Abstract
CTCF is a transcription factor with highly versatile functions ranging from gene activation and repression to the regulation of insulator function and imprinting. Although many of these functions rely on CTCF-DNA interactions, it is an emerging realization that CTCF-dependent molecular processes involve CTCF interactions with other proteins. In this study, we report the association of a subpopulation of CTCF with the RNA polymerase II (Pol II) protein complex. We identified the largest subunit of Pol II (LS Pol II) as a protein significantly colocalizing with CTCF in the nucleus and specifically interacting with CTCF in vivo and in vitro. The role of CTCF as a link between DNA and LS Pol II has been reinforced by the observation that the association of LS Pol II with CTCF target sites in vivo depends on intact CTCF binding sequences. “Serial” chromatin immunoprecipitation (ChIP) analysis revealed that both CTCF and LS Pol II were present at the β-globin insulator in proliferating HD3 cells but not in differentiated globin synthesizing HD3 cells. Further, a single wild-type CTCF target site (N-Myc-CTCF), but not the mutant site deficient for CTCF binding, was sufficient to activate the transcription from the promoterless reporter gene in stably transfected cells. Finally, a ChIP-on-ChIP hybridization assay using microarrays of a library of CTCF target sites revealed that many intergenic CTCF target sequences interacted with both CTCF and LS Pol II. We discuss the possible implications of our observations with respect to plausible mechanisms of transcriptional regulation via a CTCF-mediated direct link of LS Pol II to the DNA.
CTCF, or CCCTC binding factor, is an 11-Zn-finger transcription factor with highly versatile functions and a candidate tumor suppressor (30, 42). CTCF is localized to the nucleus and is ubiquitous and highly conserved. CTCF regulates transcription in diverse modes, such as promoter activation and repression, silencing, and constitutive- and methylation-dependent chromatin insulation; CTCF also organizes epigenetically controlled chromatin insulators that regulate imprinted genes in soma (30, 42). The characterized genes regulated by CTCF include c-myc (16, 31), chicken lysozyme (7), BRCA1 (8), hTERT (49), IRAK2 (35), amyloid beta-protein precursor (APP) (62), and other genes (42). Among vertebrate insulators controlled by CTCF are the β-globin (3) and the H19 imprinting control region (ICR) (42, 44) insulators. In our previous report, the number of CTCF binding sites in the mouse genome was estimated as ∼4,000 (40) but the real number may be much higher (∼30,000 in the human genome), as suggested in a more recent publication (61). Many of these sites are methylation sensitive and map to promoter, inter- and intragenic regions, and introns; some sites contain Alu-like repeated elements (40, 61).
Posttranslational modifications of CTCF were found to be involved in the regulation of CTCF function(s). Thus, specific phosphorylation of CTCF by the protein kinase CK2 (former casein kinase II) affects CTCF function in transcriptional regulation (15, 29). Poly(ADP-ribosyl)ation is another recently discovered modification of CTCF that is important for insulator function (27, 67) and nucleolar transcription (60). Posttranslational modifications of CTCF have also been implicated in human myeloid cell differentiation (14).
CTCF association with other proteins is also important for the regulation of CTCF-dependent molecular processes. Thus, CTCF interactions with Sin3 (37) and YB-1 (10, 28) are shown to modulate CTCF function as a transcriptional repressor. The cooperation of CTCF with nucleophosmin (68), Kaiso (13), and helicase protein CHD8 (22) has been linked to the control of insulator function of CTCF and epigenetic regulation.
In this report, we describe the interaction of CTCF with RNA polymerase II (Pol II). The eukaryotic Pol II enzyme transcribes all protein-coding genes and also noncoding regulatory RNAs (e.g., snRNA and microRNA) (52). The Pol II enzyme is composed of 12 subunits (termed Rpb1 to Rpb12) (66). Rpb1, the largest subunit of Pol II (LS Pol II), is highly conserved among eukaryotic RNA polymerases. Its characteristic feature is the carboxyl-terminal domain (CTD), which contains multiple copies of the heptapeptide repeat Tyr-Ser-Pro-Thr-Ser-Pro-Ser. The CTD can be modified by phosphorylation which results in the appearance of two forms of LS Pol II: hypophosphorylated (LS Pol IIa), migrating at 220 kDa, and hyperphosphorylated (LS Pol IIo), migrating at 240 kDa. The LS Pol IIa has been associated with the initiation complex, whereas the LS Pol IIo has been found in elongating complexes (12).
Accurate initiation of transcription by Pol II can be directed by the TATA box, INR, and possibly other less-characterized promoter elements. The mechanisms of TATA-mediated transcription initiation are very well understood. The TATA binding protein (TBP) subunit of the TFIID complex is necessary for the recognition of the TATA box and accurate initiation of transcription by Pol II (19, 57). Very little, however, is known at present about the mechanisms of transcription initiation mediated by other promoter elements, in particular, proteins that recognize these elements and aid Pol II (19).
The views on how the transcriptional machinery is assembled and targeted to specific promoters do not harmonize. Thus, a “stepwise assembly” model proposes a coordinated step-by-step recruitment of the proteins in the transcription preinitiation complex. The alternative “preassembly complex” model suggests the recruitment of a preassembled Pol II complex for transcription initiation (36). In both models, general transcription factors are required to form a stable initiation complex at promoters, and mediators and coactivators are necessary to communicate signals from transcriptional activators and repressors (39).
In this report, we describe the association of a subpopulation of CTCF with the Pol II protein complex. A component of this complex, the LS Pol II protein, has been identified as a protein-interacting partner with CTCF. We demonstrate that CTCF is associated in vivo with LS Pol II at the selected known CTCF target sequences (CTSs). Furthermore, we show that a single CTCF binding site is sufficient to activate the transcription of the reporter gene in a stably transfected cell line, which is likely to occur through the interaction between CTCF and LS Pol II. A genome-wide analysis of CTCF and LS Pol II interactions indicates that CTCF may recruit LS Pol II to a certain subpopulation of CTSs. These findings may provide a basis to link the transcriptional machinery directly to CTSs on the DNA with various potential functional implications.
MATERIALS AND METHODS
Cell lines, stable transfections, and luciferase assay.
Human HeLa (cervical carcinoma), MCF7 (breast carcinoma), and K562 (myeloid leukemia) cells were maintained in RPMI 1640 medium supplemented with HEPES, GlutaMAX, sodium bicarbonate, 50 μg/ml gentamicin, and 10% fetal calf serum (all from Life Technologies). Chicken erythroblast HD3 cells were grown in Dulbecco's modified Eagle's medium supplemented with 50 μg/ml gentamicin, 8% fetal calf serum, and 2% chicken serum. Cells were induced to differentiate according to the protocol previously described by Nicolas et al. (41). Briefly, 107 cells from a logarithmically growing culture were plated at 1 × 106 cells/ml in the above medium containing 10 mM HEPES, pH 8, and 20 μM protein kinase C inhibitor H7 and incubated at 42°C for 2 days. Staining with benzidine (5) was performed to confirm differentiation; cultures with >80% of benzidine-positive cells were used in the experiments. Human choriocarcinoma JEG-3 cells were propagated as described previously (17). DNA transfection into JEG-3 cells was performed using the calcium phosphate method (51); 106 cells were transfected with 5 μg of plasmid DNA in 10-cm plates. NIH 3T3 mouse fibroblasts were maintained in Dulbecco's modified Eagle's medium supplemented with 10% donor serum and 50 μg/ml gentamicin. Growth arrest of NIH 3T3 was induced by serum starvation (0.05%) for 48 h.
To generate pN-Myc plasmids or deficient-for-CTCF-binding pN-Sac-Myc mutant (mut) plasmids (pN-Myc-Luc wild type [wt] and pN-MycLuc mut) for stable transfection in NIH 3T3 cells, the 90-bp N-Myc and N-Myc-Sac mutant fragments were excised by HindIII from pBend-N-Myc and pBend-N-Myc-Sac, respectively (38). The fragments were then cloned into the HindIII-digested dephosphorylated pGL2 basic promoterless plasmid. For stable transfections, the FuGENE 6 reagent (Roche Applied Science) was used. Three micrograms of pMyc-N or mutant pMyc-N-Sac was mixed with 0.3 μg of pCIIN containing the neomycin resistance selection marker, and 106 cells were used for each transfection. For the selection of transfectants, cells were incubated with 500 μg/ml of G-418 for 2 weeks, followed by the subcloning of single cells. The colonies found to be positive in a luciferase assay were pooled and grown as a mass culture.
Luciferase assay.
For the luciferase reporter assay, cells were grown in six-well plates for 24 h; the luciferase activity was measured using a luciferase reporter assay system kit (Promega) according to the manufacturer's instructions. The luciferase activities were normalized to transgene copy number, which was estimated by comparing the band intensities of the ClaI/XhoI genomic fragments from pN-Myc-Luc wt and pN-MycLuc mut cells (see “Southern blot procedures” below). All assays were performed in triplicate.
Immunoprecipitation.
HeLa cells (2 × 106) were collected, washed twice with ice-cold phosphate-buffered saline (PBS), and lysed in 500 μl of high-salt radioimmunoprecipitation assay (RIPA) buffer as previously described (10, 26). A panel of antibodies at a final concentration of 5 μg/ml was used for immunoprecipitations (IP). They included rabbit polyclonal antibodies anti-LS Pol II (N-20), Sp1 (PEP2), and TBP (SI-1), all from Santa Cruz, and monoclonal antibodies anti-LS Pol II (8WG16) raised against the 220-kDa form of Pol II (hypophosphorylated, Pol IIa) and anti-LS Pol II (H14) raised against the 240-kDa form (hyperphosphorylated, Pol IIo), both from Covance Research Products. The anti-histone H2A and-histone H3 antibodies were kindly provided by S. Muller and J. Dadoune. The anti-CTCF rabbit polyclonal antibodies were raised against the bacterially expressed CTCF N-terminal domain and are able to recognize CTCF in different species. We also used mouse monoclonal antibody Rb1 (Ab-6) and p53 (Ab1 through Ab6 from the sampler kit) from Oncogene Research Products. For the neutralization of the antibody, we followed the protocol developed at Santa Cruz. The fivefold excess of the N-20 peptide (250 μg/ml) was added to 100 μl of the anti-LS Pol II antibody diluted to 50 mg/ml in 1× PBS and incubated for 2 h at room temperature. The blocked antibodies were then used at 5 μg/ml in co-IP. In some experiments, RNase-free DNase I (Roche Diagnostics) was added to the extracts at 300 U/ml for 50 min at 32°C prior to immunoprecipitation.
CTCF expression in the baculovirus system.
The CTCF-producing recombinant SF9 cells were grown following the manufacturer's instructions (BacVector system manual; Novagen). The CTCF protein (termed “baculoCTCF”) was purified to 80 to 90% purity from infected SF9 cells using Ni affinity chromatography with a linear gradient of imidazole for elution and subsequent gel filtration on an S-200 column.
Purification of the Pol II and TFIIH complexes.
The Pol II and TFIIH complexes were prepared from HeLa cells as previously described (25, 64, 65).
Expression of the His-tagged N-terminal, Zn finger, and C-terminal domains of CTCF in the bacterial system.
Preparation of the vectors expressing the His-tagged N-terminal, Zn finger, and C-terminal domains of CTCF was described in detail in our previous report (10); their detailed maps are available on request. To generate proteins in a bacterial system, transformants carrying the plasmids expressing the His-tagged N-terminal, Zn finger, and C-terminal domains of CTCF were grown in LB medium supplemented with ampicillin (50 μg/ml) for 3 h at 37°C. Protein expression was induced by the addition of 0.4 mM isopropyl-β-D-thiogalactoside (IPTG), with further incubation for 3 h at 37°C. For the purification of each of the desired proteins, the bacterial cells were collected by centrifugation, and washed twice with 0.1 volume of cold phosphate-buffered saline, followed by lysis in 0.1 volume of the original culture in the cold freshly prepared lysis buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl, pH 8.0). The lysates were then subjected to immobilized metal ion affinity chromatography for further purification. For this purpose, the total bacterial lysates were supplemented with 20 mM imidazole and then loaded onto the nickel-charged His-Bind resin (R&D Systems, Europe Ltd.), washed with 1 bed volume of the washing buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl, pH 8.0, and 20 mM imidazole), and finally, eluted with 10 ml of the elution buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl, pH 8.0, and 0.5 M imidazole).
Production of GST-LS Pol II (bactPol II) from Escherichia coli.
The construct for the expression of LS Pol II in E. coli contained the cDNA of LS Pol II (hRPB1) cloned as a glutathione S-transferase (GST) fusion using bacterial IPTG-inducible pGEX-2T vector (1). To produce the GST-LS Pol II protein in E. coli DH5α, we used the standard protocols (54, 63) with some modifications to solubilize bactPol II, which was extracted from the inclusion bodies by incubation in HEPES-guanidine buffer (50 mM HEPES, pH 7.5, 6 M guanidine HCl, 25 mM dithiothreitol [DTT]) and renaturing in ice-cold folding buffer (50 mM HEPES, pH 7.5, 0.2 M NaCl, 1 mM DTT, 1 M NDSB201 [3(1-pyridinio)-1-propane sulfonate (Fluka Chem.)], followed by dialysis against 100 volumes of the dialysis buffer (50 mM HEPES, pH 7.5, 0.2 M NaCl, 1 mM DTT, and 10% glycerol) at +4°C overnight.
Immobilization of the bacterially expressed proteins on the matrix.
To generate matrices for interaction assays, the in vitro-produced proteins were immobilized onto cystamine-coupled Sepharose 4B. Cystamine was first converted into aminoethylthiol after a reduction reaction with 50 mM dithiothreitol in TE buffer (50 mM Tris-HCl, 1 mM EDTA, pH 8.3) for 30 min at room temperature and then treated with 5 mM 2,2-dipyridyldisulfide for 2 h. The activated matrix was washed with the TE buffer. Each protein was reduced by incubation with 5 mM dithiothreitol for 1 h at room temperature, desalted through a G50 column equilibrated with TE, and then incubated with the activated matrix overnight at +4°C (protein-to-Sepharose [vol/wt] ratio was 5 mg/1 ml). The amounts of protein retained on the matrix were monitored by a protein assay (Bio-Rad) according to the manufacturer's instructions. The protein-Sepharose conjugates were finally washed with the TE buffer to remove nonincorporated materials and stored in the buffer containing 20% glycerol, 50 mM KH2PO4, pH 7.0, and 0.2% Na3N.
The interaction (pull-down) assay.
Fifty microliters of the Sepharose suspension carrying each of the different CTCF protein domains produced in E. coli or in bovine serum albumin (BSA) as a control was mixed with 1 ml of K562 cell lysate in 0.25 M RIPA buffer and incubated for at least 6 h on a rotating platform. Each suspension was then washed six times with 0.25 M RIPA buffer, boiled in sodium dodecyl sulfate (SDS) sample-loading solution for 5 min, run on 10% SDS-polyacrylamide gel electrophoresis (PAGE), and analyzed by a Western blot assay. The presence of LS Pol II was detected with the anti-LS Pol II (N-20) antibody.
Western blot analysis.
Proteins were resolved, blotted, and visualized as previously described by Chernukhin et al. (10). The primary anti-CTCF antibody was used at a 1:300 dilution, the anti-LS Pol II antibodies at 1:100 (1-μg/ml final concentration), and the anti-His tag monoclonal antibodies (Sigma) at 1:500. The secondary anti-rabbit peroxidase-conjugated (Abcam) or anti-mouse peroxidase-conjugated (Abcam) antibodies were used at 1:10,000 dilutions. Before reprobing, a membrane was stripped in a buffer containing 100 mM mercaptoethanol, 2% SDS, and 62.5 mM Tris HCl, pH 6.7, incubated twice at 55°C for 15 min and then rinsed three times for 15 min at room temperature in PBS (pH 7.5) supplemented with 0.1% Tween 20. The quantification of the bands was performed by using Image J software (http://rsb.info.nih.gov/ij/).
Mass spectrometry.
The protein bands were subjected to in-gel digestion, and peptide mass fingerprint analysis using a matrix-assisted laser desorption-time of flight mass spectrometry system (Bruker Daltonics Reflex 4) was performed as previously described by Chernukhin et al. (10). The obtained spectra were interpreted using Bruker Daltonics FlexAnalysis 2.0 software, and the sequence retrieval was performed with the Mascot peptide mass fingerprint search engine (Matrix Science)using the Swiss-Prot database.
Chromatin immunopurification (ChIP) assay and “serial” ChIP assays.
Harvested cells were cross-linked with formaldehyde according to the method of Kuo and Allis (34), and the DNA concentration was adjusted to 100 μg/ml. DNA-protein complexes were immunopurified using anti-CTCF or anti-Pol II antibodies (N-20; Santa Cruz) and protein A 4 Fast Flow Sepharose beads (Sigma).
The “serial” ChIP assay is a modification of the standard ChIP assay designed to assess the DNA occupancy by two protein molecules simultaneously. In this assay, the formaldehyde cross-linked DNA-protein complexes are first passed through the matrix linked with the antibody against one of the partner proteins; after elution, the retained complexes are subjected to the subsequent IP with the second partner antibody. The resulting complexes therefore contain DNA fragments associated with the two partner proteins. The matrices containing the conjugated anti-CTCF, anti-LS Pol II antibody (N-20), or preimmune serum were prepared as previously described (9). The DNA purified from ChIP assays was measured, and 1 to 10 μl of the DNA was used for PCR amplification. The primers and conditions for PCR are described in Table 1. Full protocols for ChIP and serial ChIP assays used in these experiments are available on request.
TABLE 1.
PCR primers used in ChIP analysis and generation of probes for hybridization
| Type of primer | Clone no. or gene element | GenBank accession no. | Primer (5′-3′)
|
PCR fragment size (bp) | PCR cycle conditionsa (×30) | |
|---|---|---|---|---|---|---|
| Forward | Reverse | |||||
| Microarray | 6 | AY457178 | TGCAGGAGAAGCAATATTAT | TAGAAGAGGTAGAAGAGGAAG | 278 | 95°C, 30 s; 50°C, 30 s; 72°C, 20 s |
| 116 | AY457216 | AGGTTTTCAGGCTAGACAGA | TCTTCTGGTCTTCTCTGAATG | 96 | 95°C, 30 s; 53°C, 30 s; 72°C, 20 s | |
| 265 | AY457268 | TCTCAGTGGAGAAAAACTTTGC | GTCGACAAATCAGAAGCTGA | 270 | 95°C, 30 s; 50°C, 30 s; 72°C, 20 s | |
| 267 | AY457269 | GACCTATTGAGAATGCTCACA | ATGATTGTTACCTCTCTTTG | 235 | 95°C, 30 s; 50°C, 30 s; 72°C, 20 s | |
| 293 | AY457285 | CCACTAAACCTCCTTCTCCA | AAGAAGGCTGTAGGTGGCTGT | 127 | 95°C, 30 s; 63°C, 30 s; 72°C, 15 s | |
| 294 | AY457286 | CCAGCAAGCCCTTTAGGAACA | TACTCCTACATCCTGAAAGTG | 124 | 95°C, 30 s; 59°C, 30 s; 72°C, 20 s | |
| 396 | AY457330 | CTAATCCTTATTGTACAGGA | CATGGAAATTCTACTTTGAA | 264 | 95°C, 30 s; 50°C, 30 s; 72°C, 20 s | |
| 513 | AY457372 | CCCTTGCTCCATCTTTTGG | GTCTGCAGAAGCACTTGAAG | 94 | 95°C, 30 s; 53°C, 30 s; 72°C, 20 s | |
| 717 | AY457431 | GACTAATATTGAAAAATGTAGC | ATAGGCATTCTGGCCTTCTGAG | 190 | 95°C, 30 s; 50°C, 30 s; 72°C, 20 s | |
| 794 | AY457460 | AAGTATTGAATTTTAGGATT | TTGAGAACCACTGCTCTAAC | 234 | 95°C, 30 s; 48°C, 30 s; 72°C, 20 s | |
| 1031 | AY457551 | TCCTCTCGGGGTTTTCTCCA | AGGAAAAGACAAAAATAACCC | 138 | 95°C, 30 s; 50°C, 30 s; 72°C, 20 s | |
| For ChIP assays in HD3 cells | β-Globin insulator | E02199 | GAGCTCACGGGGACAGCCCC | GATCCCGTGCCACCTTCCCC | 174 | 94°C, 1 min; 60°C, 1 min; 72°C, 4 min |
| β-Actin promoter | E02199 | CGCTCCGAAAGTTTCCTTTT | AGAAAAGAAACGAGCCGTCA | 238 | 94°C, 1 min; 60°C; 1 min; 72°C, 4 min | |
| CTCF exon 8 | Z22605 | CGAGTTTTATGATACAGAAGTGGAAG | AGTTATTTACAAGCTTGACCATTACAG | 318 | 94°C, 1 min; 55°C, 1 min; 72°C, 4 min | |
| For hybridization (nested) | β-Globin insulator | E02199 | CCCAAAGCCCCCAGGGATGTAAT | CCCGGGCTGTCCCCGCACGCT | 130 | 94°C, 1 min; 60°C, 1 min; 72°C, 4 min |
| β-Actin promoter | E02199 | CCTTTTATGGCGAGGCGGCGGCGG | GGGCGAAGGCAACGCAGCGACTCC | 84 | 94°C, 1 min; 60°C, 1 min; 72°C, 4 min | |
| CTCF exon 8 | Z22605 | CGAGTTTTATGATACAGAAGTGGAAG | AGTTATTTACAAGCTTGACCATTACAG | 301 | 94°C, 1 min; 55°C, 1 min; 72°C, 4 min | |
| For ChIP assays in | N-Myc site | X00364 | ACCTGACCCCCGCCCTCGTTGA | CTCTACTGGCAGCAGAGATCAT | 58 | 95°C, 30 s; 65°C, 30 s; 72°C, 20 s |
| transgenic NIH 3T3 | GAPDH (exon 1) | X55448 | ATCATGGCAGAGCAGGTGGC | GATGCACCCATGATGATAA | 122 | 95°C, 30 s; 58°C, 30 s; 72°C, 20 s |
| cells | GAPDH (promoter) | X55448 | TCCTGCAATGATAGACTAG | CTGCCAAACACGTTCACAGA | 158 | 95°C, 30 s; 55°C, 30 s; 72°C, 20 s |
| For ChIP assays of wt and mut H19 ICR | AF049091 | TCCCTTTGGTCACTGAACC | AATCCCTATTTGGGTGACCC | 319 | 94°C 30 s; 50°C 45 s; 72°C 50 s | |
PCR conditions for all cycles were initial denaturation, 94°C and 5 min, and final elongation, 72°C and 10 min.
ChIP-on-ChIP analysis.
ChIP samples were prepared from 5 × 106 NIH 3T3 cells as described in the previous section. A ChIP-on-ChIP hybridization assay was performed using microarrays of a library of CTCF target sites derived from a ChIP of mouse fetal liver (40). Briefly, the ChIP samples were amplified with SR1/SR2 primers (40) and PCR labeled with Cy3/Cy5 dyes (Amersham) using the nested primers. Labeled targets were purified with the QIAquick PCR purification kit (QIAGEN) and eluted in hybridization solution (GlassHyb hybridization solution; Clontech). The Cy3- and Cy5-labeled targets were denatured and incubated at 45°C for 1 h in the presence of 100 μg of Cot-1 DNA (Clontech). The samples were pooled and hybridized to borohydride-pretreated slides. Following a washing procedure according to the user manual of the GlassHyb hybridization solution kit (Clontech), the slides were scanned using ScanArray 4000 and analyzed with ScanArray Express 3.0 (Packard Biosciences).
Indirect immunofluorescence and analysis of colocalization.
For indirect immunofluorescent staining, the original protocol of Harlow and Lane (20) was used with an additional modification (53). Cells were incubated at +4°C overnight with anti-CTCF rabbit polyclonal antibody (Abcam) (dilution, 1:5) and the anti-Pol II CTD monoclonal antibody 7G5 (4) (dilution, 1:50), followed by the subsequent incubation with the secondary antibodies: swine anti-rabbit fluorescein isothiocyanate (Dako) and rabbit anti-mouse tetramethyl rhodamine isocyanate (Dako), both diluted 1:50. The cells were visualized using confocal laser scanning microscopy (Bio-Rad). Images were obtained using a Bio-Rad Radiance 2000 confocal unit on an Olympus IX70 microscope. The fluorescein labels were illuminated by a 488-nm laser line and detected via a 500- to 560-nm band-pass filter, while the rhodamine probes were excited by a 543-nm laser with a 570-nm long-pass filter; transmission images were also recorded. The colocalization of the two probes was analyzed by in-house software using the methods of Costes et al. (11) to estimate the background intensity. Bleed-through of rhodamine signals into the fluorescein images was measured to be 17.5% using single-labeled samples and was corrected for in the colocalization analysis.
Southern blot procedures.
DNA was extracted from NIH 3T3 stable transfectant cells using the DNeasy tissue kit (QIAGEN) and digested with ClaI/XhoI restriction enzymes, followed by electrophoresis in a 0.8% agarose gel. DNA was then transferred to a HybondN+ membrane (Amersham Biosciences). Blots were probed with a 32P-labeled luciferase cDNA probe (ClaI-XhoI fragment) synthesized using a random priming labeling kit (Roche Applied Science). Membranes were hybridized at 68°C for 4 to 6 h in a buffer containing 0.5 M sodium phosphate, pH 6.8, 1 mM EDTA, 7% SDS, and 0.2 mg/ml herring sperm DNA. Following hybridization, the membranes were washed twice for 10 min in a 5% SDS, 0.04 M sodium phosphate, pH 6.8, 1 mM EDTA solution and then four times for 10 min in the same solution containing 1% SDS and exposed to X film (Kodak) for 24 h. The quantification of the bands was performed using the ImageQuant 5.0 software program. Similar blotting and hybridization procedures were used to analyze PCR products obtained in ChIP experiments from HD3 cells. The primers and conditions for PCR are described in Table 1.
RESULTS
CTCF is a component of the Pol II protein complex.
Potential CTCF-Pol II interactions were first hinted at when one of the CTCF binding sites in human and mouse MYC promoters was found to map precisely within the region of Pol II pausing and release (33, 56) (CTCF site A) (see Fig. 4A). In our ensuing experiments to isolate proteins interacting with CTCF, affinity chromatography on a matrix with immobilized purified recombinant CTCF was employed. Routinely, a doublet of two proteins of about 200 kDa and 240 kDa, reminiscent of two differentially phosphorylated forms of the LS Pol II (LS Pol IIa and LS Pol IIo), was retained by CTCF from nuclear extracts of different cellular origins (10). Based on these observations, we hypothesized that CTCF could be a part of the Pol II complex and interact with the large proteins from this complex, such as LS Pol II. This supposition was further examined by biochemical analyses.
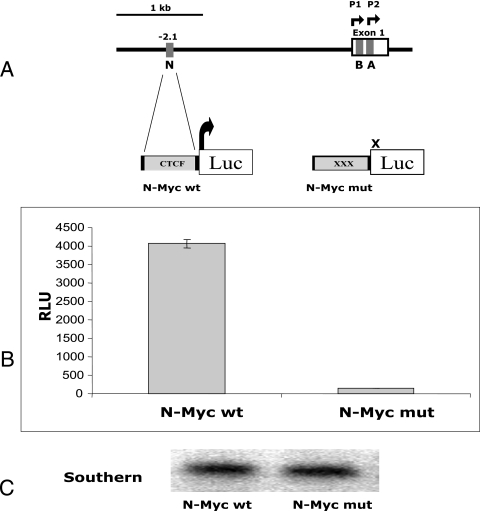
FIG. 4.
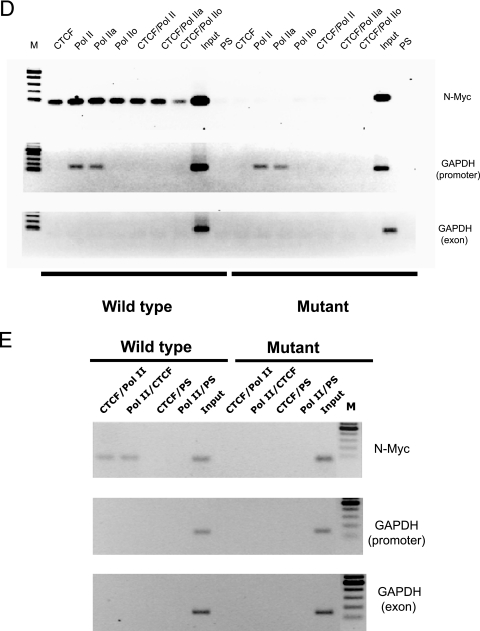
CTCF and LS Pol II are associated with wild-type N-Myc, which alone can activate transcription from the luciferase reporter gene. (A) Cartoon illustration of the 5′ noncoding region of the human c-myc gene promoter (16). Gray boxes depict the CTCF binding sites A, B, and N (38). (B) The wild-type N-Myc sequence activates the luciferase reporter gene. The NIH 3T3 cells stably transfected with pN-Myc-Luc wt and pN-MycLuc mut were harvested and assayed for luciferase activity as described in Materials and Methods. The luciferase activity normalized to the plasmid copy number is shown in relative luciferase units (RLU). Each bar represents an average of three experiments performed in triplicate. The error bar indicates standard deviation. Panel A shows the structure of the two plasmids, pN-Myc-Luc wt and pN-MycLuc mut. Gray boxes depict N-Myc sites. Luc, luciferase reporter gene. (C) Southern blot analysis of the DNA extracted from NIH 3T3 cells (pN-Myc-Luc wt and pN-MycLuc mut). Genomic DNA was extracted, digested with ClaI/XhoI, blotted, and hybridized as described in Materials and Methods. (D) CTCF and LS Pol II are associated with the wild-type N-Myc site in stably transfected NIH 3T3 cells. Standard ChIP and serial ChIP assays were performed to assess the in vivo occupancies by CTCF and Pol II at the N-Myc target sites. The antibodies used in ChIP and serial ChIP are indicated above the corresponding lanes as follows: CTCF, ChIP with the anti-CTCF antibody; Pol II, ChIP with the anti-LS Pol II antibody; Pol IIa, ChIP with the anti-LS Pol IIa antibody (hypophosphorylated form); Pol IIo, ChIP with the anti-LS Pol IIo antibody (hyperphosphorylated form); CTCF/Pol II, serial ChIP with the anti-CTCF antibody, followed by the anti-LS Pol II antibody; CTCF/Pol IIa, serial ChIP with the anti-CTCF antibody, followed by the anti-LS Pol IIa antibody; CTCF/Pol IIo, serial ChIP with the anti-CTCF antibody, followed by the anti-LS Pol IIo antibody; PS, ChIP with preimmune serum; input, DNA from NIH 3T3 cell lysates. DNA prepared from these samples was amplified using corresponding pairs of primers as described in Materials and Methods and in Table 1. The PCR products were resolved in a 1% agarose gel. M, DNA marker (100-bp DNA ladder). (E) CTCF and LS Pol II association with the wild-type N-Myc site in stably transfected NIH 3T3 cells is specific. The serial ChIP assays were performed to further assess the specificity of the in vivo occupancies by CTCF and Pol II at the N-Myc target sites. The antibodies used in ChIP and serial ChIP are indicated above the corresponding lanes as follows: CTCF/Pol II, serial ChIP with the anti-CTCF antibody, followed by the anti-LS Pol II antibody; Pol II/CTCF, serial ChIP with the anti-LS Pol II antibody, followed by the anti-CTCF antibody; CTCF/PS, serial ChIP with the anti-CTCF antibody, followed by PS; Pol II/PS, serial ChIP with the anti-LS Pol II antibody, followed by PS. Input, DNA from NIH 3T3 cell lysates. DNA prepared from these samples was amplified using corresponding pairs of primers as described in Materials and Methods and in Table 1. The PCR products were resolved in a 1% agarose gel. M, DNA marker (100-bp ladder).
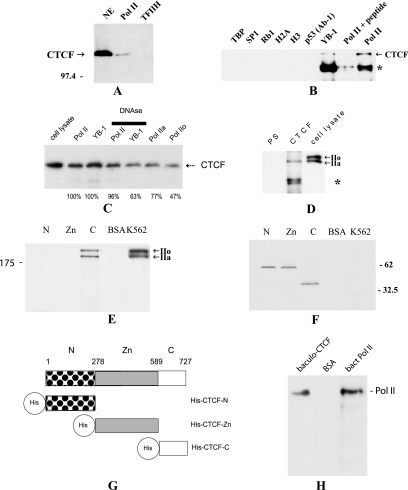
First we tested whether CTCF may be a part of the Pol II protein complex. In these experiments, the Pol II complex was purified from a cell line conditionally expressing the FLAG-tagged RPB9 subunit of human Pol II (64) and the TFIIH complex was obtained from a cell line conditionally expressing the p62 subunit of human TFIIH (25). These complexes had previously been purified and characterized (65); the same preparations were used in this study. When purified, the Pol II and TFIIH complexes were resolved by SDS-PAGE and then subjected to Western blot analysis with the anti-CTCF antibody, and the band specific for CTCF appeared in the Pol II complex but not in the TFIIH complex (Fig. 1A), thus confirming that this association is specific for the Pol II complex. The amount of CTCF associated with the Pol II protein complex in cell extracts is relatively small. This indicates that only a proportion of CTCF in the nucleus may exist in a complex with Pol II and/or this interaction in vivo is not strong, with CTCF being lost from the complex in a process of lengthy purification. The former explanation is consistent with the partial overlap of CTCF and Pol II staining in the K562 and HeLa cells' nuclei (Fig. 2); however, the latter cannot be ruled out.
FIG. 1.
CTCF is associated with LS Pol II in vivo and in vitro. (A) CTCF is a part of the Pol II protein complex. The Pol II and TFIIH complexes were purified, resolved by 10% SDS-PAGE, transferred onto a membrane, and then probed with the anti-CTCF antibody. The band specific for CTCF (indicated) can be seen in the nuclear extract (NE) and in the Pol II complex but not in the TFIIH complex. The position of the molecular marker is indicated on the left. (B) Analysis of the in vivo interactions between CTCF and LS Pol II by coimmunoprecipitation with the anti-LS Pol II antibody. The co-IP reactions were performed with a series of antibodies shown on top of the image; lysates from 5 × 105 HeLa cells were used in each reaction. The arrow signals the position of CTCF coimmunoprecipitated with anti-LS Pol II antibody (N-20) and anti-YB-1 antibody. Preincubation with peptide N-20 can block co-IP with the anti-LS Pol II antibody. On the other hand, CTCF does not coimmunoprecipitate with TBP, Sp1, Rb1, histone H2A, histone H3, or p53 (Ab-1). An ∼85-kDa protein, indicated by the asterisk, is most likely partially reduced IgG (21). (C) Analysis of the interactions between CTCF and LS Pol II pretreated with DNase I and interactions between CTCF, LS Pol IIa, and LS Pol IIo. The antibodies used for co-IP are shown at the top of the image. The anti-LS Pol II antibodies were as follows: the anti-LS Pol II (N-20) antibody that detects both forms of Pol II, the anti-LS Pol II (8WG16) that recognizes the hypophosphorylated LS Pol II (LS Pol IIa), and the anti-LS Pol II (H14) that recognizes the hyperphosphorylated LS Pol II (Pol IIo). Lysates from 5 × 105 HeLa cells were used for each reaction. Samples were electrophoretically separated, blotted, and probed with the anti-CTCF antibody. The arrow signals the position of CTCF. The developed films were scanned, and images were quantified. Levels of CTCF precipitated by the anti-Pol IIa and anti-Pol IIo antibodies and anti-Pol II treated with DNase I prior to co-IP are presented as a percentage from the co-IP reactions with the anti-Pol II (N-20) antibody (designated as 100%). An amount of CTCF precipitated by the anti-YB-1 antibody after treatment with DNase I is presented as a percentage from the co-IP reaction with the anti-YB-1 antibody (designated as 100%). Numbers below the lanes represent these results. (D) Analysis of the in vivo interactions between CTCF and LS Pol II by immunoprecipitation with the anti-CTCF antibodies. A Western blot assay with anti-LS Pol II antibody (N-20) was performed after co-IP from HeLa lysates with PS or anti-CTCF antibody (CTCF); 5 × 105 HeLacells were used for each reaction. The immunocomplexes were resolved by SDS-PAGE and blotted, and the membrane was probed with the anti-LS Pol II antibody N-20. Arrows on the right indicate the positions of the hypophosphorylated LS Pol II (IIa), sized 220 kDa, and the hyperphosphorylated LS Pol II (IIo), sized 240 kDa. An ∼85-kDa protein (depicted by the asterisk) is most likely partially reduced immunoglobulin G (21). (E) The C-terminal domain of CTCF interacts with LS Pol II in vitro. The three domains of CTCF (CTCF-N [N], CTCF-Zn [Zn], and CTCF-C [C]) expressed in E. coli and BSA (control) were coupled to the matrix and incubated with the whole lysate from K562 cells and washed with 0.25 M RIPA buffer, and the retained proteins were analyzed by a Western blot assay with the anti-LS-Pol II antibody. Arrows indicate the positions of two forms of the LS Pol II. K562, 20 μl of K562 cell lysate used in the assay. The position of the molecular marker is indicated on the left. (F) Analysis of the proteins used in the interaction assay. The membrane utilized in the experiment described for panel E was stripped and subsequently probed with the anti-His tag antibodies. The positions of the molecular markers are indicated on the right. (G) Three-domain structure of CTCF. The three domains of CTCF are depicted as follows: N, N-terminal domain (patterned box); Zn, 11-Zn-finger domain (gray box); and C, C-terminal (open box) domain. The His tags are shown as open circles. Amino acids are numbered as in Filippova et al. (16). (H) The full-length CTCF and LS Pol II interact directly in vitro. The complete peptides of CTCF (baculoCTCF) and LS Pol II (bactLS Pol II) were generated in vitro using baculoviral and bacterial systems, respectively. baculoCTCF and BSA were coupled to the matrix, incubated with the lysate containing bactLS Pol II, and washed with 0.25 M RIPA buffer, and the retained proteins were analyzed by a Western blot assay with anti-LS-Pol II antibody. The position of the bactLS Pol II is shown.
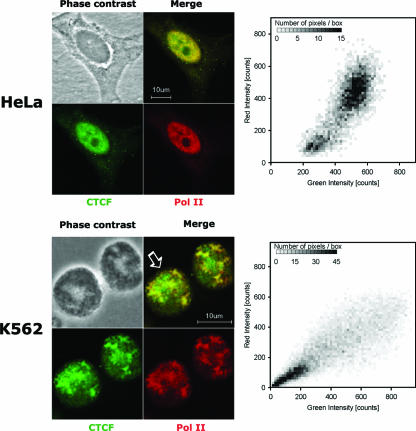
FIG. 2.
Confocal analysis of CTCF and LS Pol II in HeLa and K562 cells. HeLa and K562 cell lines were prepared and immunostained as described in Materials and Methods. The endogenous CTCF and LS Pol II proteins are extensively colocalized in the nucleus in both cell lines (HeLa, upper panel, and K562, lower panel) as shown by the merge of the CTCF (fluorescein isothiocyanate; green) and Pol II (tetramethyl rhodamine isocyanate; red) staining and colocalization analysis using the methods of Costes et al. (11). The typical two-dimensional histograms of the fluorescence for a K562 cell (indicated by the arrow) and a HeLa cell are shown.
CTCF interacts with the largest subunit of Pol II in vivo.
Next we carried out a series of co-IP assays with a panel of antibodies against proteins known to be associated with the Pol II protein complex and also proteins known to form functional interactions important for transcriptional regulation. Figure 1B shows that while the anti-LS Pol II and anti-YB-1 antibodies were able to co-IP CTCF from cell extracts, CTCF was absent from the complexes precipitated with the anti-TBP, Sp1, Rb1, histone H2A, and histone H3 antibodies. No CTCF was observed with any of six anti-p53 antibodies (Fig. 1B and data not shown). Our earlier study revealed no association between CTCF and other nuclear factors, such as p21, the ubiquitous nuclear receptor UR, thyroid receptor TRα, hTAFII130, and MYC (10). The CTCF-LS Pol II association was abolished when the peptide N-20, originally employed to raise the anti-LS Pol II antibodies, was preincubated with the anti-RNA-Pol II antibody (Fig. 1B).
CTCF was also coimmunoprecipitated by the anti-LS Pol II (8WG16) that recognizes predominantly the hypophosphorylated LS Pol II and the anti-LS Pol II (H14) that is specific to only the hyperphosphorylated LS Pol II (6) (Fig. 1C). Interestingly, CTCF was 1.6-fold more efficiently coimmunoprecipitated with the anti-LS Pol IIa antibody than with the anti-LS Pol IIo antibody. Treatment with DNase has not significantly changed the interaction between CTCF and LS Pol II (96%), which rules out possible contamination by chromatin fragments (Fig. 1C). On the other hand, interaction between CTCF and YB-1 decreased to a higher degree (63%).
The specificity of this association was further corroborated by our observation that the anti-CTCF antibody coimmunoprecipitated LS Pol II from HeLa cell extracts, while there was no LS Pol II band in coimmunoprecipitates from the preimmune serum (Fig. 1D). Notably, in cell lysates the hypophosphorylated form of Pol II (Pol IIa) was precipitated more efficiently, thus confirming previously made observations (Fig. 1C). This may reflect the nature of the interaction between CTCF and LS Pol II in vivo when posttranslational modifications of CTCF and LS Pol II or the presence of other proteins in the complex may be important in the establishment of the specific protein association. Our preliminary data indeed show that the phosphorylation of CTCF results in decreased binding to LS Pol II (I. Chernukhin and S. Shamsuddin, unpublished observations). Results similar to those described above, showing interaction between CTCF and LS Pol II, were obtained when the lysates from other cell lines (K562 and NIH 3T3) were used in co-IP experimentations (data not shown).
The two proteins sized ∼220 kDa and ∼240 kDa were also observed after a preparative immunoprecipitation with the anti-CTCF antibody (data not shown). These bands were excised and subjected to the “in-gel” digestion, and peptides were analyzed by matrix-assisted laser desorption-time of flight mass spectrometry. The database interrogation revealed the presence of peptides matching the DNA-directed RNA polymerase II largest subunit, RPB1 (Swiss-Prot protein database accession number P24928). These data complement the co-IP results showing that LS Pol II is a protein interacting with CTCF.
CTCF-LS Pol II interaction is mediated via the C-terminal domain of CTCF and is direct.
To define which portion of the CTCF protein is involved in the interaction with LS Pol II, the three His-tagged domains of CTCF (N, Zn, and C; a map is shown in Fig. 1G) were generated in a bacterial system, coupled to the matrix, and employed in the interaction assay. Figure 1E shows that the K562 cell-derived LS Pol II interacts with the CTCF C-terminal domain immobilized on the matrix. As in Fig. 1C, two bands of the LS Pol II, the hypophosphorylated LS Pol IIa, and the hyperphosphorylated LS Pol IIo were observed. However, in this case, both bands were retained efficiently by the C-terminal domain of CTCF, which may be due to the absence of the posttranslational modifications in the CTCF-C. No LS Pol II was seen with the CTCF-N, CTCF-Zn, or BSA. Equal loading of the proteins was verified by the subsequent probing of the membrane with the anti-His tag antibodies (Fig. 1F).
The directness of the association between CTCF and LS Pol II was further confirmed in the interaction assay between the in vitro-generated CTCF and LS Pol II. In this experiment, the full-length recombinant CTCF produced in the baculovirus system (baculoCTCF) was immobilized on the matrix, whereas the LS Pol II protein produced in E. coli (bactLS Pol II) was present in the solution. As shown in Fig. 1H, in this experiment, bactLS Pol II was retained after the incubation of the lysate containing bactLS Pol II with the matrix conjugated with baculoCTCF. On the other hand, no bactLS Pol II was observed in the control experiment when the matrix contained immobilized BSA.
CTCF and LS Pol II are significantly colocalized in the nucleus.
Interaction between CTCF and LS Pol II was confirmed by imaging techniques, such as immunofluorescent staining, using the anti-CTCF polyclonal antibody and anti-LS Pol II monoclonal antibody. For immunofluorescent staining, HeLa and K562 cell lines were chosen because of their differences in CTCF distribution, uniform in HeLa and patchy in K562. The staining revealed that CTCF and LS Pol II proteins are significantly colocalized in the nucleus (typical images are shown in Fig. 2). This was confirmed by further analysis of the merged images shown in the two-dimensional histograms of the fluorescence (Fig. 2, right panels). Signals were corrected for background and bleed-through (11), and the Pearson coefficient was found; the average for three sets of images of HeLa cells was 0.83, and that for three sets of images of K562 cells was 0.85, which shows good correlation between the LS Pol II and CTCF staining patterns. However, these results also indicate that there are pools of CTCF and LS Pol II which are not colocalized and therefore may not be involved in the interaction.
Analysis of the in vivo distribution of CTCF and LS Pol II at the chicken β-globin insulator in proliferating and differentiated HD3 cells.
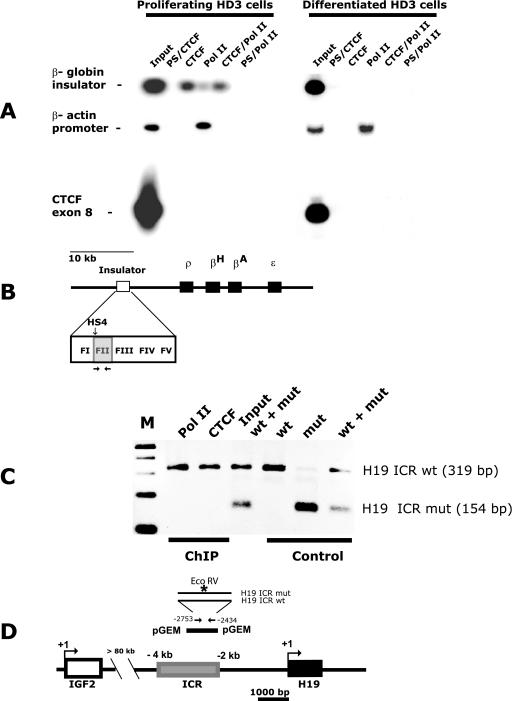
To explore the possibility that the interaction of DNA-bound CTCF and LS Pol II occurs in vivo at the β-globin insulator (site FII) (Fig. 3B) and may be important for the regulation of gene activity, we chose the erythroblast cell line HD3. In proliferating HD3 cells, the globin genes are inactive and in differentiated HD3 cells, ρ- and βA-globin genes are transcriptionally active (41), prompting a hypothesis that LS Pol II might be held by CTCF at the insulator in nonglobin-synthesized cells. To assess the simultaneous presence of CTCF and LS Pol II at CTCF binding sites, we developed the standard ChIP assay into a modified version, which we termed the serial ChIP assay. In this assay, two subsequent IP reactions of formaldehyde cross-linked DNA-protein complexes were performed to specifically precipitate the DNA-CTCF-LS Pol II complexes. The input samples were first passed through the matrix with the immobilized anti-CTCF antibodies or with the preimmune serum as a control. The advantage of using the anti-CTCF antibody covalently bound to the matrix was that only retained protein complexes, not the unwanted free antibodies, were anticipated after the first IP. After a subsequent IP with the anti-LS Pol II antibody, the composition of the resulting complexes was expected to be DNA-CTCF-LS Pol II. The fragment sizes of the sonicated DNA were 300 to 400 bp on average; hence it is very unlikely that CTCF would be associated with the same DNA fragment as LS Pol II, unless they form a protein-protein complex, because the β-globin FII site is located more than 10 kb upstream from the first transcription start site (50) (Fig. 3B).
FIG. 3.
CTCF and LS Pol II interact in vivo at the β-globin insulator and the H19 ICR. (A) CTCF and LS Pol II are associated at the β-globin insulator in proliferating HD3 cells as shown by ChIP and serial ChIP assays. Nuclear extracts were prepared from 5 × 106 of proliferating and differentiated HD3 cells; the standard ChIP assay was performed to assess the in vivo occupancies at the DNA target sites, and the serial ChIP assay was performed to assess the simultaneous presence of CTCF and LS Pol II at the β-globin insulator. PCR products were resolved by a 1% agarose gel, and a Southern blot assay was performed with the 32P-labeled β-globin insulator FII probe. PCR and hybridization with the CTCF exon 8 and chicken β-actin probes were used as a background control and as a LS Pol II loading control, respectively (see Table 1 for details of the hybridization probes). The antibodies used in ChIP and serial ChIP assays are indicated above the corresponding lanes as follows: PS/CTCF, serial ChIP with PS, followed by the anti-CTCF antibody; CTCF, ChIP with the anti-CTCF antibody; Pol II, ChIP with the anti-LS Pol II antibody; CTCF/Pol II, serial ChIP with the anti-CTCF antibody, followed by the anti-LS Pol II antibody; PS/Pol II, serial ChIP with PS, followed by the anti-LS Pol II antibody; input, DNA from HD3 cell lysates. (B) Cartoon illustration of the chicken β-globin domain (3, 46). The 1.2-kb insulator core element is shown as an open box; the detailed structure is represented in the enlarged image. CTCF binds to the 42-bp F II region within the insulator (gray box). The four β-globin genes are shown as black boxes. The hypersensitive site HS4 is indicated with a vertical arrow. Primers used for amplification of the FII are shown by horizontal arrows (the sequences of the primers are given in Table 1). (C) LS Pol II association with the H19 ICR requires functional CTCF target sites. pGEM vectors containing the wt and mut 1.2-kb H19 ICRs were transfected into JEG-3 cells individually or mixed together (+) as indicated. The image shows DNA amplified from ChIP material pulled down by CTCF and LS Pol II antibodies or control material, not subjected to ChIP, digested with EcoRV. The antibodies used in the assay are indicated above the corresponding lanes. The PCR products were resolved by a 1% agarose gel. M, DNA marker (100-bp DNA ladder). (D) Cartoon illustration of the IGF2-H19 locus (2). The positions of IGF2 (white box) and H19 (black box) genes are shown. The 2.4-kb H19 ICR element (gray box) is located −2 kb to −4.4 kb relative to the transcription start site of H19 (58). The IGF2 and H19 ICRs are separated by more than 80 kb of intervening sequences. Transcription start sites of IGF2 and H19 are presented by bent arrows. The 1.2-kb H19 ICR fragment cloned into pGEM vector is shown as a black bar. Primers used for H19 ICR amplification are denoted by straight arrows (sequences of the primers are given in Table 1). The sequence recognized by EcoRV is specific for the mutated CTCF target site 3 (indicated by an asterisk) (44).
As shown in Fig. 3A, in proliferating HD3 cells, the binding of CTCF to the DNA at the β-globin insulator can be detected after ChIP with anti-CTCF and anti-LS Pol II antibodies. Importantly, the globin insulator could be specifically amplified in the samples subjected to the serial ChIP. The interpretation of this result is that LS Pol II can interact with the β-globin insulator either directly or indirectly via CTCF in these cells. The level of β-globin sequences precipitated with the anti-LS Pol II antibody is lower than that with anti-CTCF (Fig. 3A); this result can be explained by the relative inefficiency of formaldehyde in protein-protein cross-linking in HD3 cells.
In differentiated HD3 cells, no signal was detected in samples precipitated with the anti-CTCF, anti-LS Pol II antibodies and also after the serial ChIP, suggesting that CTCF and LS Pol II were no longer associated with the β-globin insulator after the induction of HD3 cells. In both nondifferentiated and differentiated HD3 cells, no amplification was seen when the preimmune serum (PS) was used for precipitation and when primers from a region from exon 8 of the chicken CTCF gene lacking CTCF binding sites were employed for PCR. The same ChIP samples were subjected to amplification with primers designed to overlap the TATA box within the promoter region of the chicken β-actin gene, which served as a control for LS Pol II loading. The PCR products could be seen in only the samples precipitated with the anti-LS Pol II antibodies prepared from both proliferating and differentiated HD3 cells.
From these data, we conclude that CTCF and LS Pol II epitopes colocalize to the β-globin insulator, despite the absence of any known transcriptional unit at this domain.
Association of the LS Pol II to the H19 ICR requires functional CTCF target sites.
The H19 ICR is devoid of any promoter except for the H19 promoter separated from the H19 ICR by 2 kb (24) (Fig. 3D). To ascertain that the CTCF-LS Pol II signal depended on the CTCF target sites within the H19 ICR, we used previously described vectors (67) carrying a 1.2-kb region of the H19 ICR. This region contains CTCF target sites 3 and 4 in the wild type, or with mutations in the CTCF binding sites, in a pGEM vector devoid of any known eukaryotic regulatory cis elements. We have previously characterized these mutations in the H19 ICR (44) and shown that they abolished CTCF binding in vivo and in vitro (43, 44). The vectors with the wild-type and mutant H19 ICRs were mixed in equimolar amounts and transfected in JEG-3 cells, followed by ChIP analysis using CTCF or LS Pol II antibodies. The ChIP material was subsequently amplified and analyzed by using a diagnostic EcoRV restriction site which is present in only the mutated H19 ICR allele (Fig. 3D). These experiments revealed that by using CTCF target site 3 as a diagnostic marker, the sequences pulled down by the CTCF and LS Pol II antibodies exclusively contained the wild-type H19 ICR sequences, while both the wild-type and mutant H19 ICR sequences were present in the input DNA extracted from the cell lysates used for ChIP (Fig. 3C). Therefore, we conclude that the association of the LS Pol II to the H19 ICR requires functional CTCF target sites.
A single CTCF binding site is sufficient to activate a reporter gene.
One possible function of the interaction between CTCF and LS Pol II could be the activation of transcription, followed by the recruitment of LS Pol II by CTCF at the CTCF target site. To test this hypothesis and explore the functional dimension of this interaction, we prepared two vectors containing a CTCF binding site, N-Myc, and its mutated variant incapable of CTCF binding fused to the promoterless luciferase reporter gene (pN-Myc-Luc wt and pN-MycLuc mut, respectively) (Fig. 4A and B). The N-Myc site is located 2.1 kb downstream from the P1 promoter of the human c-myc gene (38) and was chosen randomly. The NIH 3T3 cell lines containing stably integrated constructs N-Myc-Luc wt and N-Myc-Luc mut were then generated.
When assessed for luciferase activity normalized to the integrated vector copy number, the cells containing the wild-type N site fused with the luciferase reporter showed significantly higher levels of luciferase activity than did the cells containing the mutant element deficient for CTCF binding (Fig. 4B and C). This implies that CTCF bound to DNA may recruit LS Pol II and factors associated with LS Pol II, which could be sufficient to initiate transcription in the absence of the promoter elements.
To confirm that both CTCF and LS Pol II are present at the wild-type N-Myc site, we used transgenic pN-Myc-Luc wt NIH 3T3 and and pN-MycLuc mut NIH 3T3 cells to perform a series of “single” and “serial” ChIP assays with the anti-CTCF and anti-LS Pol II antibodies. In these experiments, in addition to the anti-LS Pol II antibody that detects both forms of Pol II (N-20), we also tested the anti-LS Pol II (8WG16) that recognizes predominantly the LS Pol IIa and the anti-LS Pol II (H14) that is specific only to the Pol IIo (6). This analysis demonstrated that only the wild-type N site sequences could be detected after ChIP with all of these antibodies individually and together, whereas no DNA was detected in ChIP samples from cells containing the mutant N site (Fig. 4D). The same ChIP samples were amplified with primers designed to overlap the TATA box within the promoter region of the mouse GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene used as a control for LS Pol II loading. Intriguingly, both forms of LS Pol II, hyperphosphorylated and hypophosphorylated, were present at the N site, whereas only the hypophosphorylated form of LS Pol II was detected in the control (GAPDH promoter). This is likely to reflect the fact that the nonelongating Pol II is associated with the GAPDH promoter, some distance away from the elongating RNA Pol II complex. On the other hand, the presence of both forms of Pol II at the N-Myc site may indicate that nonelongating Pol II and elongating Pol II are confined to closely located promoter regions. No amplification was seen when the PS was used for precipitation and when primers from a region from exon 1 of the mouse GAPDH gene lacking CTCF binding sites were employed for PCR. Additional control experiments with the anti-Pol II (N-20) as the first antibody in the serial ChIP assay confirmed the simultaneous presence of CTCF and LS Pol II at the-N-Myc site, whereas no signal was detected when the preimmune serum was used as the second antibody in the serial ChIP with both anti-CTCF and anti-LS Pol II used as the first antibodies (Fig. 4D).
From these experiments, we conclude that a single CTCF binding site is sufficient to activate a reporter gene in the transgenic context and the site occupancy of the N-Myc site by CTCF and LS Pol II depends on functional CTCF sequences. The presence of both CTCF and elongating Pol II at the wild-type N-Myc site indicates that CTCF may be responsible for recruiting Pol II to the site which then can lead to the transcription of the reporter gene.
DNA-bound CTCF and the largest subunit of Pol II simultaneously interact genome-wide to a subset of CTCF binding sites.
To gain insight into a more genome-wide perspective of this association, we utilized a ChIP-on-ChIP hybridization assay using microarrays of a library of CTSs derived from a ChIP of mouse fetal liver. This library has been characterized in terms of both patterns of CTCF occupancy and DNA methylation status in mouse fetal liver as well as its ability to prevent enhancer-promoter communications (40). Although it represents only a proportion of total CTSs, the library gives a genome-scale impression of the occupancy of binding sites. For our experimentations, proliferating and resting mouse NIH 3T3 cells were used to prepare DNA samples from a standard ChIP or a serial ChIP assay for the hybridization to the CTS microarrays. Following amplification and labeling with Cy3/Cy5, the ChIP samples were hybridized to the target microarray.
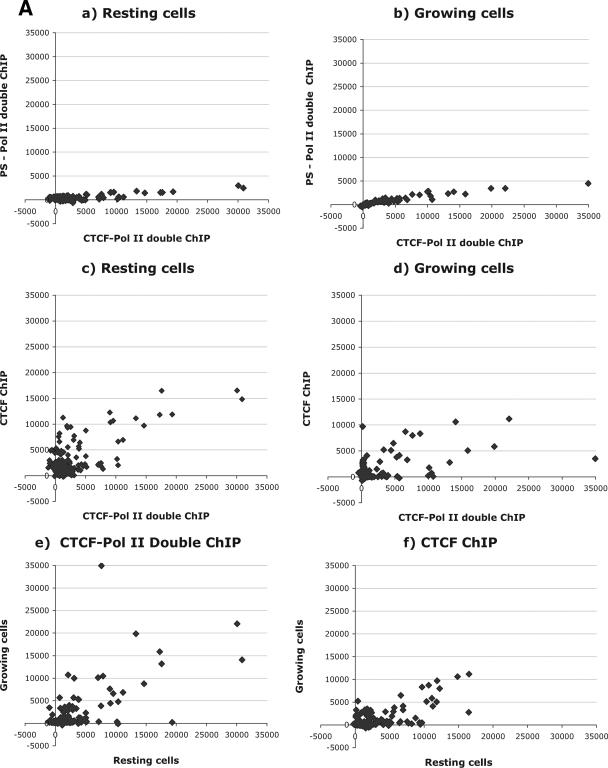
To determine the specificity of the assay, we first compared the CTCF-LS Pol II and preimmune serum-LS Pol II serial ChIP samples in resting and growing cells (Fig. 5A, panels a and b). The hybridization signals of 266 different CTCF target sites were quantified and represented on a scatter plot diagram. The signal intensities in the compared samples in both cases were low, indicating the nonspecific or background nature of the signals.
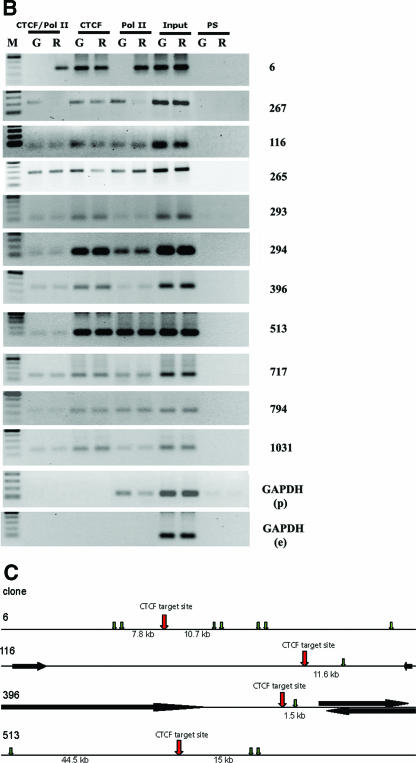
FIG. 5.
Genome-wide interaction between CTCF and LS Pol II. (A) ChIP-on-ChIP hybridization analysis revealing the simultaneous presence of CTCF and LS-Pol II epitopes genome-wide. DNA samples from the standard ChIP or serial ChIP assays from proliferating and resting mouse NIH 3T3 cells were prepared and hybridized to CTCF target site microarrays. Hybridization signals are expressed in relative fluorescence units; the results of analyses are presented in scatter plots as follows. (a) Comparison of hybridization data between serial ChIP samples CTCF-Pol II and preimmune serum-Pol II in resting cells. (b) Comparison of hybridization data between serial ChIP samples CTCF-Pol II and preimmune serum-Pol II in growing cells. (c) Comparison of the CTCF ChIP with the CTCF-Pol II serial ChIP signals in resting cells. (d) Comparison of the CTCF ChIP with the CTCF-Pol II serial ChIP signals in growing cells. (e) Comparison between serial ChIP CTCF-Pol II samples in resting and growing cells. (f) Comparison between single CTCF ChIP/CTCF ChIP signals in resting and growing cells. (B) Analysis of the 11 sequences in growing (G) or resting (R) NIH 3T3 identified by screening of the CTCF target site microarrays demonstrating the simultaneous presence of CTCF and LS-Pol II. Proliferating and resting mouse NIH 3T3 cells were used to perform the standard ChIP or serial ChIP assays. The antibodies used in ChIP and serial ChIP assays are indicated above the corresponding lanes as follows: CTCF/Pol II, serial ChIP with the anti-CTCF antibody, followed by the anti-LS Pol II antibody; CTCF, ChIP with the anti-CTCF antibody; Pol II, ChIP with the anti-LS Pol II antibody (Pol II); PS, ChIP with the preimmune serum. Input, DNA from NIH 3T3 cell lysates. DNA prepared from these samples was amplified using corresponding pairs of primers as described in Materials and Methods and Table 1, and resolved by a 1% agarose gel. M, DNA marker (100-bp DNA ladder); GAPDH (p), promoter region of GAPDH; GAPDH (e), exon 1 region of GAPDH. (C) A gene map depicting the location of transcriptional units of identified genes (black arrows) or ESTs (green arrows). The numbers below each row indicate the distance between the CTCF target site and the closest known transcriptional unit. Additional sequences are described in Table 3.
Having established the background levels for hybridizations, we examined the CTCF-LS Pol II serial ChIP samples from both resting and growing NIH 3T3 cells. The analysis of hybridizations revealed highly specific signals, with at least eightfold enrichment over the preimmune serum-LS Pol II serial ChIP samples (Fig. 5A, panels c and d). This is a conservative estimate as multiplex PCR of the original serial ChIP samples revealed minimally a 10-fold enrichment (data not shown). Of note, in this and all subsequent analyses, all sequences harboring repeat elements were excluded to avoid ambiguity.
Next we evaluated the serial CTCF-LS Pol II ChIP samples with CTCF occupancy, as determined by the single CTCF ChIP samples. As shown in Fig. 5A, panels c and d, in both resting and growing NIH 3T3 cells, only a subpopulation of CTCF target sites, approximately 10%, was pulled down with the LS Pol II antibody.
To characterize these sequences further, we compared the hybridization signals between serial ChIP samples derived from resting and growing NIH 3T3 cells. A summary of the results of the serial ChIP assay in growing and resting cells is given in Fig. 5A, panels e and f, and Tables 2 and 3. The scatter plot analyses reveal that while a majority of the sequences interact with both LS Pol II and CTCF in resting cells and growing cells, a subset of the sequences were present in the serial ChIP material from primarily resting cells. The finding that a subpopulation of CTCF target sites is occupied by CTCF and LS Pol II in only resting cells may be linked to the nature of CTCF as an inhibitor of cell growth and proliferation (47, 48, 59). It is also in agreement with the fact that CTCF interacts with low-affinity sites just downstream of each of the three MYC promoters in only resting B cells (V. Lobanenkov et al., unpublished data). The hypothesis that CTCF may sequester LS Pol II at such sites and thus support the establishment and maintenance of transcriptional repression or pausing states is further examined in Discussion.
TABLE 2.
Summary of number of intragenic, intergenic, and unidentified sequences interacting with both CTCF and the LS Pol II in growing and resting cellsa
| Interaction location | Intra | Inter | Unident |
|---|---|---|---|
| Growing and resting cells | 5 | 4 | 10 |
| Growing cells | 1 | 0 | 0 |
| Resting cells | 0 | 1 | 5 |
Intra, intragenic; inter, intergenic; and unident, unidentified.
TABLE 3.
Summary of the identified CTCF target sites in growing and resting cells
| Target site | Clone no.a | Gene | Location | Neighboring gene(s)/product
|
|
|---|---|---|---|---|---|
| Upstream | Downstream | ||||
| Intronic/exonic CTCF | 265 | GDP-mannose dehydratase related | Intron 7 | FOXc1 | MAP/CAM kinase |
| 294 | Putative prostate cancer suppressor | Intron 1 | Gene similar to cyclophilin A pseudogene | 60S ribosomal protein pseudogene | |
| 717 | Ahi1 isoform 1 | Exon 20 | Myeloblastis oncogene | ||
| 794 | Intron 3 | HnRNP-related protein | |||
| 1031 | Krüppel-related Zn finger | Intron 1 | GAPDH pseudogene | Zinc finger protein 140 | |
| 267* | Ring finger protein 144 | Intron 3 | NADH ubiquinone oxidoreductase-related protein | Mo cofactor biosynthesis-related protein | |
| Intergenic CTCF | 6** | 60S acidic ribosomal protein | Cbp/p300-interacting transactivator | ||
| 116 | Basic helix-loop-helix | ADP ribosylation-related protein | |||
| 293 | Adenomatosis polyposis coli binding protein; Dnmt3b | ||||
| 396 | Membrane-associated guanylate kinase superfamily | ||||
| 513 | Cadherin-like protein | ||||
*, present in only growing cells; **, present in only resting cells.
The simultaneous binding of CTCF and LS Pol II to the 11 identified targets in growing or resting NIH 3T3 cells was further confirmed by ChIP and serial ChIP. In these experiments, DNA material retained by IP with the anti-CTCF, anti-LS Pol II (single ChIP) or subsequent IP with the immobilized anti-CTCF antibody and then with the anti-LS Pol II antibody (serial ChIP) was amplified with the primers specific for each target. In almost all cases, the DNA sequences were precipitated individually by the anti-CTCF and anti-LS Pol II antibodies and also by both antibodies in serial ChIP assays (see below). The intensities of the signals in these assays differed, which may reflect the differences of the individual targets in the affinities to CTCF-LS Pol II. These experiments confirm the simultaneous presence of CTCF and LS Pol II on the identified microarray sites in growing and resting cells (Fig. 5B). In agreement with the microarray hybridization data, sequence 267 was not present in the DNA sample immunoprecipitated with the anti-LS Pol II antibody or with both antibodies from the resting cells. Similarly, sequence 6 was not present in the DNA sample immunoprecipitated with these antibodies from growing cells. Thus, although both of these sites are occupied by CTCF in growing and resting cells, the interaction with the LS Pol II at these sites may depend on the functional state of the cells.
The same ChIP samples were subjected to amplification with primers designed to overlap the TATA box within the promoter region of the mouse GAPDH gene, which served as a control for LS Pol II loading. The PCR products could be seen in only the samples precipitated with the anti-LS Pol II antibodies prepared from both resting and growing cells. No amplification was seen when PS was used for precipitation and when primers from a region from exon 1 of the mouse GAPDH gene lacking CTCF binding sites were employed for PCR.
Fifteen of the 26 different sequences that interact with both CTCF and LS Pol II could not be identified in the mouse genome database (http://www.ensembl.org/Mus_musculus/),which currently contains almost exclusively euchromatic sequence (Table 2). Taken together with the absence of known expressed sequence tags (ESTs), this observation points to a heterochromatic origin of these sequences. The striking conclusion that nontranscribed sequences nonetheless interact with LS Pol II can be extended to several intergenic sequences. Figure 5C identifies four such clones that contain a CTCF target site that is pulled down with the LS Pol II antibody. In all of these instances, there are one or several ESTs separated from the CTCF target site by 1.5 to 15 kb. This observation prompts the proposal that CTCF recruits Pol II to a subset of the CTCF target sites and that these complexes remain intact until the signal for the release of Pol II is received. We speculate that posttranslational modifications of CTCF (29, 67) may lead to the release of the Pol II, with ensuing activation of transcription from neighboring cryptic promoters.
DISCUSSION
In this report, we have investigated the interaction between CTCF and Pol II and discovered that LS Pol II can be physically associated with CTCF. Both proteins are ubiquitous and essential for cell viability (32; Lobanenkov et al., unpublished). While the LS Pol II is an important subunit of the Pol II complex, which is an essential component of the transcriptional machinery, CTCF is a multivalent, versatile factor that activates or represses gene transcription in various modes, including chromatin insulation. Given the biological importance of the two proteins, one can envisage that their interaction may have important functional implications.
In this study, we first documented that CTCF is a component of the Pol II protein complex and this association is specific since CTCF is not part of the TFIIH complex. Next, in a series of IP experiments, the LS Pol II was identified as the protein interacting with CTCF. The specific nature of this interaction is evident because (i) CTCF can be precipitated from cell lysates with at least three different anti-LS Pol II antibodies; (ii) conversely, the anti-CTCF antibody can coimmunoprecipitate both isoforms of LS Pol II; (iii) the CTCF-LS Pol II co-IP reaction can be blocked by peptide N-20, originally employed to raise the anti-LS Pol II antibody N-20; (iv) neither CTCF nor LS Pol II was retained when preimmune serum was used in co-IP; (v) CTCF was not detected in the co-IP reactions with a large panel of the antibodies against various proteins, nuclear and cytoplasmic; (vi) CTCF and LS Pol II could be immunoprecipitated from cell extracts treated with DNase; and (vii) peptides matching LS Pol II were detected in the high-molecular-weight bands obtained after co-IP with the anti-CTCF antibody. Notably, CTCF-LS Pol II complexes were detected in various cell types, thus pointing to the “universal” functions for the association of these two ubiquitous proteins.
Further in vitro binding analyses revealed that the interaction between CTCF and LS Pol II is direct because CTCF and LS Pol II produced and purified from the baculoviral and bacterial systems are still able to interact in vitro. Since this interaction occurs via its C-terminal portion, CTCF can be subjected to regulatory influences while bound to DNA. This may be achieved, for example, by reversible posttranslational modifications of CTCF. We previously reported the presence of several functional phosphorylation sites for protein kinase CK2 within the C-terminal domain (29), and our preliminary results show that the phosphorylation of the C-terminal domain in vitro with protein kinase CK2 decreases the binding of the LS Pol II (Chernukhin and Shamsuddin, unpublished). It is therefore conceivable that the phosphorylation of CTCF may be important for the regulation of the CTCF-LS Pol II interaction in vivo. Similar mechanisms may be involved in the regulation of CTCF and Kaiso interaction, which also occurs via the C-terminal domain of CTCF (13). On the other hand, these mechanisms may differ from those relying on CTCF interactions with YB-1 (10, 28), Sin3A (37), and the helicase protein CHD8 (22), which occur through the DNA binding zinc finger domain of CTCF.
The interaction between CTCF and LS Pol II was reinforced by the finding that CTCF and the LS Pol II significantly colocalize in the nucleus, which indicates that the subpopulations of these two proteins may be involved in the execution of the same biological processes. We hypothesized that if this were the case, then CTCF and LS Pol II could be found in vivo in association with the same functional element of DNA (insulator or promoter) via a CTCF binding site. To assess the simultaneous presence of two proteins at the same DNA sequence, we developed a serial ChIP assay. In this assay, the DNA-protein complexes are first passed through the matrix linked with the antibody against one of the partner proteins. This modification has advantages over subsequent IP in solution as only protein complexes, which are retained on the matrix, not the unwanted free antibodies, will be involved in the subsequent IP with the second partner antibody. The analysis of the in vivo occupancies of the CTCF binding sites at the β-globin insulator by CTCF and LS Pol II, using this approach, revealed that in nondifferentiated HD3 cells with no globin expression, CTCF and LS Pol II are associated with the β-globin insulator. Since FII is positioned more than 10 kb upstream of the transcription start site and the likelihood of precipitation of the same fragment with the two individual antibodies is very small, the serial ChIP assay data also point out that LS Pol II is associated with CTCF bound to the FII site.
Although these results suggested that the association of CTCF with LS Pol II depends on functional CTCF target sites, possible indirect or nonspecific effects still could not be ruled out. The cotransfection of plasmids containing the wild-type and CTCF target site-mutated H19 ICRs, followed by ChIP assays, showed that both the anti-CTCF and anti-Pol II antibodies retained the wild-type but not the mutated allele (Fig. 3C). Furthermore, in the chromatin context, only the wild-type CTCF binding site, N-Myc, but not its mutated variant deficient for CTCF binding, could be precipitated by the anti-CTCF and anti-LS Pol II antibodies, individually or in a serial ChIP format (Fig. 4D and E). These results further confirm that LS Pol II is associated with CTCF via the CTSs.
A genome-wide screen of the CTS microarray subsequently revealed that the majority of the sequences that were pulled down with both the CTCF and the LS Pol II antibodies are not transcribed. We interpret this information to mean that a CTCF-LS Pol II complex is recruited to these CTCF target sites in a transcription-independent manner. In some instances where the sequences could be identified, those CTCF target sites map in the relative vicinity of ESTs, i.e., at a distance from 1.5 to 15 kb (Fig. 5C). Given that the sonicated fragments were less than 1 kb on average, it is less likely that the low-abundant ESTs signified an interaction between CTCF and LS Pol II in a transcription-dependent manner. Similarly, it remains intriguing how LS Pol II can be associated with the murine β-globin locus positioned far from promoters in a transcription-independent manner (23). We speculate that such association could be mediated by CTCF. The formation of the locus control region-promoter loops known to exist within the β-globin locus (55) may be responsible for bringing LS Pol-II and CTCF in direct physical proximity.
A common theme of this report is that the abundant and ubiquitous CTCF may provide a novel pathway to recruit transcription complexes to particular targets. In this scenario, noncoding transcripts known to originate throughout the genome might be transcribed in an enhancer-independent manner. We thus hypothesize that the release of the LS Pol II from a DNA-bound CTCF complex might transcriptionally activate nearby cryptic promoters, or alternatively, the CTCF site itself could act as a promoter in a certain genomic context. As a result, the genome may be represented by low-abundant noncoding transcripts in a manner that is dictated by CTCF target site occupancy. Indeed, we demonstrate here that only the wild-type version of the CTCF binding site (N-Myc) fused with the luciferase reporter gene was able to activate the reporter gene efficiently. As in vivo binding of CTCF and LS Pol II to the wild-type N-Myc site was verified by ChIP and serial ChIP assays, it is possible that the binding of the CTCF-LS Pol II complex to this site was sufficient to activate the transcription from this “artificial” promoter. Another and not mutually exclusive possibility is that the DNA-bound CTCF-LS Pol II complex might be mistaken for a promoter by nearby enhancers. Such promoter decoys have been proposed to provide one of several essential mechanisms by which chromatin insulators block enhancer-promoter communications (18).
The identification of intronic/exonic sequences that simultaneously interact with CTCF and LS Pol II hints at another possibility: the tracking Pol II encounters the DNA-bound CTCF stalling the transcriptional elongation process. Such pause elements have previously been observed downstream of each of the MYC promoters (33, 56). Intriguingly, these pause elements map to or are identical with CTCF target sites, which are occupied in growth-arrested cells (45). It is therefore conceivable that the dissociation of the CTCF-DNA complex during G0/G1 transition might subsequently release LS Pol II to complete the transcriptional elongation process. The commonality of such a scenario is indicated by our demonstrations here that CTCF is dissociated from a significant subpopulation of CTCF target sites in growing cells but not in resting NIH 3T3 cells.
Although the CTS microarray has been a valuable tool in this investigation, its limitations should be acknowledged. First, this microarray represented only a relatively small subpopulation of CTCF binding sites, which is approximately 5 to 7% of all potential CTCF target sites (40). This number, however, may be lower (∼0.75 to 1%) if the criteria described by Vetchinova et al. are used for assessment (61). Second, some of the previously identified and characterized CTSs (e.g., MYC and H19 ICR) could not be assessed in the screening as they were not present on the microarray (40). Instead, a representative panel of these CTSs was investigated separately in this report. Third, the origin of cells used for microarray experimentations (NIH 3T3 fibroblasts) should be taken into consideration as CTSs in NIH 3T3 and fetal liver (source for microarray CTS) may have different occupancy patterns and only partially overlap.
Our observations indicate that only a relatively small subpopulation (approximately 10%) of CTSs, as determined in the microarray screening, can simultaneously interact with CTCF and LS Pol II. However, given the abundance of these two proteins in the nucleus, it is likely that the number of specific CTCF-LS Pol II complexes associated with CTSs may be sufficient to meet the functional requirements of cells. On the other hand, as the immunostaining reveals (Fig. 2), the actual number of CTCF-LS Pol II complexes in the nucleus may be greater as some of these complexes may not be linked to the CTSs. Of note, the association of CTCF with other interacting proteins (e.g., helicase protein CHD8 and YB-1) has also been shown to be partial (22; S. Shamsuddin and F. Docquier, unpublished data). We hypothesize that different subpopulations of CTCF may generate different specific complexes with various protein partners as a means of creating molecular and functional diversity.
Based on the experimental data presented in this study, we suggest several possible functions of CTCF interaction with LS Pol II. First, CTCF may engage RNA polymerase II to potentially generate the “storage” of proteins necessary for transcription in promoter-proximal positions, thereby modulating transcription in response to a stimulus. Data in Fig. 3A demonstrating the presence of CTCF and LS Pol II at the CTCF binding site at the insulator in nondifferentiated chicken HD3 cells and the absence of CTCF and LS Pol II at this site in globin-producing differentiated HD3 cells support this case. Second, CTCF may “piggyback” LS Pol II to a certain set of DNA targets to establish an appropriate configuration for pausing of transcriptional elongation once a CTCF target site has been recognized. Alternatively, CTCF may play a role of a functional equivalent of TBP, allowing accurate initiation of transcription at some promoters. In our model system, just the presence of a single CTCF binding site was sufficient to activate transcription from the adjacent luciferase gene (Fig. 4B). Finally, we have here also discussed the potential roles of CTCF-LS Pol II complexes in the expression of intergenic, noncoding transcripts and chromatin insulation. Irrespective of these considerations, the marriage between a versatile chromatin insulator protein, CTCF, and a Pol II enzyme complex constitutes a novel angle on the genome-wide regulation of gene transcription.
Acknowledgments
We thank S. Muller and J. Dadoune for anti-histone H2A and histone H3 antibodies. We are also grateful to M. Metodiev, A. Harrison, A. Akoulitchev, and A. Ramadass for helpful discussions and P. O'Toole for assistance with confocal microscopy. We gratefully acknowledge the assistance of A. Isaksson and the Wallenberg microarray platform at the Rudbeck laboratory.
This research was supported by the Association for International Cancer Research (I.C. and E.K.), the Breast Cancer Campaign (F.D. and E.K.), the Medical Research Council (D.F. and E.K.), the Research Promotion Fund from University of Essex (E.K.), the National Institutes of Health grant CA103867 (C.-M.C.), a scholarship from the Malaysian Government and Fundamental Research Grant Scheme from the Malaysian Government (S.S.), the Swedish Science Research Council (R.O.), the Juvenile Diabetes Research Foundation International (R.O.), the Swedish Cancer Research Foundation (R.O.), the Swedish Pediatric Cancer Foundation (R.O.), the Wallenberg and Lundberg Foundations (R.O.), Stiftelsen Wenner-Grenska Samfundet (R.O.), and the Intramural Research Program of the NIH, NIAID (V.L., D.L., and Y.-W.K.).
Footnotes
Published ahead of print on 8 January 2007.
REFERENCES
- 1.Acker, J., M. de Graaff, I. Cheynel, V. Khazak, C. Kedinger, and M. Vigneron. 1997. Interactions between the human RNA polymerase II subunits. J. Biol. Chem. 272:16815-16821. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. [DOI] [PubMed] [Google Scholar]
- 3.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 4.Besse, S., M. Vigneron, E. Pichard, and F. Puvion-Dutilleul. 1995. Synthesis and maturation of viral transcripts in herpes simplex virus type 1 infected HeLa cells: the role of interchromatin granules. Gene Expr. 4:143-161. [PMC free article] [PubMed] [Google Scholar]
- 5.Beug, H., G. Doederlein, C. Freudenstein, and T. Graf. 1982. Erythroblast cell lines transformed by a temperature-sensitive mutant of avian erythroblastosis virus: a model system to study erythroid differentiation in vitro. J. Cell. Physiol. Suppl. 1:195-207. [DOI] [PubMed] [Google Scholar]
- 6.Bregman, D. B., L. Du, S. van der Zee, and S. L. Warren. 1995. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burcin, M., R. Arnold, M. Lutz, B. Kaiser, D. Runge, F. Lottspeich, G. N. Filippova, V. V. Lobanenkov, and R. Renkawitz. 1997. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol. Cell. Biol. 17:1281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butcher, D. T., D. N. Mancini-DiNardo, T. K. Archer, and D. I. Rodenhiser. 2004. DNA binding sites for putative methylation boundaries in the unmethylated region of the BRCA1 promoter. Int. J. Cancer 111:669-678. [DOI] [PubMed] [Google Scholar]
- 9.Chernukhin, I. V., and E. M. Klenova. 2000. A method of immobilization on the solid support of complex and simple enzymes retaining their activity. Anal. Biochem. 280:178-181. [DOI] [PubMed] [Google Scholar]
- 10.Chernukhin, I. V., S. Shamsuddin, A. F. Robinson, A. F. Carne, A. Paul, A. I. El-Kady, V. V. Lobanenkov, and E. M. Klenova. 2000. Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1, and the multivalent zinc finger factor, CTCF. J. Biol. Chem. 275:29915-29921. [DOI] [PubMed] [Google Scholar]
- 11.Costes, S. V., D. Daelemans, E. H. Cho, Z. Dobbin, G. Pavlakis, and S. Lockett. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86:3993-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahmus, M. E. 1996. Phosphorylation of mammalian RNA polymerase II. Methods Enzymol. 273:185-193. [DOI] [PubMed] [Google Scholar]
- 13.Defossez, P. A., K. F. Kelly, G. J. Filion, R. Perez-Torrado, F. Magdinier, H. Menoni, C. L. Nordgaard, J. M. Daniel, and E. Gilson. 2005. The human enhancer blocker CTC-binding factor interacts with the transcription factor Kaiso. J. Biol. Chem. 280:43017-43023. [DOI] [PubMed] [Google Scholar]
- 14.Delgado, M. D., I. V. Chernukhin, A. Bigas, E. M. Klenova, and J. Leon. 1999. Differential expression and phosphorylation of CTCF, a c-myc transcriptional regulator, during differentiation of human myeloid cells. FEBS Lett. 444:5-10. [DOI] [PubMed] [Google Scholar]
- 15.El-Kady, A., and E. Klenova. 2005. Regulation of the transcription factor, CTCF, by phosphorylation with protein kinase CK2. FEBS Lett. 579:1424-1434. [DOI] [PubMed] [Google Scholar]
- 16.Filippova, G. N., S. Fagerlie, E. M. Klenova, C. Myers, Y. Dehner, G. Goodwin, P. E. Neiman, S. J. Collins, and V. V. Lobanenkov. 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franklin, G. C., M. Donovan, G. I. Adam, L. Holmgren, S. Pfeifer-Ohlsson, and R. Ohlsson. 1991. Expression of the human PDGF-B gene is regulated by both positively and negatively acting cell type-specific regulatory elements located in the first intron. EMBO J. 10:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geyer, P. K. 1997. The role of insulator elements in defining domains of gene expression. Curr. Opin. Genet. Dev. 7:242-248. [DOI] [PubMed] [Google Scholar]
- 19.Gross, P., and T. Oelgeschlager. 2006. Core promoter-selective RNA polymerase II transcription. Biochem. Soc. Symp. 73:225-236. [DOI] [PubMed] [Google Scholar]
- 20.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Hay, F. C., O. M. R. Westwood, P. N. Nelson, and L. Hudson. 2002. Practical immunology, 4th ed. Blackwell Science, Malden, MA.
- 22.Ishihara, K., M. Oshimura, and M. Nakao. 2006. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell 23:733-742. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, K. D., J. A. Grass, C. Park, H. Im, K. Choi, and E. H. Bresnick. 2003. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol. Cell. Biol. 23:6484-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C. F. Qi, A. Wolffe, R. Ohlsson, and V. V. Lobanenkov. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10:853-856. [DOI] [PubMed] [Google Scholar]
- 25.Kershnar, E., S. Y. Wu, and C. M. Chiang. 1998. Immunoaffinity purification and functional characterization of human transcription factor IIH and RNA polymerase II from clonal cell lines that conditionally express epitope-tagged subunits of the multiprotein complexes. J. Biol. Chem. 273:34444-34453. [DOI] [PubMed] [Google Scholar]
- 26.Klenova, E., I. Chernukhin, T. Inoue, S. Shamsuddin, and J. Norton. 2002. Immunoprecipitation techniques for the analysis of transcription factor complexes. Methods 26:254-259. [DOI] [PubMed] [Google Scholar]
- 27.Klenova, E., and R. Ohlsson. 2005. Poly(ADP-ribosyl)ation and epigenetics. Is CTCF PARt of the plot? Cell Cycle 4:96-101. [DOI] [PubMed] [Google Scholar]
- 28.Klenova, E., A. C. Scott, J. Roberts, S. Shamsuddin, E. A. Lovejoy, S. Bergmann, V. J. Bubb, H. D. Royer, and J. P. Quinn. 2004. YB-1 and CTCF differentially regulate the 5-HTT polymorphic intron 2 enhancer which predisposes to a variety of neurological disorders. J. Neurosci. 24:5966-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klenova, E. M., I. V. Chernukhin, A. El-Kady, R. E. Lee, E. M. Pugacheva, D. I. Loukinov, G. H. Goodwin, D. Delgado, G. N. Filippova, J. Leon, H. C. Morse III, P. E. Neiman, and V. V. Lobanenkov. 2001. Functional phosphorylation sites in the C-terminal region of the multivalent multifunctional transcriptional factor CTCF. Mol. Cell. Biol. 21:2221-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klenova, E. M., H. C. Morse III, R. Ohlsson, and V. V. Lobanenkov. 2002. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin. Cancer Biol. 12:399-414. [DOI] [PubMed] [Google Scholar]
- 31.Klenova, E. M., R. H. Nicolas, H. F. Paterson, A. F. Carne, C. M. Heath, G. H. Goodwin, P. E. Neiman, and V. V. Lobanenkov. 1993. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 13:7612-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobor, M. S., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261-275. [DOI] [PubMed] [Google Scholar]
- 33.Krumm, A., T. Meulia, M. Brunvand, and M. Groudine. 1992. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 6:2201-2213. [DOI] [PubMed] [Google Scholar]
- 34.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 35.Kuzmin, I., L. Geil, L. Gibson, T. Cavinato, D. Loukinov, V. Lobanenkov, and M. I. Lerman. 2005. Transcriptional regulator CTCF controls human interleukin 1 receptor-associated kinase 2 promoter. J. Mol. Biol. 346:411-422. [DOI] [PubMed] [Google Scholar]
- 36.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 37.Lutz, M., L. J. Burke, G. Barreto, F. Goeman, H. Greb, R. Arnold, H. Schultheiss, A. Brehm, T. Kouzarides, V. Lobanenkov, and R. Renkawitz. 2000. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 28:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lutz, M., L. J. Burke, P. LeFevre, F. A. Myers, A. W. Thorne, C. Crane-Robinson, C. Bonifer, G. N. Filippova, V. Lobanenkov, and R. Renkawitz. 2003. Thyroid hormone-regulated enhancer blocking: cooperation of CTCF and thyroid hormone receptor. EMBO J. 22:1579-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malik, S., and R. G. Roeder. 2005. Dynamic regulation of pol II transcription by the mammalian mediator complex. Trends Biochem. Sci. 30:256-263. [DOI] [PubMed] [Google Scholar]
- 40.Mukhopadhyay, R., W. Yu, J. Whitehead, J. Xu, M. Lezcano, S. Pack, C. Kanduri, M. Kanduri, V. Ginjala, A. Vostrov, W. Quitschke, I. Chernukhin, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 14:1594-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolas, R. H., G. Partington, G. N. Major, B. Smith, A. F. Carne, N. Huskisson, and G. Goodwin. 1991. Induction of differentiation of avian erythroblastosis virus-transformed erythroblasts by the protein kinase inhibitor H7: analysis of the transcription factor EF1. Cell Growth Differ. 2:129-135. [PubMed] [Google Scholar]
- 42.Ohlsson, R., R. Renkawitz, and V. Lobanenkov. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17:520-527. [DOI] [PubMed] [Google Scholar]
- 43.Pant, V., S. Kurukuti, E. Pugacheva, S. Shamsuddin, P. Mariano, R. Renkawitz, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 24:3497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pant, V., P. Mariano, C. Kanduri, A. Mattsson, V. Lobanenkov, R. Heuchel, and R. Ohlsson. 2003. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 17:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pérez-Juste, G., S. Garcia-Silva, and A. Aranda. 2000. An element in the region responsible for premature termination of transcription mediates repression of c-myc gene expression by thyroid hormone in neuroblastoma cells. J. Biol. Chem. 275:1307-1314. [DOI] [PubMed] [Google Scholar]
- 46.Prioleau, M. N., P. Nony, M. Simpson, and G. Felsenfeld. 1999. An insulator element and condensed chromatin region separate the chicken beta-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 18:4035-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qi, C. F., A. Martensson, M. Mattioli, R. Dalla-Favera, V. V. Lobanenkov, and H. C. Morse III. 2003. CTCF functions as a critical regulator of cell-cycle arrest and death after ligation of the B cell receptor on immature B cells. Proc. Natl. Acad. Sci. USA 100:633-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasko, J. E., E. M. Klenova, J. Leon, G. N. Filippova, D. I. Loukinov, S. Vatolin, A. F. Robinson, Y. J. Hu, J. Ulmer, M. D. Ward, E. M. Pugacheva, P. E. Neiman, H. C. Morse III, S. J. Collins, and V. V. Lobanenkov. 2001. Cell growth inhibition by the multifunctional multivalent zinc-finger factor CTCF. Cancer Res. 61:6002-6007. [PubMed] [Google Scholar]
- 49.Renaud, S., D. Loukinov, F. T. Bosman, V. Lobanenkov, and J. Benhattar. 2005. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 33:6850-6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saitoh, N., A. C. Bell, F. Recillas-Targa, A. G. West, M. Simpson, M. Pikaart, and G. Felsenfeld. 2000. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 19:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Saunders, A., L. J. Core, and J. T. Lis. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7:557-567. [DOI] [PubMed] [Google Scholar]
- 53.Shi, S. R., M. E. Key, and K. L. Kalra. 1991. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J. Histochem. Cytochem. 39:741-748. [DOI] [PubMed] [Google Scholar]
- 54.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 55.Splinter, E., H. Heath, J. Kooren, R. J. Palstra, P. Klous, F. Grosveld, N. Galjart, and W. de Laat. 2006. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 20:2349-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strobl, L. J., and D. Eick. 1992. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 11:3307-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas, M. C., and C. M. Chiang. 2006. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41:105-178. [DOI] [PubMed] [Google Scholar]
- 58.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12:3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torrano, V., I. Chernukhin, F. Docquier, V. D'Arcy, J. Leon, E. Klenova, and M. D. Delgado. 2005. CTCF regulates growth and erythroid differentiation of human myeloid leukemia cells. J. Biol. Chem. 280:28152-28161. [DOI] [PubMed] [Google Scholar]
- 60.Torrano, V., J. Navascues, F. Docquier, R. Zhang, L. J. Burke, I. Chernukhin, D. Farrar, J. Leon, M. T. Berciano, R. Renkawitz, E. Klenova, M. Lafarga, and M. D. Delgado. 2006. Targeting of CTCF to the nucleolus inhibits nucleolar transcription through a poly(ADP-ribosyl)ation-dependent mechanism. J. Cell Sci. 119:1746-1759. [DOI] [PubMed] [Google Scholar]
- 61.Vetchinova, A. S., S. B. Akopov, I. P. Chernov, L. G. Nikolaev, and E. D. Sverdlov. 2006. Two-dimensional electrophoretic mobility shift assay: identification and mapping of transcription factor CTCF target sequences within an FXYD5-COX7A1 region of human chromosome 19. Anal. Biochem. 354:85-93. [DOI] [PubMed] [Google Scholar]
- 62.Vostrov, A. A., and W. W. Quitschke. 1997. The zinc finger protein CTCF binds to the APBβ domain of the amyloid β-protein precursor promoter. Evidence for a role in transcriptional activation. J. Biol. Chem. 272:33353-33359. [DOI] [PubMed] [Google Scholar]
- 63.Vuillard, L., T. Rabilloud, and M. E. Goldberg. 1998. Interactions of non-detergent sulfobetaines with early folding intermediates facilitate in vitro protein renaturation. Eur. J. Biochem. 256:128-135. [DOI] [PubMed] [Google Scholar]
- 64.Wu, S. Y., and C. M. Chiang. 1998. Properties of PC4 and an RNA polymerase II complex in directing activated and basal transcription in vitro. J. Biol. Chem. 273:12492-12498. [DOI] [PubMed] [Google Scholar]
- 65.Wu, S. Y., E. Kershnar, and C. M. Chiang. 1998. TAFII-independent activation mediated by human TBP in the presence of the positive cofactor PC4. EMBO J. 17:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young, R. A. 1991. RNA polymerase II. Annu. Rev. Biochem. 60:689-715. [DOI] [PubMed] [Google Scholar]
- 67.Yu, W., V. Ginjala, V. Pant, I. Chernukhin, J. Whitehead, F. Docquier, D. Farrar, G. Tavoosidana, R. Mukhopadhyay, C. Kanduri, M. Oshimura, A. P. Feinberg, V. Lobanenkov, E. Klenova, and R. Ohlsson. 2004. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 36:1105-1110. [DOI] [PubMed] [Google Scholar]
- 68.Yusufzai, T. M., H. Tagami, Y. Nakatani, and G. Felsenfeld. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13:291-298. [DOI] [PubMed] [Google Scholar]