Abstract
Recent research has identified critical roles for microRNAs in a large number of cellular processes, including tumorigenic transformation. While significant progress has been made towards understanding the mechanisms of gene regulation by microRNAs, much less is known about factors affecting the expression of these noncoding transcripts. Here, we demonstrate for the first time a functional link between hypoxia, a well-documented tumor microenvironment factor, and microRNA expression. Microarray-based expression profiles revealed that a specific spectrum of microRNAs (including miR-23, -24, -26, -27, -103, -107, -181, -210, and -213) is induced in response to low oxygen, at least some via a hypoxia-inducible-factor-dependent mechanism. Select members of this group (miR-26, -107, and -210) decrease proapoptotic signaling in a hypoxic environment, suggesting an impact of these transcripts on tumor formation. Interestingly, the vast majority of hypoxia-induced microRNAs are also overexpressed in a variety of human tumors.
MicroRNAs represent approximately 1% to 2% of the eukaryotic transcriptome and have been shown to play critical roles in the coordination of cell differentiation, proliferation, death, and metabolism (1, 3, 6, 7, 17, 24) and more recently in tumorigenesis (2, 5, 6, 10, 13, 20, 21). Indeed, a significant percentage of microRNA-encoding genes are located at fragile sites, minimal loss-of-heterozygosity regions, minimal regions of amplification, or common breakpoint regions in cancers.
Moreover, global microRNA expression changes have been described to occur in human cancers and in some cases shown to correlate with the clinico-pathological features of the tumor (2, 13, 29). However, no mechanism has been proposed to date for these profile alterations.
Despite this wealth of data, relatively little is known about microRNA regulation and response to microenvironmental factors. One mechanism involves the activation of specific signal transduction pathways that in turn promote the transcription of certain microRNAs. For example, it was reported that the miR-1 genes are targets of serum response factor, a converging downstream effector for a variety of oncoproteins and growth factors (30). Another transcription factor, the c-myc oncogene product, was also found to activate the expression of a microRNA cluster (25).
Hypoxia is an essential feature of the neoplastic microenvironment. Tumors with widespread low oxygenation tend to exhibit increased invasion and resistance to conventional therapy (9). The molecular mechanisms responsible for the hypoxic survival of neoplastic cells are not fully characterized, and a better understanding of this process may lead to novel strategies for pharmacological intervention.
Our data indicate that hypoxia leaves a specific mark on microRNA profiles in a variety of cell types, with a critical contribution of the hypoxia-inducible factor (HIF). Moreover, at least a subgroup of these hypoxia-regulated microRNAs (HRMs) seem to play a role in cell survival in a low-oxygen environment.
Finally, by comparing hypoxia-associated microRNA spectra with published data from a large number of tumors (28), we propose that cancer-associated microRNA profiles exhibit a hypoxic signature.
MATERIALS AND METHODS
Cell culture and growth conditions.
Colon (HT29 and HCT116) and breast cancer (MCF7 and MDA-MB231) cell lines were obtained from Phil Hinds (Tufts-New England Medical Center, Boston, MA). HT29 and HCT116 cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and MCF7, and MDA-MB231 cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum. Hypoxic conditions were maintained in an InVivo200 hypoxia workstation (Biotrace International, Ruskinn Life Sciences, United Kingdom) with oxygen maintained at 0.2%. Normoxic controls were propagated at 37°C and 5% CO2.
Plasmids.
The mutant versions of HIF-1α and HIF-2α with double proline-to-alanine substitutions have been described previously (14, 15). The three-HRE-thymidine kinase (tk)-luciferase reporter is a HIF-responsive construct containing a tandem of hypoxia-responsive elements from the erythropoietin promoter. Both plasmids were provided by William Kaelin (Dana-Farber Cancer Institute, Boston, MA).
MicroRNA microarray analysis.
RNA was extracted by using TRIzol (Invitrogen, Carlsbad, CA), according to the manufacturer's protocol. RNA was labeled and hybridized on microRNA microarray chips as previously described (20). Briefly, 5 μg of RNA from each sample was biotin labeled during reverse transcription using random hexamers. Hybridization was carried out on the second version of our microRNA chip (ArrayExpress accession number A-MEXP-258), which contains 381 probes for mature and precursor human microRNAs and 393 probes for mouse microRNAs, together with controls. There are four spot replicates for each probe on the chip. Hybridization signals were detected by biotin binding of a Streptavidin-Alexa 647 conjugate using a GenePix 4000B scanner (Axon Instruments). Images were quantified using the GenePix Pro 6.0 apparatus (Axon Instruments). We used the microRNA nomenclature according to the microRNA Registry (miRBase http://microrna.sanger.ac.uk/sequences/) at the Sanger Institute and the Genome Browser (http://genome.ucsc.edu).
Analysis of microarray data.
Raw data were normalized and analyzed by GeneSpring GX software version 7.3 (Agilent Technologies). The GeneSpring software generated a unique value for each microRNA, determining the average of the results from the four spots. Samples were normalized using the on-chip median normalization feature of GeneSpring software; the result for each cell line time course experiment was normalized to that at time zero. MicroRNAs showing an altered expression across the time course were identified using the filter of the n-fold-change tool. In detail, a GeneSpring GX filter with the n-fold-change tool was used to point out the microRNAs that, for at least two cell lines, were 1.5-fold upregulated at 24 h compared to expression levels at time zero. The 1.5-fold-change threshold was chosen on the basis of its use in previously published articles employing these particular types of microarrays. Analysis of variance (ANOVA) with the Benjamini and Hochberg correction for false-positive reduction was used to compare the average values obtained with microRNA probes in all cell lines at 24 and 48 h with values at time zero. Hierarchical cluster analysis was performed using average linkage and Pearson correlation as a measure of similarity.
Transfections.
Plasmid transfections were performed using Lipofectamine 2000 (Invitrogen) as per the manufacturer's protocol. Fresh medium was added after 5 h of transfection, and RNA and protein were harvested 24/48 h posttransfection. Transfection of mature microRNAs or inhibitors (Ambion, Inc.) was performed using siPORT NeoFX transfection agent (Ambion, Inc.) according to the manufacturer's protocol. Transfection complexes were added to cells at a final oligonucleotide concentration of 20 nM, and medium was replaced 24 h later.
Luciferase assays using reporters based on HRM promoter fragments.
We started by amplifying HRM promoter fragments predicted to encompass HIF sites, as shown in Table S1b in the supplemental material. Thus, the genomic region 4.9 kb immediately upstream of miR-24-1 was amplified using primers 5′ATACTCGAGCTGCTAGGCCATGCGTGTCC3′ (forward) and 5′ATTAAGCTTCAAGAGAGAGTTCACCGCGC3′ (reverse) (underlining indicates restriction sites inserted). For miR-181c, a region of approximately 2.0 kb immediately upstream was PCR amplified using 5′ATAGGTACCCACTCCACAGCCTGAATG3′ (forward) and 5′TATAAGCTTGGTGGGGTAGGTGGCAGGGAAC3′ (reverse). For miR-26b, a region situated approximately 3 kb upstream of the 5′ end (and encompassing a cluster of four predicted HIF sites) was PCR amplified using the following primers: 5′ATAGCTAGCGAGACAGATGTCCCGCTCCCAG3′ (forward) and 5′ATCGCTAGCACGCTCTTGAATGGGACGG3′ (reverse). Due to the size and CG contents, PCR amplification was done using the Herculase II fusion DNA polymerase (Stratagene). The resulting fragments were cloned in the pGL3 basic vector (Promega) (miR-24-1 and -181c) or pGL3-tk-luciferase vector (from David Fisher, DFCI) (miR-26b). MCF7 or HT29 cells were cotransfected using Lipofectamine 2000 (Invitrogen) with the reporter plasmids and either of the HIF mutants or the empty vector. Luciferase assays were performed 24 h posttransfection by following the manufacturer's protocol (luciferase assay system; Promega). Luciferase activity (measured as relative light units) was scored using a Femtomaster FB12 luminometer (Zylux Corporation) and normalized to that of an equal protein concentration in all samples.
Apoptosis assays.
Cells were plated in triplicate in a 96-well plate, transfected with microRNA precursors or inhibitors (Ambion, Inc.) as described above, and scored for caspase activation 72 h posttransfection using an Apo-ONE homogeneous caspase-3/7 assay kit (Promega) according to manufacturer's instructions. The protein concentration was determined using Bradford's method, and caspase activities for all samples were normalized to that of an equal protein amount. The data are presented as means plus standard deviations from three independent experiments performed in triplicate.
Northern hybridization.
Total RNA isolation was performed using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA samples (20 μg each) were run on 15% acrylamide-denaturing (urea) Criterion precast gels (Bio-Rad Laboratories, Hercules, CA) and then transferred onto a Hybond-N+ membrane (Amersham Pharmacia Biotech). The hybridization with [α-32P]ATP was performed at 42°C in 7% sodium dodecyl sulfate (SDS)-0.2 M Na2PO4 (pH 7.0) overnight. Membranes were washed at 42°C, twice in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])-0.1% SDS and twice with 0.5× SSPE-0.1% SDS. Blots were stripped by boiling them in 0.1% aqueous SDS-0.1× SSC for 10 min and were reprobed several times. As a loading control, we used 5S rRNA stained with ethidium bromide or probing for the U6 snRNA.
TaqMan RT-PCR.
The method was optimized for microRNA, and reagents, primers, and probes were obtained from Applied Biosystems. Human 18S rRNA was used to normalize all RNA samples. Reverse transcriptase (RT) reactions and real-time PCR (PCR) were performed according to manufacturer protocols. RNA concentrations were determined with a NanoDrop apparatus (NanoDrop Technologies, Inc.), and 1 nanogram per sample was used for the assays. All RT reaction mixtures, including no-template controls and RT-minus controls, were run in duplicate in an Applied Biosystems 9700 thermocycler. Gene expression levels were quantified using the ABI Prism 7900HT sequence detection system (Applied Biosystems). Analysis was performed in duplicate, including with no-template controls. Relative expression was calculated using the comparative cycle threshold method.
ChIP.
HT29 cells (60% confluent) were exposed to hypoxia (0.2% O2) or normoxia (21% O2) for 24 h. Cross-linking was performed using 1% (vol/vol) formaldehyde for 10 min, and the reaction was stopped with 125 mM glycine for 5 min. Cells were washed twice with ice-cold 1× phosphate-buffered saline and collected by scraping them in 1 ml of 1× phosphate-buffered saline supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), followed by centrifugation and lysis in 400 μl of buffer (1% SDS, 10 mM EDTA, 50 mM Tris-Cl, pH 8.0, protease inhibitor cocktail, and 1 mM PMSF). The resulting lysate was incubated on ice for 10 min, followed by sonication for DNA shearing to fragments between 200 bp and 1 kb. The supernatant was recovered and diluted in chromatin immunoprecipitation (ChIP) dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-Cl, pH 8.0, protease inhibitor cocktail, 1 mM PMSF), followed by immunoprecipitation overnight at 4°C with a polyclonal anti-HIF-1α antiserum (ab2185; Abcam) or whole rabbit antiserum (immunoglobulin G [IgG] control). Immunocomplexes were recovered by the addition of a 50% slurry of salmon sperm DNA-protein A-agarose (Upstate) to the samples and sequentially washed for 4 min each in buffer I (20 mM Tris, pH 8.0, 200 mM NaCl, 0.5% Triton X-100, 0.05% deoxycholate, 0.5% NP-40, 1 mM PMSF), buffer II (20 mM Tris, pH 8.0, 500 mM NaCl, 0.5% Triton X-100, 0.05% deoxycholate, 0.5% NP-40, 1 mM PMSF), buffer III (10 mM Tris, pH 8.0, 250 mM LiCl, 1% Triton X-100, 0.1% deoxycholate, 0.5% NP-40, 0.5 mM EDTA, 1 mM PMSF), and buffer IV (10 mM Tris, pH 8.0, 5 mM EDTA). The immunoprecipitated DNA was retrieved from the beads by incubation in elution buffer (10 mM Tris, pH 8.0, 5 mM EDTA, 1% SDS) at 65°C for 1 h. Cross-linking was reversed using 200 mM NaCl at 65°C overnight followed by proteinase K digestion at 47°C for 2 h. DNA was then purified using a PCR purification kit (QIAGEN), and PCR was performed using primers spaced approximately 15 to 250 bp apart and encompassing predicted HIF binding sites in miR-210 and -26a-2 promoters (see below).
5′GGAGCCTTGACGGTTTGACC 3′ (forward) and 5′CGAGGACCAGGGTGACAGTG3′ (reverse) were used to PCR amplify the miR-210 promoter fragment (210-A) containing predicted HIF binding sites located at positions −1720 and −1822. For the 210-B fragment containing a predicted HIF consensus at position −1166, the following pair was used: 5′GGTCGTGAAGGAGCCTCTAAG3′ (forward) and 5′GGACCATCCCACTCACTACAG3′ (reverse). Finally, the miR-26a-2 promoter fragment (26-A), encompassing a predicted HIF binding site at position −2860, was amplified using 5′CCAAGGACTATGCACATCACC3′ (forward) and 5′GGAAAGGCAGTGAGATGGCAC3′ (reverse). The following primers were used to amplify a region in the promoter of miR-130b (negative control, hypoxia nonresponsive): 5′GCGAAACCCCAGCTCTACTA3′ (forward) and 5′ACACTCTCACTCTGTCGCCC3′ (reverse). For HIF site localization and sequences, see Table S1b in the supplemental material. The PCR products were resolved on a 1.5% agarose gel and visualized by ethidium bromide staining.
RESULTS
MicroRNA expression changes in hypoxia.
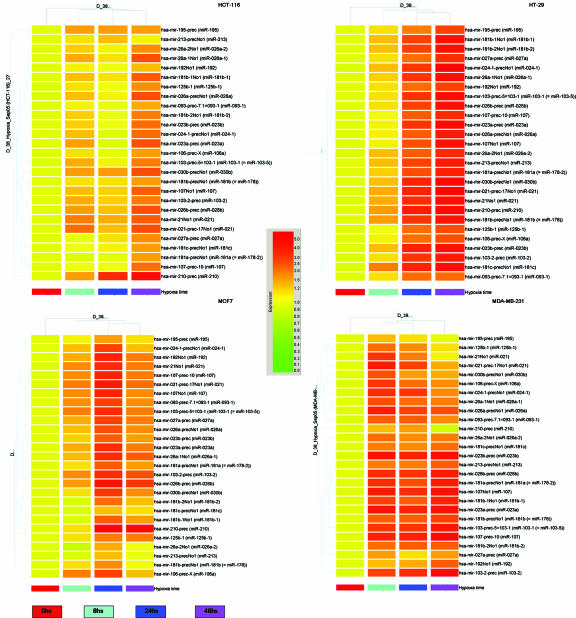
We hypothesized that microRNAs could be involved in hypoxia response and began addressing this hypothesis using a microarray-based screening. We first analyzed a panel of four human cancer cell lines (MDA-MB231, MCF7, HT29, and HCT116), which were subjected to time course exposure to 0.2% oxygen, a concentration often present in the hypoxic regions of tumors. RNA was extracted at specific time points (0, 8, 24, and 48 h), and microRNA expression profiling was done subsequently. Twenty-seven microRNA probes were identified to be at least 1.5-fold upregulated at 24 h in at least two cell lines. The ANOVA, with the Benjamini and Hochberg correction for false-positive reduction, was employed to compare the microRNA probes average values in all cell lines at 24 and 48 h versus values at time zero, yielding in all cases significant P values (<0.05) (see Table 1 for a summary of levels of induction at 24 and 48 h in all cell lines versus those at 0 h and corresponding P values). A cluster analysis of time points for each cell line is shown in Fig. 1. These probes correspond to 23 distinct mature microRNAs, which will henceforth be termed HRMs.
TABLE 1.
Summary of hypoxia-regulated microRNAsa
| MicroRNA | Symbol | MicroRNA expression ratio in indicated cell line
|
ANOVA P value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HT29
|
HCT116
|
MCF7
|
MDA-MB231
|
|||||||
| 24 h/0 h | 48 h/0 h | 24 h/0 h | 48 h/0 h | 24 h/0 h | 48 h/0 h | 24 h/0 h | 48 h/0 h | |||
| miR-103-1 | MIRN103-1 | 2.33 | 3.46 | 1.11 | 1.71 | 2.56 | 1.72 | 2.38 | 3.19 | 0.00228 |
| miR-103-2 | MIRN103-2 | 2.31 | 2.91 | 1.22 | 1.70 | 2.58 | 2.07 | 2.6 | 3.76 | 0.00228 |
| miR-106a | MIRN106A | 1.76 | 2.03 | 1.01 | 1.41 | 2.18 | 1.26 | 1.84 | 1.52 | 0.00241 |
| miR-107 | MIRN107 | 1.94 | 2.61 | 1.07 | 1.75 | 2.65 | 1.66 | 2.76 | 3.90 | 0.00231 |
| miR-107 | MIRN107 | 2.04 | 3.26 | 1.22 | 1.86 | 2.72 | 1.60 | 2.47 | 3.37 | 0.00228 |
| miR-125b-1 | MIRN125B1 | 1.83 | 2.07 | 1.07 | 1.52 | 1.71 | 1.64 | 1.51 | 1.13 | 0.00234 |
| miR-181a-1 | MIRN213 | 2.29 | 3.02 | 1.17 | 1.44 | 1.61 | 1.05 | 1.87 | 2.24 | 0.00398 |
| miR-181a-2 | MIRN181A | 2.01 | 2.73 | 1.01 | 1.44 | 1.87 | 1.47 | 2.13 | 2.84 | 0.00241 |
| miR-181b-1 | MIRN181B1 | 2.17 | 3.38 | 1.17 | 1.96 | 1.97 | 1.47 | 1.97 | 2.66 | 0.00228 |
| miR-181b-2 | MIRN181B2 | 2.17 | 3.33 | 1.04 | 1.66 | 1.73 | 1.28 | 1.95 | 1.98 | 0.00268 |
| miR-181c | MIRN181C | 2.74 | 3.33 | 1.11 | 1.48 | 1.26 | 1.17 | 1.6 | 1.75 | 0.0083 |
| miR-192 | MIRN192 | 1.75 | 2.19 | 1.07 | 1.30 | 2.7 | 1.63 | 1.33 | 1.86 | 0.00262 |
| miR-195 | MIRN195 | 1.45 | 2.26 | 1.51 | 1.51 | 1.59 | 1.15 | 1.17 | 1.02 | 0.00622 |
| miR-21 | MIRN21 | 2.81 | 4.00 | 1.26 | 2.09 | 2.86 | 1.79 | 2.28 | 1.34 | 0.00242 |
| miR-21 | MIRN21 | 2.62 | 3.51 | 1.34 | 1.92 | 2.6 | 1.66 | 1.71 | 1.12 | 0.00231 |
| miR-210 | MIRN210 | 2.92 | 3.85 | 2.47 | 3.73 | 4.33 | 3.77 | 1.45 | 0.94 | 0.00241 |
| miR-213-5p | MIRN213 | 2.14 | 2.90 | 1.3 | 1.10 | 1.25 | 1.04 | 1.64 | 1.86 | 0.00961 |
| miR-23a | MIRN23A | 2.42 | 3.73 | 0.97 | 2.02 | 2.5 | 1.71 | 2.71 | 3.41 | 0.00231 |
| miR-23b | MIRN23B | 2.7 | 3.39 | 1.05 | 1.82 | 1.87 | 1.45 | 2.76 | 3.57 | 0.00241 |
| miR-24-1-3p | MIRN24-1 | 2.44 | 4.09 | 0.98 | 1.75 | 2.22 | 1.30 | 2.33 | 1.52 | 0.00483 |
| miR-26a | MIRN26A1 | 1.91 | 2.87 | 1.12 | 1.99 | 2.06 | 1.58 | 2.21 | 2.04 | 0.00228 |
| miR-26a-1 | MIRN26A1 | 2.3 | 3.46 | 1.14 | 2.06 | 2.5 | 1.88 | 1.81 | 1.72 | 0.00228 |
| miR-26a-2 | MIRN26A2 | 1.94 | 2.62 | 1.1 | 1.57 | 1.37 | 1.07 | 1.54 | 1.66 | 0.0046 |
| miR-26b | MIRN26B | 2.21 | 3.24 | 1.29 | 2.04 | 3.23 | 2.40 | 2.32 | 2.96 | 0.00213 |
| miR-27a | MIRN27A | 1.77 | 2.48 | 1.13 | 1.51 | 2.14 | 1.65 | 1.18 | 1.44 | 0.00241 |
| miR-30b | MIRN30B | 2.74 | 3.96 | 1.52 | 2.11 | 2.38 | 1.54 | 1.5 | 1.40 | 0.00234 |
| miR-93-1 | MIRN93 | 1.84 | 1.79 | 1.02 | 1.34 | 2.18 | 1.54 | 1.86 | 1.56 | 0.00231 |
MicroRNA expression numbers represent the ratio of each microarray probe expression value at 24 or 48 h to that at time zero. The P value was obtained by performing an ANOVA statistical comparison between the mean expression levels at 24 and 48 h versus that at time zero in all cell lines.
FIG. 1.
Coordinated hypoxic changes of microRNA expression in colon and breast cancer cell lines. Cluster analysis of four cell lines according to the expression of microRNAs upregulated by hypoxia in at least two cell lines. Expression data were normalized to expression at time zero.
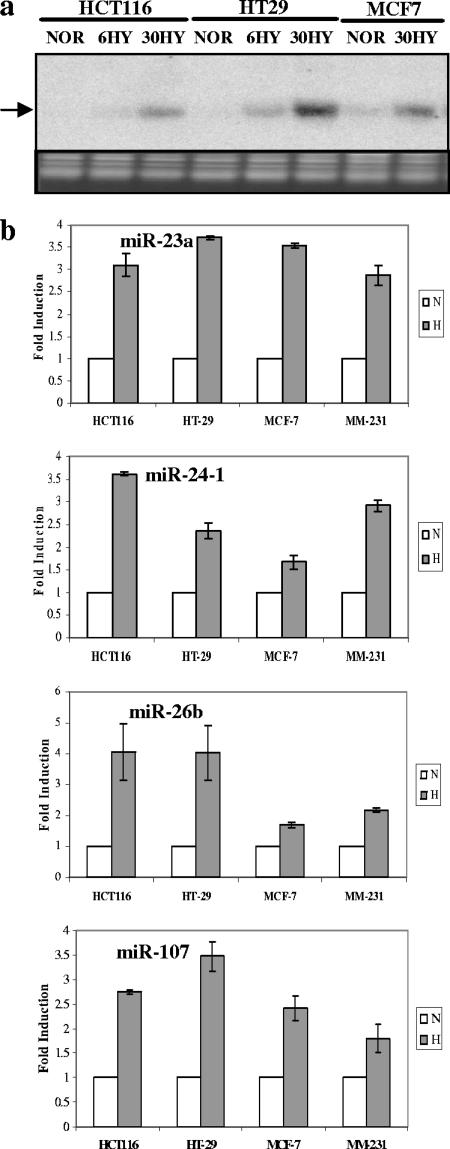
We went on to confirm the microarray data using Northern analysis (Fig. 2a for miR-210) or quantitative RT-PCRs for miR-21, -23, -24, -26, -27, -181, -103, -107, -125, -210, and -213 (see Fig. 2b for a selection; the rest can be found in Fig. S1a in the supplemental material). Priority was given to HRMs that were subjected to subsequent functional analyses. A Northern blot showing a lack of induction of miR-7-3 (not a member of the HRM list) served as negative control (see Fig. S1b in the supplemental material).
FIG. 2.
Confirmation of HRM induction by hypoxia by Northern analysis or quantitative RT-PCR. (a) Northern blotting confirmation of miR-210 induction under hypoxia (the mature form is indicated). Lanes NOR, 6HY, and 30HY show results under normoxia and at 6 h and 30 h under hypoxia, respectively. An ethidium bromide-stained gel picture is shown as a loading control. (b) Quantitative RT-PCR confirmation of HRM induction by 24-hour hypoxia (H) compared with HRM induction for normoxic controls (N). Bars indicate means from two independent experiments. I bars indicate standard deviations.
Evidence for the role of HIFs in HRM regulation.
We next addressed the role of HIFs in HRM regulation. HIFs are well-documented master regulators of the hypoxia response and transcription factors, which are stabilized during hypoxia and coordinate the transcription of a wide variety of genes that are critical for survival under conditions of low oxygen (9).
In order to investigate whether HRMs exhibit a significant enrichment in predicted HIF binding sites, we performed an in silico search upstream of the genomic sequences that encode all the known microRNAs (1,039 sequences, experimentally demonstrated and predicted; http://microrna.sanger.ac.uk/sequences/ftp/genomes/hsa.gff).
Since only a few microRNA promoters have been identified experimentally, a 6-kb region (5 kb upstream and 1 kb downstream region of the 5′ end of the annotated microRNA) was designated as a putative promoter sequence. Indeed, most of the experimentally confirmed transcription factor binding sites are located within the kb −5 region of the transcription start site. Predicted HIF binding sites were analyzed using the MATCH program and the V$HIF1_Q3 (GNNKACGTGCGGNN; boldfacing indicates the core HIF consensus), V$HIF1_Q5 (NGTACGTGCNGB), and V$HIF1_Q6 (NRCGTGNGN) position weight matrices from the TRANSFAC database (version 9.1) (23). The position weight matrices describe the position preferences of different nucleotides in the HIF binding site. We scanned regions around the transcription start sites of all the microRNAs from kilobase positions −5 to +1 using the “minFP_good91.prf” profile (the profile of cutoff values with a minimum number of false-positive predictions) of MATCH, similarly to searches described previously (16, 26, 27).
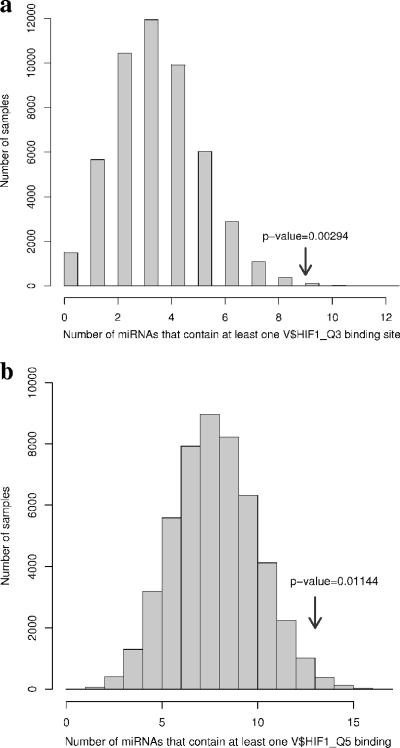
We then tested whether these consensus sequences are significantly more abundant in the promoters of the 23 HRMs (target set) than in the rest of microRNA-ome. Thus, we generated 50,000 groups, each consisting of 23 promoters randomly selected from the 1,039 microRNAs and calculated the number of promoters that contain at least one predicted HIF binding site in both target and random sets of promoters. The search was performed separately for the HIF_Q3 and HIF_Q5 consensus sequences. The selection was performed using the function “sample,” which is part of the random module in the Python 2.4 programming language (http://www.python.org/), followed by data analysis using the R programming language (http://www.r-project.org/). The results of the analysis are summarized in Fig. 3 and Table S1a in the supplemental material, indicating a highly significant enrichment of the HIF binding consensuses in the HRM group (P = 0.00294 for HIF_Q3; P = 0.011 for the HIF_Q5 consensus). The search based on the HIF_Q6 matrix did not yield a significant P value (not shown), which is not surprising, given the high probability of such a short and degenerate sequence arising by chance very often in the genome.
FIG. 3.
Distribution of random 23-microRNA groups (samples) based on HIF1_Q3 (a) or HIF1_Q5 (b) binding sites. The arrow indicates where the experimental data (HRMs) fall within the random sample population, with the corresponding P value.
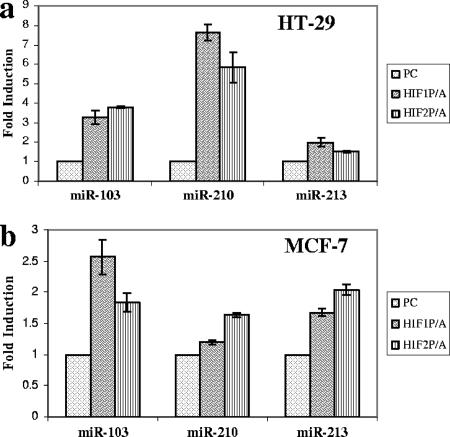
The next step was to obtain experimental evidence for the role of HIF in HRM regulation. First, we transduced constitutively stable HIF-1 and -2α subunits versus a vector-only control (pcDNA3.1) in HT29 and MCF7 cell lines under normoxia. HIF stabilization was achieved by substituting the two prolines (at positions 564 and 402 in the case of HIF-1) in the alpha subunits that are subject to oxygen-dependent hydroxylation and proteasomal degradation via VHL-dependent ubiquitylation (15, 16, 22). The activity of exogenous HIFs was confirmed by cotransfection with an HRE-tk-luciferase reporter (containing three hypoxia response elements) followed by standard luciferase assay (Fig. S2a in the supplemental material). Expression of miR-103, -210, and -213 was measured following HIF transfection using a modified real-time RT-PCR protocol which revealed a robust and reproducible increase in the expression of all three transcripts (Fig. 4).
FIG. 4.
Effect of HIF on specific microRNA expression. The impact of exogenous constitutively active HIF-1α (HIF1P/A) and HIF-2α (HIF2P/A) subunits on miR-103, -210, and -213 expression was determined by quantitative RT-PCR in HT29 (a) and MCF7 (b) cells. The control was the pcDNA3.1 empty vector (PC). Bars indicate means from two independent experiments. I bars indicate standard deviations.
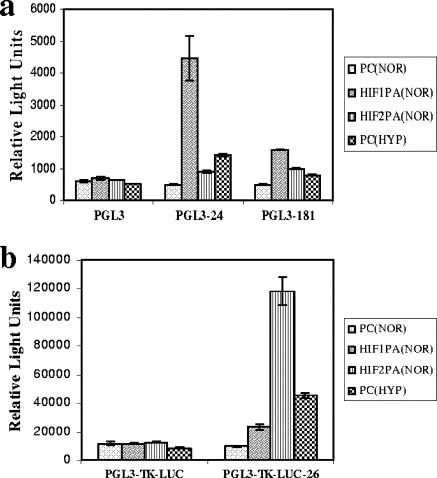
We then addressed whether HIF has a direct impact on the putative promoter regions of several HRMs (miR-24, -26, -181, and -210). To this end, we amplified 5 kb and 2 kb of the genomic regions upstream of miR-24-1 and miR-181c, respectively. We also amplified a 1.2-kb region approximately 3 kb upstream of the 5′ end of miR-26b (for details, see Materials and Methods). These regions were chosen based on the predicted HIF binding sites identified by in silico analysis; in order to minimize the chance of missing potentially important sites, we used a “less stringent search,” by considering all the position weight matrices (HIF_Q3, _Q5, and _Q6) (see Table S1b in the supplemental material). The promoter fragments were subcloned into the pGL3-luciferase (for miR-24 and -181) or, as an enhancer, into the pGL3-tk-luciferase (for miR-26) vector. HT29 cells were cotransfected with these constructs or the control construct pGL3-luc/pGL3-tk-luciferase and constitutively active HIF constructs (HIF1P/A or HIF2P/A) or the empty vector pcDNA3.1 (PC), followed by incubation under normoxia or hypoxia for 24 h. Standard luciferase assays showed that HIF transduction or hypoxia induced a robust activation of all the promoter-luciferase constructs, supporting a direct role of HIF in HRM upregulation (Fig. 5). Interestingly, HIF-1 had a more robust impact on the promoters of miR-24 and miR-181c than HIF-2, which was more efficient in activating miR-26. This is unlikely to be attributed to differences in transfection efficiency, as parallel controls using a three-HRE (Epo)-luciferase reporter cotransfected with the HIF plasmids showed comparable levels of luciferase induction in response to both isoforms (Fig. S2b in the supplemental material). While these differences between HIF-1 and HIF-2 were not tested at the level of full HRM induction, recent data argue for specific targets and biological effects of the two forms (12).
FIG. 5.
Direct effect of HIF in the up-regulation of select HRMs. Relative luciferase activities of HRM promoter reporter constructs in HT29 cells. (a) miR-24-1 and -181c promoters in a pGL3 context; (b) miR-26b fragment in a pGL3-tk context. The constructs were cotransfected with constitutively active HIF-1α (HIF1P/A), HIF-2α (HIF2P/A), or the empty vector pcDNA3.1 (PC) and incubated under normoxia (NOR). The effect of hypoxia (HYP) on the reporter is also shown. Bars indicate means from three independent experiments. I bars indicate standard deviations.
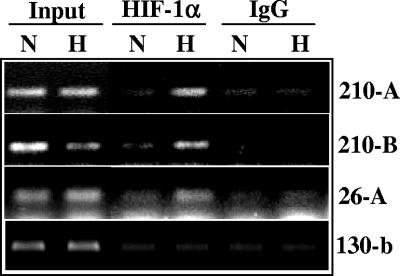
Additionally, for miR-26a-2 and miR-210, we confirmed the dynamic recruitment of HIF to the promoter by ChIP. As shown in Fig. 6, the HIF-1α antibody (but not the control IgG antibody) immunoprecipitated the miR-210 and miR-26a-2 promoter fragments in hypoxic HT29 cells but very little in the normoxic controls. A similar assay performed for a region upstream of miR-130b (which is not an HRM), did not reveal measurable recruitment of HIF upon hypoxia exposure.
FIG. 6.
Direct recruitment of HIF on the miR-210 and miR-26 promoters under hypoxia. Chromatin was immunoprecipitated from HT29 cells using a HIF-1α antibody or an IgG control, and the enriched genomic fragment was amplified using primers spanning the candidate HREs located at positions −1720 and −1822 (210-A); −1166 (210-B) of the miR-210 promoter; or −2860 for the miR-26a-2 promoter (26-A). A fragment of the miR-130b promoter was used as a negative control.
Roles of select HRMs in cell survival.
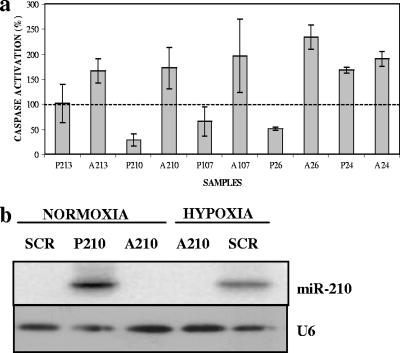
The impact of HRMs on cell survival was addressed by direct transfection of mature microRNAs or their antisense inhibitors (4) in human cell lines under hypoxic conditions. We transduced miR-24, -26, -107, -210, and -213 into MCF7 cells and then incubated them under hypoxia for 48 h. The proapoptotic response was interrogated by a caspase-3/7 assay (Fig. 7a). Efficient transfer and blockade of microRNAs were confirmed by Northern blotting (Fig. 7b). The controls employed in these experiments were pre-mir miRNA precursor molecules and negative control 1 (Ambion, Inc.).
FIG. 7.
Antiapoptotic effect of select microRNAs under hypoxia. Blockade of miR-26, -107, and -210 with antisense inhibitors leads to an increased apoptotic response in three independent experiments (each performed in triplicate). In contrast, an excess of sense microRNAs decreases the apoptotic response. The dotted line represents the apoptotic caspase-3/7 baseline activity in response to negative-control microRNA under hypoxia (P, precursor [sense]; A, antisense). (b) Northern blot confirmation of efficient transduction or blockade of miR-210 in MCF7 cells. MCF7 cells were transfected with the precursor or antisense miR-210 or the scramble control (SCR). U6 snRNA is shown as the loading control.
The antisense experiment is of particular importance since it is aimed at blocking the “natural” induction in response to hypoxia. The overexpression of the sense microRNA is anticipated to accentuate the effect of the “physiologic” induction of HRMs. Moreover, sense and antisense microRNAs should exhibit opposite effects on cell death compared to the baseline caspase activity associated with a scrambled control microRNA. At least with respect to miR-26, -107, and -210, we reproducibly noticed caspase inhibition in three independent experiments, which points towards a decrease in central components of apoptotic signaling. Of note, the mature forms of miR-103 and -107 are almost identical and their molecular targets and biological effects are predicted to be highly similar. Comparable effects on caspase activation were elicited in HT29 cells under hypoxia using the same microRNAs (not shown).
Correlations between cancer and hypoxia-specific microRNA profiles.
Recent investigations have dissected a large number of human neoplasms for microRNA expression (13, 21, 28) and identified specific alterations from normal cells; however, the mechanism and biological impact of these changes remain elusive. In order to address a potential correlation between the pattern of microRNAs altered in solid cancers and under hypoxia, we took advantage of the largest genome-wide microarray profiling study published to date, including 540 tumor samples from six types of solid cancer (breast, lung, colon, stomach, and pancreatic endocrine tumors and prostate carcinomas) and corresponding normal samples (28). From the 228 microRNAs, 137 exhibiting expression values above threshold in at least 90% of samples were retained, which generated a “common signature” of abnormally expressed microRNAs (presented in Fig. 2B and supplementary Tables 10 and 11 in reference 28). Of note, the microarray search for HRMs was performed using the same profiling technology.
The vast majority of HRMs identified by our study (Table 2 ) are also overexpressed in at least some types of tumors, suggesting that hypoxia may represent a key contributing “trigger” for microRNA alterations in cancer.
TABLE 2.
Solid correlations of microRNAs with cancersa
| MicroRNA | Expression in cancer cellsb |
|---|---|
| hsa-miR-21 | Upregulated in general |
| hsa-miR-23a | Upregulated in some cancers (pancreas, colon) |
| hsa-miR-23b | Downregulated in most cancers (upregulated in pancreas) |
| hsa-miR-24-1 | Upregulated in general |
| hsa-miR-26b | Upregulated in general (except breast) |
| hsa-miR-27a | Upregulated in general |
| hsa-miR-30b | Upregulated in most cancers (downregulated in lung and breast) |
| hsa-miR-93-1 | Upregulated in general |
| hsa-miR-103-2 | Upregulated in general (except breast) |
| hsa-miR-103-1 | Upregulated in general (except breast) |
| hsa-miR-106a | Upregulated in general |
| hsa-miR-125b-1 | Upregulated in general (except breast) |
| hsa-miR-181a-2 | Upregulated in some cancers (especially breast) |
| hsa-miR-181c | No evidence for altered expression |
| hsa-miR-195 | Upregulated in general (supplemental Table 10 in reference 27) |
| hsa-miR-210 | Upregulated in general |
| hsa-miR-213-5p | Upregulated in general (especially breast) |
| hsa-miR-26a | Upregulated in general (except breast) |
| hsa-miR-181b-1 | Upregulated in general (except breast) |
| hsa-miR-26a-1 | Upregulated in general (except breast) |
| hsa-miR-192 | No evidence for altered expression |
| hsa-miR-107 | Upregulated in general |
| hsa-miR-181b-1 | Upregulated in general |
DISCUSSION
Our work identifies a group of microRNAs (termed HRMs) which are induced in a hypoxic environment. To our knowledge, this is the first report establishing a link between a tumor-specific stress factor and microRNAs and extends the response to low oxygen beyond the “classic” translated genes.
We are fully aware that the microarray-based strategy leaves open the possibility that other microRNAs may respond to hypoxia and were simply not detected by the screen. Indeed, with consideration of the well-recognized technical limitations of microarrays, the microRNAs' world is still expanding and it is conceivable that real HRMs were not present on the chip at that stage.
On the other hand, it is possible that some of the microRNAs identified as upregulated by microarray analysis, but not validated independently by quantitative PCR, are false positives. However, given the extremely high confirmation rate of the candidates tested, it is conceivable that high proportions of the remaining candidates indeed respond to oxygen deprivation and represent bona fide HRMs.
The direct contribution of HIFs to the upregulation of HRMs was dissected for select microRNAs using a combination of luciferase-based reporters (containing fragments of microRNA promoters) and chromatin immunoprecipitation. A large variety of direct HIF target genes have been reported, and an indirect, microRNA-mediated component could further add to the complexity of the molecular response orchestrated by these transcription factors. Several in silico methods for target gene prediction have been developed and are publicly available, including PicTar (pictar.bio.nyu.edu), TargetScan 3.0. (http://www.targetscan.org/), and miRBase (http://microrna.sanger.ac.uk/sequences/). For a given microRNA, these utilize different algorithms and ranking criteria (8, 18, 19) and are known to produce a partially overlapping set of candidates, making the search for targets a complex endeavor. Using these three programs, we performed in silico searches for HRM targets, which revealed a highly complex spectrum, including genes involved in apoptosis and proliferation. This could indeed be highly significant for the response to low oxygen, since hypoxia is known to have an impact on both processes.
We show experimental evidence that caspase activation is inhibited by several HRMs during hypoxia. Interestingly, and in keeping with this, our searches with the above-mentioned programs predict that several HRMs target core components of apoptosis: BAK1 (miR-26), BIM (miR-24 and -181), BID (miR-23), caspase-7 (miR-23), CASP3 (miR-30), APAF1 (miR-27), and NIX/BNIP3L (miR-23). In the case of miR-26, we demonstrated a direct antiapoptotic effect with oxygen depletion, increasing the possibility that BAK1 (a proapoptotic protein) is a relevant target.
With regard to miR-210, another HRM with antiapoptotic effect, in silico searches did not reveal candidate targets that are part of the apoptotic machinery. However, a large variety or other genes can influence apoptosis in response to specific stresses. For example, one putative target revealed by PicTar is neuronal pentraxin 1 (11), which has been shown to mediate apoptosis in ischemic neurons, and miR-210 could help neutralize such an effect. Whether such a gene could play a role in apoptosis in nonneuronal cells in low oxygen is not known.
Another process known to be affected by hypoxia is proliferation, with many cell types undergoing cell cycle arrest during oxygen deprivation. HRMs could contribute to this process via predicted targets, such as cdc25A (miR-21, miR-103, and miR-107), cyclin D2 (miR-26, miR-103), cyclin E1 (miR-26), cyclin H (miR23), and cdk6 (miR-26, miR-103). Interestingly, and similarly to the case of apoptosis, several cell cycle genes are predicted to be targeted by multiple HRMs, thereby increasing the chance of efficient downregulation.
Since the hypoxic response described in this paper involves a multitude of microRNAs, it is conceivable that manipulation of any individual HRM could fail to fully capture the phenotypic impact of this mechanism in low oxygen. The concerted induction of these HRMs could therefore have a much more robust impact on apoptotic/proliferative behavior when oxygen is limiting for extended periods, such as in cancer. One could speculate that differences between HRM induction in various cell types could contribute to a variability in the response to hypoxia, with important consequences for cancer progression and response to therapy.
Our analysis shows that a surprisingly high proportion of HRMs are overexpressed in human tumors. The alterations of microRNAs in various cancer types is conceivably the sum of a variety of factors (including oncogene signaling, paracrine factors, and pH alterations), but hypoxia could have a significant impact, setting in motion microtranscripts with biological impact on survival and/or proliferation.
Supplementary Material
Acknowledgments
This work was supported by NIH grant P30 DK-34928 and an AACR/PanCan career development award to M.I.; a Kimmel Scholar award to G.A.C.; grants from the Italian Ministry of Public Health, the Italian Association for Cancer Research (AIRC), and Comitato Sostenitori Progetto CAN-2006 to M.N.; and grants from the National Cancer Institute to C.M.C. M.F. is a recipient of a fellowship from Fondazione Italiana per la Ricerca sul Cancro (FIRC).
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 28 December 2006.
REFERENCES
- 1.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 2.Calin, G. A., M. Ferracin, A. Cimmino, G. Di Leva, M. Shimizu, S. E. Wojcik, M. V. Iorio, R. Visone, N. I. Sever, M. Fabbri, R. Iuliano, T. Palumbo, F. Pichiorri, C. Roldo, R. Garzon, C. Sevignani, L. Rassenti, H. Alder, S. Volinia, C. G. Liu, T. J. Kipps, M. Negrini, and C. M. Croce. 2005. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 353:1793-1801. [DOI] [PubMed] [Google Scholar]
- 3.Calin, G. A., C. G. Liu, C. Sevignani, M. Ferracin, N. Felli, C. D. Dumitru, M. Shimizu, A. Cimmino, S. Zupo, M. Dono, M. L. Dell'Aquila, H. Alder, L. Rassenti, T. J. Kipps, F. Bullrich, M. Negrini, and C. M. Croce. 2004. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. USA 101:11755-11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, A. M., M. W. Byrom, J. Shelton, and L. P. Ford. 2005. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 33:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croce, C. M., and G. A. Calin. 2005. miRNAs, cancer, and stem cell division. Cell 122:6-7. [DOI] [PubMed] [Google Scholar]
- 6.Farh, K. K. 2005. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310:1817-1821. [DOI] [PubMed] [Google Scholar]
- 7.Filipowicz, W., L. Jaskiewicz, F. A. Kolb, and R. S. Pillai. 2005. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 15:331-341. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths-Jones, S., R. J. Grocock, S. van Dongen, A. Bateman, and A. J. Enright. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34:D140-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris, A. L. 2002. Hypoxia—a key regulatory factor in tumour growth. Nat. Rev. Cancer 2:38-47. [DOI] [PubMed] [Google Scholar]
- 10.He, L., J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon, and S. M. Hammond. 2005. A microRNA polycistron as a potential human oncogene. Nature 435:828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hossain, M. A., J. C. Russell, R. O'Brien, and J. Laterra. 2004. Neuronal pentraxin 1: a novel mediator of hypoxic-ischemic injury in neonatal brain. J. Neurosci. 24:4187-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, C. J., S. Iyer, A. Sataur, K. L. Covello, L. A. Chodosh, and M. C. Simon. 2006. Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1α) and HIF-2α in stem cells. Mol. Cell. Biol. 26:3514-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iorio, M. V., M, Ferracin, C. G. Liu, A. Veronese, R. Spizzo, S. Sabbioni, E. Magri, M. Pedriali, M. Fabbri, M. Campiglio, S. Menard, J. P. Palazzo, A. Rosenberg, P. Musiani, S. Volinia, I. Nenci, G. A. Calin, P. Querzoli, M. Negrini, and C. M. Croce. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65:7065-7070. [DOI] [PubMed] [Google Scholar]
- 14.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIF-alpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 15.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 16.Jin, V. X., Y. W. Leu, S. Liyanarachchi, H. Sun, M. Fan, K. P. Nephew, T. H. Huang, and R. V. Davuluri. 2004. Identifying estrogen receptor alpha target genes using integrated computational genomics and chromatin immunoprecipitation microarray. Nucleic Acids Res. 32:6627-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karp, X., and V. Ambros. 2005. Developmental biology. Encountering microRNAs in cell fate signaling. Science 310:1288-1289. [DOI] [PubMed] [Google Scholar]
- 18.Krek, A., D. Grun, M. N. Poy, R. Wolf, L. Rosenberg, E. J. Epstein, P. MacMenamin, I. da Piedade, K. C. Gunsalus, M. Stoffel, and N. Rajewsky. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495-500. [DOI] [PubMed] [Google Scholar]
- 19.Lewis, B. P., I. H. Shih, M. W. Jones-Rhoades, D. P. Bartel, and C. B. Burge. 2003. Prediction of mammalian microRNA targets. Cell 115:787-798. [DOI] [PubMed] [Google Scholar]
- 20.Liu, C. G., G. A. Calin, B. Meloon, N. Gamliel, C. Sevignani, M. Ferracin, C. D. Dumitru, M. Shimizu, S. Zupo, M. Dono, H. Alder, F. Bullrich, M. Negrini, and C. M. Croce. 2004. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA 101:9740-9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, J., G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz, and T. R. Golub. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834-838. [DOI] [PubMed] [Google Scholar]
- 22.Masson, N., C. William, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20:5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matys, V., E. Fricke, R. Geffers, E. Gossling, M. Haubrock, R. Hehl, K. Hornischer, D. Karas, A. E. Kel, O. V. Kel-Margoulis, D. U. Kloos, S. Land, B. Lewicki-Potapov, H. Michael, R. Munch, I. Reuter, S. Rotert, H. Saxel, M. Scheer, S. Thiele, and E. Wingender. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31:374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miska, E. A. 2005. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 15:563-568. [DOI] [PubMed] [Google Scholar]
- 25.O'Donnell, K. A., E. A. Wentzel, K. I. Zeller, C. V. Dang, and J. T. Mendell. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839-843. [DOI] [PubMed] [Google Scholar]
- 26.Palaniswamy, S. K., V. X. Jin, H. Sun, and R. V. Davuluri. 2005. OMGProm: a database of orthologous mammalian gene promoters. Bioinformatics 21:835-836. [DOI] [PubMed] [Google Scholar]
- 27.Sun, H., S. K. Palaniswamy, T. T. Pohar, V. X. Jin, T. H. Huang, and R. V. Davuluri. 2006. MPromDb: an integrated resource for annotation and visualization of mammalian gene promoters and ChIP-chip experimental data. Nucleic Acids Res. 34:D98-D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volinia, S., G. A. Calin, C. G. Liu, S. Ambs, A. Cimmino, F. Petrocca, R. Visone, M. Iorio, C. Roldo, M. Ferracin, R. L. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C. C. Harris, and C. M. Croce. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 103:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanaihara, N., N. Caplen, E. Bowman, M. Seike, K. Kumamoto, M. Yi, R. M. Stephens, A. Okamoto, J. Yokota, T. Tanaka, G. A. Calin, C. G. Liu, C. M. Croce, and C. C. Harris. 2006. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9:189-198. [DOI] [PubMed] [Google Scholar]
- 30.Zhao, Y., E. Samal, and D. Srivastava. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436:214-220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.