Abstract
The core promoter is a critical DNA element required for accurate transcription and regulation of transcription. Several core promoter elements have been previously identified in eukaryotes, but those cannot account for transcription from most RNA polymerase II-transcribed genes. Additional, as-yet-unidentified core promoter elements must be present in eukaryotic genomes. From extensive analyses of the hepatitis B virus X gene promoter, here we identify a new core promoter element, XCPE1 (the X gene core promoter element 1), that drives RNA polymerase II transcription. XCPE1 is located between nucleotides −8 and +2 relative to the transcriptional start site (+1) and has a consensus sequence of G/A/T-G/C-G-T/C-G-G-G/A-A-G/C+1-A/C. XCPE1 shows fairly weak transcriptional activity alone but exerts significant, specific promoter activity when accompanied by activator-binding sites. XCPE1 is also found in the core promoter regions of about 1% of human genes, particularly in poorly characterized TATA-less genes. Our in vitro transcription studies suggest that the XCPE1-driven transcription can be highly active in the absence of TFIID because it can utilize either free TBP or the complete TFIID complex. Our findings suggest the possibility of the existence of a TAF1 (TFIID)-independent transcriptional initiation mechanism that may be used by a category of TATA-less promoters in higher eukaryotes.
Transcriptional initiation is a key regulatory step of gene expression. Regulation of transcriptional initiation is carried out by a complex network of interactions between cis-acting DNA elements and DNA-binding transcription factors and also among many transcription factors, including sequence-specific DNA-binding transcriptional regulators, coregulators, and general (basal) transcriptional machinery (i.e., general transcription factors [GTFs] and RNA polymerase [pol]) (22, 26, 33-35, 41, 51, 54).
The DNA regions that specify the transcriptional program of each gene contain two functionally distinct regions, the core promoter (for reviews, see references 9, 61, and 62) and the regulatory regions (e.g., enhancers, silencers, etc.). The core promoter is the minimum essential region necessary for accurate transcription. The core promoter typically comprises about 40 nucleotides (nt) and contains functional subregions called core promoter elements. When transcription starts, the core promoter elements are recognized by some of the GTFs or by other factors that trigger the assembly of a large protein complex that consists of GTFs and RNA polymerase (i.e., the transcriptional preinitiation complex [PIC]) at the core promoter. The transcriptional initiation complex positions RNA polymerase at the correct site and makes it possible to start RNA synthesis accurately. Thus, core promoter elements play an essential role in specifying transcription start sites. Core promoter elements also determine the specific transcriptional properties of each core promoter, dictating (i) which RNA polymerase among the class I, II, and III systems should be used and (ii) which enhancers can act to regulate transcriptional activity, thereby helping to specify a temporal and spatial regulation pattern of a gene. Unlike core promoters, regulatory regions contain binding sites for sequence-specific DNA-binding transcriptional regulators that can control levels of transcription but cannot promote transcriptional initiation by themselves. Some DNA-binding transcriptional regulators (e.g., enhancer-binding factors) can also interact with the components of the general transcription machinery directly and help to control recruitment of RNA polymerase to the vicinity of the core promoters, thereby activating or repressing transcription of a gene. Other DNA-binding transcriptional regulators interact with transcriptional coregulators that modulate the structure of the transcriptional template which normally exists, in vivo, in the form of a highly ordered structure of a DNA-histone complex called chromatin. Changes in chromatin structure result in an increase or decrease of accessibility of the core promoter to the transcriptional machinery, which acts to enhance or repress transcriptional initiation. Thus, the precise temporal and spatial regulation of transcription is accomplished through the regulation of the core promoter activity by the interplay between the regulatory regions and the core promoter. Consequently, characterization of different core promoters and core promoter elements is an integral and essential component of studies of transcriptional regulation.
Several core promoter elements have previously been identified for the RNA polymerase II system. These elements include the TATA box (consensus, TATAWAWR; located about 25 to 30 bp upstream of the transcriptional start site), the initiator (Inr; consensus, YYA+1NWYY for mammals, TCA+1KTY for Drosophila melanogaster; the A residues at position +1 correspond to the start sites), the downstream promoter element (DPE; consensus, RGWYV(T); located between positions +28 and +33), the TFIIB recognition element (BRE; consensus, SSRCGCC; located immediately upstream of some TATA boxes), the motif ten element (MTE; consensus, CSARCSSAACGS; located between positions +18 and +29), and the downstream core element (DCE; consensus, CTTC…CTGT…AGC; located around positions +6 to +35) (2, 9, 31, 39, 49, 61, 62) (degenerate nucleotides are designated according to the IUPAC code; also see Table 2).
TABLE 2.
Estimated frequency of XCPE1 and previously known core promoter elements
| Core promoter element (consensus sequence: position) | % Estimated frequencya |
|---|---|
| XCPE1 (DSGYGGRASM: −8 to +2) | 0.72 (110/15,262) |
| TATA box (TATAWAWR: −31 to −24, 1-bp mismatch allowed) | 19.6 (2,986/15,262) |
| DPE (RGWYVT: +28 to +33 or RGWYV: +28 to +32) | 18.9 (2,886/15,262) or 74.3 (11,337/15,262) |
| MTE (CSARCSSAACGS: +18 to +29) | <0.01 (1/15,262) |
| Initiator (YYANWYY: −2 to +5) | 40.1 (6,115/15,262) |
| XCPE1 + TATA box | 0.02 (3/15,262) |
| XCPE1 + DPE (RGWYVT or RGWYV) | 0.05 (8/15,262) or 0.24 (37/15,262) |
| XCPE1 + TATA box + DPE | <0.01 (1/15,262) |
| TATA box + DPE (RGWYVT or RGWYV) | 3.4 (521/15,262) or 10.1 (1,541/15,262) |
| Initiator + TATA box | 3.87 (591/15,262) |
| Initiator + DPE (RGWYVT or RGWYV) | 3.4 (522/15,262) or 11.7 (1,787/15,262) |
| TATA box + DPE + initiator | 1.00 (152/15,262) |
Number of genes containing the element at correct positions divided by the total number of genes in the database. The regions searched for each element included 10-bp upstream and 5-bp downstream margins. The lower half of the table shows frequencies of genes that have transcription start sites that appear to be driven by the indicated combinations of core promoter elements. Degenerate nucleotides are designated according to IUPAC code. D: A, T, or G; S: C or G; Y: C or T; R: A or G; M: C or A; W: T or A; V: A, C, or G.
The first and best characterized of these elements is the TATA box. In the classic model of transcriptional initiation (6, 12, 78), the first step involves recognition of the TATA box by the multisubunit, general transcription factor TFIID. After binding to the TATA element, TFIID nucleates a PIC that subsequently recruits RNA polymerase II to the promoter. However, this classic model does not explain transcription from TATA-less promoters which drive transcription for as many as 68% of human protein-coding genes (65) or that for 57% of Drosophila protein-coding genes (29).
The presence of a second group of more recently identified core promoter elements (Inr, DPE, and MTE), which have been found in a subset of both TATA-containing and TATA-less promoters, helps to explain transcription from some TATA-less genes. While the DNA sequences of these core promoter elements are not similar to the TATA box, TFIID can recognize and bind directly to Inr and DPE elements by using the TAF (TATA-binding protein [TBP]-associated factor) subunits [7, 72]). Although TFIID does not directly bind to the BRE or the MTE motif, these elements are found only in promoters that contain other TFIID-dependent elements. For example, BRE is found only in a subset of TATA-containing promoters, and MTE is found only in a subset of Inr-containing promoters (30, 39). Thus, the BRE- and MTE-containing promoters are also TFIID dependent. These observations have led to the idea that TFIID is the key factor for promoter recognition and PIC formation not only for TATA-containing promoters but also for Inr-, DPE-, BRE-, and MTE-containing promoters. However, a significant number of genes do not contain any of these core promoter elements (29, 62; this study), and thus, it is not yet clear whether TFIID is universally important for RNA polymerase II promoter recognition. Therefore, it is important to identify additional core promoter elements for those genes that do not fall into the known classes and to determine mechanisms of transcriptional initiation from these novel types of promoters. In this report, we describe the identification and characterization of a novel core promoter element that is present in the hepatitis B virus (HBV) X gene promoter and the cellular promoters and that utilizes both TFIID-dependent and TFIID-independent mechanisms.
Our motivations for characterizing the HBV X gene promoter were twofold. First, the X gene could be a good model system for studying general mechanisms of transcription from a novel type of TATA-less promoter, since the core promoter driving transcription of the X gene contained no recognizable core promoter elements. Second, the X gene may be involved in HBV-induced hepatocarcinogenesis (1, 55). X gene expression has been observed in most, if not all, HBV-infected liver tissues at both the mRNA and protein levels (52, 63). However, the regulatory mechanism of X gene expression is complicated and remains poorly understood. For example, Su et al. (63) detected X protein preferentially in hepatocellular carcinoma and the surrounding parenchyma but only in a small number of parenchymal and malignant cells. Why only some of the malignant cells expressed the X gene or why levels of X gene expression in individual cells varied considerably was not clear. Previous investigators (52, 63) also observed no clear synchronization of X gene expression with other HBV genes, despite the fact that all HBV genes are regulated by the same two HBV enhancers. Therefore, it appeared that the key to understanding the mechanisms of X gene-specific regulation must lie in the interplay between the HBV enhancers and the X gene core promoter. Identification of the core promoter element and determination of some of the transcription factors required for X gene transcription described here provide the first step toward clarification of the X gene regulation.
MATERIALS AND METHODS
HBV plasmids.
All the HBV plasmids used in this study are derived from an HBV strain (subtype adr) reported by Kobayashi and Koike (27). Nucleotide position 1 of this strain corresponds to nt 127 of the strain whose EcoRI site is designated nt 1. The plasmid pHBVX-1, which contains the HBV enhancer 1 core, the X promoter region, and the X open reading frame (ORF) through the poly(A) addition signal, has been described previously (70). The X gene template, pBS-HBXB, was constructed by inserting an HBV DNA fragment (nt 123 [XbaI site] to nt 1858 [BglII site]), which contains the whole HBV enhancer 1, the X promoter region, and the X ORF through the poly(A) addition signal, into the pBluescript vector. A chloramphenicol acetyltransferase (CAT) reporter plasmid, pXStNcCAT, and some of its derivatives have been previously described (15). The other deletion mutants and point mutants of the enhancer 1-X promoter region were constructed by standard molecular biological methods.
In vitro transcription.
Nuclear extracts from HeLa cells and HepG2 cells were prepared essentially as described previously (13). The standard transcription reaction mixtures contained 15 mM HEPES (pH 7.6), 5% glycerol, 6 mM MgCl2, 60 mM KCl, 1 mM dithiothreitol, 75 μg nuclear extract, 0.2 μg of a supercoiled template DNA, and 0.5 mM ribonucleotides. The transcripts were detected by primer extension using a 32P-labeled primer, HB1252/1229, corresponding to nt 1252 to 1229 of the HBV DNA sequence surrounding the first methionine codon of the X protein, or a CAT primer corresponding to the region near the beginning of the CAT protein coding region. For verification of the transcriptional start sites, another HBV primer, “DR2,” corresponding to nt 1473 to 1450, was also used. The resultant 32P-labeled cDNAs were analyzed by electrophoresis on 6% acrylamide gels containing 7 M urea. Sequence ladders were made by the dideoxy termination method using the same sets of primers and template plasmids as used in the primer extension reactions to determine the transcriptional start sites.
Analysis of HBV RNAs from tissue specimens.
Frozen liver tissue samples from HBV-infected patients were homogenized in TRIzol reagent (Invitrogen), and total RNA was extracted according to the manufacturer's instructions. Poly(A)+ RNA was selected using oligo(dT) beads (QIAGEN), and the X mRNA start sites were determined by primer extension analyses.
Site-directed mutagenesis of the X gene core promoter 1.
For extensive mutagenesis of the X core promoter 1, PCRs were performed using pBS-HBXB as the template, with mutated oligonucleotides corresponding to the core promoter region as the primers. After PCR, the template plasmid was selectively removed by DpnI digestion, and the PCR products were introduced into Escherichia coli. The sequences of the mutated plasmids were verified by DNA sequencing.
Antibodies.
To generate anti-human TBP antibody, rabbits were immunized and boosted with recombinant human TBP expressed and purified from E. coli. To affinity purify anti-TBP antibody, the antiserum was first precleared with a glutathione S-transferase (GST) affinity column and then applied to an affinity column made of purified GST-TBP fusion proteins cross-linked to Affi-Gel 10 gel. Anti-human TAF1 antibody was raised by immunizing rabbits with His-tagged protein fragments corresponding to amino acids 1371 to 1629 of human TAF1. The anti-TAF1 antibody was affinity purified using the TAF1 fragment fused to the intein and the chitin-binding domain (IMPACT system; NEB) that was immobilized on chitin beads. Anti-human MED26 was raised by immunizing a rabbit with His-tagged full-length MED26 proteins and affinity purified using maltose binding protein-MED26 fusion proteins cross-linked to Affi-Gel 10 gel.
Immunodepletion analysis.
Affinity-purified antibodies were first bound to protein A-Sepharose beads (Amersham Pharmacia) and then mixed with nuclear extract in D-100 buffer (100 mM KCl, 20% glycerol, 20 mM HEPES [pH 7.6], 0.2 mM EDTA, 2 mM MgCl2) at 4°C for 6 to 8 h. After incubation, the beads were spun down and the supernatant was used for transcription reactions or immunoblot analyses.
Preparation of TBP, TFIID, and mediator complex for in vitro transcription assays.
A nontagged form of human recombinant TBP was expressed by E. coli and purified as described previously (53). To purify endogenous TFIID, HeLa nuclear extract was fractionated with phosphocellulose (P11) as described previously (16, 40). The fraction that was step-eluted from 0.5 M to 1 M KCl (the P1.0 fraction) was used to immunoprecipitate TFIID with an anti-hTAF4 monoclonal antibody. After extensive washing, the TFIID was eluted from the antibody beads by incubation with an excess amount of peptides that correspond to the epitope for the antibody. The eluted TFIID was further dialyzed to remove the peptides. To purify the mediator complex, nuclear extract from the HeLa cell line (57) that expresses FLAG-tagged MED26 was fractionated with P11, and the mediator complex was immunopurified using anti-FLAG antibodies from P.5 (step eluate from 0.3 M to 0.5 M KCL) and P1.0 fractions.
RESULTS
X gene transcription shows two major start sites.
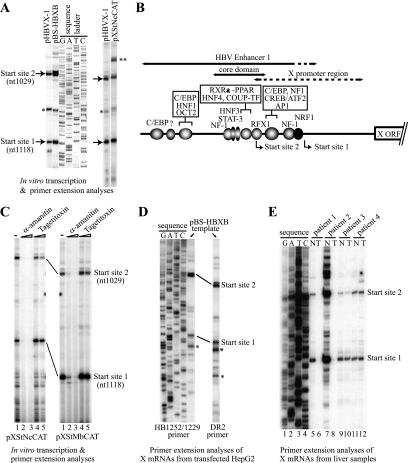
To map the start sites of the X transcripts, we performed in vitro transcription reactions with crude nuclear extracts prepared from HepG2 hepatoblastoma cells or HeLa cells. We used several different HBV template DNAs that contained the complete or core domain of enhancer 1 with or without the X gene coding region/enhancer 2. As shown in Fig. 1A and B, two major start sites (nt 1118 ± 1 and nt 1029 ± 1) were reproducibly observed under any of the conditions tested. The same start sites were also observed in primer extension analyses using two different reverse transcriptases (murine leukemia virus reverse transcriptase and avian myeloblastosis virus reverse transcriptase) and in runoff in vitro transcription assays (data not shown). We call the start site located at nt 1118 ± 1 “start site 1” and the start site located at nt 1029 ± 1 “start site 2.” Start site 1 is consistent with the start site (nt 1117 ± 3) observed by Yaginuma et al. (77) and is very near start sites nt 1123 and nt 1125 reported by Siddiqui et al. (60) and Yaginuma et al. (77), respectively. We recently reported start site 2 for the first time (70), although its signal had apparently been observed previously in tissue culture experiments described by others (60, 77). Previous investigators have also reported additional start sites downstream of start site 1 (17, 18, 60, 71). The reason for the start site difference is not completely clear, but those additional sites may reflect strain (genotype)-specific start sites, incomplete primer extension products, or degradation of the X mRNA and other HBV RNAs.
FIG. 1.
Determination of transcriptional start sites of the HBV X gene mRNA. (A) In vitro transcription analysis of the X gene. In vitro transcription reactions were performed using HeLa (left panel) or HepG2 (right panel) nuclear extract with different wild-type HBV X gene templates (pHBVX-1, pBS-HBXB, or pXStNcCAT) as described in Materials and Methods. By comparison with the sequencing ladder, two major transcriptional start sites, 1118 ± 1 (start site 1) and 1029 ± 1 (start site 2) were identified. An asterisk indicates a band that was not consistently observed. Two asterisks indicate a band that corresponds to a transcript starting from a cryptic (uncharacterized) start site in the pSV00CAT vector. (B) Schematic of the HBV enhancer 1-X promoter region. The enhancer 1, the enhancer 1 core domain, and the previously coarsely mapped X promoter region, as well as reported or potential transcription factor binding sites in enhancer 1, are shown with the two major transcriptional start sites reported in this study. (C) X gene transcription by RNA polymerase II. In vitro transcription of the X gene was carried out using HepG2 nuclear extract and enhancer 1-X promoter templates in the presence or absence of inhibitors. Lane 1, no inhibitor; lane 2, 2 μg/ml α-amanitin; lane 3, 200 μg/ml α-amanitin; lane 4, 0.5 U/ml tagetitoxin; lane 5, 1.5 U/ml tagetitoxin. For structures of pXStNcCAT and pXStMbCAT, see Fig. 2A. (D) X mRNA from transfected HepG2 cells as analyzed by primer extension with two different primers (HB1252/1229 or DR2). An asterisk (*) denotes likely degradation products of the X mRNA (see text). (E) Analysis of X mRNA from HBV-infected liver tissue samples. Poly(A)+ RNA derived from 125 μg of total RNA from each tissue was analyzed by primer extension using DR2 primer. The sequence ladder was made using pBS-HBXB (as the template) and DR2 primer. T, tumor; N, nontumor.
X RNA is transcribed by RNA polymerase II.
The eukaryotic RNA polymerases I, II, and III each function with a unique set of general (basal) transcription factors. pol II is the major polymerase for the transcription of protein-coding genes. However, under some nonphysiological in vitro conditions or under certain physiological conditions, pol III can also transcribe genes that normally are transcribed by pol II. To determine which RNA polymerase is responsible for transcription of the HBV X gene, sensitivity to α-amanitin and to tagetitoxin (a selective inhibitor of pol III) was tested. In vitro transcription with nuclear extracts was carried out in the presence of different amounts of the inhibitors. Human pol II is known to be inhibited by low concentrations (1 to 2 μg/ml) of α-amanitin; human pol I and pol III are not. At high concentrations (100 to 200 μg/ml) of α-amanitin, pol II and pol III are known to be inhibited but pol I is not. Transcription of the X gene from both start sites 1 and 2 was inhibited by 2 μg/ml of α-amanitin (Fig. 1C, lane 2) but was resistant to tagetitoxin (Fig. 1C, lanes 4 to 5). These results indicated that transcription of the X gene from the major start sites was carried out by pol II.
X transcripts produced in transfected tissue culture cells and hepatocellular carcinoma tissues show the same transcriptional start sites 1 and 2.
To verify the relevance of the start sites observed in the in vitro transcription experiments, we examined X mRNAs expressed in tissue culture cells. HepG2 cells were transiently transfected with the HBV template plasmid pBS-HBXB. Two days after transfection, cells were harvested and X mRNA was analyzed by primer extension using the same primer as in the in vitro transcription reactions (HB1252/1229) or a primer containing the direct repeat 2 (DR2) sequence at nt 1473 to 1450 of the HBV genome. With either primer, the use of both start sites 1 and 2 in tissue culture cells was confirmed (Fig. 1D). Several other downstream start sites were occasionally observed (Fig. 1D, asterisks) that were similar to those observed by others (18, 60). However, these downstream start sites most likely resulted from sample degradation for two reasons: (i) because their intensities were generally weak and varied considerably among different preparations of RNA and (ii) because previous investigators have shown that removing this downstream region had no detectable effect on the level of X gene transcription (47, 64).
X mRNAs from HBV-infected liver samples were also analyzed. Since clinical samples may include different HBV strains, we used the DR2 primer, which is specific for a sequence that is highly conserved among different HBV strains, to carry out primer extension analyses. As shown in Fig. 1E, X mRNA expression was detected in most of the tissues, with transcripts that mapped to the same transcriptional start sites 1 and 2.
Mapping of essential DNA regions for the major two transcriptional start sites.
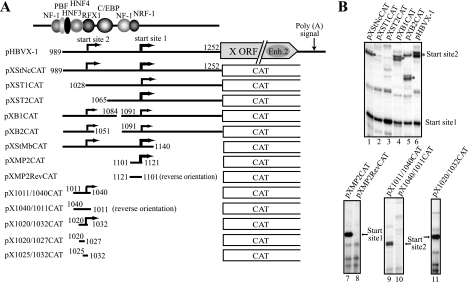
To locate the minimal DNA regions required for accurate transcription from each start site, we constructed various deletion mutants of the enhancer-X promoter region and determined their transcriptional activity by in vitro transcription and primer extension analyses. We found that the 21-bp DNA region between nt 1101 and 1121 (contained in pXMP2CAT) could promote accurate transcription from start site 1 (nt 1118 ± 1) (Fig. 2A and B). The orientation dependence of the promoter activity was confirmed by comparing the transcription activity of pXMP2CAT with that of pXMP2RevCAT, which contains the same 21-bp DNA sequence but in reverse orientation (Fig. 2B, compare lanes 7 and 8). Therefore, we concluded that the 21-bp DNA region contained a bona fide core promoter. We previously observed that various mutations (multinucleotide replacements or insertions) within the 21-bp region largely reduced or abolished the transcription from start site 1 (68, 70); thus, the 21-bp region appeared to be the minimum required region. The level of transcription from pXMP2CAT is about the same as that of adenovirus E1B basal (TATA) promoter (data not shown).
FIG. 2.
Mapping of X gene minimal promoters. (A) Deletion mutants of the enhancer 1-X promoter region were analyzed for X gene transcription by in vitro transcription assays. The results of the analyses are summarized here. Transcriptional start sites are indicated by bent arrows whose thicknesses roughly correspond to the levels of transcription. The minimal promoter for start site 1 was mapped within the nt 1101 to 1121 region. The core promoter for start site 2 was found to be present between nt 1020 and nt 1032. (B) Examples of in vitro transcription analyses. Asterisks in the upper panel show the positions of primer extension products corresponding to start site 2.
For start site 2, we localized the core promoter activity within a 13-bp DNA region between nt 1020 and 1032 (CCCCGTTGCC+1CGG) that is located between −9 and +4 relative to start site 2 (Fig. 2A and B, lane 11, pX1020/1032CAT). The accuracy of the transcription start site (nt 1029 ± 1) and the orientation dependency was also confirmed by in vitro transcription using the CAT reporter plasmids containing a 30-bp region in forward or reverse orientation (pX1011/1040CAT and pX1040-1011CAT in Fig. 2A and B, compare lanes 9 and 10).
Taken together, these data demonstrated that the 21-bp DNA region (nt 1101 to 1121) and the 13-bp region (nt 1020 to 1032) are the minimal regions necessary to function as core promoters for start sites 1 and 2, respectively.
Determination of the core promoter element for transcription from start site 1.
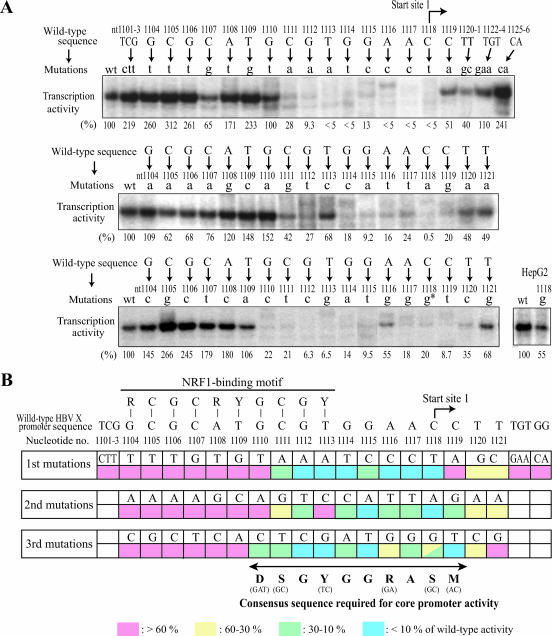
Since the promoters for start sites 1 and 2 both lacked any known core promoter elements, we were curious to determine which specific DNA sequences within these regions were critical for transcriptional activity. In addition, even though the 21-bp minimal promoter region contains the NRF1-binding site which we recently identified as an essential element for transcription from start site 1, it was still not clear whether the NRF1-binding site itself was sufficient for complete core promoter activity, since all other previously identified NRF1-binding sites had been shown to function as regulatory elements but not as core promoter elements. To examine the start site 1 core promoter in more detail, we carried out further mutagenesis of this region (nt 1101 to 1121). First, we mutated the three nucleotides TCG (nt 1101 to 1103) upstream of the NRF1 recognition sequence (TCG→CTT) and tested the activity of the mutant promoter by in vitro transcription assays. As shown in Fig. 3A (the second lane of the top panel), the mutation (TCG→CTT) at nt 1101 to 1103 did not show any adverse effect on transcription from start site 1, suggesting that nt 1101 to 1103 were not essential for the promoter activity. Therefore, the region encompassing nt 1101 to 1103 was not analyzed further. For the nt 1104 to 1121 region (the NRF1-binding site to start site 1), essentially the complete set of single nucleotide point mutations was generated. To verify the dispensability of the sequence downstream of the 21-bp minimal promoter region, two additional mutants that contained either a triple mutation at nt 1122 to 1124 (TGT→GAA) or a double mutation at nt 1125 to 1126 (GG→CA) were made. The activities of the various mutated promoters were tested by in vitro transcription assays using HeLa and HepG2 nuclear extracts. Figure 3A shows representative results of the in vitro transcription assays using the mutant templates. The activity of the mutated promoters measured with HepG2 extracts was essentially consistent with that of HeLa extracts except in one case: HeLa cells were more sensitive than HepG2 cells to the C-to-G mutation at nt 1118, as shown in Fig. 3A, bottom panel. HepG2 cells were also transfected with the mutated template plasmids, and transcription patterns were analyzed by primer extension of the cellularly produced HBV X mRNAs. Levels of transcription from the core promoter 1 with the introduced mutations, as measured from transfected cells, were consistent with the results from in vitro transcription analyses (data not shown). Figure 3B summarizes the results of the in vitro transcription and transient transfection analyses of the 56 mutated promoters. The nucleotides most critical for the core promoter 1 activity were located between nt 1110 and nt 1119, partially overlapping with the previously found NRF1-binding site (nt 1104 to 1113, as shown in Fig. 3B). The functionally tolerant range of a single-base change from the HBV sequence (i.e., the sequence that showed the same, higher, or mildly reduced [>30%] level of promoter activity relative to that of the wild-type promoter) was DSGYGGRASM (or G/A/T-G/C-G-T/C-G-G-G/A-A-G/C+1-A/C). Importantly, this 10-bp DNA sequence is highly conserved among all previously reported HBV strains (the corresponding HBV sequence is G-C-G-T/C-G-G-A-A-C+1-C), suggesting that this sequence may be essential for X gene expression and the virus life cycle. Hereinafter, we refer to this 10-bp sequence as XCPE1, for X core promoter element 1.
FIG. 3.
Determination of the core promoter element for the X gene core promoter 1. (A) Nucleotides within the 21-bp minimal promoter region were individually mutated into three other nucleotides, and the transcription activity of the mutants was assayed in vitro. Primer extension products corresponding to start site 1 are shown with the mutated nucleotides (in lowercase letters) and the nucleotide numbers of the HBV genome. The levels of transcription from mutant templates were determined by densitometry and are shown in each lane as the percentage of transcription activity relative to that of the wild-type promoter. Since the use of HeLa and HepG2 extracts produced somewhat different results for one mutation (1118 C→g), the result for that mutant with HepG2 extract is also shown separately in the bottom right panel. (B) Summary of site-directed mutagenesis. Each construct was assayed at least three times. Promoter activities of mutants are expressed as percentages of the wild-type promoter activity.
Curiously, we found that point mutations in the consensus NRF1-binding sequence did not significantly affect transcriptional activity of the HBV X gene in our assays. One possible explanation for this observation was that our point mutations were not sufficient to abrogate NRF1 binding. To determine the mutations' effects on NRF1 binding affinity, a number of electrophoretic mobility shift assays were carried out using wild-type DNA as the labeled probe and differing amounts of mutant templates as cold competitors. Most of the single-base-change mutations in the region between nt 1104 and nt 1109 (the region within the NRF1-binding sequence but not overlapping with XCPE1) could bind NRF1 with only slightly (approximately threefold) reduced affinity relative to that of the wild-type promoter (data not shown). Given our previous observation that treatment with either small interfering RNA or a dominant negative mutation of NRF1 was required to elicit NRF1's effect on X gene transcription (70), it had been shown that NRF1 was essential for transcription from start site 1 but that NRF1 was not limiting in HeLa and HepG2 cells. Therefore, the approximately threefold reduced affinity of NRF1 for the core promoter 1 may not have significantly reduced the levels of transcription. The levels of NRF1 were sufficient to compensate for the moderate loss of binding affinity.
NRF1-binding site and 10-bp XCPE1 sequence in the 21-bp minimal promoter are cooperative but separable elements.
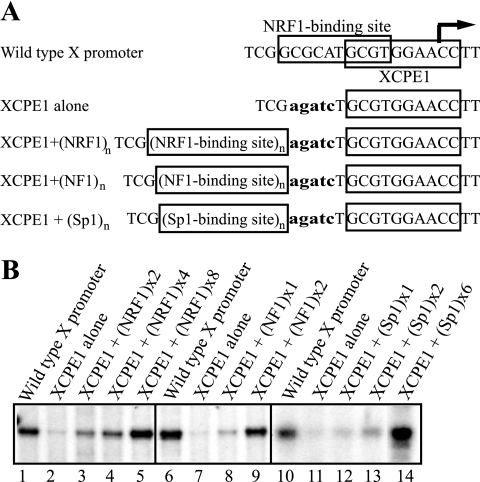
To further clarify the roles of the two DNA elements essential for promoter activity within the 21-bp minimal promoter region, another mutant construct was made in which XCPE1 was intact but the NRF1-binding site was drastically mutated (GCGCATGCGT→AGATCTGCGT), as illustrated in Fig. 4A. This mutated 21-bp minimal promoter did not bind NRF1 (tested by electrophoretic mobility shift assays [data not shown] and see reference 70), and its transcription activity was at about the same background level (Fig. 4B, lanes 2, 7, and 11 versus lanes 1, 6, and 10). To determine whether the position of the NRF1-binding site relative to the XCPE1 element was important for promoter activity, we inserted two to eight copies of tandemly repeated wild-type NRF1-binding elements at a different position relative to XCPE1. Transcription activity assays showed that insertion of the NRF1 binding sequences at this position, although not as effective as at the original position, gave rise to specific transcription at levels equivalent to or higher than those induced by the wild-type minimal promoter (Fig. 4B, lanes 1 through 5). Therefore, we concluded that the NRF1-binding site could be moved but that there were some requirements for the copy number or the distance from XCPE1 in order for maximal activation of transcription by NRF1. These results indicate that the 21-bp minimal promoter for start site 1 consists of two separable but essential elements, an NRF1-binding site and XCPE1. XCPE1 by itself is capable only of inducing a background level of promoter activity but, in cooperation with NRF1-binding sites, can drive transcription from the specific start site at a level clearly distinguishable from the background. Therefore, XCPE1 appears to be an activator-dependent core promoter (or initiator) element.
FIG. 4.
XCPE1 is an activator-dependent core promoter. (A) The sequences of the wild-type X gene core promoter 1 and the mutant promoters containing no NRF1-binding site (XCPE1 alone) or a tandem repeat of NRF1, NF1, or Sp1-binding sites are shown. (B) In vitro transcription from templates containing NRF1, NF1, or Sp1-binding sites upstream of XCPE1.
To determine whether other transcription factor binding sites might also be able to cooperate with XCPE1, Sp1-binding sites and NF1-binding sites were inserted upstream of XCPE1. Transcription from start site 1 could also be activated by the insertion of these activator-binding sites (Fig. 4B, lanes 6 through 14). These results suggest that the XCPE1 element not only can work with a NRF1-binding site but also can cooperate with other activator-binding sites to achieve high levels of transcription.
Approximately 1% of human genes contain XCPE1 in their core promoter regions.
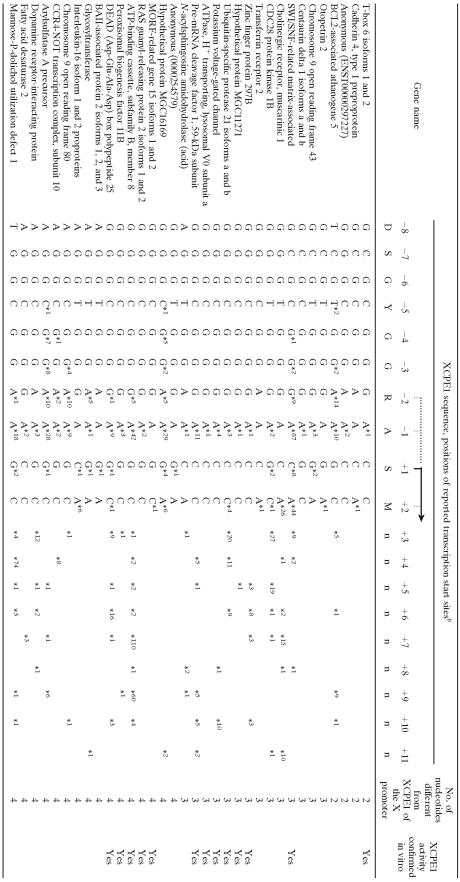
To investigate whether XCPE1 is also utilized in human gene promoters, the Ref-Full human promoter database (http://dbtss.hgc.jp/) (65, 66) was searched for the XCPE1 consensus sequence. DNA regions between −18 and +7 bp relative to the transcription start sites (5′ ends of cDNA clones) were searched for the XCPE1 sequence. This range included the expected position of the XCPE1 sequence (−8 to +2) and margins of 10 bp upstream and 5 bp downstream. The 10-bp upstream margin was used to account for any non-full-length cDNAs still present in the database, and the 5-bp downstream margin allowed for small errors in start site determination. This search identified 110 out of 15,262 genes (0.7%) in the Ref-Full database that contain the XCPE1 sequence within the specified region and in the correct orientation. Table 1 shows a partial list of the 110 genes. The complete list can be found in the table in the supplemental material.
TABLE 1.
Partial list of human genes that contain XCPE1 sequences around previously identified transcriptional start sitesa
The genes are sorted by XCPE1 sequences and shown in the order of increasing numbers of nucleotides different from the XCPE1 sequence in the HBV X promoter.
Positions of previously identified transcriptional start sites are shown by stars (*). The numbers next to the stars show how many times the start sites were mapped to the particular positions. The genes whose XCPE1 sequences have been confirmed to have specific core promoter activity by our in vitro assays (Fig. 5) are shown as “Yes” in the last column. The complete list can be found in the table in the supplemental material.
To compare the frequency of XCPE1 with those of other previously known core promoter elements, the Ref-Full database was also searched for TATA box, DPE, MTE, and Inr sequences using the same margins as those used for the XCPE1 search. In the case of the TATA box, sequences containing a one-base mismatch to the consensus sequence were also included because some TATA-like sequences can be functional (62). This level of stringency has previously been used by Kutach and Kadonaga (29) to determine the frequency of TATA-containing promoters in Drosophila. As summarized in Table 2, our search indicated that about 20% of the human genes in the Ref-Full database contained a TATA box, 19% (when RGWYVT was used as the consensus sequence for search) or 74% (when the looser consensus sequence RGWYV was used for search) contained a DPE, <0.01% contained an MTE, and 40% contained an Inr sequence within the respective search regions. We also found that only 0.02% of all genes (corresponding to 2.7% of XCPE1-containing genes) contained both the XCPE1 and the TATA elements. This result indicates that XCPE1 is preferentially found in TATA-less promoters.
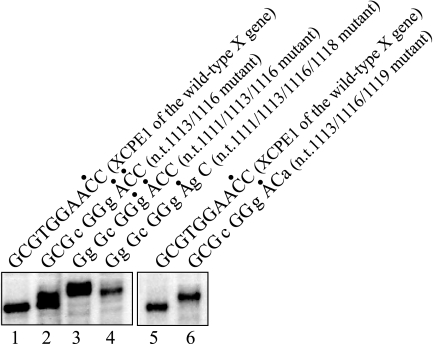
As seen in Table 1 and the table in the supplemental material, most XCPE1 sequences found in the human promoter database do not utilize exactly the same sequence as that observed in the HBV X gene promoter and often differ at more than a single nucleotide position. Since our initial mutational analyses had examined only single-base changes, the extent to which multiple base changes to the HBV XCPE1 sequence would affect the function of the core promoter element was examined. Several additional mutants were constructed that contained multiple simultaneous mutations of the original HBV sequence, and these were tested for promoter activity. As shown in Fig. 5, all of the double-, triple-, and quadruple-base-change mutations we examined retained activity, although some minor start site shifts were observed. It was not possible to test all of the XCPE1 sequences found in the cellular genes because of the overwhelming number of occurrences. However, as seen in Table 1, for many of the XCPE1-containing human promoters, transcriptional start sites have been detected at positions consistent with transcription being driven by XCPE1, suggesting an important functional contribution of XCPE1 to the transcriptional initiation of cellular genes. Most recently, we cloned several XCPE1-containing human promoters and confirmed the presence of their transcriptional start sites at the positions expected to be driven by XCPE1 by in vitro transcription assays and that mutations of the XCPE1 sequence abolished transcription from those start sites (unpublished results).
FIG. 5.
XCPE1 variants containing multiple base changes from the wild-type HBV X promoter sequence retain specific promoter activities. The XCPE1 sequences that are 2-, 3-, and 4-nucleotide divergent from the HBV sequence were made by site-directed mutagenesis of pBS-HBXB (HBV enhancer and X promoter-containing construct) and analyzed for the promoter activity. In vitro transcription was carried out with HepG2 nuclear extract. Lanes 2, 3, 4, and 6 show the results of the transcription from the templates containing base changes from the wild-type HBV X promoter sequence at nt 1113 and 1116; nt 1111, 1113, and 1116; nt 1111, 1113, 1116, and 1118; and nt 1113, 1116, and 1119, respectively. Dots above the XCPE1 sequences indicate positions of the observed start sites.
The majority of XCPE1-containing promoters also contain Sp1-, NF1-, and NRF1-binding sites.
Since XCPE1 functions with activator binding sites, including NRF1, Sp1, and NF1 (Fig. 4), we next asked if the XCPE1-containing promoters also contain binding sites for these activators. Among the 110 genes that were found to contain one or more XCPE1 sequences in their core promoter regions, 69 genes (63%) were found to contain an NRF1-binding consensus sequence (RCGCRYGCGY; 1-bp mismatch allowed per search) in their promoter regions (−1000 to +200). We also found that all of the 110 genes contained one or more (up to 24) Sp1-binding sequences (GC boxes) in the region between −100 and +50 and that 96% of the XCPE1-containing genes contained one or more NF1-binding sequences (CAAT boxes) between −100 and +50.
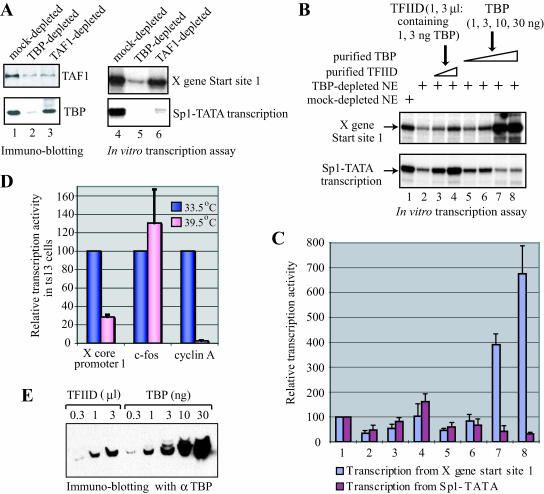
Immunodepletion experiments indicate that TBP is essential for transcription from the X gene promoter but TAF1 (TFIID) is dispensable.
TFIID has been shown to play an important role in transcriptional initiation not only from TATA-containing promoters but also from DPE- and Inr-containing promoters. However, whether TFIID is universally required for RNA polymerase II transcription in higher eukaryotes is still unclear. In order to investigate whether transcription of the X gene would require TFIID, HeLa nuclear extracts were depleted of TFIID using anti-TBP or anti-TAF1 antibodies. The transcriptional activity of the immunodepleted nuclear extracts was then examined by in vitro transcription using the X gene templates and a control template, Sp1-TATA, that is known to exhibit TFIID-dependent transcriptional activation by Sp1 (56). Extracts immunodepleted by anti-TBP antibody showed considerably reduced levels of transcription of both the X gene and Sp1-TATA (Fig. 6A, lane 5), indicating that TBP was essential for transcription of both the X gene and Sp1-TATA. Transcription of Sp1-TATA was also largely reduced by TFIID depletion with anti-TAF1 antibody (Fig. 6A, lane 6, lower panel). However, it was interesting that the extracts immunodepleted by anti-TAF1 were still able to strongly transcribe the X gene (Fig. 6A, lane 6, upper panel), indicating that transcription from the X gene did not require TAF1, i.e., the TFIID complex. The inhibition of transcription from the Sp1-TATA template by TAF1 depletion was consistent with the previous observation (56) and also confirmed that our anti-TAF1 antibody immunodepleted the functional TFIID complex. These results suggest that TBP is essential for transcription of the X gene but that TAF1 is dispensable. Since TAF1 is the unique and major subunit of TFIID, our results indicate that a free form of TBP or some TBP-containing factor other than the complete TFIID complex can be involved in transcription of the X gene.
FIG. 6.
Transcription from the HBV X gene promoter is TBP dependent, but it can use either free TBP or TFIID. (A) Transcription from the X gene is TBP dependent but not TAF1 dependent. HeLa cell nuclear extracts were immunodepleted with anti-TBP, anti-TAF1, or control (preimmune) antibody. The levels of TAF1 and TBP proteins in the depleted extracts were monitored by immunoblotting (left panel, lanes 1 to 3). The depleted nuclear extracts were also tested for activity of transcription from the X gene template (start sites 1) or a TFIID-dependent transcription template, Sp1-TATA, by in vitro transcription assays (right panel, lanes 4 to 6). Note that TBP antibody depleted TAF1 as well as TBP because the majority of TAF1 in cells is present in TFIID complexes. In contrast, TAF1 antibody depleted large amounts of TAF1 but only a small portion of TBP because TBP is also present in TBP-containing complexes other than TFIID. (B) Transcription from the X gene can utilize either a free form of TBP or TFIID. In vitro transcription of the X gene and the Sp1-TATA templates (in a single two-template reaction) was performed using control (lane 1) or TBP-depleted (lanes 2 to 8) HeLa nuclear extract in the absence (lane 2) or presence of 1 μl (lane 3) or 3 μl (lane 4) of purified TFIID or in the presence of 1 ng (lane 5), 3 ng (lane 6), 10 ng (lane 7), or 30 ng (lane 8) of purified recombinant TBP. One microliter of the TFIID contained about 1 ng of TBP (determined by quantitative Western blotting, shown in panel E). (C) Quantification of the transcription assay shown in panel B. The experiment was done twice, and the graph shows a summary of the two independently performed experiments. (D) ts13 cells were transfected with a firefly luciferase reporter plasmid driven by the X gene core promoter 1, the c-fos promoter, or the cyclin A promoter. After 16 h of incubation at the permissive (33.5°C) or nonpermissive (39.5°C) temperature, the luciferase activity in transfected cells was measured and normalized for transfection efficiency. For each reporter construct, the activity at 39.5°C compared to that at 33.5°C was calculated. (E) Determination of the concentration of TBP in the purified TFIID. Different volumes of purified TFIID and known amounts of purified TBP were loaded on a sodium dodecyl sulfate-polyacrylamide gel, and TBP contents were examined by immunoblotting using anti-TBP.
The X gene promoter can be transcribed with either free TBP or the TFIID complex.
Since the X gene promoter lacks any TATA element to which TBP could efficiently bind, TBP is probably not the XCPE1 recognition factor. However, if free TBP was present in human cells, it could be recruited to the X promoter via interaction with some other factor or factors bound to the promoter. To examine the possibility that a free form of TBP might play a role in transcription of the X gene, recombinant human TBP was expressed in E. coli, purified, and tested for its ability to restore X gene transcriptional activity to a TBP-depleted nuclear extract. For comparison, TFIID was also purified from HeLa nuclear extract as described in Materials and Methods and tested for its ability to reconstitute X gene transcriptional ability. As shown in Fig. 6B, the free form of recombinant TBP could efficiently restore X gene transcriptional activity to the TBP-depleted nuclear extracts (Fig. 6B, lanes 5 through 8, upper panel, and C). Ten nanograms of TBP (about one-third to one-half the amount of total TBP the nuclear extract contained before immunodepletion) appeared to be more than sufficient to completely restore the X gene transcription activity (Fig. 6B, lane 7). In contrast, the same amount of free TBP was not able to fully restore the transcription activity for Sp1-TATA template, and the addition of larger amounts of TBP (10 and 30 ng) instead reduced the level of transcription (Fig. 6B, lanes 5 through 8, lower panel, and C). These results suggest that transcription from the XCPE1-containing promoters can be carried out effectively with free TBP, consistent with our observation that immunodepletion of TAF1 did not significantly change transcription of the X gene (Fig. 6A). This property of the X gene promoter is quite different from that of the Sp1-TATA promoter, which is largely dependent on TFIID to exert its full transcription activity.
Even though TAF1 was dispensable for transcription from the X gene, the purified TFIID was also able to contribute to restoring transcription activity to the X gene (Fig. 6B, lanes 3 and 4, upper panel). However, interestingly, the specific activity of TFIID needed for restoring X gene transcription was at a level comparable to that of free TBP (Fig. 6B, upper panel, lanes 3 and 4 versus lanes 5 and 6). The reason that the TFIID did not show significantly higher transcriptional activity than the free TBP on the X gene template was not because the TFIID was inactive, since the same TFIID restored transcription of Sp1-TATA template much more effectively than free TBP did (Fig. 6B, lower panel, lanes 3 and 4 versus lanes 5 and 6). Our results suggest that the transcription driven by XCPE1 can be performed through two alternative transcription mechanisms: a TFIID (TAF1)-independent mechanism that utilizes a free form of TBP and a second mechanism that utilizes the TFIID complex.
To analyze TAF1-independent transcription of the X gene in vivo, ts13 cells, a baby hamster kidney cell line with a TAF1 missense mutation (20), were used (Fig. 6D). The ts13 mutant TAF1 protein functions as wild-type TAF1 at the permissive temperature (33.5°C) but becomes inactive at the nonpermissive temperature (39.5°C). The ts13 cells were transfected with a reporter plasmid driven by the X gene core promoter 1 and cultured at either 33.5°C or 39.5°C. As controls, reporter plasmids driven by either c-fos or cyclin A promoters were also analyzed. The c-fos promoter has been shown to be TAF1 independent, while the cyclin A promoter is TAF1 dependent (74). As expected, the level of cyclin A transcription was reduced about 30- to 40-fold upon inactivation of TAF1 at 39.5°C, but c-fos transcription was not reduced. Under the same experimental conditions, transcription of the X gene minimal promoter decreased but only three- to fourfold, which was an intermediate response between those observed for the c-fos and the cyclin A promoters. These results further support the notion that the X gene (XCPE1-containing) promoter can utilize TFIID but it can also be transcribed by an alternative, TAF1-independent mechanism.
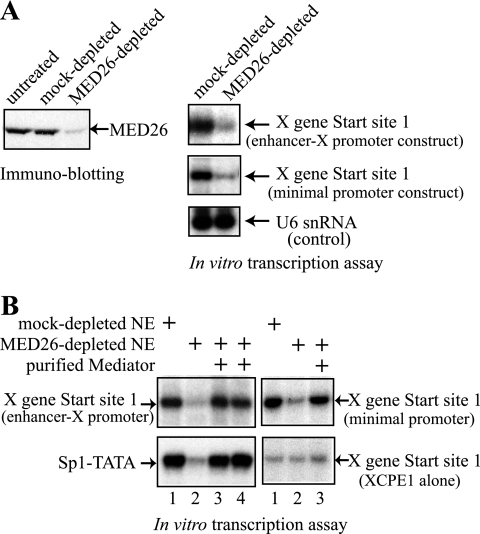
The mediator is also essential for transcription from the X gene promoter.
In other, ongoing biochemical studies, we had observed that the mediator was associated with TBP in the absence of other subunits of TFIID. This finding prompted us to test whether the mediator is important/required for transcription from the X gene. We generated antibodies against the MED26 subunit and carried out immunodepletion of the mediator complex. As shown in Fig. 7A, nuclear extract depleted with anti-MED26 showed much less X gene transcriptional activity than the control, while the same extract retained comparable transcription activity for a U6 snRNA gene that is transcribed by RNA polymerase III. These results suggest that the MED26-containing mediator complex is essential for transcription from the XCPE1-containing core promoters.
FIG. 7.
Requirement of the mediator for transcription from the X gene promoter. (A) HeLa cell nuclear extract was immunodepleted with anti-MED26 or control (preimmune) antibody. The levels of MED26 protein in the depleted extracts were assayed by immunoblotting with anti-MED26 antibody. The immunodepleted nuclear extracts were then tested for transcriptional activity of the X gene (using the enhancer-X promoter construct pBS-HBXB or using the minimal promoter construct pXMP2CAT) or for transcriptional activity of a U6 snRNA gene that is not regulated by the mediator (control). MED26 depletion specifically abolished transcription of the X gene. (B) In vitro reconstitution confirms that the mediator complex is required for transcription from the X gene. In vitro transcription of the X gene (enhancer-X promoter or X minimal promoter 1), XCPE1 alone, and the Sp1-TATA templates was performed using control (lane 1) or MED26-depleted (lanes 2 to 4) HeLa nuclear extracts (NE) in the absence (lane 2) or presence (lanes 3 and 4) of the mediator (MED26-containing) complexes purified from P.5 (lane 3) or P1.0 (lane 4) phosphocellulose fractions.
Since the mediator has well-defined activity as a coactivator (4, 5, 11, 54, 67) and since the HBV enhancer 1 contains binding sites for transcriptional activators that have been shown to be coactivated by the mediator in other genes, we next tested whether the mediator dependence of the X gene transcription could be observed even in the absence of the HBV enhancer. For this purpose, an X gene minimal promoter construct (pXMP2CAT, shown in Fig. 2) was used for the transcription assays. As shown in Fig. 7A (right, second panel), transcription from the X gene minimal promoter still required the mediator. To further confirm our results, we tested whether purified mediator complexes could restore X gene and Sp1-TATA transcription to MED26-depleted nuclear extracts. As shown in Fig. 7B, our purified mediator could completely restore transcription activity for both the X gene (either enhancer-X promoter or the X minimal promoter) and the Sp1-TATA constructs. Thus, the mediator appears to have an essential role in transcription from the X gene minimal (core) promoter 1.
Although XCPE1 does not exert clear transcription activity above background without an activator-binding site, we additionally examined whether transcription from the template containing XCPE1 alone (see Fig. 4) shows mediator dependence or not. Since the transcription level of the “XCPE1 alone” construct is very low, it was difficult to tell whether the level of transcription was decreased by mediator depletion or whether the addition of the mediator activated transcription from this template (Fig. 7B, right lower panel, compare lanes 1, 2, and 3). However, it was at least clear that the response of the “XCPE1 alone” template to mediator depletion was much smaller than that of the X minimal promoter (Fig. 7B, right, compare upper panel versus lower panel). Therefore, the mediator dependence of transcription from the X gene core promoter 1 may be largely a reflection of XCPE1's activator (i.e., NRF1) dependence. However, because of its low level of transcription activity, the possibility that the mediator also contributes to the function of XCPE1 itself still remains. To clearly address this point, in vitro reconstitution with purified general transcription factors would be necessary.
DISCUSSION
A new core promoter element, XCPE1, an activator-dependent initiator.
Recent studies have revealed an unexpectedly broad variation and flexibility of pol II transcription mechanisms. For example, the discovery of tissue-specific and cell-type-specific paralogs of GTFs is evidence that GTFs are not necessarily universal (3, 14, 21, 42, 69). Core promoters are also more diverse than previously thought. There is even evidence of TBP-independent pol II transcriptional mechanisms from the TBP knockout mouse study (43) and from studies of the TFTC (TBP-free TAF-containing) complex (19, 75). These findings suggest that there are multiple alternative pathways for the recruitment of pol II. Studies of enhancer-promoter specificity have demonstrated that different core promoters exhibit different and selective responses to enhancers (8, 10, 38, 50). These findings indicate that it is the combination of a specific promoter and an enhancer that determines the transcriptional regulation pattern for each gene and that different core promoters may utilize different sets of GTFs. Thus, it is very important to identify additional core promoter motifs and to determine which GTFs contribute to the transcription of individual genes.
To explore new core promoter elements for pol II transcription and to better understand the mechanisms of transcriptional regulation of the HBV X gene, we started dissecting the HBV X gene promoter. Using an in vitro transcription system in parallel with analyses of the X mRNAs expressed in vivo, we identified two major transcriptional start sites of X mRNA (start sites 1 and 2) and determined two independent core promoters (core promoters 1 and 2) that drive transcription from the respective two start sites. We further found that the core promoter 1 contained two DNA elements essential for the promoter activity: the NRF1-binding site and a new core promoter element called XCPE1. Transcription driven by XCPE1 alone occurs at levels close to background, but accompanied by the NRF1-binding site, XCPE1 shows specific transcription from start site 1 at a level clearly above background. XCPE1 can also be activated by other transcriptional activators. The optimum distance between activator-binding sites and XCPE1 and how many copies of the activator-binding sites are required to activate XCPE1 are points that remain to be determined. Interestingly, there are reports of cellular promoters where individual CAAT boxes or GC boxes contribute differentially to promoter activity (24, 36, 46). These observations suggest that activator-binding sites can modulate core promoter activity from various locations relative to the core promoter but that the relative locations of activator-binding sites to core promoters can strongly determine how effectively the activators can activate transcription.
Why does XCPE1 need an activator(s)? It may be that the interactions between XCPE1 and XCPE1 recognition factors or between the XCPE1 recognition factor(s) and other general transcription factors are specific but weak, and therefore, the recruitment of general transcription factors requires additional bridging interactions. For example, NRF1 interacts with PGC-1 (PPARγ coactivator-1). PGC-1 has been shown to interact with both pol II (45) and MED1 (a subunit of mediator) (73). Therefore, NRF1/PGC-1 interaction may bridge XCPE1 interactions with pol II.
Mechanisms of transcriptional initiation from XCPE1-containing promoters.
Which factors are required to initiate transcription from the X gene and XCPE1-containing promoters? Our data suggest that X gene transcription requires, at a minimum, TBP, the mediator complex, an activator, and pol II. Interestingly, the X gene exhibits functional plasticity and can be transcribed using two alternative pathways. The X gene can be transcribed by using either free TBP in a TAF1-independent mechanism or TFIID in a TAF1-dependent mechanism. As a result for the X gene, TAF1 is not essential for transcription. There have been a few previous reports of TAF-independent transcriptional activation occurring for viral and mammalian promoters (48, 74, 76), but to date, little is known about the mechanisms of TAF-free transcription in higher eukaryotes.
However, there are a number of different complexes that have been implicated in TFIID-free transcription for other systems that are good candidates for the factors driving the TAF-independent transcription of the X gene. The TFTC complex and a multifunctional transcriptional coactivator, SAGA (Spt-Ada-Gcn5 acetyltransferase), both share multiple TAF subunits with TFIID, and in Saccharomyces cerevisiae, SAGA has been reported to interact with TBP. It will be worth examining whether STAGA (human SAGA) or TFTC is involved in the TAF1 (TFIID)-independent transcription of the XCPE1-containing genes.
In contrast to the situation in higher eukaryotes, the presence of TAF-independent promoters has been well described for yeast (28, 32, 37, 58). Different promoters in yeast have different levels and repertoires of TAF requirements. Interestingly, the TFIID-specific subunit TAF1 has been shown to be necessary for the expression of only 16 to 27% of yeast genes (23, 32). Paradoxically, genome-wide expression analyses of TAF mutant strains and chromatin immunoprecipitation of TFIID and SAGA (25) have shown that the regulation of about 90% of the yeast genome is TFIID dominated, about 10% is SAGA dominated, and at some promoters, SAGA and TFIID contributions are more or less equivalent. However, both of these observations are consistent with the notion that the individual yeast genes do not exclusively depend on either TFIID or SAGA, i.e., many genes can be transcribed through either a TFIID-dependent or a SAGA-dependent (TFIID-independent) mechanism. This may be a situation similar to our observations of transcription of the HBV X gene promoter that showed that either TFIID or free TBP could be utilized.
What determines whether TFIID is required for a particular promoter? Studies of yeast have shown that the main determinants of TAF requirements are the core promoters but not upstream activating sequences (10, 59), although in some cases, activators play a part in TFIID recruitment through direct activator-TAF interactions (44). Recent bioinformatics studies have shown that, as a general tendency, TATA box-containing genes preferentially utilize SAGA rather than TFIID and that TATA-less genes tend to be TAF dependent and utilize TFIID (2). Identification of additional determinants for TFIID utilization or SAGA utilization or other combinations of transcription factors that replace TFIID or SAGA function will help to gain an understanding of transcription mechanisms for individual genes. Shen and Green (59) suggested that in addition to the canonical TATA sequence, the region surrounding the TATA box was also important for TAF1 dependence. Since the XCPE1 element is found mainly in TATA-less promoters but may not require TAF1 for activity, XCPE1 promoters appear to be additional examples of promoters where the presence or absence of the TATA box is not the only criterion by which to determine TAF dependence. An interesting remaining question to be determined is which mechanism, TAF1 dependence or TAF1 independence, predominates for XCPE1-containing genes in vivo. Our present work at least suggests that XCPE1-containing genes may represent a class of promoters that can utilize either mechanism. This behavior is very different from the properties of the Sp1-TATA promoters, the Inr-containing promoters, the DPE-containing promoters, or the MTE-containing promoters, which are all clearly TFIID dependent.
In order to further clarify what other transcription factors are necessary for transcription from the X gene core promoters, in vitro reconstitution of the transcription reaction with highly purified factors will be essential. In addition, identification of the recognition factor for XCPE1 will be particularly important for determining the mechanisms of pol II recruitment and for understanding how NRF1 and other activators activate transcription from the XCPE1 promoters.
XCPE1 is also found in human TATA-less promoters.
In light of our database search, we estimated that XCPE1 sequences are present in the core promoter regions of about 1% of human protein-encoding genes, particularly TATA-less genes. The estimated frequency of XCPE1 appears significant but is not high enough to account for the majority of TATA-less, DPE-less, and MTE-less promoters. Thus, there must still be many additional core promoter elements that remain to be identified. Our observation of a 20% frequency for TATA box-containing genes is lower than the previous report (32%) for human promoters (65). The differences between our results and those from previous searches can be attributed to two factors. The first factor is the difference in search conditions. In the previous search, more than one mismatch was allowed and a wider region was searched (−90 to +27, whereas our search region was −41 to −19). The second factor may reflect a more comprehensive sampling of genes in the database. The current Ref-Full database contains almost 15 times more genes than previously. In Drosophila, the most recent analysis of promoter database (containing 1,941 genes) estimated the frequency of TATA-containing genes (in the region between −45 and −15) to be 28.3% (49). The bioinformatics study of yeast genes that carefully defined functional TATA box consensus sequences found that 19% of the yeast genes contained a TATA box, a frequency very similar to our result for human TATA promoters. In any case, in order to more accurately determine the frequency of the TATA box and other elements, more-complete information describing transcriptional start sites and a rigorous examination of consensus sequences of core promoter elements will be required. Taken together, our results suggest the presence of a new core promoter element, XCPE1, that may be utilized not only by HBV X promoter 1 but also by many cellular gene promoters. Our study of transcription driven by the XCPE1 element may help explain general mechanisms of transcription from a subset of the human TATA-less promoters.
The HBV X gene as a model system for studying TATA-less, DPE-less promoters.
To understand the role of the X protein in the transformation of hepatocytes, it is essential to understand both the regulatory mechanisms of gene expression and the function of the gene product. Our extensive characterization of the X gene core promoter and its required general transcription factors and cofactors is the first step toward understanding the regulatory mechanisms of X gene expression. At the same time, the X gene can serve as a general model system for studying transcription from TATA-less promoters since RNA transcription of the HBV genes utilizes only the cellular transcriptional machinery.
Recent analyses of the human genome sequence have revealed a substantial number of genes that are regulated by as-yet-unidentified core promoter elements. This category of genes includes (but is not restricted to) many cell growth-related genes and housekeeping genes, as well as mitochondrial function-related genes. The knowledge we have obtained from our research should help us to understand not only the basis of HBV-induced liver diseases but also the transcription of important classes of TATA-less genes in eukaryotic cells.
Supplementary Material
Acknowledgments
We thank Robert Tjian and James Goodrich for providing the anti-hTAF4 monoclonal antibody; Naoko Tanese for the nontagged human TBP expression vector; Joan Conaway, Shigeo Sato, and Chieri Sato for the MED26 cell line; Katsuro Koike for many HBV DNA constructs; David Graham (Texas Gulf Coast Digestive Disease Center, Study Design and Clinical Specimen Core Facility) for liver tissue samples; and Edith Wang for c-fos and cyclin A reporter plasmids and ts13 cells. We also thank Yutaka Suzuki for helping us search the DBTSS database.
This study was supported by an institutional research grant from the M. D. Anderson Cancer Center, by an American Gastroenterological Association/Elsevier Research Initiative Award, and by a grant (AI057504) from the NIH to S.T. DNA sequencing was supported by the DNA core facility at the M. D. Anderson Cancer Center.
Footnotes
Published ahead of print on 8 January 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arbuthnot, P., and M. Kew. 2001. Hepatitis B virus and hepatocellular carcinoma. Int. J. Exp. Pathol. 82:77-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basehoar, A. D., S. J. Zanton, and B. F. Pugh. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116:699-709. [DOI] [PubMed] [Google Scholar]
- 3.Bell, B., and L. Tora. 1999. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp. Cell Res. 246:11-19. [DOI] [PubMed] [Google Scholar]
- 4.Blazek, E., G. Mittler, and M. Meisterernst. 2005. The mediator of RNA polymerase II. Chromosoma 113:399-408. [DOI] [PubMed] [Google Scholar]
- 5.Bourbon, H. M., A. Aguilera, A. Z. Ansari, F. J. Asturias, A. J. Berk, S. Bjorklund, T. K. Blackwell, T. Borggrefe, M. Carey, M. Carlson, J. W. Conaway, R. C. Conaway, S. W. Emmons, J. D. Fondell, L. P. Freedman, T. Fukasawa, C. M. Gustafsson, M. Han, X. He, P. K. Herman, A. G. Hinnebusch, S. Holmberg, F. C. Holstege, J. A. Jaehning, Y. J. Kim, L. Kuras, A. Leutz, J. T. Lis, M. Meisterernest, A. M. Naar, K. Nasmyth, J. D. Parvin, M. Ptashne, D. Reinberg, H. Ronne, I. Sadowski, H. Sakurai, M. Sipiczki, P. W. Sternberg, D. J. Stillman, R. Strich, K. Struhl, J. Q. Svejstrup, S. Tuck, F. Winston, R. G. Roeder, and R. D. Kornberg. 2004. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14:553-557. [DOI] [PubMed] [Google Scholar]
- 6.Buratowski, S. 1994. The basics of basal transcription by RNA polymerase II. Cell 77:1-3. [DOI] [PubMed] [Google Scholar]
- 7.Burke, T. W., and J. T. Kadonaga. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler, J. E., and J. T. Kadonaga. 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15:2515-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler, J. E., and J. T. Kadonaga. 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16:2583-2592. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, J. X., M. Floer, P. Ononaji, G. Bryant, and M. Ptashne. 2002. Responses of four yeast genes to changes in the transcriptional machinery are determined by their promoters. Curr. Biol. 12:1828-1832. [DOI] [PubMed] [Google Scholar]
- 11.Conaway, J. W., L. Florens, S. Sato, C. Tomomori-Sato, T. J. Parmely, T. Yao, S. K. Swanson, C. A. Banks, M. P. Washburn, and R. C. Conaway. 2005. The mammalian mediator complex. FEBS Lett. 579:904-908. [DOI] [PubMed] [Google Scholar]
- 12.Conaway, R. C., and J. W. Conaway. 1993. General initiation factors for RNA polymerase II. Annu. Rev. Biochem. 62:161-190. [DOI] [PubMed] [Google Scholar]
- 13.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dikstein, R., S. Zhou, and R. Tjian. 1996. Human TAFII 105 is a cell type-specific TFIID subunit related to hTAFII130. Cell 87:137-146. [DOI] [PubMed] [Google Scholar]
- 15.Fukai, K., S. Takada, O. Yokosuka, H. Saisho, M. Omata, and K. Koike. 1997. Characterization of a specific region in the hepatitis B virus enhancer I for the efficient expression of X gene in the hepatic cell. Virology 236:279-287. [DOI] [PubMed] [Google Scholar]
- 16.Ge, H., E. Martinez, C. M. Chiang, and R. G. Roeder. 1996. Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 274:57-71. [DOI] [PubMed] [Google Scholar]
- 17.Guo, W. T., J. Wang, G. Tam, T. S. Yen, and J. S. Ou. 1991. Leaky transcription termination produces larger and smaller than genome size hepatitis B virus X gene transcripts. Virology 181:630-636. [DOI] [PubMed] [Google Scholar]
- 18.Gustin, K., M. Shapiro, W. Lee, and R. D. Burk. 1993. Characterization of the role of individual protein binding motifs within the hepatitis B virus enhancer I on X promoter activity using linker scanning mutagenesis. Virology 193:653-660. [DOI] [PubMed] [Google Scholar]
- 19.Hardy, S., M. Brand, G. Mittler, J. Yanagisawa, S. Kato, M. Meisterernst, and L. Tora. 2002. TATA-binding protein-free TAF-containing complex (TFTC) and p300 are both required for efficient transcriptional activation. J. Biol. Chem. 277:32875-32882. [DOI] [PubMed] [Google Scholar]
- 20.Hayashida, T., T. Sekiguchi, E. Noguchi, H. Sunamoto, T. Ohba, and T. Nishimoto. 1994. The CCG1/TAFII250 gene is mutated in thermosensitive G1 mutants of the BHK21 cell line derived from golden hamster. Gene 141:267-270. [DOI] [PubMed] [Google Scholar]
- 21.Hiller, M. A., T. Y. Lin, C. Wood, and M. T. Fuller. 2001. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 15:1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochheimer, A., and R. Tjian. 2003. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17:1309-1320. [DOI] [PubMed] [Google Scholar]
- 23.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 24.Hu, Q., C. Bhattacharya, and S. N. Maity. 2002. CCAAT binding factor (CBF) binding mediates cell cycle activation of topoisomerase IIalpha. Conventional CBF activation domains are not required. J. Biol. Chem. 277:37191-37200. [DOI] [PubMed] [Google Scholar]
- 25.Huisinga, K. L., and B. F. Pugh. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13:573-585. [DOI] [PubMed] [Google Scholar]
- 26.Kadonaga, J. T. 2004. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116:247-257. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, M., and K. Koike. 1984. Complete nucleotide sequence of hepatitis B virus DNA of subtype adr and its conserved gene organization. Gene 30:227-232. [DOI] [PubMed] [Google Scholar]
- 28.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 29.Kutach, A. K., and J. T. Kadonaga. 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20:4754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagrange, T., A. N. Kapanidis, H. Tang, D. Reinberg, and R. H. Ebright. 1998. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, D.-H., N. Gershenzon, M. Gupta, I. P. Ioshikhes, D. Reinberg, and B. A. Lewis. 2005. Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol. Cell. Biol. 25:9674-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 33.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 34.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 35.Lewis, B. A., and D. Reinberg. 2003. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 116:3667-3675. [DOI] [PubMed] [Google Scholar]
- 36.Li, F., and D. C. Altieri. 1999. Transcriptional analysis of human survivin gene expression. Biochem. J. 344:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, X. Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 38.Li, X. Y., S. R. Bhaumik, X. Zhu, L. Li, W. C. Shen, B. L. Dixit, and M. R. Green. 2002. Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr. Biol. 12:1240-1244. [DOI] [PubMed] [Google Scholar]
- 39.Lim, C. Y., B. Santoso, T. Boulay, E. Dong, U. Ohler, and J. T. Kadonaga. 2004. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 18:1606-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maldonado, E., R. Drapkin, and D. Reinberg. 1996. Purification of human RNA polymerase II and general transcription factors. Methods Enzymol. 274:72-100. [DOI] [PubMed] [Google Scholar]
- 41.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 42.Martianov, I., S. Brancorsini, A. Gansmuller, M. Parvinen, I. Davidson, and P. Sassone-Corsi. 2002. Distinct functions of TBP and TLF/TRF2 during spermatogenesis: requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development 129:945-955. [DOI] [PubMed] [Google Scholar]
- 43.Martianov, I., S. Viville, and I. Davidson. 2002. RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science 298:1036-1039. [DOI] [PubMed] [Google Scholar]
- 44.Mencia, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras, and K. Struhl. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9:823-833. [DOI] [PubMed] [Google Scholar]
- 45.Monsalve, M., Z. Wu, G. Adelmant, P. Puigserver, M. Fan, and B. M. Spiegelman. 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6:307-316. [DOI] [PubMed] [Google Scholar]
- 46.Muller, C., R. Yang, L. Beck-von-Peccoz, G. Idos, W. Verbeek, and H. P. Koeffler. 1999. Cloning of the cyclin A1 genomic structure and characterization of the promoter region. GC boxes are essential for cell cycle-regulated transcription of the cyclin A1 gene. J. Biol. Chem. 274:11220-11228. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura, I., and K. Koike. 1992. Identification of a binding protein to the X gene promoter region of hepatitis B virus. Virology 191:533-540. [DOI] [PubMed] [Google Scholar]
- 48.Oelgeschlager, T., Y. Tao, Y. K. Kang, and R. G. Roeder. 1998. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol. Cell 1:925-931. [DOI] [PubMed] [Google Scholar]
- 49.Ohler, U., G. C. Liao, H. Niemann, and G. M. Rubin. 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol 3:RESEARCH0087. http://genomebiology.com/2002/3/12/research/0087. [DOI] [PMC free article] [PubMed]
- 50.Ohtsuki, S., M. Levine, and H. N. Cai. 1998. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 12:547-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 52.Paterlini, P., K. Poussin, M. Kew, D. Franco, and C. Brechot. 1995. Selective accumulation of the X transcript of hepatitis B virus in patients negative for hepatitis B surface antigen with hepatocellular carcinoma. Hepatology 21:313-321. [PubMed] [Google Scholar]
- 53.Pugh, B. F. 1995. Purification of the human TATA-binding protein, TBP. Methods Mol. Biol. 37:359-367. [DOI] [PubMed] [Google Scholar]
- 54.Roeder, R. G. 2005. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579:909-915. [DOI] [PubMed] [Google Scholar]
- 55.Rossner, M. T. 1992. Review: hepatitis B virus X gene product: a promiscuous transcriptional activator. J. Med. Virol. 36:101-117. [DOI] [PubMed] [Google Scholar]
- 56.Ryu, S., S. Zhou, A. G. Ladurner, and R. Tjian. 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397:446-450. [DOI] [PubMed] [Google Scholar]
- 57.Sato, S., C. Tomomori-Sato, T. J. Parmely, L. Florens, B. Zybailov, S. K. Swanson, C. A. Banks, J. Jin, Y. Cai, M. P. Washburn, J. W. Conaway, and R. C. Conaway. 2004. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14:685-691. [DOI] [PubMed] [Google Scholar]
- 58.Shen, W. C., S. R. Bhaumik, H. C. Causton, I. Simon, X. Zhu, E. G. Jennings, T. H. Wang, R. A. Young, and M. R. Green. 2003. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 22:3395-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen, W. C., and M. R. Green. 1997. Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90:615-624. [DOI] [PubMed] [Google Scholar]
- 60.Siddiqui, A., S. Jameel, and J. Mapoles. 1987. Expression of the hepatitis B virus X gene in mammalian cells. Proc. Natl. Acad. Sci. USA 84:2513-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smale, S. T. 2001. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 15:2503-2508. [DOI] [PubMed] [Google Scholar]
- 62.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 63.Su, Q., C. H. Schroder, W. J. Hofmann, G. Otto, R. Pichlmayr, and P. Bannasch. 1998. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology 27:1109-1120. [DOI] [PubMed] [Google Scholar]
- 64.Sugata, F., H. S. Chen, S. Kaneko, R. H. Purcell, and R. H. Miller. 1994. Analysis of the X gene promoter of woodchuck hepatitis virus. Virology 205:314-320. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki, Y., T. Tsunoda, J. Sese, H. Taira, J. Mizushima-Sugano, H. Hata, T. Ota, T. Isogai, T. Tanaka, Y. Nakamura, A. Suyama, Y. Sakaki, S. Morishita, K. Okubo, and S. Sugano. 2001. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 11:677-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki, Y., R. Yamashita, S. Sugano, and K. Nakai. 2004. DBTSS, DataBase of Transcriptional Start Sites: progress report 2004. Nucleic Acids Res. 32:D78-D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taatjes, D. J., M. T. Marr, and R. Tjian. 2004. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 5:403-410. [DOI] [PubMed] [Google Scholar]
- 68.Takada, S., N. Kaneniwa, N. Tsuchida, and K. Koike. 1996. Hepatitis B virus X gene expression is activated by X protein but repressed by p53 tumor suppressor gene product in the transient expression system. Virology 216:80-89. [DOI] [PubMed] [Google Scholar]
- 69.Takada, S., J. T. Lis, S. Zhou, and R. Tjian. 2000. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell 101:459-469. [DOI] [PubMed] [Google Scholar]
- 70.Tokusumi, Y., S. Zhou, and S. Takada. 2004. Nuclear respiratory factor 1 plays an essential role in transcriptional initiation from the hepatitis B virus X gene promoter. J. Virol. 78:10856-10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Treinin, M., and O. Laub. 1987. Identification of a promoter element located upstream from the hepatitis B virus X gene. Mol. Cell. Biol. 7:545-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Verrijzer, C. P., J. L. Chen, K. Yokomori, and R. Tjian. 1995. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 81:1115-1125. [DOI] [PubMed] [Google Scholar]
- 73.Wallberg, A. E., S. Yamamura, S. Malik, B. M. Spiegelman, and R. G. Roeder. 2003. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell 12:1137-1149. [DOI] [PubMed] [Google Scholar]
- 74.Wang, E. H., and R. Tjian. 1994. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science 263:811-814. [DOI] [PubMed] [Google Scholar]
- 75.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393:187-191. [DOI] [PubMed] [Google Scholar]
- 76.Wu, S.-Y., T. Zhou, and C.-M. Chiang. 2003. Human mediator enhances activator-facilitated recruitment of RNA polymerase II and promoter recognition by TATA-binding protein (TBP) independently of TBP-associated factors. Mol. Cell. Biol. 23:6229-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yaginuma, K., I. Nakamura, S. Takada, and K. Koike. 1993. A transcription initiation site for the hepatitis B virus X gene is directed by the promoter-binding protein. J. Virol. 67:2559-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zawel, L., and D. Reinberg. 1992. Advances in RNA polymerase II transcription. Curr. Opin. Cell Biol. 4:488-495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.