Abstract
Although the actin cytoskeleton and the translation machinery are considered to be separate cellular complexes, growing evidence supports overlapping regulation of the two systems. Because of its interaction with actin, the eukaryotic translation elongation factor 1A (eEF1A) is proposed to be a regulator or link between these processes. Using a genetic approach with the yeast Saccharomyces cerevisiae, specific regions of eEF1A responsible for actin interactions and bundling were identified. Five new mutations were identified along one face of eEF1A. Dramatic changes in cell growth, cell morphology, and actin cable and patch formation as well as a unique effect on total translation in strains expressing the F308L or S405P eEF1A mutant form were observed. The translation effects do not correlate with reduced translation elongation but instead include an initiation defect. Biochemical analysis of the eEF1A mutant forms demonstrated reduced actin-bundling activity in vitro. Reduced total translation and/or the accumulation of 80S ribosomes in strains with either a mutation or a null allele of genes encoding actin itself or actin-regulating proteins Tpm1p, Mdm20p, and Bnirp/Bni1p was observed. Our data demonstrate that eEF1A, other actin binding proteins, and actin mutants affect translation initiation through the actin cytoskeleton.
Actin, an essential component of the cell cytoskeleton, is responsible for the regulation of the structure of eukaryotic cells. Actin organization in the yeast Saccharomyces cerevisiae produces two major types of filament-based structures: patches and cables. These provide the structural basis for cell morphology, polarity, and endocytosis. Both structures are well defined and differentially regulated. Actin cortical patches are discrete cytoskeletal bodies associated with invaginations of the plasma membrane (47) and are clustered near regions of exocytosis in growing cells (2). The assembly and regulation of the cortical patches is in part controlled by the actin-related protein 2/3 complex. Actin cables are long bundles of actin filaments oriented along the mother-bud axis which guide the majority of polarizing events, such as mRNA localization and organelle inheritance, in yeast. Numerous actin binding proteins including fimbrin (Sac6p) (1), two tropomyosin isoforms (Tpm1p and Tpm2p) (17, 42), and the mitochondrial disruption and morphology protein (Mdm20p) (25) have been reported to participate in the elaboration and composition of the actin cytoskeleton. Because these highly dynamic actin structures are subject to a very strict balance between stabilization and disassembly, the overexpression of actin or actin-related proteins is detrimental to yeast cell viability (18, 43, 56), suggesting that the correct stoichiometry of cytoskeletal components is crucial. One such factor whose overexpression has been shown to specifically affect the actin cytoskeleton in yeast is the eukaryotic translation elongation factor 1A (eEF1A; formerly known as EF-1α) (24, 48).
eEF1A is a highly abundant 52-kDa protein whose canonical function is the delivery of aminoacyl-tRNA to the elongating ribosome. The last decade, however, has seen the discovery of other functions for eEF1A outside of its essential role in protein elongation. eEF1A has been shown to play roles in the quality surveillance of newly synthesized proteins (31), ubiquitin-dependent degradation (13, 23), and viral functions (reviewed in reference 37). Reports have also proposed a role for eEF1A in facilitating apoptosis (12, 19, 39). The most abundant source of information available regarding a noncanonical function is the association of eEF1A with the cytoskeleton. Since the first report demonstrating the interaction of eEF1A with the actin cytoskeleton in Dictyostelium amoebae (64), this association has been established across species from yeast to mammals (20, 24, 48, 58). The prediction that a high percentage of eEF1A is associated with the actin cytoskeleton and that the actin binding function is a universal property of eEF1A implies that actin is important for eEF1A functions and/or vice versa (14). It has been proposed that eEF1A cross-links actin filaments via a unique bonding rule that excludes other F-actin cross-linking proteins (50). Because other findings implicated eEF1A in microtubule binding, bundling, or severing (45, 46, 60) and an association with the centrosome and the mitotic apparatus (38), current models propose that eEF1A is a key factor in regulating cytoskeleton organization. These models have been further strengthened by our recent work demonstrating that mutations in eEF1A that reduce its actin-bundling activity result in aberrant actin cytoskeletons of yeast cells in vivo (24). The fact that other components of the protein synthesis machinery, such as aminoacyl-tRNA synthetases (15, 44), eukaryotic initiation factors (32), and the elongation factors eEF1Bβ (22) and eEF2 (9), are reported to associate with the actin cytoskeleton suggests that these two different systems are intrinsically connected and may show reciprocal regulation. A model has emerged in which actin and the cytoskeleton may have important regulatory functions in protein synthesis, providing a scaffold for translational components including polyribosomes, translation factors, and mRNAs. Direct evidence of the regulation of protein synthesis by actin cytoskeleton components, especially in vivo, remains missing.
Although the eEF1A-actin interaction has been extensively documented, the location, functional consequences, potential regulation of association, and effect on other actin binding proteins are not well understood. Using a unique genetic screen (24), we identified eEF1A mutations that affect the bundling or binding of actin in order to gain insight into the regions of eEF1A essential for such properties. In this work, we demonstrate that five newly identified eEF1A mutations that suppress the overexpression phenotype are clustered into two regions on a single face of eEF1A. While the genetic screen yielded mutants with various levels of suppression of the overexpression phenotype, all mutant proteins were functional as the only form of eEF1A. The two eEF1A mutations that most efficiently suppressed the eEF1A overexpression phenotypes induced dramatic changes in cell morphology and the disappearance of actin cables and increased actin patches when the mutant protein was expressed as the only form of eEF1A. The eEF1A mutant strains also had a reduction in total protein synthesis and, surprisingly, a polyribosome defect consistent with reduced initiation. Strikingly, translation initiation defects were also seen in strains with mutant alleles of the genes encoding several other actin-regulating proteins and actin. Our data present novel effects of altered actin and actin binding proteins, including eEF1A, through the cytoskeleton to alter translation initiation.
MATERIALS AND METHODS
Strains and media.
S. cerevisiae strains used in this study are described in Table1). The disruption of TPM1 (YNL079C) or MDM20 (YOL076W) in strain MC214 was obtained by PCR of genomic DNA from the Open Biosystem deletion set (Open Biosystem, AL) using primers 200 nucleotides 5′ and 3′ of the open reading frame and transformation of the PCR fragments by using the Frozen-EZ Yeast Transformation II kit (Zymo Research, CA). Cells in which in vivo recombination had occurred were selected on medium containing 200 μg/ml G418 sulfate. Strain TKY803 was transformed with plasmids expressing the mutant forms of eEF1A, and the loss of the TEF1 LYS2 plasmid was monitored by the ability to grow on media containing α-aminoadipate. Yeast cells were grown in either yeast extract-peptone-dextrose (YEPD; 1% [wt/vol] Bacto yeast extract, 2% [wt/vol] peptone, 2% [wt/vol] dextrose) or defined synthetic complete medium (C) supplemented with 2% (wt/vol) dextrose as a carbon source. Cells were transformed with yeast plasmids by the lithium acetate method (33). Growth assays were performed by spotting cells as 10-fold serial dilutions onto C-Trp or YEPD medium and incubating the cells at 30 and 37°C for 2 or 3 days, respectively.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| MC214 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 met2-1 his4-713 tef1::LEU2 tef2Δ pTEF2 TRP1 | 55 |
| TKY259 | MATaura3-52 leu2 his3-Δ200 trp1-Δ100 ade2-10 | 48 |
| TKY621 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 MET2 his4-713 tef1::LEU2 tef2Δ pTEF2 TRP1 | 3 |
| TKY622 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 MET2 his4-713 tef1::LEU2 tef2Δ pTEF2 E286K TRP1 | 3 |
| TKY702 | MATα leu2-3,112 ura3-52 trp1-7 yef3::LEU2 lys2-20 his4-713 met2-1 pYEF3 TRP1 | 3 |
| TKY707 | MATα leu2-3,112 ura3-52 trp1-7 yef3::LEU2 lys2-20 his4-713 met2-1 pYEF3 F650S TRP1 | 3 |
| TKY803 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 met2-1 his4-713 tef1::LEU2 tef2Δ pTEF1 LYS2 | This study |
| TKY880 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 met2-1 his4-713 tef1::LEU2 tef2Δ pTEF1 TRP1 | This study |
| TKY881 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 met2-1 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 | This study |
| TKY882 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 met2-1 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 N329D Y355C | This study |
| TKY883 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 met2-1 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 K333E | This study |
| TKY885 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 met2-1 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 H294A Q296R | This study |
| TKY886 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 met2-1 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 F308L | This study |
| TKY888 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 met2-1 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 S405P | This study |
| TKY892 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 MET2 his4-713 tef1::LEU2 tef2Δ pTEF1 LYS2 | 24 |
| TKY895 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 MET2 his4-713 tef1::LEU2 tef2Δ pTEF1 TRP1 | 24 |
| TKY896 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 MET2 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 | 24 |
| TKY897 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 MET2 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 N329D Y355C | This study |
| TKY898 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 MET2 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 K333E | This study |
| TKY900 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 MET2 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 H294A Q296R | This study |
| TKY901 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 MET2 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 F308L | This study |
| TKY903 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 MET2 his4-713 tef1::LEU2 tef2Δ pTEF1-URA3 TRP1 S405P | This study |
| TKY1056 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 met2-1 his4-713 tef1::LEU2 tef2Δ pTEF2 TRP1 tpm1Δ::KanMX6 | This study |
| TKY1057 | MATα ura3-52 leu2-3,112 trp1-Δ1 lys2-20 met2-1 his4-713 tef1::LEU2 tef2Δ pTEF2 TRP1 mdm20Δ::KanMX6 | This study |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0Δura3Δ1 | Open Biosystem |
| YNL079C | MATahis3Δ1 leu2Δ0 met15Δ0Δura3Δ1 tpm1Δ::KanMX6 | Open Biosystem |
| YOL076W | MATahis3Δ1 leu2Δ0 met15Δ0Δura3Δ1 mdm20Δ::KanMX6 | Open Biosystem |
| Y1239 | MATahis3Δ1 met15Δ0 leu2Δ0 ura3Δ0Δ | 21 |
| Y3024 | MATahis3Δ1 met15Δ0 leu2Δ0 ura3Δ0 bni1-11::URA3 bnr1Δ::KanMX6 | 21 |
| AAY1453 | MATa/MATα ura3-52/ura3-52 trp1/trp1 his3-Δ200/his3-Δ200 pep4::HIS3/pep4::HIS3 prb11.6R/prb11.6R can1/can1 leu2-Δ1/leu2-Δ1 GAL/GAL | 34 |
| IGY191 | MATα barΔ::LYS2 ura3-52 his3-Δ200 lys2-801 leu2-3,112 ade2 ACT1 | 63 |
| IGY58 | MATα barΔ::LYS2 ura3-52 his3-Δ200 lys2-801 leu2-3,112 ade2 act1-122::HIS3 | 63 |
| IGY88 | MATα barΔ::LYS2 ura3-52 his3-Δ200 lys2-801 leu2-3,112 ade2 act1-20::HIS3 | 63 |
| IGY116 | MATα barΔ::LYS2 ura3-52 his3-Δ200 lys2-801 leu2-3,112 ade2 act1-2::HIS3 | 63 |
DNA manipulation.
Restriction endonucleases and DNA-modifying enzymes were obtained from Roche or Stratagene. All manipulations of the eEF1A- or eEF1A-Ura3p-encoding genes were performed using plasmid pTKB731 or pTKB744, respectively (24). Preparation of the eEF1A-Ura3p fusion protein and PCR mutagenesis for the genetic screen were performed as described previously (24). The eEF1A-Ura3p E286K mutant was prepared by site-directed mutagenesis using the QuikChange system (Stratagene). Plasmid pTKB975 was generated from pRS426 by the insertion of HpaI and AgeI sites 5′ and 3′, respectively, of the URA3 open reading frame. Following digestion, the MET2 open reading frame prepared by PCR amplification from wild-type genomic DNA was inserted. pRS426 (URA3 2μm)-based plasmids expressing TPM1 (pTKB1005) or MDM20 (pTKB998) were prepared by PCR of genomic DNA from wild-type strain BY4741 by using primers corresponding to regions 1,000 nucleotides 5′ and 3′ of the open reading frame containing restriction sites for NotI and KpnI in MDM20 and EcoRI in TPM1.
Actin phalloidin staining.
Yeast strains were grown in YEPD or an appropriate defined synthetic complete medium for 16 h in log phase by continual dilution at 30°C. Strains were transferred to the appropriate temperature for the time indicated in the figure legends as required. For strains grown in synthetic medium, cells were shifted to YEPD for 4 h prior to staining. Fixation, staining, and sample preparation were performed as previously described (24). Images were captured with an IX70 inverted fluorescence microscope (Olympus) equipped with a HiQ fluorescein filter set (excitation wavelength, 450 to 492 nm), a planapochromatic 100× oil immersion objective lens, and a 100-W Hg lamp. Images were collected and analyzed with a Princeton Instruments 5-MHz MicroMax cooled-charge-coupled-device camera, shutter, and controller unit and IPLab software (version 3.5; Scanalytics). Cell size was determined and actin cables and patches were quantitated for a minimum of 100 cells counted from five different fields, and results were plotted as the average number of cells per size or the average number of actin cables or patches per cell.
In vivo [35S]methionine incorporation.
Strains containing each eEF1A mutant protein were prepared in the MET2 strain TKY892 by plasmid shuffling. The yeast strains TKY1056 (tpm1Δ), TKY1057 (mdm20Δ), and MC214 (wild type) were transformed with the MET2 plasmid pTKB975 to allow growth in C-Met medium. The three yeast strains were also transformed with plasmids expressing TPM1 (pTKB1005) or MDM20 (pTKB998) or with pRS426. Liquid cultures (100 ml) were grown in either C-Met or C-Ura-Met at 30°C to an A600 of 0.5 to 0.7, and experiments were performed with collections of aliquots in triplicate at 15-min intervals as previously described (24).
Protein purification and actin-eEF1A binding and -bundling assays.
Wild-type actin was purified from strain AAY1453 as described previously (24). eEF1A, eEF1A-Ura3p, and the eEF1A-Ura3p mutants were purified by the method of Cavallius et al. (10) with the adjustments described in reference 24. Actin binding and -bundling assays were performed by using previously described procedures (41, 48) with the following adaptations. eEF1A was dialyzed into cosedimentation assay buffer [20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 7.2), 2 mM EGTA, 1 mM dithiothreitol, 1 mM ATP, 2 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, and 0.25 mM GDP] for 4 h at 4°C. Dialyzed eEF1A and purified G-actin were clarified by centrifugation at 130,000 × g in a Sorvall Discovery M120SE tabletop ultracentrifuge for 40 min at 4°C. G-actin (3 μM) was added to cosedimentation assay buffer, followed by the addition of 0.125 μM eEF1A, eEF1A-Ura3p, or eEF1A-Ura3p mutant forms in a total volume of 100 μl. The mixture was incubated for 18 to 20 h at 4°C to allow equilibration and divided into Hitachi high-walled 500-μl tubes for centrifugation in a Sorvall Discovery M120SE at low speed (50,000 × g; 36,000 rpm) for 2 min at 4°C to assay bundling or high-speed (130,000 × g; 60,000 rpm) for 40 min at 4°C after a first low-speed centrifugation cycle to assay actin binding. Supernatants and pellets were separated and solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer. Densitometry analysis was performed using ImageQuant 5.2 (Molecular Dynamics).
Ribosome extraction and polyribosome profile analysis.
Yeast polyribosome preparation was performed as previously described (7) with the following modifications. Growth took place either in YEPD or in C-Ura medium when the expression of either TPM1 or MDM20 was required. Yeast cultures were grown at 30°C or shifted to 37°C for the time indicated in the figure legends, divided, and extracted with or without cycloheximide added to the cells (100 μg/ml) and lysis buffer (80 μg/ml). Cell extracts (A260, 25) were layered on a 35-ml 7 to 47% (wt/vol) sucrose gradient and centrifuged for 4 h at 23,000 rpm in a Surespin630 rotor. The A254 was monitored and recorded using a model 185 density gradient fractionator (ISCO, Inc., Lincoln, NE). Quantification of the 80S/polyribosome ratio and polyribosome areas was performed through measurement of the areas for different peak populations in a minimum of three replicate experiments using the ImageJ software 1.36b (Wayne Rasband, National Institutes of Health). The polyribosome area values were standardized relative to the value for the wild-type control for each experiment and are presented as percentages of the control value.
RESULTS
Several classes of mutations suppress eEF1A overexpression phenotypes.
Although the eEF1A-actin protein-protein interaction has been extensively studied in vitro, the location, functional consequences, and potential regulation of this association are not well understood. Using a unique genetic screen, we previously identified two mutations, N305S and N329S, that specifically inhibit the actin-bundling function of eEF1A without altering total protein translation in vivo (24). The positions and locations of these two amino acids indicated that the N-terminal region of domain III of eEF1A plays an essential function in its bundling interaction with actin.
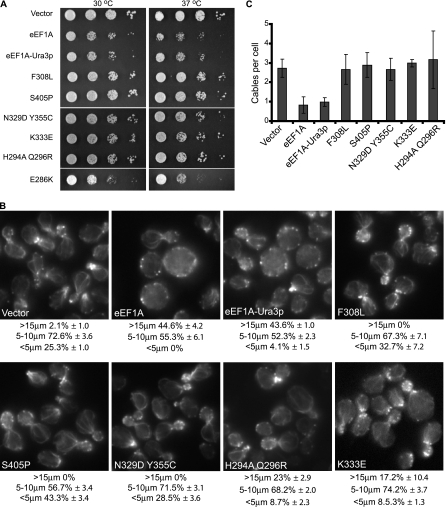
Extension of this genetic approach led to the identification of five additional mutant forms of the eEF1A-Ura3p fusion protein that no longer induce a severe growth defect when overexpressed (Fig. 1A). Two mutant forms (those with the F308L or S405P mutation) were found to fully suppress the slow-growth phenotype while the other three mutants (those with Y355C N329D, K333E, and H294A Q296R mutations) only partially suppressed the effect (Fig. 1A). Suppression of the slow growth was not due to reduced expression, as all mutant proteins were expressed at similar levels in the cells (data not shown). Because mutant forms of eEF1A that alter its intrinsic function in translation elongation could potentially affect the overexpression growth phenotype (see below), an E286K mutant form of eEF1A previously shown to significantly reduce the protein's activity in translation was analyzed (3, 55). Overexpression of the eEF1A-Ura3p E286K mutant form conferred a growth defect similar to that conferred by the overexpression of eEF1A (Fig. 1A), indicating that lowering the intrinsic activity of eEF1A in translation elongation is not sufficient to diminish the actin-dependent slow-growth phenotype.
FIG. 1.
eEF1A-Ura3p mutations suppress the overexpression phenotypes. (A) eEF1A-Ura3p mutants do not show an overexpression-induced slow-growth phenotype. TKY259 cells overexpressing eEF1A (pTKB731), the fusion eEF1A-Ura3p (pTKB744), or the F308L (pTKB884), S405P (pTKB886), K333E (pTKB881), H294A Q296R (pTKB883), N329D Y355C (pTKB880), or E286K (pTKB945) mutant forms of eEF1A-Ura3p were spotted as 10-fold serial dilutions onto C-Trp media and incubated at 30 or 37°C for 2 or 3 days, respectively. (B) Actin staining is restored in strains overexpressing eEF1A-Ura3p mutant forms. Cells described in the legend to panel A were further grown in YEPD medium for 4 h and stained with rhodamine phalloidin prior to mounting. Images were captured with an IX70 Olympus inverted fluorescence microscope equipped with a planapochromatic 100× oil immersion objective lens. Cells were scored as large (>15 μm), medium (5 to 10 μm), and small (<5 μm), and results are presented as percentages of the total population. (C) The overexpression of eEF1A-Ura3p mutants restored actin cable numbers. Cells from the experiment described in the legend to panel B were scored for numbers of actin cables per cell, and the averages of results were plotted.
We next determined whether cell morphology and the actin cytoskeleton were improved when the mutant forms of the eEFA-Ura3p fusion protein, compared to the wild-type protein, were overexpressed. We have previously shown that eEF1A overexpression results in dramatic changes in both the actin cytoskeleton and cell size (24, 48). eEF1A and eEF1A-Ura3p overexpression led to similar effects, mainly an increase in cell size such that about 50% of the population was >15 μm in diameter (Fig. 1B) and the loss of actin cables throughout the cells (<1 cable per cell) (Fig. 1C) as determined by actin staining using rhodamine phalloidin. Overexpression of the five mutant forms led to various levels of amelioration of both cell size and actin structural defects (Fig. 1B and C). All mutants showed a restoration of actin cable formation (Fig. 1B and C). Cell size and morphology were also improved when the different mutants were overexpressed (Fig. 1B). Cells overexpressing the F308L, S405P, or Y355C N329D mutant form presented normal morphology and cell sizes, with 100% of the cells <15 μm in diameter (Fig. 1B) compared to approximately 45% with eEF1A overexpression. The overexpression of the K333E or H294A Q296R mutant form led to 17.2 or 23% of the population's measuring >15 μm in diameter, respectively (Fig. 1B). However, the size distribution was closer to that found among wild-type cells than was that found among cells overexpressing eEF1A or eEF1A-Ura3p, suggestive of partial suppression of the actin-dependent phenotype. Overexpression of the eEF1A-Ura3p E286K mutant form induced slow growth and disorganization of the actin cytoskeleton, similar to the overexpression of eEF1A or eEF1A-Ura3p (data not shown), demonstrating that inhibiting the eEF1A function in protein synthesis does not necessarily affect its actin-associated properties.
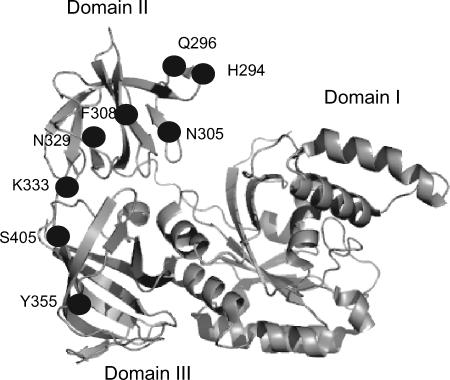
All the mutations identified via this genetic screen (this study and reference 24) were mapped on the structure of eEF1A. Strikingly, all localized on one face of the protein (Fig. 2). The mutations clustered into two different folds of the protein, on the tip of domain II between amino acids 290 and 310 (H294A Q296R, N305S, and F308L) and the strand connecting domains II and III (N329S, N329D Y355C, and K333E). The S405P mutation is relatively far away when considering the linear sequence; however, it clusters near those located on the connecting strand of the domains.
FIG. 2.
The structure of eEF1A (5) is shown with the mutations clustering on one face of the protein.
eEF1A-Ura3p F308L and S405P mutant forms affect growth, cell morphology, and the actin cytoskeleton.
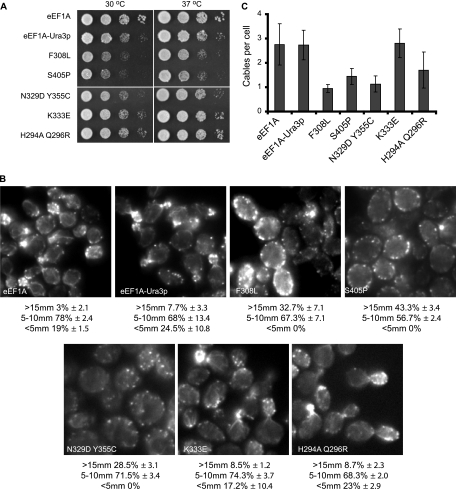
The five different mutated forms of the eEF1A-Ura3p fusion were all able to function as the only form of the eEF1A protein as determined by plasmid shuffling (Fig. 3A). The eEF1A-Ura3p protein was functional as the only form and was found to slightly reduce cell growth (24). F308L and S405P mutant strains presented pronounced growth defects at 30°C and, more significantly, at 37°C (Fig. 3A). The strains expressing the N329D Y355C, K333E, and H294A Q296R mutant proteins grew similarly to the strain expressing the wild-type eEF1A-Ura3p fusion protein.
FIG. 3.
eEF1A-Ura3p mutant proteins are functional as the only form of eEF1A, and strains expressing them demonstrate altered cell growth, morphology, and actin cytoskeletal organization. (A) eEF1A-Ura3p mutant strains show differential growth effects. Strain TKY803 containing the wild-type TEF1 LYS2 plasmid and deletions of the two genes encoding eEF1A (tef1::LEU2 and tef2Δ) was transformed with the plasmids expressing the eEF1A-Ura3p mutant forms described in the legend to Fig. 1, and the loss of wild-type eEF1A was monitored on α-aminoadipate. Strains expressing eEF1A (TKY880), eEF1A-Ura3p (TKY881), or the five mutant forms of eEF1A (TKY882, TKY883, TKY885, TKY886, and TKY888) were diluted to an A600 of 1.0, spotted as 10-fold serial dilutions onto YEPD plates, and incubated at 30 and 37°C for 2 and 3 days, respectively. (B) Loss of actin organization and cell size in eEF1A-Ura3p mutants. Cells described in the legend to panel A were grown in YEPD medium and stained with rhodamine phalloidin prior to mounting. Cells were scored as large (>15 μm), medium (5 to 10 μm), and small (<5 μm), and results are presented as percentages of the total population. (C) Actin cable numbers are generally reduced when eEF1A-Ura3p mutants are expressed as the only form of eEF1A. Cells described in the legend to panel B were scored for numbers of actin cables per cell, and the averages of results were plotted.
Changes to the cell morphology and actin cytoskeletons of strains expressing only the mutant eEF1A-Ura3p fusion forms were determined by rhodamine phalloidin staining (Fig. 3B). Cells expressing eEF1A, eEF1A-Ura3p, or the K333E mutant form demonstrated similar actin staining patterns, cell sizes, and morphologies (Fig. 3B) and comparable numbers of actin cables (Fig. 3C). The H294A Q296R mutant strain did not show any significant changes in overall actin organization or cell size, although slightly fewer actin cables per cell were seen. The numbers of actin cables were reduced in the N329D Y355C, F308L, and S405P mutant strains to an average of approximately 1 per cell (Fig. 3C). An increase in cell size was pronounced among the F308L, N329D Y355C, and, most significantly, S405P eEF1A-Ura3p mutant strains (Fig. 3B). The N329D Y355C mutant strain showed changes in overall actin organization and size similar to those in the previously identified N329S mutant strain (24). The loss of a defined actin cytoskeleton and significant changes in the numbers of actin patches localized around the cell periphery were most dramatic in the F308L and S405P eEF1A-Ura3p mutant strains (24). Taken together, these data demonstrate that a series of strains expressing mutant eEF1A-Ura3p forms show differential disruption of both cell and actin organization in living cells, allowing for the analysis of the effects of changes in the actin-directed functions of eEF1A.
eEF1A-Ura3p F308L and S405P mutant forms demonstrate deficient actin bundling in vitro.
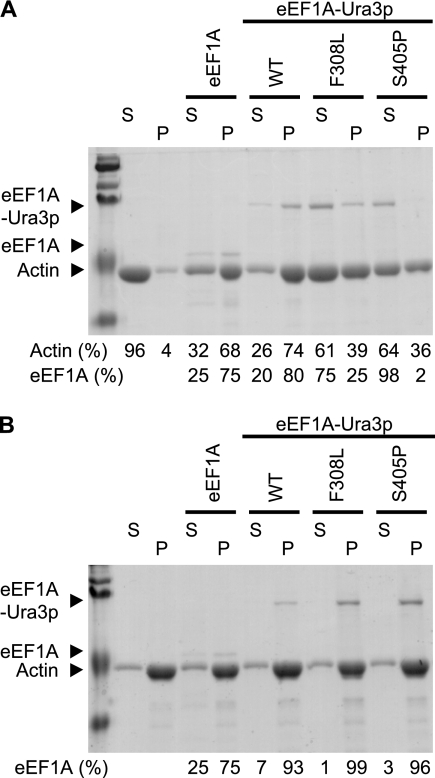
Because of the nature of the genetic screen and the actin cytoskeletal disorganization in some yeast strains expressing a mutant form of eEF1A-Ura3p as the only copy, we investigated whether the mutant proteins demonstrated any alteration in their actin-bundling activities. We chose to analyze the F308L and S405P mutants because of the clear inability to disrupt the actin cytoskeleton when overexpressed (Fig. 1), as well as the deficiency in actin organization when the mutant fusion proteins were expressed as the only form (Fig. 3B). Their ability to both bind and bundle actin in vitro was determined using a cosedimentation assay (Fig. 4). Purified actin, at the concentration used here, was unable to bundle and consequently pellet in a low-speed spin (4% of total actin). The addition of purified eEF1A, or eEF1A-Ura3p to a slightly lower extent, was sufficient to induce the polymerization of a complex that pelleted after centrifugation, yielding 68% or 74% of the actin in the pellet, respectively (Fig. 4A). F308L and S405P mutant proteins showed an approximately 50% reduction in the ability to bundle and thus pellet actin. The eEF1A mutant forms were preferentially recovered in the supernatant along with actin, at 75% (F308L) and 98% (S405P), compared to eEF1A-Ura3p and eEF1A (25% and 20%, respectively) (Fig. 4A). These data correlate with the loss of actin disorganization observed when these proteins were overexpressed (Fig. 1) or when the eEF1A mutant proteins were the sole form (Fig. 3). No significant changes in actin binding in any of the mutants could be observed when they were assayed for their abilities to pellet during high-speed centrifugation (Fig. 4B).
FIG. 4.
The F308L and S405P eEF1A-Ura3p mutants are deficient in bundling actin in vitro. Actin-bundling (A) and binding (B) assays were performed with purified yeast eEF1A, eEF1A-Ura3p, or the different eEF1A-Ura3p mutants. Actin polymerized in the presence of eEF1A, eEF1A-Ura3p, or the eEF1A-Ura3p mutants was collected by low-speed centrifugation for the bundling assay (A) or high-speed centrifugation for the binding assay (B). Supernatants (S) and pellets (P) were resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Gelcode blue. The positions of yeast actin, eEF1A, and eEF1A-Ura3p are indicated. Densitometry analysis was performed using ImageQuant 5.2, and the intensities of the signals are represented as percentages of the total (supernatant and pellet). WT, wild type.
Strains carrying F308L and S405P eEF1A-Ura3p mutant forms show a reduction in total protein translation.
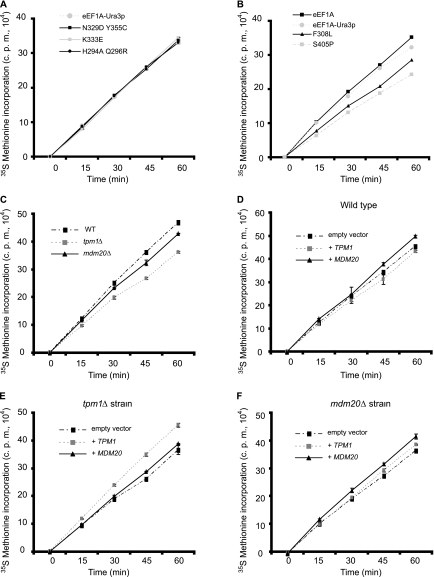
Although the integrity of the actin cytoskeleton and cell morphology were severely affected in strains expressing the different eEF1A mutants, it was unlikely that these phenotypes were solely responsible for the slow growth observed. Previous analysis of an N305S eEF1A mutant strain also revealed the disruption of the actin cytoskeleton but no significant differences in growth (24). It was possible, however, that these eEF1A mutations might also affect the canonical activity of the factor in translation elongation and, consequently, growth rates. To test this hypothesis, total translation was monitored by in vivo [35S]methionine incorporation (Fig. 5A and B). Compared to a wild-type-eEF1A strain, no significant differences in levels of total protein synthesis in the strains expressing the eEF1A-Ura3p fusion protein or the K333E, H294R Q296A, or N329D Y355C mutant form were found (Fig. 5A). The F308L and S405P mutant strains, however, demonstrated a significant reduction in [35S]methionine incorporation (Fig. 5B). A correlation between cell growth and total translation was observed, as the slowest growing strain carrying the S405P mutation reduced translation by about 20 to 25%. The F308L mutant strain, which grew slightly better than the S405P strain, reduced incorporation by 15%. Taken together, these data demonstrate that two specific mutations in eEF1A that affect its actin-bundling activities in vitro, as well as cell organization, the actin cytoskeleton, and cell growth, also lead to reductions in total protein synthesis.
FIG. 5.
A subset of eEF1A-Ura3p and actin-bundling protein mutant strains show reduced total translation. Strain TKY892 containing the wild-type TEF1 LYS2 plasmid and deletions of the two genes encoding eEF1A (tef1::LEU2 and tef2Δ) was transformed with the plasmids expressing the eEF1A-Ura3p mutant forms described in the legend to Fig. 1, and the loss of wild-type eEF1A was monitored on α-aminoadipate. N329D Y355C, K333E, and H294A Q296R (TKY897, TKY898, and TKY900) (A) or F308L and S405P (TKY901 and TKY903) (B) mutant strains and the strains expressing eEF1A (TKY895) and eEF1A-Ura3p (TKY896) were grown in C-Met to mid-log phase, and total protein synthesis was measured by trichloroacetic acid precipitation of [35S]methionine-labeled proteins. The strains from which TPM1 (TKY1056) and MDM20 (TKY1057) had been deleted and the corresponding wild-type strain (MC214) were transformed with pTKB975 to allow growth in C-Met (C). Strains were grown and the assay was performed as described in the legend to panel A. Strains MC214 (D), TKY1056 (E), and TKY1057 (F) were transformed with plasmids expressing TPM1 (pTKB1005) or MDM20 (pTKB998) or the corresponding empty vector (pRS426). Strains were grown in C-Met-Ura and the assay was performed as described in the legend to panel A.
Inhibiting translation elongation is not sufficient to affect actin cytoskeleton organization.
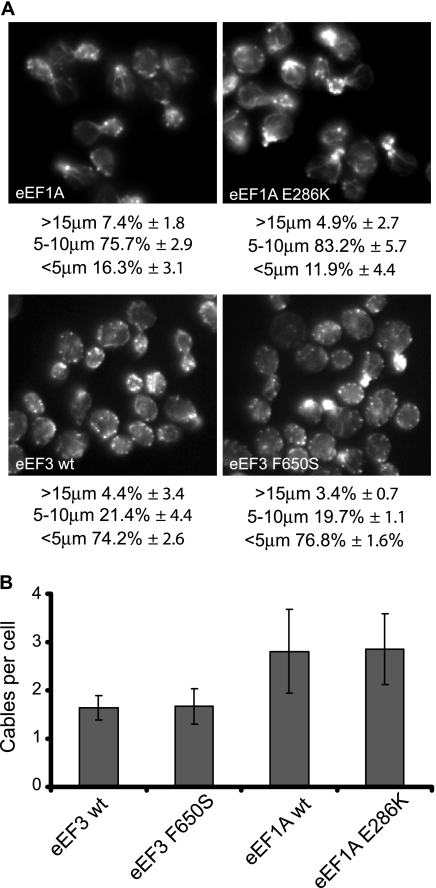
Since a subset of eEF1A mutations that alter the actin cytoskeleton also reduce total translation, it is possible that the inhibition of protein synthesis alone could be responsible for the changes seen in actin cytoskeleton organization in the eEF1A mutant strains. To rule out this possibility, we analyzed the organization of the actin cytoskeleton in two different translation elongation factor mutant strains in which total protein synthesis was reduced (Fig. 6A). Actin staining of strains expressing either the eEF1A E286K or eEF3 F650S mutant form, both of which lead to the inhibition of protein synthesis by 50% (3), was analyzed and compared to that of the isogenic wild-type strain. As shown in Fig. 6, while the mutant proteins were expressed in two different strain backgrounds with slight differences between the cytoskeletons of the wild-type strains, no significant changes in actin cable numbers in the strains expressing the mutated forms of the elongation factor were observed compared to those in their respective wild-type strains. The eEF1A strains contained an average of 2.8 cables per cell, while the eEF3 strains showed about 1.4 cables per cell (Fig. 6B). Similar results were obtained for cell size and morphology (Fig. 6A), as no significant differences between wild-type and mutant strains could be seen. Taken together, these data indicate that the changes in the actin cytoskeleton and cell morphology observed with eEF1A mutants are due not to the defective rates of elongation but more likely to the reduced actin-bundling activity of the protein.
FIG. 6.
eEF1A or eEF3 mutant strains with reduced total translation do show not altered cell morphology or actin cytoskeletal organization. (A) Cells expressing eEF1A (TKY621), the eEF1A E286K mutant (TKY622), wild-type EF3 (eEF3 wt; TKY702), or the eEF3 F650S mutant (TKY707) were grown in YEPD medium and stained with rhodamine phalloidin prior to mounting. Cells were scored as large (>15 μm), medium (5 to 10 μm), and small (<5 μm), and results are presented as percentages of the total population. (B) eEF1A or eEF3 mutant strains show no changes in the numbers of actin cables per cell. Cells described in the legend to panel A were scored for the numbers of actin cables per cell, and averages of the results were plotted.
Strains carrying F308L and S405P mutant forms of eEF1A-Ura3p show an initiation defect.
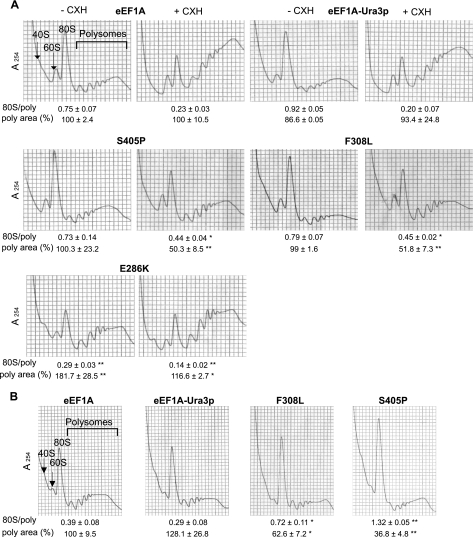
The reduction in total protein synthesis in strains carrying the F308L or S405P mutant form of eEF1A raises the possibility that the translation activity of the factor was affected, even though reduced translation elongation is not sufficient to account for the actin phenotypes observed. To determine whether these mutations affected the rate of elongation, we prepared extracts from strains expressing eEF1A, eEF1A-Ura3p, or F308L or S405P mutant forms of the eEF1A-Ura3p fusion for polyribosome profile analysis. In a strain in which elongation is reduced, extracts made in the absence of cycloheximide showed an accumulation of polyribosomes. This accumulation was observed in an E286K eEF1A mutant strain (Fig. 7A) in which the translation elongation activity is reduced (3). The 80S/polyribosome ratio for this strain demonstrated a significant retention of the ribosome population in polyribosomes (0.29 ± 0.03; P < 0.001) compared to that in either the eEF1A or eEF1A-Ura3p fusion strain (0.75 ± 0.07 or 0.92 ± 0.05, respectively). Strikingly, no increases in polyribosomes were observed in the extracts from strains expressing the F308L or S405P mutant forms of eEF1A-Ura3p in the absence of cycloheximide, as seen from the 80S/polyribosome ratio (0.79 ± 0.07 or 0.73 ± 0.14, respectively). Although unexpected, these data suggest that the reductions in protein translation seen in strains expressing the F308L or S405P eEF1A mutant forms are not due to an altered elongation function of the eEF1A protein.
FIG. 7.
F308L and S405P eEF1A-Ura3p mutant strains show an initiation defect. (A) Ribosome extracts of F308L and S405P eEF1A-Ura3p mutant strains (TKY901 and TKY903, respectively) and strains expressing eEF1A (TKY895), eEF1A-Ura3p (TKY896), or the eEF1A E286K mutant (TKY622) were grown at 30°C and prepared and analyzed in the presence (+CHX) or absence (−CHX) of cycloheximide by using 7 to 47% sucrose gradients. (B) Cells were prepared as described in the legend to panel A with cycloheximide after being shifted to 37°C for 6 h. Significant differences in 80S/polyribosome ratios compared to that for the wild type are indicated by either one asterisk (P < 0.05) or two asterisks (P < 0.001; Student's t test). Significant differences in the polyribosome areas, standardized into percentages relative to the value for the wild-type control for each experiment, are indicated by either one asterisk (P < 0.05) or two asterisks (P < 0.001; Student's t test).
An analysis of the polyribosome profiles of cells expressing either eEF1A, eEF1A-Ura3p, or the E286K eEF1A mutant protein in the presence of cycloheximide revealed significant retention of polyribosomes in the heavy fraction of the sucrose gradient (Fig. 7A). The 80S/polyribosome ratios were found to decrease significantly, by three- to fourfold. The accumulation of polyribosomes, however, was significantly reduced in extracts when strains expressing the F308L or S405P mutant forms were prepared and analyzed in the presence of cycloheximide, where the 80S/polyribosome ratio was decreased by less than 1.7-fold. Such a phenomenon could be due to either a reduction in cytoplasmic mRNA abundance, an acquired resistance to cycloheximide, or a slowdown of the initiation step. To eliminate the first possible explanation, the polyribosome profiles from the different eEF1A strains were analyzed to determine the sizes of the polyribosome fractions. A reduced abundance of cytoplasmic mRNAs would result in an increase in the area under the 80S peak with no changes in the areas under the polysome peaks. Average areas for the different polyribosome fractions were calculated, and the values were standardized into percentages of the wild-type control value for each experiment (Fig. 7). These data correlate the 80S/polyribosome ratio with a decrease in polyribosomal areas and are consistent with a block in initiation for the eEF1A mutants which is not due to a limitation of the mRNA cytoplasmic pool. Acquired cycloheximide resistance was tested by performing a growth inhibition assay (Table 2). Strains expressing the eEF1A-Ura3p fusion forms of eEF1A were found to be more sensitive to cycloheximide than a strain expressing eEF1A (P < 0.05), suggesting that the presence of the Ura3p tag slightly affects the properties of the eEF1A protein. Interestingly, however, the two mutant strains that showed a strong reduction of total translation, those expressing F308L and S405P mutant forms of eEF1A, demonstrated the greatest sensitivity to cycloheximide compared to the strains expressing either eEF1A or the eEF1A-Ura3p fusion (P, <0.001 or <0.05, respectively). This indicates that increased resistance to the drug cannot explain the absence of polyribosomes.
TABLE 2.
Drug sensitivities of the different eEF1A-Ura3p mutant strainsa
| Strain | Diam (mm) of area of inhibition |
|---|---|
| Strain expressing eEF1A | 11 ± 1.15 |
| Strain expressing eEF1A-Ura3p | 13 ± 0.95b |
| F308L mutant | 16.5 ± 1.00b,c |
| S405P mutant | 19.25 ± 1.25b,c |
| Y355C N329D mutant | 15.25 ± 0.94b |
| K333E mutant | 15.5 ± 1.29b |
| H294A Q296R mutant | 14 ± 0.81b |
Cells described in the legend to Fig. 3 were grown in YEPD medium and diluted to an A600 of 0.5 before being spread on YEPD. Cell sensitivity was determined by measuring the diameter of the area of growth inhibition around a filter containing 0.7 mM cycloheximide.
P, <0.05 for comparison with the strain expressing eEF1A.
P, <0.05 for comparison with the strain expressing the eEF1A-Ura3p fusion.
Because the strains expressing the mutant forms of eEF1A presented severe morphological and actin organization defects, we sought to exacerbate these phenotypes by shifting the strains for 6 h at 37°C, a temperature which also reduces their growth. These conditions led to the deterioration of both the organization of the actin cytoskeleton and cellular morphology, as strains expressing the F308L or S405P eEF1A-Ura3p mutant form demonstrated a significant increase in cell size (data not shown). Since the actin cytoskeleton in yeast is highly regulated and organized to adjust to external conditions, stresses such as heat shock have been reported to depolarize and disorganize the actin cytoskeleton (11, 16). Thus, as a control the eEF1A and eEF1A-Ura3p strains were subjected to the same temperature shift. The temperature shift reduced the number of actin cables in the eEF1A strain from an average of 2.8 to fewer than 1.5 cables per cell, modestly increased the average cell size, and reduced the number of polarized cells (data not shown). The temperature shift caused similar changes in the eEF1A-Ura3p strain in actin organization and cell morphology; however, a third of the population increased to a diameter of >15 μm, suggesting that the Ura3p tag may affect eEF1A function under stress.
Cell extracts were prepared after the temperature shift and analyzed by using polyribosome gradients in the presence of cycloheximide. A small but significant increase in the 80S peak (Fig. 7B), as well as a corresponding augmentation in the 80S/polyribosome ratio, was observed in either the eEF1A or the eEF1A-Ura3p strain after the temperature shift compared to the same strains grown at 30°C (Fig. 7A). Strains carrying the S405P or F308L eEF1A-Ura3p mutant forms, however, demonstrated a clear block in initiation, as a majority of the ribosomes were found in the 80S peak. The 80S/polyribosome ratio demonstrated that the F308L mutant strain had a more than twofold increase in the population of ribosomes in the 80S peak (0.72 ± 0.11 compared to 0.39 ± 0.08 for the wild type; P < 0.05). The S405P mutant strain presented a dramatic 3.5-fold increase in the 80S population (1.32 ± 0.05 compared to 0.39 ± 0.08 for the wild type; P < 0.001). Determination of the average polyribosome fraction areas also demonstrated a reduction in the amount of polysomes, consistent with a reduced initiation that is not due to a limitation of the mRNA cytoplasmic pool. These data suggest that specific eEF1A mutations that alter the actin-bundling activity of eEF1A in vitro and actin organization in vivo reduce translation via reduced initiation.
Disruption of the actin cytoskeleton by deletion or mutation of actin binding proteins leads to a block in initiation.
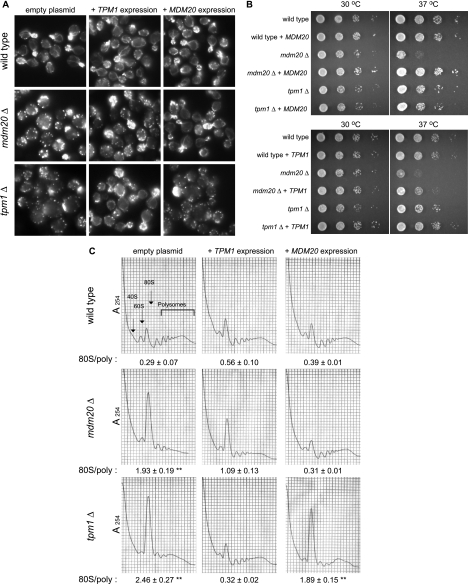
The above-described data suggested a link between actin organization and translation initiation independent of eEF1A's function in translation elongation. Because of the general loss of actin cables and the enhanced presence of actin patches in eEF1A mutant strains with reduced translation, strains carrying a single deletion of other well-characterized proteins involved in actin organization were analyzed. The tropomyosin isoform Tpm1p (17, 42) and the mitochondrial disruption and morphology protein (Mdm20p) (25) have been reported to participate in the elaboration and composition of the actin cytoskeleton. Initially, the effect of the loss of either protein was monitored with the Open Biosystem deletion library strains, in which clear effects on cell size, the actin cytoskeleton, and translation were observed (data not shown). To ensure a clean genetic background and allow direct comparison to the effects of eEF1A mutant forms, the genes encoding these proteins were deleted in strain MC214 by in vivo recombination. Actin cytoskeleton organization in the control strain demonstrated normal-sized and -shaped cells (Fig. 8A, top panel), with numerous actin cables generating from the buds and elongating throughout the cell body and clustered actin patches. Interestingly, the presences of a high-copy-number plasmid expressing Mdm20p or Tpm1p in the wild-type strain did not induce any changes in the integrity of the actin cytoskeleton (Fig. 8A, top panel).
FIG. 8.
The loss of TPM1 or MDM20 reduces protein synthesis at the step of initiation. The wild-type strain (MC214) and isogenic strains in which TPM1 (TKY1056) or MDM20 (TKY1057) had been deleted were transformed with a plasmid expressing TPM1 (pTKB1005) or MDM20 (pTKB998) or the corresponding empty vector (pRS426). Strains were grown in C-Ura. TPM1 and MDM20 expression complemented the loss of actin organization in (A) and the growth defect of (B) tpm1Δ and mdm20Δ strains, respectively. (A) Strains were grown in C-Ura for 16 h and further incubated in YEPD medium for 4 h at 30°C and stained with rhodamine phalloidin prior to mounting. (B) Cells were diluted to an A600 of 1.0, spotted as 10-fold serial dilutions onto C-Ura plates, and incubated at 30 and 37°C for 2 and 3 days, respectively. (C) The loss of TPM1 or MDM20 induces an accumulation at the 80S peak. Ribosome extracts of the different strains were prepared and analyzed in the presence of cycloheximide by using 7 to 47% sucrose gradients. Significant differences in the 80S/polyribosome ratios compared to that for the wild type are indicated by either one asterisk (P < 0.05) or two asterisks (P < 0.001; Student's t test).
As previously reported (25, 53), the tpm1 and mdm20 deletion strains showed a similar loss of actin cables and complete disruption of the actin cytoskeleton structure, as seen in the eEF1A mutant strains, and were confirmed in this strain background (Fig. 8A). The expression of Tpm1p or Mdm20p in the corresponding deleted strain compensated for the loss of the protein and reestablished proper actin organization. Interestingly, the overexpression of Tpm1p was able to partially suppress the actin phenotype seen in the mdm20Δ strain (Fig. 8A, middle panel), with clear amelioration of the formation of actin structures, mainly cables. Mdm20p overexpression, however, did not have any effects on actin organization in the tpm1Δ strain (Fig. 8A, bottom panel). Similar observations on complementation and suppression were made in analyzing the growth of the different strains (Fig. 8B). The deletion of either TPM1 or MDM20 in the MC214 strain induced a slight mutant growth phenotype at 30°C (for either strain) and at 37°C (for the mdm20Δ strain), which was complemented by the expression of the corresponding protein. The overexpression of Tpm1p in the mdm20Δ strain also partially suppressed the growth phenotype, especially at 37°C (Fig. 8B, right panel).
The analysis of polyribosome profiles of extracts from the tpm1Δ and mdm20Δ strains indicated a significant correlation between the disorganization of the actin cytoskeleton and an increased 80S peak (Fig. 8C). A reproducible increase in the 80S/polyribosome ratio for the mdm20Δ (1.93 ± 0.19; P < 0.05) and tpm1Δ (2.46 ± 0.27; P < 0.001) strains compared to that for the wild-type strain (0.29 ± 0.07) was observed. The changes in the average polyribosome fraction area were 43.4 ± 4.6 for the mdm20Δ strain and 43.4 ± 4.6 for the tpm1Δ strain relative to the average area for the wild-type strain (100 ± 10.1). These data demonstrate that a significant reduction in the polyribosomal pool was seen in both the mdm20Δ and tpm1Δ strains, consistent with reduced initiation and not a lower number of cytoplasmic mRNAs. The expression of the corresponding protein in the mdm20Δ and tpm1Δ strains resulted in complementation and gave ribosome profiles similar to that of the wild type. Similar to the observations made when we studied actin staining or growth, we found that the overexpression of Tpm1p in the mdm20Δ strain partially suppressed the accumulation of the 80S peak and reduced the 80S/polyribosome ratio (1.09 ± 0.13; P < 0.05) to be closer to that of the wild type (0.29 ± 0.07). The overexpression of Tpm1p in the wild-type strain also demonstrated a small but significant accumulation at the 80S peak (0.56 ± 0.10; P < 0.05) compared to that in the wild type (0.29 ± 0.07).
To see whether the accumulation at the 80S peak in the tpm1Δ and mdm20Δ strains related to altered total protein synthesis, the level of in vivo [35S]methionine incorporation was determined (Fig. 5C). Compared to the wild-type strain, the mdm20Δ and tpm1Δ deletion strains demonstrated a 5 to 10% and a 10 to 15% reduction in [35S]methionine incorporation, respectively. Total translation was most affected in the tpm1Δ strain, correlating with the differences seen in the polyribosome profiles. The expression of either Tpm1p or Mdm20p in the corresponding deletion strain compensated for the loss of the gene and restored wild-type levels of [35S]methionine incorporation (Fig. 5E and F). The overexpression of Tpm1p in the mdm20Δ strain (Fig. 5F) restored total translation to an extent similar to that of the effect seen in the polyribosome profile analysis (Fig. 8C), cell morphology (Fig. 8A), and growth (Fig. 8B).
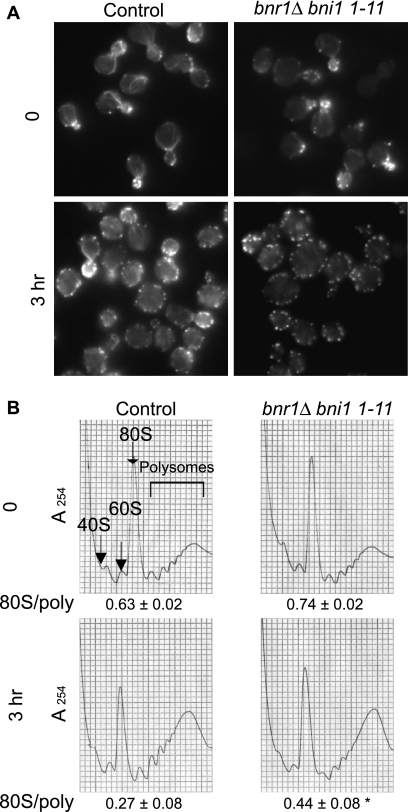
Another set of essential proteins that regulate actin cable organization in yeast is the formin protein family, bud neck-involved protein 1 (Bni1p) and bud neck-related protein 1 (Bnr1p). The deletion of BNR1 in a strain harboring the bni1-11 (D1511G K1601R) allele leads to the loss of function of the protein Bni1p after a shift to the restrictive temperature of 37°C (21). The bnr1Δ bni1-11 strain was shifted to 37°C for 3 h before cell staining or extract preparation for polyribosome analysis (Fig. 9), a time frame during which no significant changes in growth rates were observed (data not shown). Incubation at 37°C led to the rapid loss of actin organization as previously observed (21), and this loss was more pronounced after 3 h (Fig. 9B). Polyribosome profiles and the 80S/polyribosome ratio were similar in the wild-type and bnr1Δ bni1-11 strains when the strains were grown at a permissive temperature of 20°C. Following incubation at 37°C, a clear accumulation at the 80S peak and, thus, an increase in the 80S/polyribosome ratio was observed for the bnr1Δ bni1-11 strain (0.44 ± 0.08 compared to 0.27 ± 0.08 for the wild type; P < 0.05).
FIG. 9.
Mutations in the formins Bni1p and Bnr1p lead to a block in initiation. (A) Loss of actin organization in bnr1Δ bni1-11 strains. Y1239 (wild-type) and Y3024 (bnr1Δ bni1-11) strains were grown in YEPD medium at 22°C before being shifted to 37°C for 3 h before staining with rhodamine phalloidin prior to mounting. (B) A bnr1Δ bni1-11 strain shows an accumulation at the 80S ribosome peak. Ribosome extracts from the strains described in the legend to panel A were prepared and analyzed in the absence of cycloheximide by using 7 to 47% sucrose gradients. Significant differences in 80S/polyribosome ratios compared to that for the wild type are indicated by an asterisk (P < 0.05; Student's t test).
Disruption of the actin cytoskeleton in certain actin mutant strains leads to a block in initiation.
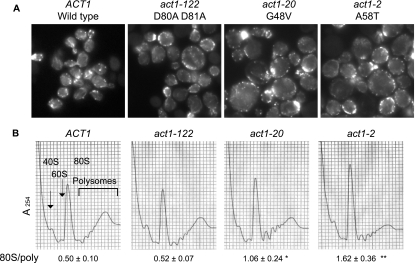
Our data indicate that correct actin organization through the role of different actin binding proteins (Tpm1p, Mdm20p, formins, and eEF1A) has direct consequences on the regulation of the initiation step. To see whether actin itself could also be responsible for a possible control of translation, polyribosome profile analysis of several actin mutant strains was performed (Fig. 10). The control strain presented normal-sized and -shaped cells (Fig. 10A, left panel), with numerous actin cables generating from the buds and elongating throughout the cell body. Both the polyribosome profile and the 80S/polyribosome ratio indicated normal physiological distribution. Actin mutants selected for this study were previously reported to affect translation and drug sensitivity to various extents (35). Actin organization and cell size were affected in strains expressing the actin mutants act1-2, act1-20, and act1-122 (Fig. 10A). Loss of actin structures, mainly cables, was seen throughout the cells. Interestingly, however, translation initiation as assessed through polyribosome analysis was differentially affected in these strains in a manner that correlated with the extent of the increase in cell size (Fig. 10A) and the reduction in [35S]methionine incorporation (35). The act1-122 (D80A D81A) strain showed no effect on the distribution of polyribosomes and an 80S/polyribosome ratio similar to that of the wild type (0.52 ± 0.07 compared to 0.50 ± 0.10). In contrast, the act1-20 (G48V) mutant (1.06 ± 0.24; P < 0.05) and, more significantly, the act1-2 mutant (1.62 ± 0.36; P < 0.001) showed an increase in the 80S peak and the 80S/polyribosome ratio compared to the wild type (0.50 ± 0.10). These two mutants showed the greatest increase in cell size and the most significant reduction in total protein synthesis. Thus, similar to those in eEF1A, a series of mutations within the same protein yield differential effects on translation but subtle but distinct cytoskeletal phenotypes and altered cell size. Taken together, these data demonstrate that inhibiting proper actin organization, through either the reduction of eEF1A actin-bundling activities, the deletion of TPM1 or its regulator MDM20, the loss of formin function, or specific actin mutations, leads to a general limitation of translation initiation.
FIG. 10.
Loss of cytoskeletal organization in actin mutant strains can induce a block in initiation. (A) Loss of cytoskeletal organization in actin mutant strains. The ACT1 (wild-type; IGY191) strain and act1-122 (D80A D81A; IGY58), act1-20 (G48V; IGY88), and act1-2 (A58T; IGY116) mutant strains were grown in YEPD medium at 30°C for 16 h before staining with rhodamine phalloidin prior to mounting. (B) Specific mutations in actin lead to an accumulation at the 80S ribosome peak. Ribosome extracts from the different strains grown as described in the legend to panel A were prepared and analyzed in the absence of cycloheximide by using 7 to 47% sucrose gradients. Significant differences in 80S/polyribosome ratios compared to that for the wild type are indicated by either one asterisk (P < 0.05) or two asterisks (P < 0.001; Student's t test).
DISCUSSION
Since the first report demonstrating the interaction of eEF1A with the actin cytoskeleton (64), the consequences of their association have generated great interest. Further evidence of direct connections between the translation machinery and the actin cytoskeleton has accumulated. The binding of eEF1A to actin is now regarded as a function that may link two distinct cellular processes, cytoskeletal organization and gene expression. Although eEF1A-actin interactions in vitro have been extensively studied (40, 41), the location and the potential regulation of eEF1A-actin association, especially in vivo, are not well understood. Using a unique genetic screen, we have identified seven mutations that reduce the actin disorganization induced by the overexpression of eEF1A. This analysis mapped the regions and residues of eEF1A essential for such properties (Fig. 2) (24). The first cluster of mutations is located on the tip of domain II between amino acids 290 and 310 (H294A Q296R, N305S, and F308L). The crystal structure of the EF-Tu-Phe-tRNAPhe-GDPNP complex of the prokaryotic homolog of eEF1A indicates that H273, H274, and R300 recognize the aminoacyl-tRNA (49). The equivalent H293, H294, and R320 residues on eEF1A are proposed to similarly bind aminoacyl-tRNA and contact eEF1Bα (4). The striking similarity of regions linked to aminoacyl-tRNA, eEF1Bα, and actin binding may not have been obtained by mere coincidence. Previous work in vitro has demonstrated that actin binding and bundling by eEF1A are significantly reduced in the presence of aminoacyl-tRNA (41). The possibility that actin bundling-defective mutant forms of eEF1A may affect aminoacyl-tRNA or eEF1Bα binding is therefore feasible. However, such changes would likely affect the intrinsic activity of eEF1A in elongation, as seen with the E286K mutant form of eEF1A. Although the inhibition of total protein synthesis in the F308L eEF1A-Ura3p mutant strain was observed (Fig. 5), the cause of such a defect is most likely an initiation defect. Additionally, the E286K form of eEF1A does not affect the actin-dependent phenotypes. Thus, while a related site in eEF1A likely contributes to the binding of eEF1Bα, aminoacyl-tRNA, and actin, unique aspects of each interaction are clear from specific eEF1A mutants. The H294A Q296R and N305S mutations clearly did not alter eEF1A elongation activity, with the mutant strain showing a wild-type level of total translation (Fig. 5) (24) and no dramatic changes in polyribosome accumulation (Fig. 7; data not shown) (24).
The other set of mutations identified through the screen were either localized on the strand connecting domains II and III or clustered near the connecting strand (N329S, N329D Y355C, K333E, and S405P) (Fig. 2). It is therefore possible that the connecting strand between these two domains, or the orientation of the domains themselves, plays an important regulatory function in eEF1A-actin complex formation.
Interestingly, none of the mutations identified, except for S405P, were actually located in domain III. Although this screen was originally based on data showing that domain III truncations and fragments affect the actin-bundling phenotypes (24, 40), the nature of the assay itself may have prevented such an outcome. First, eEF1A interacts with actin through several regions of domain III, as indicated by our data and those of others (24, 40). It was not sufficient to remove this specific domain, since such a mutation only partially suppresses the eEF1A overexpression phenotype (24). Second, the addition of the Ura3p fusion at the C terminus of eEF1A may also have affected the results. Although indispensable for an efficient screen, the addition of a 25-kDa polypeptide appears to slightly affect the actin-bundling properties of eEF1A. eEF1A-Ura3p showed a slight reduction in bundling efficiency in the cosedimentation assay (Fig. 4), and when this fusion protein was overexpressed, the strain showed slightly less growth inhibition. Last, although indiscernible in vivo at permissive conditions (Fig. 3), the cell morphology of an eEF1A-Ura3p strain was slightly affected at the restrictive temperatures. The absence of changes in either total protein synthesis (Fig. 5) or polyribosome profiles of the eEF1A-Ura3p strain compared to those of its wild-type counterpart (Fig. 7), however, suggests that the intrinsic function of the protein in elongation has not been affected.
Most interestingly, F308L and S405P eEF1A-Ura3p mutant strains had disrupted cytoskeletons (Fig. 3), with the disappearance of actin cables, increased cell size, and reduced total protein synthesis (Fig. 5). These reductions correlate with an effect on the initiation step of translation as demonstrated by polyribosome analysis (Fig. 7). This effect was not exclusive to the subset of eEF1A mutant strains, as the deletion or mutation of other actin-bundling proteins, namely, Tpm1p, Mdm20p, and the formins Bni1p and Bnrp, also led to the inhibition of translation initiation and the accumulation of 80S monoribosomes. In all mutants tested, the degree of 80S complex accumulation, and therefore initiation inhibition, correlated with the degree of actin disorganization. It is thought that 25 to 40% of the polyribosome population is associated with the actin cytoskeleton (28, 54, 62). Perturbations of the F-actin cytoskeleton in mammalian cells have been shown to induce profound effects on protein synthesis (57). Cases in which the inhibition of translation initiation is accompanied by the depolymerization of the actin cytoskeletons of yeast cells in vivo following glucose deprivation have been reported previously (6, 61). Although the actin cytoskeleton is believed to provide a scaffold for the translational apparatus, it is unclear how such depolymerization would result in a reduction in translation, particularly the accumulation of the 80S monoribosome complex. The fact that the different constituents of the translation machinery interact with the actin cytoskeleton suggests that this effect could be due to steric inhibition. Translation initiation factors such as eukaryotic translation initiation factor 2 (eIF2), eIF3, eIF4A, and eIF4B have been shown to associate with the cytoskeleton (26, 27, 29, 30, 32, 59, 65). It is therefore conceivable that upon actin depolymerization, one or all of these constituents loose their spatial arrangement and cannot properly interact with the ribosome.
The comparison of eEF1A mutant strains identified in this screen raised another question (this work and reference 24). Why do the F308L and S405P eEF1A-Ura3p mutations lead to a translation initiation block and reduced total translation while the N329S and N305S mutations that affect actin organization do not? We find that an apparent similarity in the loss of actin organization does not always induce a comparable block in initiation. However, the correlation is greatest with those eEF1A mutant forms that induce the smallest growth defect when overexpressed. The different actin mutant strains used in this study also presented severe actin defects; however, the consequences on translation initiation also differed. An act1-122 (D80A D81A) mutant strain did not show any accumulation at the 80S peak and therefore no initiation block. On the other hand, the act1-20 (G48V) strain and, more significantly, the act1-2 (A58T) strain showed 80S accumulation similar to that in the eEF1A mutant strains. While TPM1 or MDM20 deletion resulted in the loss of actin cables and cell integrity, translation rates and the inhibition of initiation were differentially affected, with the more than twofold reduction for the tpm1Δ strain consistent with the more direct role of Tpm1p in actin binding. In parallel, while the loss of formins also resulted in the disruption of the actin cytoskeleton, a more modest change in 80S peak accumulation was seen. The one general trend is that those strains with mutations in eEF1A, actin, or actin binding proteins that demonstrate the greatest increase in cell size show the largest reduction in total translation. Several hypotheses can be postulated to begin to explain these differences. These studies quantitated actin cables, patches, and cell morphology. Previous studies have shown that even when the formation of actin cables is apparently abolished, leading to the observation of cableless cells, some advanced imaging techniques detect truncated or very fine cables (36, 51-53). Thus, different levels of fine and truncated cables may exist among the different eEF1A, actin binding protein, and actin mutant strains. This statement is strengthened by the fact that the tropomyosin family of proteins have been shown to be essential constituents of actin cables, as they initiate cable formation while Mdm20p is thought to regulate the actin-tropomyosin interactions. It is thought that the loss of these proteins leads to the severe loss of cables (25, 53) (Fig. 8). The deletion of TPM1 was found to induce a more severe initiation block than the deletion of MDM20, arguing that the effects reported here are indeed due to the inability to induce proper cable formation. The partial suppression of the defect in an mdm20Δ strain by TPM1 further supports the key role of Tpm1p in the phenotypes observed.
Also, it is still unclear how the translation machinery associates with the cytoskeleton in yeast. Actin cables are essential in the majority of polarizing events, such as mRNA localization and organelle inheritance, in yeast. It is possible that polyribosomes associate along with mRNAs into more detailed and finer structures of the actin network. In fact, reports have postulated that eEF1A may localize at actin-filament intersections with ribosomes and mRNA (8, 41). Further analysis of changes in these substructures and functions of the different classes of eEF1A-Ura3p or actin binding protein mutant strains will give greater insight into the direct consequences of such regulation.
Acknowledgments
We acknowledge the assistance of the Robert Wood Johnson Medical School DNA core facility sequencing laboratory and the fluorescence microscopy laboratory in the Robert Wood Johnson Medical School Department of Pharmacology. Special thanks to Charles Boone for strains Y1239 and Y3024 and Paul Copeland for helpful comments.
This research was supported by grants from the National Institutes of Health (GM62789 and GM57483) to T.G.K.
Footnotes
Published ahead of print on 18 December 2006.
REFERENCES
- 1.Adams, A. E., D. Botstein, and D. G. Drubin. 1991. Requirement of yeast fimbrin for actin organization and morphogenesis in vivo. Nature 354:404-408. [DOI] [PubMed] [Google Scholar]
- 2.Adams, A. E., and J. R. Pringle. 1984. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98:934-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand, M., K. Chakraburtty, M. J. Marton, A. G. Hinnebusch, and T. G. Kinzy. 2003. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J. Biol. Chem. 278:6985-6991. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, G. R., L. Pedersen, L. Valente, I. Chatterjee, T. G. Kinzy, M. Kjeldgaard, and J. Nyborg. 2000. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Bα. Mol. Cell 6:1261-1266. [DOI] [PubMed] [Google Scholar]
- 5.Andersen, G. R., L. Valente, L. Pedersen, T. G. Kinzy, and J. Nyborg. 2001. Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex. Nat. Struct. Biol. 8:531-534. [DOI] [PubMed] [Google Scholar]
- 6.Ashe, M. P., S. K. De Long, and A. B. Sachs. 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11:833-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baim, S. B., D. F. Pietras, D. C. Eustice, and F. Sherman. 1985. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol. Cell. Biol. 5:1839-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassell, G. J., C. M. Powers, K. L. Taneja, and R. H. Singer. 1994. Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J. Cell Biol. 126:863-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bektas, M., R. Nurten, Z. Gurel, Z. Sayers, and E. Bermek. 1994. Interactions of eukaryotic elongation factor 2 with actin: a possible link between protein synthetic machinery and cytoskeleton. FEBS Lett. 356:89-93. [DOI] [PubMed] [Google Scholar]
- 10.Cavallius, J., A. P. Popkie, and W. C. Merrick. 1997. Site-directed mutants of post-translationally modified sites of yeast eEF1A using a shuttle vector containing a chromogenic switch. Biochim. Biophys. Acta 1350:345-358. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee, I., S. R. Gross, T. G. Kinzy, and K. Y. Chen. 2006. Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression. Mol. Genet. Genomics 275:264-276. [DOI] [PubMed] [Google Scholar]
- 12.Chen, E., G. Proestou, D. Bourbeau, and E. Wang. 2000. Rapid up-regulation of peptide elongation factor EF-1alpha protein levels is an immediate early event during oxidative stress-induced apoptosis. Exp. Cell Res. 259:140-148. [DOI] [PubMed] [Google Scholar]
- 13.Chuang, S. M., L. Chen, D. Lambertson, M. Anand, T. G. Kinzy, and K. Madura. 2005. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol. Cell. Biol. 25:403-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Condeelis, J. 1995. Elongation factor 1a, translation and the cytoskeleton. Trends Biochem. Sci. 20:169-170. [DOI] [PubMed] [Google Scholar]
- 15.Dang, C. V., D. C. Yang, and T. D. Pollard. 1983. Association of methionyl-tRNA synthetase with detergent-insoluble components of the rough endoplasmic reticulum. J. Cell Biol. 96:1138-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delley, P. A., and M. N. Hall. 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147:163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drees, B., C. Brown, B. G. Barrell, and A. Bretscher. 1995. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J. Cell Biol. 128:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drubin, D. G., K. G. Miller, and D. Botstein. 1988. Yeast actin-binding proteins: evidence for a role in morphogenesis. J. Cell Biol. 107:2551-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duttaroy, A., D. Bourbeau, X. L. Wang, and E. Wang. 1998. Apoptosis rate can be accelerated or decelerated by overexpression or reduction of the level of elongation factor-1 alpha. Exp. Cell Res. 238:168-176. [DOI] [PubMed] [Google Scholar]
- 20.Edmonds, B. T., J. Wyckoff, Y. G. Yeung, Y. Wang, E. R. Stanley, J. Jones, J. Segall, and J. Condeelis. 1996. Elongation factor-1 alpha is an overexpressed actin binding protein in metastatic rat mammary adenocarcinoma. J. Cell Sci. 109:2705-2714. [DOI] [PubMed] [Google Scholar]
- 21.Evangelista, M., D. Pruyne, D. C. Amberg, C. Boone, and A. Bretscher. 2002. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4:260-269. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa, R., T. M. Jinks, T. Tishgarten, M. Mazzawi, D. R. Morris, and M. Fechheimer. 2001. Elongation factor 1beta is an actin-binding protein. Biochim. Biophys. Acta 1527:130-140. [DOI] [PubMed] [Google Scholar]
- 23.Gonen, H., C. E. Smith, N. R. Siegel, C. Kahana, W. C. Merrick, K. Chakraburtty, A. L. Schwartz, and A. Ciechanover. 1994. Protein synthesis elongation factor EF-1a is essential for ubiquitin-dependent degradation of certain Na-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc. Natl. Acad. Sci. USA 91:7649-7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross, S. R., and T. G. Kinzy. 2005. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 12:772-778. [DOI] [PubMed] [Google Scholar]
- 25.Hermann, G. J., E. J. King, and J. M. Shaw. 1997. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J. Cell Biol. 137:141-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hesketh, J. E., G. P. Campbell, and P. F. Whitelaw. 1991. c-myc mRNA in cytoskeletal-bound polysomes in fibroblasts. Biochem. J. 274:607-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hesketh, J. E., Z. Horne, and G. P. Campbell. 1991. Immunohistochemical evidence for an association of ribosomes with microfilaments in 3T3 fibroblasts. Cell Biol. Int. Rep. 15:141-150. [DOI] [PubMed] [Google Scholar]
- 28.Hesketh, J. E., and I. F. Pryme. 1988. Evidence that insulin increases the proportion of polysomes that are bound to the cytoskeleton in 3T3 fibroblasts. FEBS Lett. 231:62-66. [DOI] [PubMed] [Google Scholar]
- 29.Hesketh, J. E., and I. F. Pryme. 1991. Interaction between mRNA, ribosomes and the cytoskeleton. Biochem. J. 277:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heuijerjans, J. H., F. R. Pieper, F. C. Ramaekers, L. J. Timmermans, H. Kuijpers, H. Bloemendal, and W. J. Van Venrooij. 1989. Association of mRNA and eIF-2 alpha with the cytoskeleton in cells lacking vimentin. Exp. Cell Res. 181:317-330. [DOI] [PubMed] [Google Scholar]
- 31.Hotokezaka, Y., U. Tobben, H. Hotokezaka, K. Van Leyen, B. Beatrix, D. H. Smith, T. Nakamura, and M. Wiedmann. 2002. Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. J. Biol. Chem. 277:18545-18551. [DOI] [PubMed] [Google Scholar]
- 32.Howe, J. G., and J. W. Hershey. 1984. Translational initiation factor and ribosome association with the cytoskeletal framework fraction from HeLa cells. Cell 37:85-93. [DOI] [PubMed] [Google Scholar]
- 33.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones, E. W. 1991. Tackling the protease problem in Saccharomyces cerevisiae, p. 428-453. In C. Guthrie and G. R. Fink (ed.), Guide to yeast genetics and molecular biology. Academic Press, New York, NY. [DOI] [PubMed]
- 35.Kandl, K. A., R. Munshi, P. A. Ortiz, G. R. Andersen, T. G. Kinzy, and A. E. M. Adams. 2002. Identification of a role for actin in translational fidelity in yeast. Mol. Gen. Genet. 268:10-18. [DOI] [PubMed] [Google Scholar]
- 36.Karpova, T. S., J. G. McNally, S. L. Moltz, and J. A. Cooper. 1998. Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J. Cell Biol. 142:1501-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinzy, T. G., and E. Goldman. 2000. Non-translational functions of the translational apparatus, p. 973-997. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Kuriyama, R., P. Savereide, P. Lefebuve, and S. Dasgupta. 1990. The predicted amino acid sequence of a centrosphere protein in dividing sea urchin eggs is similar to elongation factor (EF-1a). J. Cell Sci. 95:231-236. [DOI] [PubMed] [Google Scholar]
- 39.Lamberti, A., M. Caraglia, O. Longo, M. Marra, A. Abbruzzese, and P. Arcari. 2004. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis. Amino Acids 26:443-448. [DOI] [PubMed] [Google Scholar]
- 40.Liu, G., W. M. Grant, D. Persky, V. M. Latham, Jr., R. H. Singer, and J. Condeelis. 2002. Interactions of elongation factor 1alpha with F-actin and beta-actin mRNA: implications for anchoring mRNA in cell protrusions. Mol. Biol. Cell 13:579-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, G., J. Tang, B. T. Edmonds, J. Murray, S. Levin, and J. Condeelis. 1996. F-actin sequesters elongation factor 1a from interaction with aminoacyl-tRNA in a pH-dependent reaction. J. Cell Biol. 135:953-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, H. P., and A. Bretscher. 1989. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell 57:233-242. [DOI] [PubMed] [Google Scholar]
- 43.Magdolen, V., D. G. Drubin, G. Mages, and W. Bandlow. 1993. High levels of profilin suppress the lethality caused by overproduction of actin in yeast cells. FEBS Lett. 316:41-47. [DOI] [PubMed] [Google Scholar]
- 44.Mirande, M., D. Le Corre, D. Louvard, H. Reggio, J. P. Pailliez, and J. P. Waller. 1985. Association of an aminoacyl-tRNA synthetase complex and of phenylalanyl-tRNA synthetase with the cytoskeletal framework fraction from mammalian cells. Exp. Cell Res. 156:91-102. [DOI] [PubMed] [Google Scholar]
- 45.Moore, R. C., and R. J. Cyr. 2000. Association between elongation factor-1alpha and microtubules in vivo is domain dependent and conditional. Cell Motil. Cytoskelet. 45:279-292. [DOI] [PubMed] [Google Scholar]
- 46.Moore, R. C., N. A. Durso, and R. J. Cyr. 1998. Elongation factor-1alpha stabilizes microtubules in a calcium/calmodulin-dependent manner. Cell Motil. Cytoskelet. 41:168-180. [DOI] [PubMed] [Google Scholar]
- 47.Mulholland, J., D. Preuss, A. Moon, A. Wong, D. Drubin, and D. Botstein. 1994. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J. Cell Biol. 125:381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munshi, R., K. A. Kandl, A. Carr-Schmid, J. L. Whitacre, A. E. Adams, and T. G. Kinzy. 2001. Overexpression of translation elongation factor 1alpha affects the organization and function of the actin cytoskeleton in yeast. Genetics 157:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nissen, P., M. Kjeldgaard, S. Thirup, G. Polekhina, L. Reshetnikova, B. F. C. Clark, and J. Nyborg. 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270:1464-1472. [DOI] [PubMed] [Google Scholar]
- 50.Owen, C. H., D. J. DeRosier, and J. Condeelis. 1992. Actin crosslinking protein EF-1a of Dictyostelium discoideum has a unique bonding rule that allows square-packed bundles. J. Struct. Biol. 109:248-254. [DOI] [PubMed] [Google Scholar]
- 51.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. J. Cell Sci. 113:571-585. [DOI] [PubMed] [Google Scholar]
- 52.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113:365-375. [DOI] [PubMed] [Google Scholar]
- 53.Pruyne, D. W., D. H. Schott, and A. Bretscher. 1998. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 143:1931-1945. [DOI] [PubMed] [Google Scholar]
- 54.Ramaekers, F. C., E. L. Benedetti, I. Dunia, P. Vorstenbosch, and H. Bloemendal. 1983. Polyribosomes associated with microfilaments in cultured lens cells. Biochim. Biophys. Acta 740:441-448. [DOI] [PubMed] [Google Scholar]
- 55.Sandbaken, M. G., and M. R. Culbertson. 1988. Mutations in elongation factor EF-1a affect the frequency of frameshifting and amino acid misincorporation in Saccharomyces cerevisiae. Genetics 120:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sandrock, T. M., S. M. Brower, K. A. Toenjes, and A. E. M. Adams. 1999. Suppressor analysis of fimbrin (Sac6p) overexpession in yeast. Genetics 151:1287-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stapulionis, R., S. Kolli, and M. P. Deutscher. 1997. Efficient mammalian protein synthesis requires an intact F-actin system. J. Biol. Chem. 272:24980-24986. [DOI] [PubMed] [Google Scholar]
- 58.Suda, M., M. Fukui, Y. Sogabe, K. Sato, A. Morimatsu, R. Arai, F. Motegi, T. Miyakawa, I. Mabuchi, and D. Hirata. 1999. Overproduction of elongation factor 1alpha, an essential translational component, causes aberrant cell morphology by affecting the control of growth polarity in fission yeast. Genes Cells 4:517-527. [DOI] [PubMed] [Google Scholar]
- 59.Toh, B. H., S. J. Lolait, J. P. Mathy, and R. Baum. 1980. Association of mitochondria with intermediate filaments and of polyribosomes with cytoplasmic actin. Cell Tissue Res. 211:163-169. [DOI] [PubMed] [Google Scholar]
- 60.Ueno, H., K. Gonda, T. Takeda, and O. Numata. 2003. Identification of elongation factor-1alpha as a Ca2+/calmodulin-binding protein in Tetrahymena cilia. Cell Motil. Cytoskelet. 55:51-60. [DOI] [PubMed] [Google Scholar]
- 61.Uesono, Y., M. P. Ashe, and E. A. Toh. 2004. Simultaneous yet independent regulation of actin cytoskeletal organization and translation initiation by glucose in Saccharomyces cerevisiae. Mol. Biol. Cell 15:1544-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vedeler, A., I. F. Pryme, and J. E. Hesketh. 1991. Compartmentalization of polysomes into free, cytoskeletal-bound and membrane-bound populations. Biochem. Soc. Trans. 19:1108-1111. [DOI] [PubMed] [Google Scholar]
- 63.Whitacre, J. L., D. A. Davis, K. A. Toenjes, S. M. Brower, and A. E. M. Adams. 2001. Generation of an isogenic collection of yeast actin mutants and identification of three interrelated phenotypes. Genetics 157:533-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, F., M. Demma, V. Warren, S. Dharmawardhane, and J. Condeelis. 1990. Identification of an actin-binding protein from Dictyostelium as elongation factor 1α. Nature 347:494-496. [DOI] [PubMed] [Google Scholar]
- 65.Zumbe, A., C. Stahli, and H. Trachsel. 1982. Association of a Mr 50,000 cap-binding protein with the cytoskeleton in baby hamster kidney cells. Proc. Natl. Acad. Sci. USA 79:2927-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]