Abstract
Structural studies of GTP-binding proteins identified the Switch I and Switch II elements as contacting the γ-phosphate of GTP and undergoing marked conformational changes upon GTP versus GDP binding. Movement of a universally conserved Gly at the N terminus of Switch II is thought to trigger the structural rearrangement of this element. Consistently, we found that mutation of this Gly in the Switch II element of the eukaryotic translation initiation factor 5B (eIF5B) from Saccharomyces cerevisiae impaired cell growth and the guanine nucleotide-binding, GTPase, and ribosomal subunit joining activities of eIF5B. In a screen for mutations that bypassed the critical requirement for this Switch II Gly in eIF5B, intragenic suppressors were identified in the Switch I element and at a residue in domain II of eIF5B that interacts with Switch II. The intragenic suppressors restored yeast cell growth and eIF5B nucleotide-binding, GTP hydrolysis, and subunit joining activities. We propose that the Switch II mutation distorts the geometry of the GTP-binding active site, impairing nucleotide binding and the eIF5B domain movements associated with GTP binding. Accordingly, the Switch I and domain II suppressor mutations induce Switch II to adopt a conformation favorable for nucleotide binding and hydrolysis and thereby reestablish coupling between GTP binding and eIF5B domain movements.
GTP-binding (G) proteins include members of the Ras family, heterotrimeric G proteins, and translation factors. These proteins regulate many cellular pathways including signal transduction, transport, and protein synthesis. The core of the G proteins is the GTP-binding domain, which can be easily identified by the conservation of specific amino acid sequence motifs (25). Comparison of the structures of several G proteins in their active, GTP-bound and inactive, GDP-bound forms identified two regions that undergo marked conformational changes upon GTP versus GDP binding (25, 27). The Switch I region contains the conserved G domain sequence motif G-2 and is marked by an essential Thr residue (Fig. 1A) whose main-chain NH group contacts the γ-phosphate of GTP and whose side chain helps coordinate the Mg2+ required for GTP binding and hydrolysis. The Switch II region contains the G-3 sequence motif D-X-X-G (Fig. 1A). The main-chain NH of the Gly in G-3 also contacts the γ-phosphate of GTP. It is thought that the structural changes in Switch I and Switch II upon GTP binding and hydrolysis govern the interaction of the G proteins with specific downstream effectors (27). Thus, conformational switches in the G proteins are thought to function as activity switches regulating various cellular pathways. It is noteworthy that structural studies of several G proteins revealed conformational flexibility of the Switch I and Switch II elements and detailed the interaction of these structural elements with guanine nucleotides (see references 24 and 27).
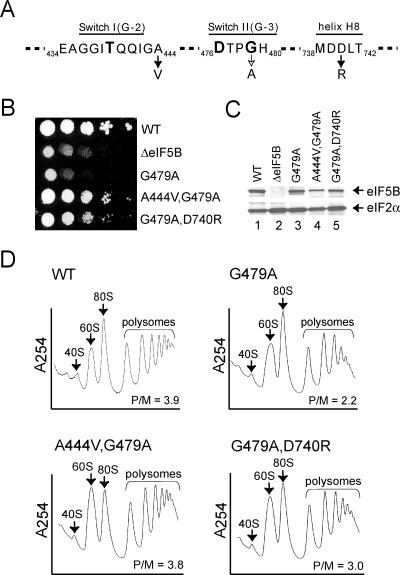
FIG. 1.
Intragenic suppressors of the eIF5B G479A mutation. (A) Amino acid sequences of the Switch I (G-2 sequence motif), Switch II (G-3 sequence motif), and helix H8 elements in yeast eIF5B. The G479A mutation and the A444V and D740R suppressor mutations are shown, and the position numbers in yeast eIF5B are indicated. The invariable residues Thr439 in Switch I and Asp476 and Gly479 in Switch II are shown in boldface. (B) Growth rate analysis of yeast expressing WT and mutant forms of eIF5B. The ΔeIF5B strain J111 was transformed with the empty vector YCplac33 (ΔeIF5B) or the same plasmid containing the indicated WT or mutant eIF5B genes. Transformants were grown to saturation, and 4 μl of serial dilutions (at optical densities at 600 nm of 1.0, 0.1, 0.01, 0.001, and 0.0001) was spotted onto synthetic dextrose medium supplemented with the required nutrients and incubated at 30°C for 4 days. (C) Western blot analysis of eIF5B expression. Whole-cell extracts prepared from transformants described in the legend for panel B were subjected to immunoblot analysis using anti-eIF5B or anti-eIF2α antiserum as described previously (7). Immune complexes were visualized using enhanced chemiluminescence. (D) Analysis of polysome profiles for strains expressing eIF5B, eIF5B-G479A, eIF5B-A444V,G479A, and eIF5B-G479A,D740R. Whole-cell extracts from yeast strain J111 expressing the indicated eIF5B WT or mutant protein were resolved by velocity sedimentation in 7 to 47% sucrose gradients. Gradients were fractionated while scanning at A254, and the positions of the 40S and 60S subunits, 80S ribosomes, and polysomes are indicated. P/M ratios were calculated by measuring the areas under the peaks representing the polysome fractions and the 80S peak.
The eukaryotic translation initiation factor 5B (eIF5B), present in all eukaryotes, is an ortholog of the bacterial translation initiation factor 2 and the archaeal initiation factor 5B (aIF5B), and in vitro eIF5B facilitates ribosomal subunit joining, the final step of eukaryotic translation initiation (6, 20). Structural studies revealed that Methanothermobacter thermoautotrophicus aIF5B contains a classical G domain, which forms part of the cup of the chalice-shaped protein (21). Comparison of aIF5B structures bound to GTP and GDP revealed that modest repositioning of the Switch II element upon GTP binding triggers lever-type domain movements that are amplified by the fulcrum arm “stem” of the chalice, resulting in an ∼5-Å displacement of the base (domain IV) of the factor (21). In previous mutational studies of eIF5B, encoded in Saccharomyces cerevisiae by FUN12, we showed that the factor must bind GTP to promote subunit joining and that GTP versus GDP binding to eIF5B governs the ribosome affinity of the factor (18, 20, 23). The substitution of Ala for the conserved Thr in eIF5B Switch I severely impairs both yeast cell growth and the ribosome-dependent GTPase activity of the factor but does not impair guanine nucleotide binding or ribosome subunit joining activity in vitro (23). An intragenic suppressor of the eIF5B Switch I mutation restores protein synthesis, but not eIF5B GTPase activity, by lowering the ribosome affinity of the factor (23). Thus, we proposed that GTP hydrolysis by eIF5B activates a regulatory switch required for eIF5B release from the ribosome following subunit joining.
In most G proteins, including Ras, Giα1, and the translation elongation factor Tu (EF-Tu), the Gly residue in the D-X-X-G G-3 sequence motif resides at the N terminus of an α-helix in the Switch II region. It has generally been thought that the movement of this Gly residue is critical for the structural transition of Switch II during GTP binding and hydrolysis (13, 15, 25). Consistent with this idea, the substitution of Ala for Gly60 in Ras does not affect GTP binding but blocks Ras function presumably by preventing the conformational changes normally induced upon GTP binding (26). Mutation of the corresponding Gly226 in the heterotrimeric Gsα protein decreases the affinity for Mg2+ approximately 100-fold and prevents the conformational changes required to release Gsα from the Gβγ complex (17, 19). Analysis of the same mutation in Giα1 (Gly203 to Ala) revealed only a sevenfold decrease in Mg2+-binding affinity, and structural studies showed an altered Switch II helix that, based on the presence of both GDP and Pi in the crystal, was proposed to reflect the structure of an intermediate in the GTP hydrolysis reaction pathway (4). In EF-Tu, the replacement of the corresponding Gly83 with Ala decreases the GTP-binding affinity threefold, increases the dissociation rate constant for aminoacyl-tRNA, impairs the binding of EF-Tu-GTP-aminoacyl-tRNA ternary complexes to the ribosome, and abolishes ribosome-dependent GTP hydrolysis activity (12, 16). The impaired function of the EF-Tu G83A mutant is consistent with the idea that dramatic movement of this Gly residue, which is dependent on the unique peptide bond flexibility associated with Gly, is critical for the reorientation of the Switch II helix α2 and the coupled movements of Switch I and EF-Tu domains II and III (13, 15).
In contrast to Ras, Giα, and EF-Tu, the Switch II element in aIF5B does not adopt an α-helical conformation. Comparison of the structures of the aIF5B-GTP and aIF5B-GDP complexes revealed a modest ∼2-Å movement of Switch II upon GTP binding, which is much less than the ∼7.5-Å movement observed in EF-Tu Switch II upon GTP binding (13, 21). Moreover, the Switch II Gly79 residue in M. thermoautotrophicus aIF5B (21) does not directly contact GTP as do the corresponding residues in Ras and EF-Tu. To gain further insights into the eIF5B GTP-binding properties and regulatory switch, and by extension the switch and guanine nucleotide-binding behaviors of other G proteins, we conducted a mutational and suppressor analysis of the conserved Switch II Gly479 residue (G-3 motif) of yeast eIF5B. Our genetic studies identified intragenic suppressor mutations mapping to the Switch I element and to domain II and, together with further biochemical analyses, provide both in vivo and in vitro support for the idea that a conformational change in Switch II triggered through the conserved Gly residue is critical for the GTP-binding, GTP hydrolysis, and translation stimulatory properties of eIF5B.
MATERIALS AND METHODS
Mutagenesis and screening.
The plasmid pC1285 carrying FLAG-tagged eIF5B with the deletion of N-terminal residues 28 to 396 [ΔN(28-396)-eIF5B] is a derivative of the vector YCplac33. Site-directed mutagenesis was used to introduce the G479A mutation, generating plasmid pC1780. To screen for intragenic suppressors, pC1780 was randomly mutated by passage through the error-prone Escherichia coli strain XL1-Red (Stratagene). A pool of mutated plasmids was introduced into S. cerevisiae strain J111 lacking eIF5B (MATα ura3-52 leu2-3 leu2-112 fun12Δ) (7), and transformants growing faster than controls transformed with the empty vector YCplac33 or unmutated pC1780 were selected. From ∼3 × 104 yeast transformants, two fast-growing revertants were identified. The eIF5B plasmids were isolated from the revertants, used to transform strain J111 to confirm the phenotype, and sequenced to identify the mutations.
Purification of proteins and ribosomes.
The purification of wild-type (WT) and mutant forms of fusion proteins consisting of glutathione S-transferase and eIF5B residues 396 to 1002 (eIF5B396-1002) expressed in yeast and the preparation of 80S ribosomes were performed as previously described (23).
Ribosomal 40S and 60S subunits were purified from S. cerevisiae strain F353 (MATα trp1 leu2-Δ1 his3-Δ200 pep4::HIS3 prb1-Δ1.6 GAL+) as described previously (1) with minor modifications. The 40S and 60S fractions obtained after subjection to sucrose gradients were collected and pelleted by centrifugation for 17 h at 35,000 rpm in a Beckman Type 45 rotor. Finally, the ribosomal subunit pellets were dissolved in ribosome storage buffer (20 mM HEPES-KOH [pH 7.4], 100 mM potassium acetate, 2.5 mM magnesium acetate, 250 mM sucrose, and 2 mM dithiothreitol [DTT]).
Measurement of Kd values for guanine nucleotide binding to eIF5B.
Fluorescence intensities of N-methylanthraniloyl (Mant)-labeled GDP (Molecular Probes and Invitrogen) were measured with a Floromax-3 steady-state fluorometer. Competition experiments were set up by first mixing 1.5 μM eIF5B with 500 nM Mant-GDP in 1× binding buffer (30 mM HEPES-KOH [pH 7.4], 100 mM potassium acetate, 2 mM DTT, and 3 mM magnesium acetate). Samples were excited at 360 nm, and fluorescence at 445 nm was then monitored as a function of the concentrations of various unlabeled nucleotides. Reaction mixtures equilibrated within 1 min after the addition of the competitor, and the change in intensity (the intensity of the bound sample minus the intensity of the free sample) was plotted as a function of competitor (GTP or GDP) concentration. The data were fit with the expression 1 − [GTP]/(Kd + [GTP]) by nonlinear regression using KaleidaGraph. The assumption implicit in this model that [eIF5B] was much greater than [eIF5B-Mant-GDP] was reasonable because the Mant-GDP concentration was 20-fold less than the lowest Kd measured.
80S formation assay.
Reconstitution 80S formation assays were performed as previously described (2, 23). Briefly, eIF2 was mixed with saturating amounts of GTP for 10 min to allow the exchange of eIF2-bound GDP for GTP, followed by the addition of [35S]Met-tRNAiMet. After 5 min of incubation to allow ternary complex formation, 80S, eIF1, eIF1A, eIF5, mRNA, and eIF5B were added simultaneously to initiate 80S complex formation. Following incubation at 26°C for up to 30 min, aliquots were mixed with 10× loading dye and loaded directly onto a running 4% polyacrylamide gel.
Ribosome-binding assay.
Ribosome-binding assays were performed as described previously (23). Briefly, wild-type or mutant forms of eIF5B were mixed with 80S ribosomes in the presence of GTP, the nonhydrolyzable GTP analog GDPNP, GDP, or no nucleotide. The mixtures were then layered on top of ice-cold 10% sucrose cushions, and the ribosomes were pelleted by centrifugation. Samples of the supernatant and pellet fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue, and the amount of eIF5B was quantified by densitometry using NIH Image software (version 1.62).
Trypsin proteolysis.
Purified eIF5B (3.5 μM) was digested with trypsin (100 nM; proteomics grade [Sigma-Aldrich]) in reaction buffer containing 20 mM HEPES (pH 7.4), 100 mM potassium acetate, 3 mM magnesium acetate, and 2 mM DTT. Reactions were performed at 25°C and stopped by freezing in dry ice. Samples were analyzed by SDS-PAGE on either 4 to 20% polyacrylamide gels using Tris-glycine buffer or 10% NuPAGE bis-Tris gels using MOPS (morpholinepropanesulfonic acid) buffer. For N-terminal sequencing of fragments obtained by trypsin cleavage, gels were blotted onto polyvinylidene difluoride membrane (Invitrogen) by using N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) buffer (pH 11.0). Peptide sequencing was performed by the Protein Chemistry Laboratory (SAIC-Frederick, Inc.).
RESULTS
Mutation of the invariable Gly residue in the G domain Switch II element impairs eIF5B function.
To address the importance of the universally conserved Switch II Gly residue in eIF5B, the corresponding Gly479 in yeast eIF5B was mutated to Ala (Fig. 1A). As the N-terminal region (residues 1 to 395) of yeast eIF5B is not required in vivo or in vitro (7), an N-terminally truncated form of eIF5B (eIF5B396-1002) was used for all experiments. Plasmids expressing eIF5B or an eIF5B G479A mutant (eIF5B-G479A) were introduced into a strain lacking the FUN12 gene encoding yeast eIF5B (a ΔeIF5B strain). Yeast expressing eIF5B-G479A as the sole source of eIF5B exhibited a severe slow-growth phenotype, similar to that of a ΔeIF5B strain, with a doubling time 2.5-fold higher than that of the WT (6.6 h versus 2.8 h) (Fig. 1B). Immunoblot analysis revealed that eIF5B-G479A was expressed at a level similar to wild-type eIF5B (Fig. 1C, compare lanes 1 and 3), indicating that the null phenotype was due not to poor protein expression but rather to the loss of catalytic function. Consistent with the notion that the Switch II mutation impairs eIF5B translational activity, polysome profile analyses revealed a significant loss of polysomes in the eIF5B-G479A-expressing strain. The polysome/monosome (P/M) ratio of 3.9 in the WT strain was reduced to 2.2 in the eIF5B-G479A-expressing strain (Fig. 1D). As the P/M ratio reflects the distribution of ribosomes on all translated mRNAs, the decreased P/M ratio indicates that this eIF5B mutation impaired translation initiation on many, if not all, of the mRNAs in the cell. Thus, the invariable Switch II Gly residue is critical for eIF5B function in vivo.
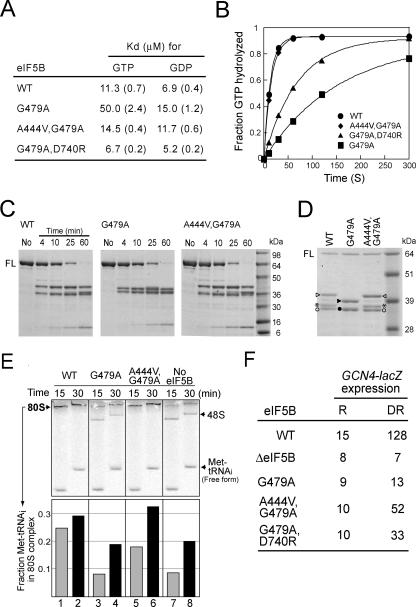
As the conserved Switch II Gly residue is part of the Walker B box and the G-3 sequence motif, the eIF5B G479A mutation was predicted to impair GTP binding. To test this possibility, we examined the binding of GTP and GDP to eIF5B and eIF5B-G479A in competition assays with a fluorescent Mant analog of GDP as described in Materials and Methods. The Kd for GTP and eIF5B was ∼11 μM, which was similar to the Kd for eIF5B and GDP (7 μM) (Fig. 2A). In contrast, eIF5B-G479A bound GTP approximately fivefold more weakly, with a Kd of 50 μM (Fig. 2A). The G479A mutation in eIF5B modestly increased the Kd for GDP, about twofold (Fig. 2A). These data revealed, as predicted, that the invariable Gly479 in eIF5B is critical for guanine nucleotide binding. The G479A mutation also significantly impaired the single-round eIF5B ribosome-dependent GTPase activity. In the presence of 40S and 60S ribosomal subunits, wild-type eIF5B hydrolyzed GTP with a rate constant of 0.076 ± 0.014 s−1 whereas the rate constant for eIF5B-G479A was 0.0064 ± 2 × 10−4 s−1 (Fig. 2B). It is noteworthy that increasing the GTP concentration up to 1 mM (20-fold over the Kd) failed to restore the GTPase activity of eIF5B-G479A (data not shown). Thus, the G479A mutation impaired not only GTP binding but also the ribosome-dependent GTPase activity of eIF5B.
FIG. 2.
Biochemical analysis of eIF5B-G479A Switch II mutant and eIF5B-A444V,G479A and eIF5B-G479A,D740R suppressor mutants. (A) Summary of guanine nucleotide Kd values for eIF5B mutants. Values shown in parentheses are standard errors. (B) Results from a ribosome-dependent GTPase assay. Reaction mixtures containing 1 μM eIF5B, 0.4 μM 40S and 60S ribosomal subunits, and 50 nM [γ-33P]GTP were incubated at 30°C, and samples were quenched at the indicated times. Data were fit to the single exponential expression A[1 − exp(−kt)], in which A is the amplitude, k is the rate constant, and t is time. Fits were performed using KaleidaGraph (Synergy Software). The data presented are representative of results from at least three independent experiments. (C) Trypsin cleavage analysis of eIF5B. Recombinant WT or mutant forms of eIF5B396-1002 (3.5 μM) were digested with trypsin (100 nM), and reactions were stopped at the indicated times. Digestion products were analyzed by 4 to 20% SDS-PAGE (Tris-glycine buffer). Molecular mass markers are shown on the right. No, intact eIF5B396-1002 prior to trypsin cleavage; FL, full-length eIF5B396-1002. Results shown are representative of results from three independent experiments. (D) Trypsin cleavage analysis of eIF5B. Reactions were performed for 25 min as described for panel C, and products were analyzed by SDS-PAGE using 10% NuPAGE bis-Tris gels with MOPS buffer. N-terminal sequencing revealed that the peptide fragments marked by the triangles (closed or open) start at residue L689 (located in domain II), fragments marked by the asterisks start at residue L721 (located at the C-terminal end of domain II), and fragments marked by the circles (closed or open) start at the N terminus of eIF5B396-1002. Refer to Fig. 4 (upper panel) for the locations of the cleavage sites. (E) Results from the 80S complex formation assay. (Upper panel) Phosphorimage of a native gel for examining the ability of eIF5B to stimulate 80S complex formation. The progress of 80S complex formation was monitored in reactions with mixtures containing WT eIF5B, eIF5B-G479A, eIF5B-A444V,G479A, or no eIF5B by stopping the reactions at 15 and 30 min. The staggering of the bands is due to the fact that the samples were loaded at different times onto a running gel. The positions of 80S and 48S complexes and free [35S]Met-tRNAiMet are indicated. The data presented are representative of results from at least three independent experiments. (Lower panel) The amount of [35S]Met-tRNAiMet that was free or bound to 48S or 80S complexes was quantified, and the fraction of Met-tRNAiMet present in the 80S complexes relative to the total Met-tRNAiMet (80S + 48S + free) was calculated. (F) Analysis of GCN4-lacZ expression. The GCN4-lacZ plasmid p180 (10) was introduced into derivatives of strain J111 expressing WT eIF5B, eIF5B-G479A, eIF5B-A444V,G479A, or no eIF5B (ΔeIF5B). Cells were grown and β-galactosidase activity was determined as described previously (10), except that longer growth periods were required for sufficient quantities of cells from the slow-growing ΔeIF5B- and eIF5B-G479A-expressing strains. R, cells were grown under nonstarvation conditions in SD minimal medium where GCN4 expression is repressed; DR, cells were grown under amino acid starvation conditions (SD medium + 10 mM 3-aminotriazole) where GCN4 expression is normally derepressed. The β-galactosidase activities are the averages from three independent transformants and have standard errors of 30% or less.
To assess the impact of the G479A mutation on eIF5B structure, WT eIF5B and eIF5B-G479A were subjected to limited proteolysis with trypsin. Recombinant eIF5B396-1002 was incubated with trypsin, and aliquots were taken at various times and analyzed by SDS-PAGE. As shown in Fig. 2C (left and middle panels), the digestion of WT eIF5B and eIF5B-G479A resulted in the rapid and stable appearance of several products with masses around 39 kDa. Interestingly, the mobilities of these prominent digestion products appeared to be different for WT eIF5B and eIF5B-G479A. To better resolve these fragments, the products from a reaction mixture incubated for 25 min were separated on a 10% gel (Fig. 2D). Three fragments were readily detected in the digest of WT eIF5B, whereas only two fragments were detected in the digest of eIF5B-G479A. N-terminal sequencing of the digestion products revealed that the 64-kDa fragment as well as the ∼36-kDa fragment initiated at the N terminus of eIF5B396−1002. The 64-kDa fragment likely represents full-length eIF5B396-1002, and based on its molecular mass the 36-kDa fragment is likely to result from cleavage in domain II. Sequencing of the ∼41-kDa fragment from WT eIF5B and the ∼39-kDa fragment from eIF5B-G479A (Fig. 2D) revealed the same N-terminal residue, L689 in domain II (see Fig. 4, top panel). Based on the different masses of these two products, we predicted that the ∼39-kDa fragment from eIF5B-G479A reflects a novel cleavage near the C terminus of the protein. Unfortunately, using matrix-assisted laser desorption ionization-time of flight mass spectrometry, we were unable to detect the C-terminal peptide fragment from either WT eIF5B or eIF5B-G479A (data not shown), so we were unable to confirm that the difference in the mobility of these fragments was due to a unique C-terminal cleavage of eIF5B-G479A. Finally, sequencing of the ∼37-kDa fragment observed in digests of WT eIF5B (Fig. 2D) and lacking in digests of eIF5B-G479A identified the N-terminal residue as L721, which is located at the C-terminal end of domain II (see Fig. 4, top panel). These changes in the trypsin cleavage pattern of eIF5B-G479A versus that of WT eIF5B are consistent with the idea that the G479A mutation alters the conformation of the Switch II element, leading to the repositioning of domains II and IV as indicated by the protection of the cleavage site at R720 (loss of one fragment as depicted in Fig. 2D) and the exposure of a cleavage site in domain IV (altered size of one fragment as depicted in Fig. 2D).
FIG. 4.
Model of the aIF5B active site depicting the interactions of Switch II with Switch I and helix H8. (Upper panels) Ribbon representation of aIF5B in complex with GTP (Protein Data Bank code, 1G7T [21]). The Switch II element is depicted in green, Switch I in blue, and helix H8 in purple. The four domains of the protein are labeled, and the N-terminal residues of trypsin cleavage fragments (L689 and L721) are indicated (Fig. 2D). The image on the right was rotated about the indicated axis to better visualize the interactions at the G domain active site. (Lower panel) Ribbon representation of the aIF5B active site in complex with GTP. As in the upper panel, the Switch II element is depicted in green, Switch I in blue, and helix H8 in purple, and GTP is depicted in gray. The side chains of key residues discussed in the text are shown, and the three residues mutated in this study (Gly79, Ala44, and Glu340) are depicted in red. The H bond or salt bridge interactions between R87 and E340, E81 and the γ-phosphate of GTP, G43 and D76, and T45 and F74 are depicted by dotted lines. The substitution of Val for A44 (A444V suppressor mutation) may create a clash (marked by the black double arrow) with I75. The image was generated using PyMOL software (8).
Isolation of intragenic suppressors of the eIF5B G479A mutation.
To gain further insight into the role of Switch II and the defect associated with the G479A mutation, we screened for intragenic suppressors of the eIF5B G479A mutation. Two suppressor mutations were identified: Ala444 to Val in Switch I and Asp740 to Arg in the α-helix H8 linking domain II to domain III (Fig. 1A and see Fig. 4). The expression of both eIF5B-G479A with the A444V mutation (eIF5B-A444V,G479A) and eIF5B-G479A with the D740R mutation (eIF5B-G479A,D740R) was less than (∼50%) the expression of WT eIF5B (Fig. 1C, lanes 1, 4, and 5), indicating that the suppression phenotype was not due to enhanced eIF5B expression. As shown in Fig. 1B, the growth rates of yeast expressing eIF5B-A444V,G479A (doubling time of 3.3 h) and eIF5B-A444V,D740R (3.6 h) were close to that of yeast expressing WT eIF5B (2.8 h) and much better than those of strains expressing eIF5B-G479A (6.4 h) or no eIF5B (6.6 h). Polysome profile analysis (Fig. 1D) revealed P/M ratios that were nearly identical for strains expressing eIF5B-A444V,G479A (3.8) and WT eIF5B (3.9) and slightly lower for the strain expressing eIF5B-G479A,D740R (3.0), consistent with the lower growth rate of the latter strain. Thus, the suppressor mutations restored general protein synthesis in the yeast cells. In contrast to its ability to suppress the slow-growth phenotype associated with the Switch II G479A mutation, the A444V mutation was unable to suppress the slow-growth phenotype caused by a Switch I mutation (T439A) or a different Switch II mutation (H480E) in eIF5B (data not shown). In addition, yeast strains expressing eIF5B with the A444V mutation only (eIF5B-A444V) or with the D740R mutation only (eIF5B-D740R) exhibited a WT phenotype (data not shown), indicating that the suppressor mutations did not significantly alter eIF5B function. Thus, the A444V and D740R mutations appear to be specific suppressors of the G479A Switch II mutation.
Suppressor mutations in eIF5B-A444V,G479A and eIF5B-G479A,D740R restore eIF5B nucleotide-binding and GTPase activities.
In order to determine how the A444V and D740R mutations suppressed the eIF5B-G479A growth defect, we first tested the guanine nucleotide-binding and GTPase activities of the suppressor mutants. As shown in Fig. 2A, eIF5B-A444V,G479A and eIF5B-G479A,D740R bound GTP and GDP with affinities similar to that of WT eIF5B. Likewise, as shown in Fig. 2B, eIF5B-A444V,G479A possessed a robust, ribosome-dependent GTPase activity with a rate constant of 0.066 ± 6 × 10−4 s−1, which was nearly the same as the rate constant of 0.076 s−1 obtained for WT eIF5B. The eIF5B-G479A,D740R suppressor mutant hydrolyzed GTP with a rate constant of 0.0145 ± 2 × 10−4 s−1, an ∼2-fold increase in the rate compared to that of eIF5B-G479A (Fig. 2B). This partial restoration of ribosome-dependent GTPase activity by the D740R mutation is consistent with the modest growth restoration observed with this mutation (Fig. 1B). Interestingly, the A444V mutation also restored a WT pattern of protease sensitivity (Fig. 2C). The protease digestion pattern of purified eIF5B-A444V,G479A resembled the pattern observed with WT eIF5B, especially as noted by the mobility of the fragment starting at residue L689 (Fig. 2D) and the reappearance of the fragment initiating at L721 (Fig. 2D). In contrast, the protease digestion pattern for eIF5B-G479A,D740R was very similar to that for eIF5B-G479A (data not shown). These results suggest that an altered conformation of Switch II and coupled changes in the structures of domains II and IV in eIF5B-G479A are restored to a WT configuration by the A444V suppressor mutation. Thus, both the Switch I A444V and the α-helix H8 D740R mutations suppressed, albeit to different extents, the guanine nucleotide binding and GTPase defects and the altered protease sensitivity associated with the Switch II G479A mutation.
To test whether the restoration of eIF5B GTPase activity was necessary for the suppression of the Switch II mutation, we introduced the Switch I mutation T439A into eIF5B-A444V,G479A. Previously, we demonstrated that this Switch I mutation eliminated eIF5B GTPase activity and could be suppressed by mutations that lowered ribosome affinity (23). The growth rate of yeast expressing the eIF5B-T439A,A444V,G479A triple mutant was similar to that of a ΔeIF5B strain (data not shown), and the eIF5B triple mutant was expressed at levels comparable to wild-type eIF5B (data not shown). These results suggest, though they do not prove, that the suppressor function of the A444V mutation is dependent on the restoration of eIF5B GTPase activity.
We next examined the ribosomal subunit joining activity of the eIF5B mutants by using a reconstituted yeast translation initiation system as described previously (2, 23). As shown in Fig. 2E, the inclusion of eIF5B (WT, lanes 1 and 2) in the subunit joining assay mixtures strongly enhanced 80S complex formation in comparison to that in assay mixtures lacking eIF5B (lanes 7 and 8). Notably, there was an apparent complete conversion of 48S complexes to 80S complexes in assay mixtures containing eIF5B versus those containing no eIF5B. In contrast, the eIF5B-G479A Switch II mutant failed to stimulate subunit joining and the assay mixture yielded about the same amount of 48S and 80S complexes as assay mixtures lacking eIF5B (Fig. 2E, lanes 3 and 4 versus lanes 7 and 8). This defect in subunit joining activity is consistent with the GTP-binding defect of the eIF5B Switch II mutant and may reflect alterations in the way eIF5B binds GTP and a resulting change in the relative positions of the eIF5B domains when eIF5B is bound to GTP. Finally, the A444V suppressor mutation in eIF5B-A444V,G479A restored 80S complex formation activity to WT levels (Fig. 2E, lanes 5 and 6). Thus, the A444V mutation appears to restore almost fully all eIF5B activities. In contrast, the eIF5B-G479A,D740R suppressor mutant failed to stimulate 80S complex formation (data not shown). As the D740R mutation was a weak suppressor of the cell growth and GTPase defects associated with the G479A mutation, the lack of activity in the 80S complex formation assay by eIF5B-G479A,D740R may reflect a higher threshold for function in this assay.
A444V and D740R suppressor mutations restore GCN4 translational control in yeast.
The observation that the suppressor mutations at least partially restored eIF5B function in the assays led us to examine the effects of the eIF5B mutations on GCN4 expression. The translation of the yeast GCN4 mRNA, encoding a transcriptional activator of genes encoding amino acid biosynthetic enzymes, is sensitive to perturbations of general translation initiation (11). Previously, we showed that the expression of GCN4 under amino acid starvation conditions was impaired in yeast lacking eIF5B or expressing GTPase-deficient forms of the factor (23). In cells containing WT eIF5B, the expression of a GCN4-lacZ reporter was low under nutrient-rich conditions and increased approximately ninefold under amino acid starvation conditions (Fig. 2F). This high-level expression of GCN4-lacZ under starvation conditions was blocked in strains lacking eIF5B or expressing eIF5B-G479A, consistent with the diminished GTPase activity of this eIF5B mutant. In contrast, GCN4-lacZ expression was induced five- or threefold under amino acid starvation conditions in strains expressing eIF5B-A444V,G479A or eIF5B-G479A,D740R, respectively (Fig. 2F). The greater derepression of GCN4-lacZ expression in strains expressing the A444V suppressor, which fully restores eIF5B GTPase function, provides additional evidence that GCN4 translational regulation in yeast is dependent on eIF5B GTPase activity.
The GTP-GDP switch governing ribosome binding is altered in the eIF5B-A444V,G479A suppressor mutant.
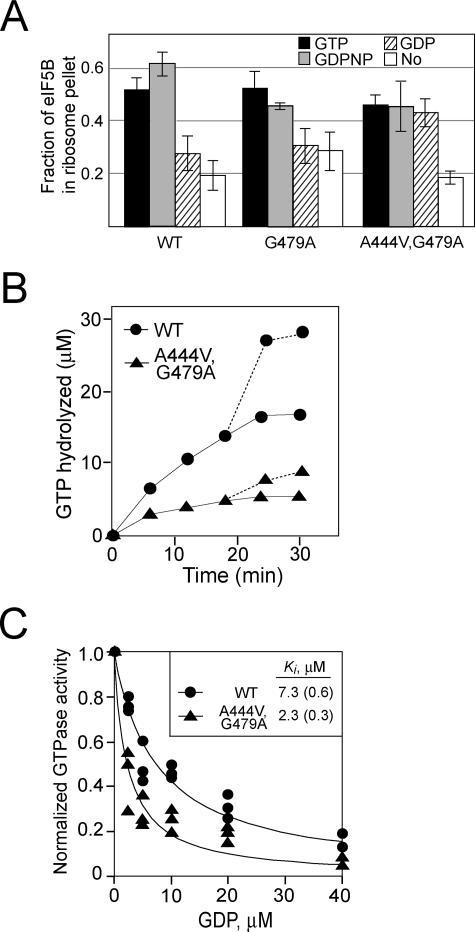
To assess ribosome binding, wild-type and mutant versions of eIF5B were mixed with purified 80S ribosomes in the presence of GTP, nonhydrolyzable GDPNP, GDP, or no nucleotide and then pelleted through a sucrose cushion. As observed previously (23), wild-type eIF5B pelleted with ribosomes in the presence of GTP or GDPNP, but the binding was significantly reduced in the presence of GDP or no nucleotide (Fig. 3A). Ribosome"T3 binding by the eIF5B-G479A mutant was similar to that by wild-type eIF5B, with good binding in the presence of GTP or GDPNP and low binding in the presence of GDP or no nucleotide (Fig. 3A). In contrast, the eIF5B-A444V,G479A suppressor mutant showed good binding to ribosomes in the presence of GTP, GDPNP, or GDP and lower binding in the absence of nucleotide. Thus, ribosome binding by eIF5B-A444V,G479A is apparently governed by a nucleotide-no nucleotide switch as opposed to the GTP-GDP switch governing ribosome binding by wild-type eIF5B.
FIG. 3.
The GTPase switch regulating ribosomal affinity is altered in the eIF5B-A444V,G479A suppressor mutant. (A) Results of a ribosome-binding assay. Purified WT eIF5B, eIF5B-G479A, or eIF5B-A444V,G479A was mixed with purified yeast 80S ribosomes in the presence of GTP, GDPNP, GDP, or no nucleotide as indicated and then loaded onto a 10% sucrose cushion. Following centrifugation, the supernatant and ribosomal pellet fractions were analyzed by SDS-PAGE. The amounts of eIF5B recovered in the supernatant and pellet fractions were determined by quantitative densitometry, and the fraction of total recovered eIF5B present in the ribosomal pellet was calculated. The data presented are the averages of results from at least three independent experiments. (B) Time course of ribosome-dependent GTPase assay. Equal amounts of purified WT eIF5B or eIF5B-A444V,G479A (0.4 μM) were incubated with 50 μM [γ-33P]GTP in the presence of purified yeast 80S ribosomes (0.1 μM). Aliquots from the reaction mixtures were analyzed at various time points by thin-layer chromatography, and the amount of phosphate released was quantified. The values were corrected by subtracting the GTPase activities observed for the proteins in the absence of ribosomes. To test whether the loss of activity by eIF5B-A444V,G479A after 10 min was due to protein instability, fresh [γ-33P]GTP (50 μM) was added at 18 min and the release of phosphate was quantified (dotted lines). Results shown are representative of results from three independent experiments. (C) GDP inhibition of eIF5B GTPase activity. Increasing amounts of GDP, as indicated, were added to GTPase reaction mixtures containing 1 μM GTP, 50 nM 80S ribosomes, and 100 nM eIF5B or eIF5B-A444V,G479A. Reaction mixtures were incubated at 30°C for 5 min, phosphate release was quantified, and the values were normalized to the amount of phosphate released in assays with mixtures lacking GDP. Results shown are from three independent experiments, and the data were fit with the following expression by nonlinear regression using KaleidaGraph: 1 − [GDP]/(Ki + [GDP]). Numbers in parentheses are errors of the fits.
Two assays were employed to further test this altered switch behavior of the eIF5B-A444V,G479A suppressor mutant. In the first experiment, the time courses of GTP hydrolysis by WT eIF5B and eIF5B-A444V,G479A were examined (Fig. 3B). We reasoned that if the GDP-bound form of eIF5B-A444V,G479A has high affinity for the ribosome, then the ribosome-dependent GTPase activity of eIF5B-A444V,G479A should be product inhibited as the concentration of GDP increases during the reaction. Whereas GTP hydrolysis by WT eIF5B increased throughout the first 25 min of the assay, the GTPase activity of eIF5B-A444V,G479A reached a plateau after 5 min (Fig. 3B). To differentiate between the product inhibition of eIF5B-A444V,G479A by GDP and the loss of activity due to enzyme instability, we added fresh GTP to the reaction mixtures after 18 min. Both WT eIF5B and eIF5B-A444V,G479A readily hydrolyzed the added GTP (Fig. 3B), indicating that the plateau observed after 5 min in the eIF5B-A444V,G479A reaction was due to GDP inhibition, probably caused by the lack of release of GDP-bound eIF5B-A444V,G479A from the ribosome. In the second experiment, we directly examined the inhibition of eIF5B GTPase activity by GDP. The GTPase assay mixtures contained a limiting amount of WT eIF5B or eIF5B-A444V,G479A, a 10-fold excess of GTP relative to eIF5B (still below the Kd), and increasing amounts of GDP. As shown in Fig. 3C, the GTPase activities of WT eIF5B and eIF5B-A444V,G479A were inhibited by the addition of GDP. Importantly, the data in Fig. 3C revealed that the apparent Ki for the inhibition of WT eIF5B GTPase activity by GDP (Ki, ∼7.3 μM) was ∼3-fold higher than the Ki for the inhibition of eIF5B-A444V,G479A (Ki, ∼2.3 μM). These results are consistent with the idea that the GDP-bound form of eIF5B-A444V,G479A is not released from the ribosome. Taken together, these results support the conclusion that whereas ribosome binding by eIF5B is governed by a GTP-versus-GDP switch, the release of eIF5B-A444V,G479A from the ribosome is triggered, not by GTP hydrolysis, but instead by nucleotide (GDP) release from the factor (Fig. 3A).
DISCUSSION
Structural studies of several G domains first identified the Switch I and Switch II elements as undergoing marked conformational changes upon GTP versus GDP binding (25, 27). The conserved Gly residue in Switch II, part of the D-X-X-G G-3 sequence motif, was proposed to function as the key switch residue, owing in part to its inherent dihedral angle flexibility (13, 14). We found that the substitution of Ala for the corresponding Gly479 in the eIF5B Switch II element altered the protease sensitivity and decreased the guanine nucleotide-binding affinity and the ribosome-dependent GTPase and subunit joining activities of the factor (Fig. 2A through E). Moreover, we identified second site suppressor mutations in Switch I and domain II that restored the guanine nucleotide-binding and GTPase activities of the factor and the ability of eIF5B to promote protein synthesis in vivo. Thus, the conserved Switch II Gly residue and the structural transitions initiated at this residue are not necessarily essential for G protein function. We propose that the suppressor mutations induce Switch II to adopt a conformation favorable for nucleotide binding and hydrolysis and thereby reestablish the coupling between GTP binding and eIF5B domain movements required for protein synthesis.
The conserved Switch II Gly residue is critical for the switch behavior of GTP-binding proteins.
In most G proteins, interactions between the γ-phosphate of GTP and the main-chain amide nitrogens of the conserved Thr in Switch I and the conserved Gly in Switch II serve to lock the Switch elements into their “on” state (27). Following GTP hydrolysis and Pi release, these contacts are lost and the Switch elements are thought to relax and assume their inactive conformations. Upon GTP binding to EF-Tu, the conserved Gly83 in Switch II repositions to contact the γ-phosphate, and this repositioning leads to the unwinding of the N terminus of an α-helix in Switch II (13). Concurrently, the Switch I element converts from a β-hairpin to a structure containing two α-helices. Significantly, the peptide flip around Gly83 in EF-Tu requires a main-chain conformational change that is possible only with Gly (and not energetically allowed by other amino acids at this position) (13). Likewise, it was proposed that the flexibility of this Gly residue in Ras initiated the restructuring of Switch II during GTP hydrolysis (9). Consistent with the notion that the flexibility of the conserved Switch II Gly residue is important for the switch behavior of a G domain, the substitution of Ala for this Gly in Ras, heterotrimeric G proteins, and EF-Tu severely impairs function (4, 19, 26). Mutation of the Switch II Gly83 in EF-Tu to Ala impairs GTP and aminoacyl-tRNA binding (12). It was proposed that the G83A mutation alters the position of helix α2 immediately C-terminal of Switch II, thereby altering the position of the water molecule that catalyzes GTP hydrolysis (15).
In contrast to these classical G domains, the Switch II element of aIF5B adopts a noncanonical position, with the conserved Gly over 7 Å from the γ-phosphate (21). Upon GTP binding to aIF5B, the Switch II element moves an average of 2 Å. This movement of Switch II triggers the rotation of domain II and is relayed through domain III and the lever arm helix H12 to reposition domain IV at the base of the protein. Based on our previous studies revealing that eIF5B has higher affinity for 80S ribosomes in the presence of GTP than in the presence GDP (23), we proposed that these domain movements govern the GTP switch that regulates eIF5B ribosome binding.
Mutation of the conserved Switch II Gly479 to Ala in yeast eIF5B significantly impaired GTP binding (Fig. 2A). Whereas in aIF5B this residue is remote from the γ-phosphate, the adjacent His80 and Glu81 (His480 and Glu481 in eIF5B) residues contact and help to stabilize the γ-phosphate of GTP (21). Thus, the eIF5B G479A mutation may impair GTP binding by indirectly affecting the interaction of His480 and Glu481 with GTP. In support of this idea, it is noteworthy that His480 in eIF5B corresponds to the catalytic Gln61 residue in Ras and that the defective GTPase activity of the eIF5B-G479A mutant was not restored at high GTP concentrations. Thus, in addition to impairing GTP binding, the G479A mutation eliminates GTP hydrolysis perhaps by altering the position of His480. As shown in Fig. 2E, the G479A mutation also impaired subunit joining, even in the presence of high GTP concentrations. As the Switch II element in aIF5B directly contacts domain II (21), the altered position of Switch II due to the G479A mutation may prevent eIF5B from achieving its active conformation necessary for subunit joining. Consistent with this proposal, it is noteworthy that the G479A mutation altered the protease sensitivity of eIF5B in both domains II and IV (Fig. 2C and D).
Intragenic suppressor mutations in Switch I and domain II may restore eIF5B function by altering the position of Switch II.
Examination of the aIF5B structure reveals contacts between Switch II and the residues mutated in the intragenic suppressors in Switch I and helix H8 in domain II (Fig. 4). Residues G43 and T45 flanking A44 in aIF5B (corresponding to A444 in eIF5B) interact with residues D76 and F74 in Switch II (Fig. 4). The carbonyl of G43 interacts via a hydrogen bond with the backbone amide of D76, and the amide and carbonyl of T45 form hydrogen bonds with the carbonyl and amide, respectively, of F74 (Fig. 4). The eIF5B A444V suppressor mutation is likely to alter the positions of G443 and T445 in Switch I, leading to the repositioning of Switch II. Moreover, the A444V mutation in Switch I may result in a direct clash with I475 in Switch II (Fig. 4). Thus, it is likely that the Switch I suppressor mutation alters the position of Switch II, enabling the active site to readily bind and hydrolyze GTP. Consistent with these proposed structural alterations, the eIF5B-A444V,G479A mutant displayed a pattern of protease sensitivity similar to that of WT eIF5B, indicating that the suppressor mutation restored a WT geometry to the eIF5B active site and reestablished the coupling between nucleotide binding and eIF5B domain movements.
The D740R intragenic suppressor mutation in eIF5B corresponds to the residue E340 in aIF5B. Interestingly, E340 in helix H8 interacts via a salt bridge with the conserved residue R87 in Switch II (corresponding to Arg487 in yeast eIF5B) (Fig. 4). The substitution of Arg for D740 would be expected to cause the repulsion of R487, and thus, like the Switch I suppressor, it is possible that the D740R mutation induces a repositioning of Switch II. In support of this repulsion model, it is noteworthy that the substitution of Ala for D740 did not suppress the G479A mutation (data not shown). Moreover, substituting Ala for Arg487 in the triple mutant eIF5B-G479A,R487A,D740R, which would disrupt the proposed repulsion between Arg487 and the Arg mutation at Asp740, blocked the suppressor phenotype (data not shown). We propose that the repulsion of Arg487 by the Arg mutation at D740 enables Switch II, whose flexibility has been impaired by the G479A mutation, to assume a conformation compatible with GTP binding, GTP hydrolysis, and the stimulation of translation initiation.
Further examination of the aIF5B structure provided a rationale for the altered switch properties governing ribosome binding by the eIF5B-A444V,G479A suppressor mutant (Fig. 3). The Asp76 residue in Switch II moves upward ∼2 Å in aIF5B-GDP versus aIF5B-GTP (21). This upward movement of Switch II is likely prevented by the eIF5B A444V mutation that creates a clash with Ile475 (Fig. 4). Thus, the Switch I suppressor mutation may prevent the local conformational change typically associated with GDP binding and lock eIF5B in a conformation resembling the GTP-bound form of the factor that has high affinity for the ribosome. This altered switch behavior of the eIF5B-A444V,G479A mutant is consistent with the presence of GDP plus Pi in the structure of the corresponding Giα1-G203A mutant (4) and with recent data indicating that Pi release rather than GTP hydrolysis is a limiting step in eIF2, Ras, Rap, and EF-G function (1, 3, 5, 22). Future structural and biochemical analyses will be useful for further understanding the mechanistic basis of the intragenic suppressors and for testing whether Pi release is a critical step for eIF5B function.
Acknowledgments
We thank our colleagues in the Dever, Lorsch, and Hinnebusch laboratories and Antonina Roll-Mecak for advice and helpful discussions and Alan Hinnebusch and Marina Rodnina for comments on the manuscript.
This work was supported in part by the Intramural Research Program of the National Institute of Child Health and Development, National Institutes of Health (T.E.D.), and by American Cancer Society grant RSG-03-156-01-GMC (J.R.L.).
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Algire, M. A., D. Maag, and J. R. Lorsch. 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 20:251-262. [DOI] [PubMed] [Google Scholar]
- 2.Algire, M. A., D. Maag, P. Savio, M. G. Acker, S. Z. Tarun, Jr., A. B. Sachs, K. Asano, K. H. Nielsen, D. S. Olsen, L. Phan, A. G. Hinnebusch, and J. R. Lorsch. 2002. Development and characterization of a reconstituted yeast translation initiation system. RNA 8:382-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allin, C., M. R. Ahmadian, A. Wittinghofer, and K. Gerwert. 2001. Monitoring the GAP catalyzed H-Ras GTPase reaction at atomic resolution in real time. Proc. Natl. Acad. Sci. USA 98:7754-7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berghuis, A. M., E. Lee, A. S. Raw, A. G. Gilman, and S. R. Sprang. 1996. Structure of the GDP-Pi complex of Gly203→Ala gialpha1: a mimic of the ternary product complex of galpha-catalyzed GTP hydrolysis. Structure 4:1277-1290. [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti, P. P., Y. Suveyzdis, A. Wittinghofer, and K. Gerwert. 2004. Fourier transform infrared spectroscopy on the Rap. RapGAP reaction, GTPase activation without an arginine finger. J. Biol. Chem. 279:46226-46233. [DOI] [PubMed] [Google Scholar]
- 6.Choi, S. K., J. H. Lee, W. L. Zoll, W. C. Merrick, and T. E. Dever. 1998. Promotion of Met-tRNAMeti binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science 280:1757-1760. [DOI] [PubMed] [Google Scholar]
- 7.Choi, S. K., D. S. Olsen, A. Roll-Mecak, A. Martung, K. L. Remo, S. K. Burley, A. G. Hinnebusch, and T. E. Dever. 2000. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol. 20:7183-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLano, W. L. 2004. The PyMOL user's manual. DeLano Scientific, San Carlos, CA.
- 9.Hall, B. E., D. Bar-Sagi, and N. Nassar. 2002. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc. Natl. Acad. Sci. USA 99:12138-12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinnebusch, A. G. 1985. A hierarchy of trans-acting factors modulate translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 5:2349-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinnebusch, A. G. 2000. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes, p. 185-243. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Kjaersgard, I. V., C. R. Knudsen, and O. Wiborg. 1995. Mutation of the conserved Gly83 and Gly94 in Escherichia coli elongation factor Tu. Indication of structural pivots. Eur. J. Biochem. 228:184-190. [DOI] [PubMed] [Google Scholar]
- 13.Kjeldgaard, M., P. Nissen, S. Thirup, and J. Nyborg. 1993. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure 1:35-50. [DOI] [PubMed] [Google Scholar]
- 14.Kjeldgaard, M., J. Nyborg, and B. F. Clark. 1996. The GTP binding motif: variations on a theme. FASEB J. 10:1347-1368. [PubMed] [Google Scholar]
- 15.Knudsen, C., H. J. Wieden, and M. V. Rodnina. 2001. The importance of structural transitions of the switch II region for the functions of elongation factor Tu on the ribosome. J. Biol. Chem. 276:22183-22190. [DOI] [PubMed] [Google Scholar]
- 16.Knudsen, C. R., I. V. Kjaersgard, O. Wiborg, and B. F. Clark. 1995. Mutation of the conserved Gly94 and Gly126 in elongation factor Tu from Escherichia coli. Elucidation of their structural and functional roles. Eur. J. Biochem. 228:176-183. [PubMed] [Google Scholar]
- 17.Lee, E., R. Taussig, and A. G. Gilman. 1992. The G226A mutant of Gs alpha highlights the requirement for dissociation of G protein subunits. J. Biol. Chem. 267:1212-1218. [PubMed] [Google Scholar]
- 18.Lee, J. H., T. V. Pestova, B. S. Shin, C. Cao, S. K. Choi, and T. E. Dever. 2002. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc. Natl. Acad. Sci. USA 99:16689-16694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, R. T., S. B. Masters, K. A. Sullivan, B. Beiderman, and H. R. Bourne. 1988. A mutation that prevents GTP-dependent activation of the alpha chain of Gs. Nature 334:712-715. [DOI] [PubMed] [Google Scholar]
- 20.Pestova, T. V., I. B. Lomakin, J. H. Lee, S. K. Choi, T. E. Dever, and C. U. T. Hellen. 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403:332-335. [DOI] [PubMed] [Google Scholar]
- 21.Roll-Mecak, A., C. Cao, T. E. Dever, and S. K. Burley. 2000. X-ray structures of the universal translation initiation factor IF2/eIF5B. Conformational changes on GDP and GTP binding. Cell 103:781-792. [DOI] [PubMed] [Google Scholar]
- 22.Savelsbergh, A., D. Mohr, U. Kothe, W. Wintermeyer, and M. V. Rodnina. 2005. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 24:4316-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin, B. S., D. Maag, A. Roll-Mecak, M. S. Arefin, S. K. Burley, J. R. Lorsch, and T. E. Dever. 2002. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell 111:1015-1025. [DOI] [PubMed] [Google Scholar]
- 24.Spoerner, M., C. Herrmann, I. R. Vetter, H. R. Kalbitzer, and A. Wittinghofer. 2001. Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc. Natl. Acad. Sci. USA 98:4944-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sprang, S. R. 1997. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 66:639-678. [DOI] [PubMed] [Google Scholar]
- 26.Sung, Y. J., M. Carter, J. M. Zhong, and Y. W. Hwang. 1995. Mutagenesis of the H-ras p21 at glycine-60 residue disrupts GTP-induced conformational change. Biochemistry 34:3470-3477. [DOI] [PubMed] [Google Scholar]
- 27.Vetter, I. R., and A. Wittinghofer. 2001. The guanine nucleotide-binding switch in three dimensions. Science 294:1299-1304. [DOI] [PubMed] [Google Scholar]