Abstract
The Sleeping Beauty (SB) transposase reconstructed from salmonid fish has high transposition activity in mammals and has been a useful tool for insertional mutagenesis and gene delivery. However, the transposition efficiency has varied significantly among studies. Our previous study demonstrated that the introduction of methylation into the SB transposon enhanced transposition, suggesting that transposition efficiency is influenced by the epigenetic status of the transposon region. Here, we examined the influence of the chromatin status on SB transposition in mouse embryonic stem cells. Heterochromatin conformation was introduced into the SB transposon by using a tetracycline-controlled transrepressor (tTR) protein, consisting of a tetracycline repressor (TetR) fused to the Kruppel-associated box (KRAB) domain of human KOX1 through tetracycline operator (tetO) sequences. The excision frequency of the SB transposon, which is the first step of the transposition event, was enhanced by approximately 100-fold. SB transposase was found to be colocalized with intense DAPI (4′,6′-diamidino-2-phenylindole) staining and with the HP1 family by biochemical fractionation analyses. Furthermore, chromatin immunoprecipitation analysis revealed that SB transposase was recruited to tTR-induced heterochromatic regions. These data suggest that the high affinity of SB transposase for heterochromatin conformation leads to enhancement of SB transposition efficiency.
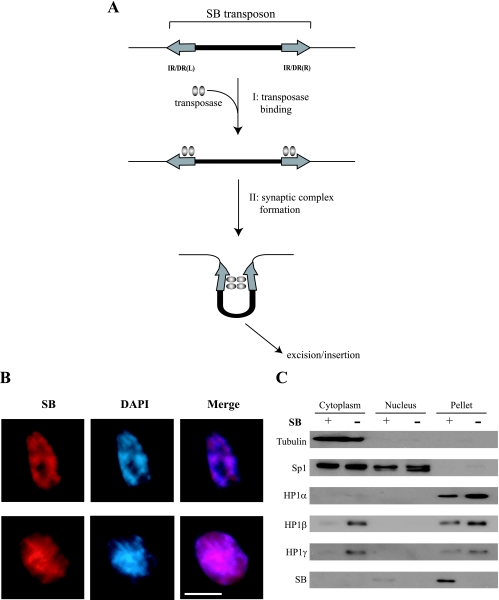
Transposable elements, constituting a large part of the mammalian genome, may have a major impact on the evolution and integrity of the genome (30). One of the most active transposases in mammalian cells, Sleeping Beauty (SB), was reconstructed from the salmonid genome and it is categorized within the Tc1/mariner superfamily of transposons (20). SB-based gene mobilization has been utilized as a tool for insertional mutagenesis (3, 4, 7, 14, 19, 24) and several gene therapy paradigms (32, 39). The SB transposon system has been developed as a nonautonomous system consisting of two independent components: transposon and transposase. The SB transposon is a DNA fragment flanked by the terminal inverted repeats (IRs) and is mobilized by SB transposase. Each IR contains two copies of a short (15- to 20-bp) direct repeat (DR), and the resultant structures are named IR/DRs. SB transposase can initiate a transposition event by binding to IR/DRs. The two IR/DRs are then probably paired through interactions of the transposase subunits (21), thereby forming a synaptic complex (Fig. 1A), followed by excision and reinsertion of the SB transposon into another locus. The excision events result in a “footprint” containing several additional base pairs at the excised site.
FIG. 1.
Schematic representation of the initial transposition reaction and localization of SB transposase. (A) Summary and schematic representation of steps occurring in the initial transposition event: step 1 (I), SB transposase binds to IR/DRs; step 2 (II), synaptic-complex formation, followed by excision and reinsertion of the transposon from the original donor site. IR/DR(R) and IR/DR(L) indicate right and left IR/DRs, respectively. (B) Colocalization of SB transposase with intense DAPI staining. After SB transposase gene transfection, ES cells were fixed, followed by staining them with anti-SB antisera (red) and counterstaining with DAPI (blue). Scale bar, 10 μm. (C) Intracellular distribution of SB and HP1 proteins. ES cells expressing exogenous SB10 or vector control were extracted with an NE-PER kit (see Materials and Methods), and the indicated fractions were examined by Western blot analysis using the indicated antibodies.
The epigenetic modification for transposable-element mobility is an important issue for understanding the relationship between host and exogenous elements, such as transposons. When the copy number of the retrotransposon is increased in Arabidopsis, the transposed retrotransposon is inactivated by DNA methylation. These retrotransposons were found to be reactivated in a ddm (for decrease in DNA methylation) mutant background (17, 29). Moreover, although expression of retrovirus was often repressed in the genome by cytosine methylation, it was transcriptionally activated in mouse embryos deficient in DNA methyltransferase-1 (38) and by treatment with 5-azacytidine, which induces genomewide demethylation (27, 36). Thus, reactivation of silent retroviral or retrotransposable elements due to the lack of DNA methylation suggests that this may have evolved as a basic defense mechanism to prevent the harmful activity of mobile elements within the mammalian genome (37, 40).
On the other hand, we demonstrated previously that CpG methylation within the transposon sequence enhances the transposition frequency of the SB transposon (42). However, that study was performed using cultured cells introduced with in vitro-methylated DNA. In order to further clarify this result, we took advantage of a tetracycline-controlled transrepressor (tTR) protein, in which the tetracycline repressor (TetR) from Escherichia coli Tn10 is fused to the Kruppel-associated box (KRAB) domain of human KOX1 (5), which can induce epigenetic gene silencing in specific regions of the genome. KRAB is a 75-amino-acid transcriptional repression domain found in many zinc finger-containing proteins (2) and can suppress in an orientation-independent manner both polymerase II- and polymerase III-mediated transcription within a distance of up to 3 kb from its binding site, presumably by triggering heterochromatin conformation change (5, 28, 34). This heterochromatic conformation induced by the KRAB domain has been found to be mitotically heritable (1). When linked to the DNA-binding domain of TetR, KRAB can modulate transcription from an integrated promoter juxtaposed with tetracycline operator (tetO) sequences (5). In the absence of doxycycline (DOX), tTR binds specifically to tetO and suppresses the activity of any nearby promoter. Conversely, in the presence of DOX, tTR is sequestered away from tetO, thus permitting gene expression (5). Using this mechanism, we performed a tTR-based study by inducing heterochromatin formation in inserted tetO sequences within or near the transposon IR/DRs and compared the efficiencies of SB transposition in vivo.
MATERIALS AND METHODS
Construction of plasmids.
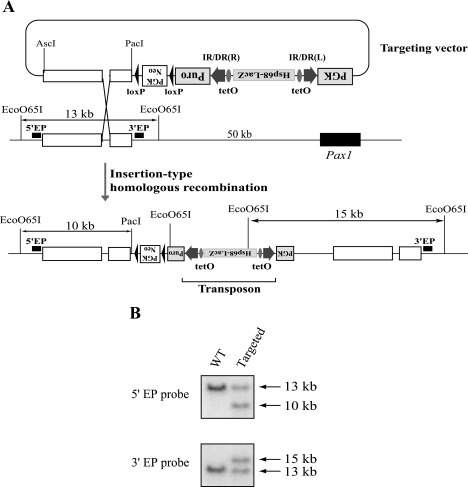
A targeting vector to introduce a single-copy SB transposon into the Pax1 locus was generated as part of a separate project in our laboratory, and full details will be described elsewhere (C. Kokubu, K. Hories, R. Ikeda, and J. Takeda, unpublished data). As shown in Fig. 2A, tetO sequences were inserted within the SB transposon immediately inside both IR/DRs. An enhancer detection cassette, consisting of the Hsp68 minimal promoter and the LacZ reporter gene, was also inserted into the transposon between the tetO elements. The resulting SB transposon was inserted between the puromycin resistance gene and the phosphoglycerate kinase (PGK) promoter. The PGK-Neo cassette, located outside the transposon, was used as a positive selection marker for genomic integration of the entire targeting vector.
FIG. 2.
Targeted insertion of a transposon into the Pax1 locus. (A) Introduction of a single SB transposon copy into the Pax1 locus by insertion-type homologous recombination. The open boxes indicate homologous sequences for recombination. An Hsp68 minimal promoter-LacZ cassette and tetO were inserted between IR/DRs of the transposon. (B) Southern blot analysis of the targeted ES cells. DNA from targeted and wild-type (WT) ES cells were double digested with EcoO65I and PacI and analyzed with two different probes. The locations of the probes are shown in panel A.
To generate the pCAG-IRES-Hygro vector, the fusion PCR product of the internal ribosome entry site (IRES)-hygromycin resistance gene was inserted downstream of the CAG promoter. This vector was used for generating control embryonic stem (ES) cell clones. To generate the tTR expression vector (pCAG-tTR-IRES-Hygro), the tTR fragment generated from pCMV-TetR(B/E)-KRAB (12) was cloned into the HpaI site of the pCAG-IRES-Hygro vector.
Cell culture and gene targeting.
Mouse ES cells were cultured in Dulbecco modified Eagle medium containing 20% fetal bovine serum, nonessential amino acids, sodium pyruvate, and leukemia-inhibitory factor (1,000 U/ml) on mitomycin C-treated (15 μg/ml for 2.5 h) mouse embryonic fibroblasts.
Targeted integration of the SB transposon at the Pax1 locus was performed by means of insertion-type homologous recombination. Briefly, 25 μg of the targeting vector was linearized at the BamHI site located in the homologous region and introduced into 107 ES cells by electroporation (240 V; 500 μF) with a Gene Pulser II (Bio-Rad, Hercules, CA). Cells were selected for 7 days with G418 (150 μg/ml), after which resistant clones were picked up, expanded, and analyzed by Southern blotting.
Generation and characterization of stably transfected cell clones.
For the generation of ES cell lines containing a stably expressed tTR, ES cells with a targeted integration of the transposon element in the Pax1 locus were transfected with either the pCAG-IRES-Hygro control or pCAG-tTR-IRES-Hygro expression vector and cultured in ES cell medium containing 200 μg/ml hygromycin B. The colonies were isolated, expanded, and tested for G418 (150 μg/ml) resistance and LacZ expression using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining 3 days after the addition of DOX.
Antibodies.
The antibodies used and their sources were as folows: anti-dimethyl H3-K4 (Upstate Biotechnology, Lake Placid, NY), anti-dimethyl H3-K9 (Abcam, Cambridge, United Kingdom), anti-trimethyl H3-K9 (Abcam, Cambridge, United Kingdom), anti-acetyl H3 (Upstate Biotechnology), anti-HP1α (Upstate Biotechnology), anti-HP1β (Chemicon International, Inc., Temecula, CA), anti-HP1γ (Upstate Biotechnology), anti-SB antisera (a gift from Zoltan Ivics), anti-tubulin (Sigma Chemical Co., St. Louis, MO), and anti-Sp1 (Sigma Chemical Co., St. Louis, MO).
ChIP assay.
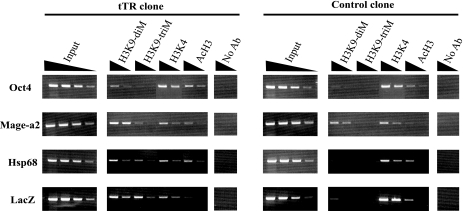
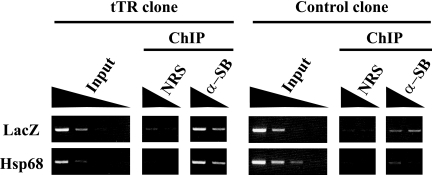
The chromatin immunoprecipitation (ChIP) experiment was performed with a ChIP assay kit using the protocol recommended by the supplier (Upstate Biotechnology) with some modifications. Briefly, 107 (for histone modification [see Fig. 4]) and 2 × 106 (for recruitment of SB transposase [see Fig. 8]) ES cells were cross-linked with 1% formaldehyde at 37°C for 10 min. The cross-linked cells were centrifuged and washed once with phosphate-buffered saline. The cells were suspended in sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl at pH 8.1) containing protease inhibitor cocktail (Sigma) and sonicated to obtain average fragment sizes of 200 to 500 bp. Solubilized chromatin was clarified by centrifugation for 10 min at 13,000 rpm at 4°C, and the supernatant was diluted 10-fold in ChIP dilution buffer (1% Triton X-100, 1 mM EDTA, 150 mM NaCl, and 15 mM Tris-HCl, pH 8.1). The diluted chromatin was precleared for 60 min with protein A (see Fig. 4) or G (see Fig. 8) agarose beads that had been blocked with salmon sperm DNA and bovine serum albumin. The precleared chromatin was incubated with anti-dimethyl H3-K4, anti-dimethyl H3-K9, anti-trimethyl H3-K9, anti-acetyl H3, and anti-SB antibodies at 4°C for 12 to 16 h. Immunocomplexes were bound to protein A or G agarose beads at 4°C for 30 min. The beads were washed once each with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris-HCl at pH 8.1), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, and 20 mM Tris-HCl at pH 8.1), and LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl at pH 8.1) and twice with Tris-EDTA buffer (1 mM EDTA and 10 mM Tris-HCl at pH 8.0). The DNA-protein immunocomplexes were eluted from the protein A or G beads with elution buffer (1% SDS and 0.1 M NaHCO3), and the eluates were treated with proteinase K for 1 h at 56°C and extracted once with phenol-chloroform, and the DNA was precipitated with ethanol. The precipitated DNA was resuspended in 50 μl of Tris-EDTA. The immunoprecipitated and input DNAs were analyzed by PCR using primer pairs specific for the Oct4 promoter sequence (5′-GGAGGTGCAATGGCTGTCTTGTCC-3′ and 5′-CTGCCTTGGGTCACCTTACACCTCAC-3′), the Mage-a2 promoter sequence (5′-TTGGTGGACAGGGAAGCTAGGGGA-3′ and 5′-CGCTCCAGAACAAAATGGCGCAGA-3′), the Hsp68 minimal promoter sequence (5′-GCGCCGCTCTGCTCTGCTTCTCTTGTCTTC-3′ and 5′-CCCCCTCAGGAATCCGTACTCTCCAGTGAA-3′), and the LacZ sequence (5′-GCGCCCATCTACACCAACGTAACCTATCCC-3′ and 5′-CGCGGCTGAAATCATCATTAAAGCGAGTGG-3′).
FIG. 4.
ChIP analyses of the transposon locus. Formaldehyde cross-linked chromatin from tTR (no. 1) and control (no. 1) ES clones cultured with DOX for 3 days to exclude tTR from tetO was immunoprecipitated without antibody (No Ab) or with anti-dimethyl H3-K9, anti-trimethyl H3-K9, anti-dimethyl H3-K4, or anti-acetyl H3 antibody. Fivefold serial dilutions of input and precipitated DNAs from tTR and control clones were used for semiquantitative PCR. Oct4 and Mage-a2 were used as controls for euchromatic and heterochromatic regions, respectively.
FIG. 8.
Recruitment of SB transposase to a tTR-induced heterochromatic region. ES cells expressing tTR (tTR clone no. 1) and control ES cells (control clone no. 1) were transfected with SB11 transposase expression vector. After 48 h, the cells were subjected to a ChIP assay using anti-SB polyclonal antibody. Fivefold serial dilutions of input and precipitated DNAs from tTR and control clones were used for semiquantitative PCR. NRS, preimmune rabbit serum.
Cell fractionation assay.
Rapidly growing ES cells stably transfected with either empty vector or SB10 transposase expression vector were harvested using the Nuclear and Cytoplasmic Extraction Reagent (NE-PER; Pierce, Rockford, IL) according to the instructions provided by the manufacturer. The respective cytoplasmic, nuclear, and pellet fractions were analyzed by Western blot analysis. Immunodetections of tubulin, Sp1, and HP1 were used as controls to identify cytoplasmic, nuclear/cytoplasmic, and pellet fractions, respectively.
Excision assay of the transposon located in the Pax1 locus.
For the transposon excision assay, 2 × 105 ES cells in either the presence or absence of 1 μg/ml of DOX were transfected with 2 μg of pCMV-SB11 (15) using TransFast (Promega, Madison, WI) and cultured for 48 h on mouse embryonic fibroblasts treated with mitomycin C (feeder cells). The transfected ES cells were harvested, and the feeder cells were removed by incubation on gelatin-coated dishes for 20 min at 37°C. Unattached ES cells were recultured on gelatin-coated dishes for another 3 days. Genomic DNA extracted from the feederless culture was used as a template in the following semiquantitative PCR analysis. Different amounts of template DNA (100 ng, 10 ng, and 1 ng) were amplified by nested PCR using the following primers: Puro-L4 (5′-GCGCGTGAGGAAGAGTTCTTGCAGCTCGGT-3′) and PGKp-U1 (5′-TGGCCTCTGGCCTCGCACACATTCCACATC-3′) for the first PCR and Puro-L3 (5′-GTCGGCGAACGCGGCGGCGAGGGTGCGTAC-3′) and PGKp-U2 (5′-CGCGCCACCTTCTACTCCTCCCCTAGTCAG-3′) for the second PCR. PCR amplifications were performed using HotStarTaq (QIAGEN, Hilden, Germany) under the following conditions: 95°C for 15 min, followed by 35 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 40 s, with a final extension at 72°C for 5 min.
For control experiments (see Fig. 6B, right), the Fas ligand locus was amplified by single-round PCR using the following primer pairs: Fex1u (5′-ATGCAAGTGAGTGGGTGTCTCACAG-3′) and Fex1l (5′-AAACTGACCCTGGAGGAGCCCAAGA-3′). PCR amplifications were performed using HotStarTaq under the following conditions: 95°C for 15 min, followed by 35 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 5 min.
FIG. 6.
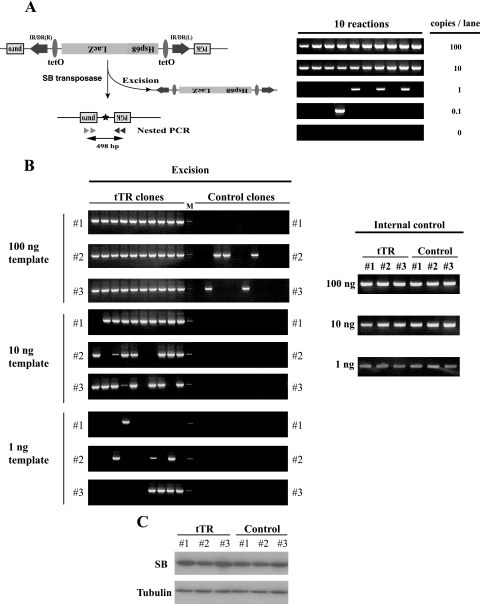
Effect of heterochromatin on the excision of the transposon by transiently transfected SB transposase. (A) Schematic representation of the excision assay and sensitivity of PCR conditions. (Left) SB transposase (pCMV-SB11) was transfected into tTR and control clones in the presence of DOX. After 48 h, feeder cells were removed, and the remaining cells were cultured for 3 days on a gelatin-coated dish; genomic DNA was extracted, and nested PCR was performed. The excised transposon locus yielded a 498-bp PCR product. Arrowheads, primers for nested PCR; asterisk, excision footprint. (Right) A single copy of the excised DNA could be detected by this nested-PCR condition. An excision-positive ES clone was obtained, and genomic DNA was prepared. The indicated copy numbers of excision-positive genomic DNAs were mixed with 100 ng of excision-negative genomic DNA. (B) High excision frequencies of the tTR-positive clones at the defined genomic locus. (Left) Genomic DNA from each clone (1, 10, and 100 ng) was used as a template for nested PCR. Ten PCR amplifications were carried out for each clone. Note that about 100-fold-higher frequency of excision events was detected in tTR-positive clones than in control clones. Two independent experiments were performed, and a representative result is presented. M, molecular marker. (Right) Control experiment to evaluate the quality of isolated DNA. The Fas ligand locus was amplified by single-round PCR. (C) Expression of SB transposase after transfection. tTR and control clones were transfected with the SB11 transposase expression vector. The Western blot analysis after 48 h posttransfection is shown. Tubulin was used as a loading control.
Immunofluorescence microscopy.
ES and NIH 3T3 cells were transfected with pCMV-SB11. After 48 h, these cells were trypsinized and seeded on a 24-well plate coated with gelatin. After 5 h, the cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 before immunoreaction with rabbit anti-SB polyclonal antiserum (dilution, 1:1,000), followed by Alexa594-conjugated donkey anti-rabbit immunoglobulin G (dilution, 1:1,000). Counterstaining was performed using 4′,6′-diamidino-2-phenylindole (DAPI). These samples were observed under fluorescence microscopy (Olympus IX70; Olympus, Tokyo, Japan).
Determination of transposon insertion sites by ligation-mediated (LM) PCR.
Genomic DNA was isolated from tTR and control clones after transfection with SB11 transposase expression vector. After 1 μg of genomic DNA was digested with a 4-base restriction enzyme (RsaI or HaeIII), splinkerettes (6) were ligated to the cleaved ends. The ligation products were purified with phenol-chloroform, followed by ethanol precipitation. Samples were digested with BglII to prevent amplification from the donor site before being used as templates for subsequent PCRs. We performed nested PCR with first primers specific for splinkerettes (Spl-P1, 5′-CGAATCGTAACCGTTCGTACGAGAA-3′) and the transposon vector (T/DR2, 5′-CTGGAATTGTGATACAGTGAATTATAAGTG-3′) using the HotStarTaq system (QIAGEN) under the following conditions: 50-μl PCR volume; 1 cycle at 95°C for 15 min, 30 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and 1 cycle at 72°C for 5 min. For the second nested PCR, we used 1 μl of the first nested-PCR product as a template under similar conditions, using second-PCR primers specific for splinkerettes (Spl-P2, 5′-TCGTACGAGAATCGCTGTCCTCTCC-3′) and the transposon vector (T/BAL, 5′-CTTGTGTCATGCACAAAGTAGATGTCC-3′). Nested-PCR product bands were purified with a Gel Extraction Kit (QIAGEN), followed by sequencing using the primer Spl-P2 or T/BAL.
RESULTS
Preferential localization of SB transposase to heterochromatin.
Transposition of DNA-type transposons generally consists of its excision and subsequent reinsertion into the host genome. Before excision occurs, two important steps are required: step 1, transposase binding, and step 2, synaptic-complex formation (Fig. 1A). We have previously shown that CpG methylation of the SB transposon enhanced transposition efficiency, and we postulated that either one of the above-mentioned steps or both are affected by CpG methylation, followed by heterochromatin formation. To investigate whether the nuclear localization of SB transposase accounts for the transposition enhancement, we transiently transfected the SB transposase gene into ES cells. As revealed by immunofluorescence analysis, SB transposase preferentially colocalized with intense DAPI staining, which is indicative of heterochromatin (25, 33) (Fig. 1B). Similar results were also observed in NIH 3T3 cells (data not shown).
To further characterize the SB transposase, we determined the intracellular distribution of SB transposase and investigated whether SB transposase was tightly bound to nuclear components, like HP1 family members (31). The transposase was first introduced into ES cells, and cytoplasmic/nuclear extracts and pellet fractions were subsequently analyzed by Western blot analysis. As shown in Fig. 1C, transcription factors, such as Sp1, could be efficiently extracted from the nuclear fraction, whereas chromatin-associated proteins, such as HP1α/β/γ, could not be extracted. A large amount of the introduced SB transposase was found in the pellet fraction, indicating that SB transposase is tightly associated with chromatin (Fig. 1C). To investigate whether SB transposase cooperatively functioned with the HP1 family in heterochromatin, we examined possible physiological interaction between SB transposase and the HP1 family using coimmunoprecipitation analysis. However, in our hands, we could not detect any interaction (data not shown).
Our immunostaining and biochemical analyses suggest that SB transposase has an affinity for the heterochromatic regions.
Generation of ES cells bearing one copy of transposon vector inserted into a defined locus.
In our previous study, we introduced an in vitro-methylated transposon into cultured cells and examined its transpositional efficiency (42). Since modification of the transposon was performed in “test tubes,” the enhanced transposition could be the result of unusual mechanisms. To investigate this phenomenon further, modification of the transposon was introduced by using an in vivo mechanism. Since the KRAB domain can recruit causative heterochromatic proteins, such as HP1 and Kap1 (1), tTR, which contains the KRAB domain, may induce heterochromatic conformation with its binding proteins around the inserted tetO sequences. We generated a transposon vector with tetO sequences positioned near the IR/DRs (Fig. 2A), and this single transposon copy was inserted into the Pax1 locus, 50 kb upstream of the first exon, by insertion-type homologous recombination. Southern analysis was performed to confirm successful insertion of a single transposon copy bearing tetO sequences in the Pax1 locus (Fig. 2B).
Gene silencing by tTR expression around the transposon insertion locus.
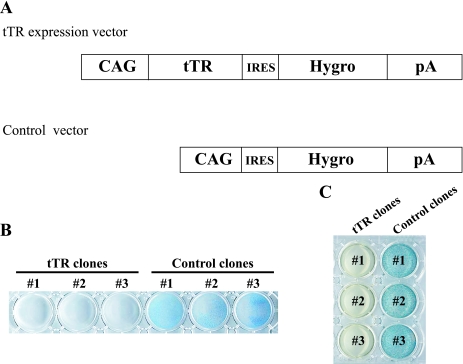
To generate ES cell lines expressing tTR, pCAG-tTR-IRES-Hygro (Fig. 3A) was transfected into ES cells bearing a single transposon copy in the Pax1 locus. As a control, we also generated stable cell lines that contained only the hygromycin resistance gene (Fig. 3A). From each transfection, three individual clones were isolated, and they were named tTR no. 1 to 3 or control no. 1 to 3 for experimentally or control-transfected clones, respectively.
FIG. 3.
Schematic representation of tTR expression vector and characterization of stably expressed tTR cells. (A) tTR expression vector under the control of the CAG promoter and its control vector. Three individual clones generated by each vector were selected for further analyses. Hygro, hygromycin. (B) Sensitivity to G418. The tTR and control clones were cultured in G418-containing medium (150 μg/ml) for 6 days. Resistant colonies were stained with Giemsa solution. Clones expressing tTR completely perished, while all control clones survived. (C) X-Gal staining. X-Gal staining showed that tTR clones were negative while control clones were positive. These data suggest that the LacZ gene was completely silenced by tTR-induced heterochromatin.
To evaluate the degree of heterochromatic change induced by tTR expression, all clones were cultured in ES medium supplemented with G418 for 6 days. Although the distance between tetO sequences and the PGK promoter of the Neo cassette is more than 3 kb, tTR clones were G418 sensitive, while control clones were G418 resistant (Fig. 3B). In addition, the X-Gal staining assay revealed no detectable LacZ gene expression in tTR clones, while control clones were all LacZ positive (Fig. 3C). Since the transcriptional activity of the Hsp68 minimal promoter is very low, the PGK promoter located immediately upstream of Hsp68 may enhance its activity. To examine whether the effect of gene silencing was continuously maintained without tTR binding to tetO, we cultured the ES cell lines expressing tTR with DOX for 3 weeks and looked for LacZ expression. The absence of any LacZ-positive colonies suggests that the gene silencing of the LacZ gene continued for at least 3 weeks without tTR binding to tetO (data not shown). These data, shown in Fig. 3B and C, suggest that the gene silencing induced by tTR expression was maintained around the transposon/tetO region.
Histone modification and DNA methylation status of the transposon locus.
tTR, consisting of TetR and the KRAB domain, can bind to tetO sequences in the genome and coordinate the machinery required for the establishment of a highly localized heterochromatin-like silenced state in euchromatic loci, which are epigenetically heritable (1). Thus, to examine heterochromatic markers at the transposon locus, we performed ChIP analysis of the targeted locus using antibodies specific to dimethylated H3-K9, trimethylated H3-K9, acetylated H3, and dimethylated H3-K4 (Fig. 4). We performed these analyses in the presence of DOX for 3 days to keep tTR away from tetO and generate more natural heterochromatin conformation in the transposon locus. The recovered DNAs from tTR and control clones were analyzed by semiquantitative PCR using gene-specific primer pairs for the Hsp68 minimal promoter, the LacZ coding region, and the internal control locus Oct4 promoter region (16) for the euchromatic region and the Mage-a2 promoter region (35) for the heterochromatic region. Although both control loci were detected at similar levels by these antibodies in all clones tested, the LacZ and Hsp68 regions were highly enriched by dimethylated H3-K9 and trimethylated H3-K9 antibodies in tTR-positive clones, but not in control clones. Furthermore, in the LacZ locus, control clones were moderately enriched by acetylated H3 antibodies and more enriched by dimethylated H3-K4 antibodies than tTR-positive clones. These results suggest that dynamic changes in histone modifications were effectively induced by tTR around the transposon/tetO region.
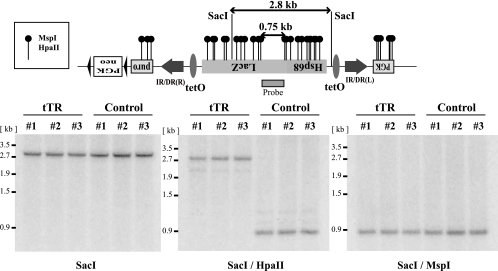
To further investigate epigenetic changes associated with heterochromatin formation, we analyzed the methylation status of CpG dinucleotides within the transposon containing tetO sequence by Southern blotting using methylation-sensitive restriction enzymes. As shown in Fig. 5, DNA from tTR clones double digested with SacI and methylation-sensitive HpaII yielded a 2.8-kb band that corresponded to the band with SacI-only digestion, while DNA from the control clones yielded a 0.75-kb band (Fig. 5, middle). DNAs from both tTR and control clones double digested with SacI and methylation-insensitive MspI always yielded a 0.75-kb band (Fig. 5, right). These results suggest that tTR effectively induced DNA methylation around the transposon/tetO region.
FIG. 5.
Analysis of methylation status within the transposon. (Top) Detailed restriction map of methylation-sensitive restriction enzymes. The LacZ probe used is shown as a box under the LacZ coding region. (Bottom) Genomic DNA from tTR-positive clones was not digested with methylation-sensitive HpaII, indicating that the region was highly methylated. On the other hand, genomic DNA from control clones was completely digested. Genomic DNAs from tTR and control clones were digested with methylation-insensitive MspI (shown on the right). The tTR-positive and control clones were cultured with DOX for 3 days.
Taken together, tTR effectively induced not only histone modifications, but also DNA methylation in the transposon locus.
Effect of heterochromatin on excision of the transposon.
To test whether heterochromatic formation influences transposition efficiency, we transiently transfected a transposase gene (SB11) into tTR and control clones and analyzed excision by nested PCR. As shown in Fig. 6A, left, a 498-bp PCR band is expected when the transposon is excised from the Pax1 locus. Before the excision analysis, we tested the sensitivity of our PCR conditions and found that they were sensitive enough to amplify a single excision event (Fig. 6A, right). Using the same PCR conditions, we performed the excision analysis in the presence of DOX for 3 days. Excision was detected with as little as 1 ng template in tTR clones. In contrast, excision in control clones was detected only at the 100-ng level (Fig. 6B, left). Therefore, the heterochromatin conformation induced by tTR resulted in approximately 100-fold-higher efficiency of excision events than that with control clones. DNA templates from tTR and control clones were of similar quality for PCR amplification (Fig. 6B, right). Expression of SB proteins was not affected by the expression of tTR (Fig. 6C). Thus, tTR can enhance the excision of transposons after introducing heterochromatin conformation change.
Analyses of reinsertion events of the SB transposon.
To evaluate transposition events, including reinsertions, puromycin selection could also be available. If the transposon mobilizes to other loci, a functional PGK-puromycin gene is generated (Fig. 6A, left). After puromycin selection, transposon reinsertion sites would be identifiable. However, the transposon locus is silenced, probably due to the recruitment of heterochromatic proteins through the tetO site (Fig. 3B and C). Therefore, even if transposition occurs, the clones would still be puromycin sensitive. To revert the heterochromatic conformation, ES cells were treated with histone deacetylase inhibitor, trichostatin A (10), and cytosine methylation inhibitor, 5-aza-dC (9). Our preliminary experiments revealed that ES cells were very sensitive to these drugs, even at very low doses, indicating that trichostatin A and 5-aza-dC could not be used to revert the heterochromatic conformation in our present experiments (data not shown).
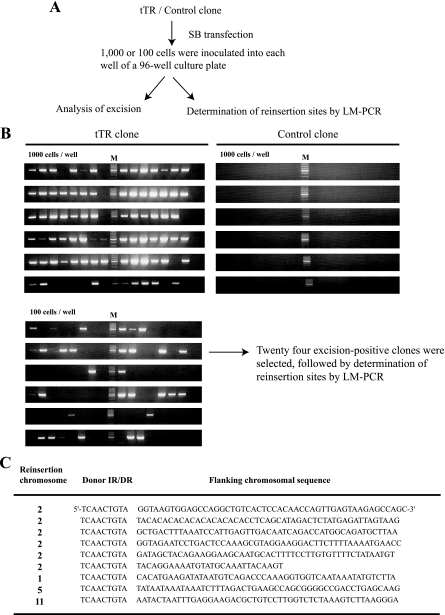
To examine reinsertion events without puromycin selection, we set up a new strategy (Fig. 7A). First, we inoculated 100 or 1,000 tTR-expressing ES cells and 1,000 control ES cells into each well of a 96-well culture plate and looked for excision events in these cells after brief expansion. As shown in Fig. 7B, 31 out of 96 wells from the 100-cell/well group were excision positive. A single excision event could be detected by our PCR conditions (Fig. 6A), suggesting that most excision-positive wells from the 100-cell/well group, if not all, had one excision event, and thus, a particular reinsertion site could be analyzed. Consistent with Fig. 6B, all wells from control ES cells were excision negative. Secondly, ES cells from excision-positive wells from the 100-cell/well group were further expanded, and reinsertion sites were determined by LM-PCR after the purification of genomic DNA. We could determine reinsertion sites from 9 out of 24 excision-positive clones (Fig. 7C). Distribution of insertion sites in tTR clones occurred mostly on the same chromosome as the donor site (Fig. 7C), which is consistent with our previous studies (19, 24). Using control ES cells, we collected excision-positive cells by puromycin selection and found that about half of the puromycin-resistant clones were reinsertion negative (Kokubu et al., unpublished), implying that approximately half of the excised transposons fail to land in the genome. This is consistent with the above-mentioned frequency of reinsertion events (9 out of 24) in tTR clones, suggesting that heterochromatic formation at the SB transposon does not influence the reinsertion process. These data demonstrate that enhanced excision efficiency by tTR-induced heterochromatin formation at the SB transposon leads to a high rate of transposition.
FIG. 7.
Analyses of reinsertion events in the presence of puromycin selection. (A) Experimental procedure to analyze for reinsertion events. After the transfection of SB transposase, tTR and control clones were inoculated into 96-well culture plates with the indicated cell numbers. Excision-positive cell populations were expanded, genomic DNA was isolated, and LM-PCR analyses were performed to identify reinsertion sites. (B) Screening for excision events. Excision-positive wells derived from 100-cell/well groups theoretically contained one excision event, and insertion sites were determined by LM-PCR. M, molecular marker. (C) Flanking transposon genomic sequences. The original donor site is located on chromosome 2, and reinsertion chromosomes are shown.
Recruitment of SB transposase to tTR-induced heterochromatinized transposons.
To directly evaluate whether SB transposase has a preference for tTR-induced heterochromatinized transposons, we performed a ChIP assay using anti-SB transposase polyclonal antibody. As shown in Fig. 8, SB transposase was clearly recruited more in the heterochromatinized transposon than in the nonheterochromatinized one. Thus, enhanced transposition by heterochromatin conformation at the SB transposon could be explained by enhanced recruitment of SB transposase to heterochromatin conformation.
DISCUSSION
Over the past few years, several studies of SB transposable elements (TEs) have been done with regard to their application for insertional mutagenesis (3, 19). However, only a few studies have been conducted to understand the exact molecular mechanisms underlying the interaction between SB transposase and its recognition sites for transposition (26, 43). A better understanding of the molecular mechanisms involved in transposition will enable researchers to further develop and improve the SB system for mutagenesis by increasing the rate of transposition and mobilization in the genome.
Comparison between our previous and present studies.
Heterochromatin and DNA methylation are generally thought to suppress the mobilization of transposable elements, both autonomous DNA transposons and retrotransposons, by inhibiting the expression of catalytic enzymes for transposition (17, 29, 38, 41). In our previous work, we found that heterochromatin formation might facilitate the transposition reaction when transposase was supplied in trans (42). However, there is an important question that needs to be addressed: whether the in vitro methylation of DNA and subsequent chromosomal integration resemble the native cellular heterochromatinization. That is, the unexpected modification might have caused a bias change in the frequency of SB transposase recruitment to IR/DRs. In order to exclude this possibility, we took advantage of a tetracycline-controlled transrepressor protein, tTR, consisting of a tetracycline repressor (TetR) fused to the KRAB domain of human KOX1, to establish highly localized heterochromatin states in the euchromatic region which are heritably and epigenetically maintained (1). Using this system in our present study, the transposition frequency with heterochromatin conformation was increased 100-fold compared to nonheterochromatin conformation (Fig. 6B). Compared with our previous results, the excision efficiency was similarly enhanced and was approximately 100-fold higher than controls. However, the minimum template DNA used for nested PCR in our previous study was 10 ng (42). In contrast, approximately 1 ng template was used in the present study (Fig. 6B). The PCR conditions in both studies were capable of detecting a single excision event (Fig. 6A) (42), suggesting that the excision efficiency was approximately 10-fold higher in the present study. This increased efficiency may indicate that recruitment of the repressor molecules caused a more natural heterochromatin conformation. Detailed analysis is necessary to clarify the difference in efficiency between the two studies. For example, the integration sites of the transposon vector, the versions of the transposase genes (SB10 versus SB11), and the cell lines used were different.
One or 10 nanograms of genomic DNA corresponds to approximately 150 or 1,500 cells, respectively. This transposition frequency is lower than that in mouse germ cells (i.e., 0.03 to 0.2 transposition/transposon/germ cell) (8, 11, 18). Our previous (42) and present studies have caused only a “local” introduction of heterochromatin conformation. In contrast, a tandem array of SB transposon transgenes would be widely heterochromatinized in mice. This different state of genome conformation might explain the effectiveness of SB transposition in mouse germ cells.
Which step of the transposition event is enhanced by heterochromatic conformation?
The initial transposition event can be divided into at least two major processes, as shown in Fig. 1A: step 1, binding of the transposase to its recognition sites within the IRs, and step 2, the formation of a synaptic complex to pair the two ends of IR/DRs and to hold them together with transposase subunits. The exact step(s) enhanced by the heterochromatin should be determined in future studies. In the present study, we demonstrated by immunostaining assay that SB transposase was colocalized with intense DAPI staining in the heterochromatic regions (Fig. 1B). Furthermore, SB transposase was recruited to the tTR-induced heterochromatic regions (Fig. 8). These findings suggest that SB transposase prefers heterochromatic to euchromatic regions. In other words, heterochromatic conformation can enhance step 1. With puromycin selection, we determined more than 100 reinsertion sites using a control clone (Kokubu et al., unpublished). Although there are many local hopping events which are commonly observed in the SB transposition (11, 13, 19, 24), we could not recognize any site preference within the local hopping regions, implying that the SB transposon would not prefer heterochromatin to euchromatin for reinsertion. However, detailed examination is required for a definitive conclusion.
It has been reported that the HMGB1 protein interacts with SB transposase and induces a local distortion of the DNA upon binding in vitro, indicating the enhancement of the above-mentioned step 2 (43). However, how the HMGB1 protein is involved in the SB transposition in vivo must be determined.
When we tried to introduce SB transposase in the absence of DOX, tTR-expressing and control clones had the same level of transposon excision efficiency (data not shown). Why did the excision efficiency change in tTR-expressing clones in the presence or absence of DOX? We hypothesized that tTR proteins and their associated proteins may accumulate around the tetO sequence, hindering the recruitment of SB proteins to the IR/DRs (step 1) because of the close proximity between tetO and IR/DRs. However, in the presence of DOX, tTR proteins and their associated proteins were dissociated from the tetO sequence, thus allowing access of the SB protein to the IR/DRs. Alternatively, the binding of tTR and associated proteins may disturb the bending of IR/DRs essential for synaptic-complex formation and thus inhibit step 2.
Taking these data together, we hypothesize that the heterochromatin conformation recruits SB transposase and enhances its binding to the SB transposon that is a prerequisite for subsequent reactions, such as excision and reinsertion.
The biological significance of these findings.
Clarification of the relationship between TEs and the host genome is becoming an important issue, because mammalian-genome-sequencing projects have revealed that approximately one-half of the total number of genomes consists of TEs (30). One can easily imagine that expansion of TEs would be deleterious to the host genome due to chromosomal rearrangement mediated by TEs. Expanded TEs reside mainly in the heterochromatin conformation (22). A possible explanation of this phenomenon is that heterochromatin causes a reduced rate of recombination. During evolution, species have selected against such high rates of recombination.
Positive roles of TEs in genome evolution have also been discussed (30). More recently, Kazazian proposed that mobile elements are drivers of genome evolution (23). Viewed in this light, the maintenance of TEs in heterochromatin might be an important feature of genome evolution. The current findings are that SB transposase has a preference for heterochromatin conformation and is able to mobilize TEs efficiently in trans when chromatin is in this conformation state. Therefore, TEs might have evolved to counteract the host genome defense strategy by accumulating in heterochromatin.
Acknowledgments
We thank Z. Ivics for providing the anti-SB antibody. We are grateful to M. Kouno, T. Hayakawa, K. Yae, S. Kouno, M. Tachibana, Y. Shinkai, and H. Tojo for their helpful advice and excellent technical support. We thank current members of our laboratory for their support and advice, particularly Y. Odan for her excellent secretarial support.
This work was supported by grants from the New Energy and Industrial Technology Development Organization of Japan; the Cosmetology Research Foundation; RIKEN, the Institute of Physical and Chemical Research; and a grant-in-aid for Science Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 18 December 2006.
REFERENCES
- 1.Ayyanathan, K., M. S. Lechner, P. Bell, G. G. Maul, D. C. Schultz, Y. Yamada, K. Tanaka, K. Torigoe, and F. J. Rauscher III. 2003. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 17:1855-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellefroid, E. J., D. A. Poncelet, P. J. Lecocq, O. Revelant, and J. A. Martial. 1991. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc. Natl. Acad. Sci. USA 88:3608-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson, C. M., A. J. Dupuy, S. Fritz, K. J. Roberg-Perez, C. F. Fletcher, and D. A. Largaespada. 2003. Transposon mutagenesis of the mouse germline. Genetics 165:243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson, C. M., J. L. Frandsen, N. Kirchhof, R. S. McIvor, and D. A. Largaespada. 2005. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc. Natl. Acad. Sci. USA 102:17059-17064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deuschle, U., W. K. Meyer, and H. J. Thiesen. 1995. Tetracycline-reversible silencing of eukaryotic promoters. Mol. Cell. Biol. 15:1907-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devon, R. S., D. J. Porteous, and A. J. Brookes. 1995. Splinkerettes—improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 23:1644-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupuy, A. J., K. Akagi, D. A. Largaespada, N. G. Copeland, and N. A. Jenkins. 2005. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436:221-226. [DOI] [PubMed] [Google Scholar]
- 8.Dupuy, A. J., S. Fritz, and D. A. Largaespada. 2001. Transposition and gene disruption in the male germline of the mouse. Genesis 30:82-88. [DOI] [PubMed] [Google Scholar]
- 9.Egger, G., G. Liang, A. Aparicio, and P. A. Jones. 2004. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429:457-463. [DOI] [PubMed] [Google Scholar]
- 10.Finnin, M. S., J. R. Donigian, A. Cohen, V. M. Richon, R. A. Rifkind, P. A. Marks, R. Breslow, and N. P. Pavletich. 1999. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401:188-193. [DOI] [PubMed] [Google Scholar]
- 11.Fischer, S. E., E. Wienholds, and R. H. Plasterk. 2001. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl. Acad. Sci. USA 98:6759-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forster, K., V. Helbl, T. Lederer, S. Urlinger, N. Wittenburg, and W. Hillen. 1999. Tetracycline-inducible expression systems with reduced basal activity in mammalian cells. Nucleic Acids Res. 27:708-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geurts, A. M., L. S. Collier, J. L. Geurts, L. L. Oseth, M. L. Bell, D. Mu, R. Lucito, S. A. Godbout, L. E. Green, S. W. Lowe, B. A. Hirsch, L. A. Leinwand, and D. A. Largaespada. 2006. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2:1413-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geurts, A. M., A. Wilber, C. M. Carlson, P. D. Lobitz, K. J. Clark, P. B. Hackett, R. S. McIvor, and D. A. Largaespada. 2006. Conditional gene expression in the mouse using a Sleeping Beauty gene-trap transposon. BMC Biotechnol. 6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geurts, A. M., Y. Yang, K. J. Clark, G. Liu, Z. Cui, A. J. Dupuy, J. B. Bell, D. A. Largaespada, and P. B. Hackett. 2003. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol. Ther. 8:108-117. [DOI] [PubMed] [Google Scholar]
- 16.Hattori, N., K. Nishino, Y. G. Ko, J. Ohgane, S. Tanaka, and K. Shiota. 2004. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 279:17063-17069. [DOI] [PubMed] [Google Scholar]
- 17.Hirochika, H., H. Okamoto, and T. Kakutani. 2000. Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation. Plant Cell. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horie, K., A. Kuroiwa, M. Ikawa, M. Okabe, G. Kondoh, Y. Matsuda, and J. Takeda. 2001. Efficient chromosomal transposition of a Tc1/mariner-like transposon Sleeping Beauty in mice. Proc. Natl. Acad. Sci. USA 98:9191-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horie, K., K. Yusa, K. Yae, J. Odajima, S. E. Fischer, V. W. Keng, T. Hayakawa, S. Mizuno, G. Kondoh, T. Ijiri, Y. Matsuda, R. H. Plasterk, and J. Takeda. 2003. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol. Cell. Biol. 23:9189-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivics, Z., P. B. Hackett, R. H. Plasterk, and Z. Izsvak. 1997. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91:501-510. [DOI] [PubMed] [Google Scholar]
- 21.Izsvak, Z., D. Khare, J. Behlke, U. Heinemann, R. H. Plasterk, and Z. Ivics. 2002. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem. 277:34581-34588. [DOI] [PubMed] [Google Scholar]
- 22.Junakovic, N., A. Terrinoni, C. Di Franco, C. Vieira, and C. Loevenbruck. 1998. Accumulation of transposable elements in the heterochromatin and on the Y chromosome of Drosophila simulans and Drosophila melanogaster. J. Mol. Evol 46:661-668. [DOI] [PubMed] [Google Scholar]
- 23.Kazazian, H. H., Jr. 2004. Mobile elements: drivers of genome evolution. Science 303:1626-1632. [DOI] [PubMed] [Google Scholar]
- 24.Keng, V. W., K. Yae, T. Hayakawa, S. Mizuno, Y. Uno, K. Yusa, C. Kokubu, T. Kinoshita, K. Akagi, N. A. Jenkins, N. G. Copeland, K. Horie, and J. Takeda. 2005. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat. Methods 2:763-769. [DOI] [PubMed] [Google Scholar]
- 25.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192-1200. [DOI] [PubMed] [Google Scholar]
- 26.Liu, G., A. M. Geurts, K. Yae, A. R. Srinivasan, S. C. Fahrenkrug, D. A. Largaespada, J. Takeda, K. Horie, W. K. Olson, and P. B. Hackett. 2005. Target-site preferences of Sleeping Beauty transposons. J. Mol. Biol. 346:161-173. [DOI] [PubMed] [Google Scholar]
- 27.Lorincz, M. C., D. Schubeler, S. C. Goeke, M. Walters, M. Groudine, and D. I. Martin. 2000. Dynamic analysis of proviral induction and de novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol. Cell. Biol. 20:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margolin, J. F., J. R. Friedman, W. K. Meyer, H. Vissing, H. J. Thiesen, and F. J. Rauscher III. 1994. Kruppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. USA 91:4509-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura, A., S. Yonebayashi, K. Watanabe, T. Toyama, H. Shimada, and T. Kakutani. 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411:212-214. [DOI] [PubMed] [Google Scholar]
- 30.Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 31.Remboutsika, E., Y. Lutz, A. Gansmuller, J. L. Vonesch, R. Losson, and P. Chambon. 1999. The putative nuclear receptor mediator TIF1α is tightly associated with euchromatin. J. Cell Sci. 112:1671-1683. [DOI] [PubMed] [Google Scholar]
- 32.Richardson, P. D., L. B. Augustin, B. T. Kren, and C. J. Steer. 2002. Gene repair and transposon-mediated gene therapy. Stem Cells 20:105-118. [DOI] [PubMed] [Google Scholar]
- 33.Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, and T. Jenuwein. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senatore, B., A. Cafieri, I. Di Marino, M. Rosati, P. P. Di Nocera, and G. Grimaldi. 1999. A variety of RNA polymerases II and III-dependent promoter classes is repressed by factors containing the Kruppel-associated/finger preceding box of zinc finger proteins. Gene 234:381-394. [DOI] [PubMed] [Google Scholar]
- 35.Tachibana, M., K. Sugimoto, M. Nozaki, J. Ueda, T. Ohta, M. Ohki, M. Fukuda, N. Takeda, H. Niida, H. Kato, and Y. Shinkai. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16:1779-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka, J., T. Ishida, B. I. Choi, J. Yasuda, T. Watanabe, and Y. Iwakura. 2003. Latent HIV-1 reactivation in transgenic mice requires cell cycle-dependent demethylation of CREB/ATF sites in the LTR. AIDS 17:167-175. [DOI] [PubMed] [Google Scholar]
- 37.Walsh, C. P., and T. H. Bestor. 1999. Cytosine methylation and mammalian development. Genes Dev. 13:26-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]
- 39.Yant, S. R., L. Meuse, W. Chiu, Z. Ivics, Z. Izsvak, and M. A. Kay. 2000. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 25:35-41. [DOI] [PubMed] [Google Scholar]
- 40.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]
- 41.Yu, F., N. Zingler, G. Schumann, and W. H. Stratling. 2001. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 29:4493-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yusa, K., J. Takeda, and K. Horie. 2004. Enhancement of Sleeping Beauty transposition by CpG methylation: possible role of heterochromatin formation. Mol. Cell. Biol. 24:4004-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zayed, H., Z. Izsvak, D. Khare, U. Heinemann, and Z. Ivics. 2003. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 31:2313-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]