Abstract
The Met receptor tyrosine kinase regulates a complex array of cellular behaviors collectively known as “invasive growth.” While essential for normal development and wound repair, this program is frequently co-opted by tumors to promote their own growth, motility, and invasion. Met is overexpressed in a variety of human tumors, and this aberrant expression correlates with poor patient prognosis. Previous studies indicate that Met receptor levels are governed in part by cbl-mediated ubiquitination and degradation, and uncoupling of Met from cbl-mediated ubiquitination promotes its transforming activity. Here we describe a novel mechanism for Met degradation. We find that the Met receptor interacts with the transmembrane protein LRIG1 independent of hepatocyte growth factor (HGF) stimulation and that LRIG1 destabilizes the Met receptor in a cbl-independent manner. Overexpression of LRIG1 destabilizes endogenous Met receptor in breast cancer cells and impairs their ability to respond to HGF. LRIG1 knockdown increases Met receptor half-life, indicating that it plays an essential role in Met degradation. Finally, LRIG1 opposes Met synergy with the ErbB2/Her2 receptor tyrosine kinase in driving cellular invasion. We conclude that LRIG1 is a novel suppressor of Met function, serving to regulate cellular receptor levels by promoting Met degradation in a ligand- and cbl-independent manner.
The Met receptor tyrosine kinase (RTK), the prototype of the scatter factor receptor family, is expressed predominantly in epithelial cells and is activated through binding of its stromal ligand hepatocyte growth factor/scatter factor (HGF/SF). Met receptor function is absolutely essential for normal development, as mice lacking either HGF or Met exhibit embryonic lethality (4, 56). Precise Met activation regulates an array of cellular behaviors, including growth, motility, invasion, and survival, which collaborate to yield an “invasive growth” program (16, 35). This program is physiologically employed during embryonic development, morphogenesis, and wound healing, but is frequently deregulated during tumor growth, progression, and metastasis (7).
Aberrant activation of the Met receptor occurs through overexpression, autocrine activation, or activating mutations of the receptor, which correlates with poor patient prognosis in a variety of tumors, including those of the lung, bladder, and breast (for an excellent summary, visit http://www.vai.org/vari/metandcancer) (3, 10, 14). Several Met mutations, first identified in both hereditary and sporadic forms of human papillary renal carcinoma (49), were later demonstrated to be oncogenic in vivo (26). These observations clearly link aberrant Met activity to human cancer.
In breast cancer, both Met and its ligand, HGF, are frequently overexpressed and correlate with decreased relapse-free and overall survival (11, 29, 31, 43, 59, 61). In addition, Met receptor overexpression is an independent predictor of poor prognosis in breast cancer (5, 19, 36). Several distinct lines of evidence have demonstrated that the mammary epithelium is exceptionally vulnerable to transformation by dysregulated Met signaling. For example, transgenic mice broadly expressing an oncogenic version of Met, Tpr-Met (39), or the ligand HGF (52) develop a predominant breast cancer phenotype. In addition, targeted expression of HGF (13) or mutationally activated Met receptor (27) in the mammary gland leads to metastatic mammary adenocarcinoma.
Met receptor down-regulation via ligand-stimulated ubiquitination is an essential negative regulatory mechanism that prevents receptor oversignaling. The ubiquitin ligase c-Cbl is recruited to the Met receptor following ligand stimulation through an atypical DpYR motif in the juxtamembrane domain of Met (46). Uncoupling of Met from c-Cbl-mediated ubiquitination either through loss of the juxtamembrane domain, as with Tpr-Met, or by mutation of Tyr1003F within the DpYR motif leads to cellular transformation (45). Restoration of the juxtamembrane domain to Tpr-Met potently suppresses Tpr-Met transforming activity (58). The fusion of monoubiquitin to the MetY1003F mutant, in essence bypassing the requirement for c-Cbl, suppresses its transforming activity by 60% (1). Currently, c-Cbl-mediated ubiquitination is the only known mechanism of Met receptor degradation.
LRIG1, a transmembrane leucine-rich repeat and immunoglobulin (Ig)-like domain-containing protein, is a newly identified negative regulator of the ErbB family of receptor tyrosine kinases. Previous work has demonstrated that LRIG1 is capable of interacting with all four ErbB receptors and enhancing both their basal and ligand-stimulated ubiquitination and degradation (21, 34). For ligand-stimulated degradation of the epidermal growth factor receptor (EGFR), LRIG1 appears to function by augmenting the amount of c-Cbl recruited to the receptor (21). The role of c-Cbl in EGF receptor degradation is well understood; however, the remaining ErbB receptors are not regulated by c-Cbl under physiological conditions (38).
Interestingly, several lines of evidence have demonstrated that ErbB receptors and Met receptor collaborate in driving tumor cell growth. For example, expression of the EGF receptor ligand transforming growth factor α in liver tumor cells leads to Met receptor phosphorylation and an enhanced response to HGF (47). In addition, the Met receptor is constitutively phosphorylated in A431 epidermoid carcinoma cells and inhibition of the EGF receptor inhibits this activation (30). Of most significance, the Met receptor and Her2 have recently been found to synergize in breakdown of cell-cell junctions and in promoting cellular invasion (32). This is particularly relevant since 50% of breast tumors that overexpress Met also overexpress Her2 (37).
In this report, we identify LRIG1 as a novel physiological negative regulator of the Met receptor, uncovering a new means of Met receptor regulation. We demonstrate that the endogenous proteins interact and that silencing of LRIG1 by RNA interference (RNAi) increases endogenous Met receptor half-life. Additionally, we find that LRIG1 decreases Met in a c-Cbl- and proteasome-independent manner. Transduction of LRIG1 into several breast cancer cell lines, as well as nontransformed Madin-Darby canine kidney (MDCK) epithelial cells, inhibits their HGF-dependent responses including growth, motility, and invasion. Finally, we demonstrate that LRIG1 opposes Her2 and Met receptor synergy in driving cellular invasion.
MATERIALS AND METHODS
Reagents and cell culture.
HGF was purchased from Fitzgerald/RDI. Human Met receptor cDNA was amplified by reverse transcription-PCR from A431 cells, and the sequence was confirmed followed by subcloning into pcDNA3.1+ plasmid. All cell lines were purchased from ATCC. HEK-293T, MCF-7, MDA-MB-231, and MDCK cells were all cultured in Dulbecco's modified Eagle's medium (DMEM)-10% fetal calf serum (FCS). CHO cells were grown in F-12K medium-10% FCS. Serum starvation medium contained only 0.1% FCS. Antibodies used here include anti-insulin receptor SC-711, anti-Met SP260 and C-12, anti c-Cbl C-15, anti-Cbl-b G-1 (Santa Cruz Biotechnology), anti-Met DO-24 and DQ-13, anti-phosphotyrosine 4G10 (Upstate), anti-myc, anti-EGFR H9B4 (Invitrogen), anti-Her2 Ab3 (Calbiochem), anti-FLAG M2, anti-α-tubulin, anti-actin AC-15 (Sigma), antihemagglutinin (HA) (Zymed), and anti-p-Erk (Cell Signaling Technologies). LRIG1-151 was a generous gift from Hakan Hedman. Transfections were performed with FuGENE6 (Roche Applied Sciences) according to the manufacturer's protocol.
Cell transductions.
The 293GPG packaging cell line and pMX-pie (pMX) cloning vector were generous gifts from Paola Marignani. The 293GPG cells were maintained in DMEM-10% ΔFCS, 100 μg/ml G418 (Invitrogen), 2 μg/ml puromycin, and 10 μg/ml tetracycline (Sigma). LRIG1-myc (21) subcloned into pMX was simultaneously transfected with pJ6Ωpuro into 293GPG cells and selected with 100 μg/ml zeocin (Invitrogen). Production of retrovirus was initiated by removal of tetracycline from the medium. Collected supernatant from pMX- and LRIG1-myc-producing cells was then added to the medium of MCF-7, MDCK, and MDA-MB-231 cells, followed by selection with 1, 1.5, and 0.5 μg/ml puromycin, respectively. All puromycin-resistant clones were pooled to avoid clonal variation.
Immunoprecipitation and Western blot analysis.
Coimmunoprecipitations of LRIG1 and Met were performed using HEK-293T cells. Cells were lysed in coimmunoprecipitation buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 1% Nonidet P-40, 10% glycerol, 1 mM Na3VO4, 1 mM NaF, 1 mM ZnCl2, 10 mM β-glycerophosphate, 5 mM tetrasodium pyrophosphate, 100 μM AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride], and 4 μg/ml each aprotinin, leupeptin, and pepstatin), and cleared lysates were precipitated with 1.2 μg of anti-LRIG1-151 or control polyclonal antibody. Precipitates were resolved by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted with either anti-LRIG1-151 or anti-Met C-12. Detection of all antibodies was carried out using horseradish peroxidase-conjugated secondary antibodies (Zymed Laboratories Inc.), followed by developing with SuperSignal West chemicals (Pierce). An Alpha Innotech imaging station with FluorChem software was used to capture images.
RNAi experiments.
To knock down LRIG1 in HEK-293T cells, Dharmacon On-Target plus SMART pool was utilized. On-Target plus siControl nontargeting pool was used as a control. Cells were transfected with 100 nM small interfering RNA (siRNA) using DharmaFect 1 (Dharmacon) per the manufacturer's instructions. The medium was replaced after 24 h, and cells were left to incubate for an additional 72 h. Lysates were collected and analyzed as described.
The RNAi sequences targeting c-Cbl (5′-TGCTCTCTTCCAAGCACTGATTCAAGAGATCAGTGCTTGGAAGAGAGCTTTTTTC) and Cbl-b (5′-TGGACAGACGAAATCTCACATTCAAGAGATGTGAGATTTCGTCTGTCCTTTTTTC) were synthesized along with their complementary strands. The oligonucleotides were annealed prior to ligation into pLentiLox3.7 digested with XhoI and HpaI, as described previously (28, 48). HEK-293T cells were transfected with either the RNAi targeting vector or empty vector, in the absence or presence of Met and LRIG1-myc. After 24 h, cells were serum starved overnight, followed by treatment without or with 10 ng/ml HGF for 60 min, before cell lysates were collected. All lysates were resolved by Western blot analysis.
Half-life analysis.
HEK-293T cells were transfected with 100 nM Dharmacon nontargeting or LRIG1 pool and allowed to incubate for 84 h. Cells were then washed twice in phosphate-buffered saline (PBS) and incubated in methionine- and cysteine-free DMEM with 5% dialyzed fetal bovine serum (FBS) for 30 min. Tran35S-label (MP Biomedical) was added at 167 μCi per ml of medium, and cells were allowed to incubate for 1 h (pulse). The medium was then chased with DMEM containing 5% dialyzed FBS and 2 mM methionine and cysteine, and samples at time points of 0, 3, 6, 12, and 18 h were collected. Cells were scraped in PBS and snap-frozen in liquid nitrogen. Immunoprecipitation of Met was conducted as described above, except that cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.5], 0.1% SDS, 1% Triton X-100, 150 mM NaCl, and 0.5 mM EDTA plus inhibitors listed above). Samples were resolved by SDS-PAGE and transferred to nitrocellulose. Western blot analysis was conducted as previously described, and the amount of Met receptor in each immunoprecipitate was quantified. Imaging of 35S was performed using a Molecular Dynamics Storm PhosphorImager, and the amount of 35S associated with Met was quantified with ImageQuant software.
Ubiquitination assay.
HEK-293T cells were transfected with HA-tagged ubiquitin and Met, with or without myc-tagged LRIG1. After 24 h, cells were serum starved overnight before being treated with 10 ng/ml of HGF for 5 min. Immunoprecipitation of Met was conducted using 1 μl of anti-DO-24 in RIPA buffer supplemented with 10 mM N-ethylmaleimide and resolved in sample buffer contained 8 M urea. For the ubiquitin knockout (Ub-KO) experiment, HEK-293T cells were transfected with Met with or without myc-tagged LRIG1 in the presence or absence of HA-tagged Ub-KO (40). The pRK5-Ub-KO plasmid was a kind gift of Ted M. Dawson of Johns Hopkins University School of Medicine and expresses a form of human ubiquitin in which all seven lysines involved in chain elongation have been mutated to arginine.
Inhibition studies.
HEK-293T cells were transfected with Met with or without LRIG1-myc. After 24 h, 10 μM MG132 or 100 nM concanamycin (CalBiochem) was added to each well and samples at various time points were collected by lysing cells with sample buffer. Samples were resolved and quantified as described above.
Construction of MetY1003F.
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. Tyrosine 1003 was converted to a phenylalanine using the 5′-AAATGAATCTGTAGACTTCCGAGCTACTTTTCCAG-3′ primer (underlining indicates nucleotides changed in going from tyrosine to phenylalanine) and its complementary pair.
MTT assay.
Transduced MCF-7 cells were plated at a density of 2.5 × 104 cells/well in 24-well Falcon plates. After settling for 16 h, cells were serum starved or treated with 25 ng/ml HGF for an additional 48 h. During the last 4 h of growth, MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) was added to the media to measure cell activity. Results were quantified by dissolving the crystals formed from the MTT in acidic isopropanol and recording absorption at 570 nm with a baseline subtraction at 655 nm. At least four points were averaged for each condition, and the experiment was repeated three times with a representative experiment selected.
Migration assay.
Cells transduced with LRIG1-myc or control vector pMX (2 × 104/well) were plated onto 24-well Boyden chambers with 8-μm-pore polycarbonate membranes (Corning) in serum starvation medium with or without the addition of HGF (10 ng/ml) in the lower chamber. After 24 h, filters were fixed and stained according to the manufacturer's instructions. Filters were then air dried and photographed using a 20× objective. Three fields of view for each well were counted, and the results were averaged among three experiments for each cell line.
Matrigel invasion assay.
Transduced MDCK cell lines stably expressing either pcDNA3.1+ (3.1) control vector or NeuT were created by transfection of transduced MDCK cells and selection with 600 μg/ml G418. For the invasion assay, MDCK cells were plated (2 × 104/well) onto 24-well Matrigel invasion chambers (BD Biosciences). All cells were grown in the presence of 10 ng/ml HGF for 15 days prior to plating (32). After 16 h in the invasion chambers, cells were fixed and stained according to the manufacturer's instructions. All cells in each chamber migrating through the Matrigel were counted. The results from three separate chambers were then averaged.
RESULTS
LRIG1 is a negative regulator of the Met receptor.
To determine whether LRIG1 is a functional regulator of the Met receptor, we cotransfected myc-tagged LRIG1 with Met into HEK-293T cells. Whole-cell lysates were probed with antibodies to Met to analyze effects of LRIG1 on Met receptor expression, as shown in the left panel of Fig. 1A. Expression of the Met receptor with LRIG1 resulted in a dramatic decrease in the Met receptor level in these cells. LRIG1 had no effect on the expression of green fluorescent protein (GFP) (right panel of Fig. 1A), indicating that it does not have a general effect on protein expression. To analyze whether this effect on the Met receptor was specific to LRIG1 or was an artifact of the coexpression of two vectors, Met was coexpressed with equivalent amounts of either LRIG1, epidermal growth factor receptor (EGFR), or Nrdp1, a ubiquitin ligase previously shown to target ErbB3 and ErbB4 (9). EGFR and Nrdp1 were expressed in the same vector as LRIG1, but only LRIG1 was able to decrease Met receptor levels, as shown in Fig. 1B. LRIG1 expression had no significant effect on Met transcript levels, as determined by real-time PCR (data not shown). Similar effects on Met receptor levels were observed in chinese hamster ovary (CHO) cell cotransfections (see Fig. 3A), demonstrating that the effect of LRIG1 on the Met receptor is cell-type independent.
FIG. 1.
LRIG1 interacts with and suppresses the Met receptor. (A) HEK-293T cells were transfected with Met (left panel) or GFP (right panel) along with vector control pcDNA3.1+ (3.1) or vector expressing myc-tagged LRIG1. Whole-cell lysates were then collected and blotted with antibodies to Met (C-12), GFP, myc, or actin. (B) HEK-293T cells were transfected with Met along with vector control, LRIG1-myc, EGFR, or Nrdp1-FLAG. Whole-cell lysates were blotted with antibodies for Met, myc, EGFR, FLAG, or actin. (C) Lysates from HEK-293T cells were immunoprecipitated with control rabbit antibody or anti-LRIG1-151 and blotted with antibodies to Met or LRIG1. IP, immunoprecipitate. (D) HEK-293T cells were transfected with nontargeting or LRIG1-targeting siRNA pools. Whole-cell lysates were collected and blotted for Met, LRIG1-151, and actin.
FIG. 3.
Met receptor degradation by LRIG1 is c-Cbl independent. (A) CHO cells were transfected with Met with or without LRIG1-myc and 70Z-Cbl-HA. Cells were treated with or without 10 ng/ml of HGF. Whole-cell lysates were collected and blotted with antibodies to Met, myc, HA, and actin. (B) The experiment was repeated as described for panel A, with the exception that a mutant form of Met receptor, MetY1003F, was used. WT, wild type. (C) HEK-293T cells were transfected with Met with or without LRIG1-myc and c-Cbl and Cbl-b RNAi vectors (Cbl-KD). After treatment with or without 10 ng/ml of HGF, whole-cell lysates were collected and blotted with antibodies to Met, myc, Cbl-b, c-Cbl, and tubulin. (D) HEK-293T cells were transfected with Met in the absence or presence of LRIG1-myc and c-Cbl-HA. Cells were treated with HGF as in panel C, and lysates were blotted for Met, myc, HA, and actin.
To determine whether endogenous LRIG1 and Met interact, a coimmunoprecipitation experiment was performed. As shown in Fig. 1C, HEK-293T cells express detectable amounts of both LRIG1 and Met. Lysates from HEK-293T cells were immunoprecipitated with equivalent amounts of either control antibody or with anti-LRIG1 antibody. As expected, endogenous LRIG1 precipitated only in the lane using the anti-LRIG1 antibody. Probing of the blot with anti-Met antibody revealed that endogenous Met was precipitated along with endogenous LRIG1 (Fig. 1C). This interaction was specific, as Met was not precipitated with the control antibody.
We next examined the effect of LRIG1 loss on Met receptor expression. In this experiment, LRIG1 was depleted in HEK-293T cells using RNAi. RNAi directed against LRIG1 significantly decreased the expression of endogenous LRIG1, to less than detectable levels. Importantly, knockdown of LRIG1 resulted in a modest but reproducible increase in endogenous Met receptor expression, with a representative experiment shown in Fig. 1D. These data indicate that LRIG1 is a physiological regulator of Met receptor and suggest that LRIG1 loss in tumors could contribute to Met receptor overexpression.
LRIG1 enhances Met receptor degradation in a lysosome-dependent manner.
As LRIG1 led to a decrease in Met receptor expression, but had a negligible effect of Met transcript levels, we measured the effect of LRIG1 loss on Met receptor half-life by pulse-chase analysis. LRIG1 was depleted from HEK-293T cells by RNAi, and endogenous Met receptor was immunoprecipitated from cells radiolabeled with [35S]Met/Cys (Fig. 2A) (60, 62). This experiment was repeated three times, and the data are quantified in Fig. 2B. In the presence of LRIG1, the half-life of Met receptor was 5.6 h, and in the absence of LRIG1, this half-life was extended twofold to 11.5 h (Fig. 2B). These data are comparable to previous research showing that disruption of the c-Cbl binding site on Met leads to an approximately twofold increase in Met receptor half-life in fibroblasts (0.7 to 1.2 h) (45). As disruption of c-Cbl binding was sufficient to mediate cellular transformation in fibroblasts, our results suggest that LRIG1 loss could contribute to Met-mediated cellular transformation.
FIG. 2.
LRIG1 enhances Met receptor degradation in a lysosome-dependent manner. In these experiments, HEK-293T cells were transfected with nontargeting or LRIG1-targeting pools of siRNA. (A) Cells were pulsed with Tran35S-label medium and chased with DMEM with excess cysteine and methionine. Samples were collected at 0, 3, 6, 12, and 18 h, and immunoprecipitations for Met with DO-24 were performed. Immunoprecipitates were resolved by SDS-PAGE and transferred to nitrocellulose. Immunoprecipitates were then blotted for Met and imaged for 35S activity with a Molecular Dynamics Storm PhosphorImager (upper panel). Lysates (lower panel) were blotted with LRIG1-151 and actin. (B) Graphic representation of three separate experiments tracking the degradation of radiolabeled Met over 18 h. The amount of Met in each immunoprecipitate was quantified for Met protein as well as 35S content. The error bars represent the standard error of three independent experiments. (C) Cells were transfected with Met in the absence or presence of HA-ubiquitin (HA-Ub) and in the absence or presence of LRIG1-myc. Cells were stimulated with or without 10 ng/ml HGF before being lysed in RIPA buffer and immunoprecipitated with anti-Met DO-24. Immunoprecipitates (IP; upper panel) were blotted with Met and HA, and lysates (lower panel) were blotted for Met, myc, and actin. (D) Cells were transfected in the absence or presence of UB-KO. Following HGF stimulation for 0 or 60 min, cells were lysed and blotted for Met, myc, and actin. (E) Cells were treated with either MG132 (left panel) or concanamycin for various time periods. Lysates were collected and blotted with Met, myc, and actin. A graphic representation of Met accumulation versus time is shown below each blot.
LRIG1 has previously been reported to enhance the ligand-stimulated ubiquitination of ErbB receptors in a c-Cbl-dependent manner (21). To examine whether LRIG1 increases ubiquitination of the Met receptor, Met was expressed in HEK-293T cells with either vector control or myc-tagged LRIG1 and an HA-tagged ubiquitin construct. Thirty-six hours after transfection, Met receptor was immunoprecipitated and probed with anti-HA to visualize ubiquitination. Receptor ubiquitination is observed as a smear and represents both polyubiquitination and multiple monoubiquitination events (6). As shown in Fig. 2C, Met ubiquitination could be observed in the absence of HGF, as previously reported (45), and was not significantly stimulated by HGF treatment. LRIG1 expression resulted in decreased Met expression, but did not appear to enhance the overall ubiquitination of remaining receptors. However, these residual receptors may represent a population that is refractory to LRIG1 action, while highly ubiquitinated receptors could be efficiently degraded throughout the time period of LRIG1-Met coexpression.
Receptor tyrosine kinases, including Met, undergo both multiple mono- and polyubiquitinations (6). Ubiquitin contains seven lysines that are known to participate in chain elongation: K6, K11, K27, K29, K33, K48, and K63. K48-linked polyubiquitination is associated with proteasomal degradation, while monoubiquitination is generally associated with receptor trafficking (42). However, Met receptor degradation has recently been found to require K48-linked polyubiquitination in a proteasome-independent manner (6). To examine whether the assembly of polyubiquitin chains on the Met receptor is necessary for LRIG1 action, Met was expressed in HEK-293T cells with either vector control or myc-tagged LRIG1 in the presence or absence of an HA-tagged form of ubiquitin in which all seven lysines have been mutated to arginine. This ubiquitin, termed Ub-KO, can participate in monoubiquitin linkages, but is unable to form polyubiquitin chains (40). Despite high levels of endogenous ubiquitin, mutant forms of ubiquitin act in a dominant-negative fashion when overexpressed and have been used to implicate specific ubiquitin linkages in receptor trafficking and degradation (6, 15). In agreement with this, the coexpression of Ub-KO with Met leads to increased Met receptor expression, in both the absence and presence of HGF. Interestingly, LRIG1-mediated Met degradation is unaffected by Ub-KO, demonstrating that LRIG1 does not require the assembly of polyubiquitin chains on Met (Fig. 2D). However, this experiment does not exclude the possible role of monoubiquitination in LRIG1-mediated Met receptor degradation.
Met receptor degradation has been found to depend on both the proteasome and lysosome, and inhibitors of either interfere with Met receptor degradation (6, 22). To examine the dependence of LRIG1-mediated Met receptor degradation on the proteasome or lysosome, Met receptor was expressed in HEK-293T cells with either vector control or myc-tagged LRIG1 as shown in Fig. 2E. Cells were then treated with either MG132, an inhibitor of the proteasome (6), or concanamycin, an inhibitor of the lysosome (6), and Met receptor accumulation was measured over a 4-h time course. In each case, the amount of Met receptor present at time zero was normalized to 1 and receptor accumulation was plotted as a function of time. Cells treated with MG132 and transfected with vector control demonstrated an accumulation of Met receptor to nearly 3-fold, while cells expressing LRIG1 had minimal accumulation, reaching only 1.5-fold of the initial levels after 4 h (left panel of Fig. 2E), indicating that LRIG1-mediated Met receptor degradation is largely independent of the proteasome. This is in agreement with the finding that LRIG1-mediated Met receptor degradation does not require the assembly of polyubiquitin chains. In cells treated with concanamycin and transfected with vector control or with LRIG1-myc, there was a comparable accumulation of Met receptor, increasing by about twofold after 4 h, indicating that LRIG1-mediated Met receptor degradation depends in large part on the lysosome. Similar results were observed in LRIG1-transduced MCF-7 cells (data not shown).
LRIG1 mediates Met receptor degradation in a c-Cbl-independent manner.
c-Cbl plays a critical role in Met receptor degradation, and uncoupling of Met from c-Cbl-mediated degradation through truncation or mutation leads to cellular transformation (23, 45). To examine whether c-Cbl is required for LRIG1-mediated degradation of the Met receptor, CHO cells were transfected with Met receptor alone or with LRIG1-myc. As seen in HEK-293T cells, coexpression of LRIG1 with Met receptor resulted in a decrease in Met receptor expression in both the absence and presence of HGF (Fig. 3A). To determine whether c-Cbl was involved in LRIG1-mediated degradation of the Met receptor, a dominant-negative version of c-Cbl, 70Z-Cbl, was employed. 70Z-Cbl is a naturally occurring mutant isolated from 70Z/3 pre-B-cell lymphomas and contains a 17-amino-acid deletion in its ring finger domain (12), rendering it inactive as a ubiquitin ligase. When HA-tagged 70Z-Cbl was transfected with Met, receptor levels dramatically increased, indicating that 70Z-Cbl was functioning in a dominant-negative fashion with respect to Met degradation. Probing of the blot with a c-Cbl antibody revealed that 70Z-Cbl was expressed at a minimum of 17-fold excess compared to endogenous c-Cbl (data not shown). Coexpression of 70Z-Cbl with LRIG1 had no effect on LRIG1's ability to destabilize the Met receptor, in either the absence or presence of HGF. Interestingly, LRIG1 was able to completely override the stabilizing effect of 70Z-Cbl on the Met receptor, indicating that LRIG1 down-regulation of Met receptor is Cbl independent.
Mutation of Y1003 within the c-Cbl docking motif on the Met receptor has been reported to uncouple Met from c-Cbl regulation and is sufficient to convert Met into a transforming protein when overexpressed. To examine whether MetY1003F is subject to LRIG1-mediated degradation, it was transfected into CHO cells in the absence and presence of LRIG1-myc. As seen in Fig. 3B, MetY1003F was significantly destabilized by LRIG1 coexpression, in both the absence and presence of HGF, indicating that the Y1003F mutation, while impairing c-Cbl-mediated degradation, does not impact LRIG1-mediated degradation. Despite being uncoupled from direct c-Cbl-mediated regulation by virtue of the Y1003F mutation, 70Z-Cbl still had a substantial stabilizing effect on MetY1003F. Cbl can also be recruited to Met indirectly via Grb2 (45). As with the wild-type receptor, 70Z-Cbl had no effect on LRIG1's ability to degrade MetY1003F and LRIG1 was able to completely override the stabilizing effect of 70Z-Cbl on MetY1003F. These results provide further evidence that LRIG1 functions to destabilize the Met receptor in a Cbl-independent manner. Similar results with 70Z-Cbl in HEK-293T cells indicate that this effect is not cell line specific (data not shown).
To further demonstrate that LRIG1 functions in a Cbl-independent manner with respect to Met receptor degradation, c-Cbl and Cbl-b, both reported to interact with the Met receptor (46), were depleted in HEK-293T cells by RNAi (25). The ability of myc-tagged LRIG1 to mediate Met receptor degradation was then examined. As shown in Fig. 3C, Cbl depletion by RNAi led to increased Met receptor expression, in agreement with the effects of dominant-negative 70Z-Cbl (Fig. 3A and B). Coexpression of myc-tagged LRIG1 with Met receptor resulted in significant destabilization of the Met receptor, in both the absence and presence of HGF. Despite the stabilizing effect of Cbl depletion on the Met receptor, Cbl depletion had no effect on LRIG1-mediated Met receptor degradation. Taken together with the results shown in Fig. 3A and B, this piece of data provides strong evidence that LRIG1 functions in a Cbl-independent manner with respect to Met receptor degradation.
Since LRIG1 and c-Cbl both act to destabilize the Met receptor, we examined the relative and combined impact of LRIG1 and c-Cbl expression on Met receptor stability. In Fig. 3D, Met receptor was expressed in HEK-293T cells with either vector control, myc-tagged LRIG1, HA-tagged c-Cbl, or with LRIG1 and c-Cbl together. As expected, LRIG1 dramatically destabilized the Met receptor regardless of its activation state. c-Cbl overexpression did not significantly affect Met receptor expression in the absence of HGF and had a modest destabilizing effect in the presence of HGF. The coexpression of c-Cbl and LRIG1 in the presence of HGF led to further destabilization of the Met receptor; however, the combination of c-Cbl and LRIG1 gave only a slight enhancement of Met degradation as compared to the effect of LRIG1 alone. There was no evidence for any synergy between LRIG1 and c-Cbl. Unexpectedly, we noted that LRIG1 decreased c-Cbl expression, both in CHO (Fig. 3A and B) and HEK-293T (Fig. 3D) cells, but had no effect on either GFP expression (Fig. 1A) or insulin receptor expression (Fig. 4A and 5A). However, endogenous c-Cbl does not appear to decrease following LRIG1 overexpression (Fig. 3C).
FIG. 4.
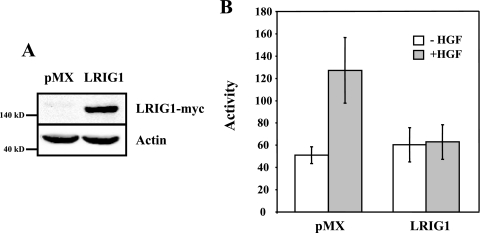
LRIG1 mediates Met receptor degradation and inhibits HGF-stimulated proliferation in MCF-7 breast cancer cells. MCF-7 cells transduced with empty vector (pMX) or LRIG1-myc were used in the following experiments. (A) Whole-cell lysates from these cells were collected and blotted with antibodies to Met, insulin receptor (IR), myc, or actin. (B) These cells were serum starved and stimulated with 10 ng/ml of HGF for various times. Whole-cell lysates were then blotted with antibodies to Met, myc, pERK, and actin. Met levels were quantified and represented graphically. (C, top panel) Following treatment with HGF as for panel B, cell lysates were immunoprecipitated with an antibody to Met (DO-24) and blotted with antibodies to phosphotyrosine (pY; 4G10) or Met. (C, bottom panel) Whole-cell lysates were blotted with pERK, myc, and tubulin antibodies. (D) Transduced MCF-7 cells were grown in either serum starvation medium or medium containing 25 ng/ml HGF. An MTT assay was employed to measure cell growth. Error bars represent the standard deviation of at least four individual readings for each condition.
FIG. 5.
LRIG1 mediates Met receptor degradation and inhibits HGF-stimulated motility in MDA-MB-231 breast cancer cells. (A) Whole-cell lysates of pMX- or LRIG1-myc-transduced MDA-MB-231 cells were collected and blotted with antibodies to Met, insulin receptor (IR), myc, and actin. (B) Motility of transduced MDA-MB-231 cells was measured by a Boyden chamber assay. Three independent ×20 fields were averaged for each chamber, and the experiment was performed in triplicate. The error bars represent the standard deviation of three independent experiments.
LRIG1 destabilizes endogenous Met receptor in human breast cancer cells and inhibits their HGF-dependent responses.
To explore whether LRIG1 could affect endogenous Met receptor expression in human breast cancer cells, MCF-7 cells were transduced using the pMX-pie retroviral system. Cells were transduced either with empty retrovirus (pMX) as a control or with retrovirus expressing LRIG1-myc. Puromycin-resistant clones were pooled to avoid artifacts due to clonal variation. As shown in Fig. 4A, stable expression of LRIG1 in MCF-7 breast cancer cells resulted in a significant decrease in the expression of endogenous Met receptor, but had no effect on endogenous insulin receptor expression. To assess the impact of LRIG1 on Met receptor signaling, transduced MCF-7 cells were stimulated with HGF over an 8-h time course (Fig. 4B). Basal Met receptor levels were decreased by approximately 60% in cells expressing LRIG1 as compared to the control pMX-transduced cells, and by 2 h of HGF treatment, very little detectable Met receptor remained. The mitogen-activated protein kinase pathway was also attenuated in LRIG1-transduced cells, demonstrating that LRIG1 impairs Met receptor downstream signaling (Fig. 4B and C). Densitometric analysis revealed that the phospho-Erk (pErk)/Met ratios were the same in pMX- and LRIG1-transduced cells, indicating that the attenuation of mitogen-activated protein kinase signaling is likely a consequence of decreased Met receptor levels. To investigate whether LRIG1 affects Met receptor phosphorylation, Met was immunoprecipitated from transduced MCF-7 cells. As shown in Fig. 4C, HGF stimulation resulted in Met phosphorylation in both cases; however densitometric analysis indicated that the phospho-Met (pMet)/Met ratio was decreased by approximately 30% following LRIG1 expression, indicating that LRIG1 decreases Met receptor activation by HGF. In agreement with this, the related leucine-rich repeat protein Kekkon-1 interferes with growth factor binding by EGF receptor (18) as does a soluble version of LRIG1 (20). The functional outcome of LRIG1 expression on HGF-dependent proliferation was measured using an MTT assay. As shown in Fig. 4D, LRIG1 significantly impaired the proliferation of MCF-7 cells in response to HGF.
To determine if LRIG1 could destabilize the Met receptor in a different breast cancer cell line, MDA-MB-231 cells were transduced as described for the MCF-7 cells. While MCF-7 cells express low levels of Met receptor, MDA-MB-231 cells express substantial amounts and are frequently used to study Met receptor signaling. As shown in Fig. 5A, stable expression of LRIG1 in MDA-MB-231 breast cancer cells resulted in a significant decrease in the expression of endogenous Met receptor, but had no effect on endogenous insulin receptor expression. To examine whether LRIG1 expression attenuated the HGF-dependent motility of MDA-MB-231 cells, a Boyden chamber assay was performed. This assay measures the migration of cells through a porous membrane toward a chemotactic gradient. As shown in Fig. 5B, MDA-MB-231 cells are highly motile and their motility can be further enhanced by HGF treatment. However, LRIG1-expressing cells show no increase in HGF-dependent motility. Cells expressing LRIG1 showed a statistically significant decrease in their HGF-dependent motility (P = 1.42 × 10−5). MDA-MB-231 cells did not proliferate in response to HGF (data not shown), in agreement with a previous report (55). Taken together, the data from the MCF-7 and MDA-MB-231 cells indicate that LRIG1 can decrease both the endogenous Met receptor levels and HGF-dependent cellular responses in breast cancer cells.
LRIG1 inhibits HGF-dependent motility in Madin-Darby canine kidney cells.
To examine HGF-dependent motility in a different cell type, MDCK cells were analyzed in a Boyden chamber assay. These cells are an immortalized, but nontransformed, kidney cell line and are frequently used to study Met receptor-mediated motility and branching morphogenesis. MDCK cells were transduced with either pMX or LRIG1-myc as described above. As shown in Fig. 6A, LRIG1 is expressed in LRIG1-myc-transduced cells but not in cells transduced with empty virus. Although we were unable to blot canine Met receptor in these cells with available antibodies, we were able to show that as with human breast cancer cells, LRIG1 expression blunts the response of these cells to HGF. Figure 6B demonstrates that LRIG1-transduced MDCK cells have reduced HGF-dependent motility. MDCK cells expressing LRIG1 showed a statistically significant decrease in their HGF-dependent motility (P = 1.69 × 10−5), once again indicating that LRIG1 inhibits HGF-dependent cellular responses. MDCK cells did not proliferate in response to HGF (data not shown), in agreement with previous reports (17, 51).
FIG. 6.
LRIG1 inhibits HGF-dependent motility of Madin-Darby canine kidney cells. (A) Whole-cell lysates of pMX- and LRIG1-myc-transduced MDCK cells were blotted for myc or actin. Canine Met could not be blotted in these cells with commercially available antibodies. (B) A Boyden chamber assay using transduced MDCK cells was performed as described for Fig. 5B. The error bars represent the standard deviation of three independent experiments.
LRIG1 opposes Her2 and Met receptor synergy in cellular invasion.
Recent studies have found that members of the ErbB receptor family, specifically EGF receptor and Her2, collaborate with Met receptor in driving tumor cell proliferation, motility, and invasion (30, 32, 47). For example, it was demonstrated that the combined activation of Her2 and Met in MDCK cells synergizes in the breakdown of cell-cell junctions and in cellular invasion (32). This finding clearly has important implications for tumors in which Her2 and Met receptor are coexpressed. Since LRIG1 is able to independently destabilize both Her2 (34) and Met receptor, we were interested in determining whether LRIG1 could oppose Met/Her2 synergy in driving cellular invasion. In the experiment shown in Fig. 7A, HEK-293T cells were cotransfected with an activated form of Her2 (NeuT), and Met receptor, in either the absence or presence of LRIG1. LRIG1 was able to significantly decrease the expression of both receptors, demonstrating that LRIG1 can simultaneously regulate both Her2 and Met.
FIG. 7.
LRIG1 simultaneously regulates Her2 and Met and opposes Met/Her2 synergy in cellular invasion. (A) HEK-293T cells were cotransfected with Met and NeuT in the absence or presence of LRIG1-myc. Whole-cell lysates were collected and blotted with antibodies to Her2, Met, myc, or actin. (B) Whole-cell lysates from transduced MDCK cells stably expressing either control vector (3.1) or NeuT were collected and blotted with antibodies to Her2, phosphotyrosine (pY; 4G10), myc, or actin. (C) Transduced MDCK cells were photographed using an inverse phase-contrast microscope under a ×10 magnification. (D) pMX-transduced MDCK cells stably expressing control vector (3.1) or NeuT were grown for 15 days in the presence of 10 ng/ml HGF, as previously described (32). Cells were plated in triplicate into Matrigel chambers with 10 ng/ml HGF in the bottom chamber. After 16 h, cells were stained and counted. The error bars represent the standard deviation of data collected from three independent experiments. (E) Transduced MDCK cells stably expressing either control vector (3.1) or NeuT were grown, plated, and counted as described for panel D.
Next, we examined the effect of LRIG1 on Her2 and Met synergy in MDCK cell invasion. MDCK cells transduced with pMX or LRIG1-myc were transfected with either empty vector (pcDNA3.1+) or NeuT, and the stable clones were pooled to avoid artifacts due to clonal variation. Importantly, there was no difference in the transfection efficiency of pMX-MDCK cells compared to LRIG1-MDCK cells as determined by transfection of glutathione S-transferase protein (data not shown). As shown in Fig. 7B, endogenous canine Her2 is detected in pMX-transduced cells (pMX-3.1), but its levels are reduced in LRIG1-transduced cells (LRIG1-3.1), demonstrating that LRIG1 is able to destabilize endogenous Her2 in these cells. Her2 levels are increased in pMX-NeuT cells, as expected, and NeuT is tyrosine phosphorylated. However, in LRIG1-transduced cells, Her2 levels are decreased with an associated reduction in phosphorylation.
MDCK cells grow in culture as organized epithelial colonies with intact cell junctions. Expression of NeuT in MDCK cells has previously been shown to promote a scattered phenotype, consistent with an epithelial-to-mesenchymal transition (32). As shown in Fig. 7C, pMX- and LRIG1-transduced MDCK cells transfected with empty vector (pcDNA3.1+) maintain an organized epithelial phenotype. pMX-transduced MDCK cells transfected with NeuT display a dramatic scattered phenotype, consistent with what has previously been reported (32). In contrast, LRIG1-transduced MDCK cells transfected with NeuT maintain an organized phenotype with intact cell junctions, comparable to wild-type MDCK cells. Taken together with data shown in Fig. 7B, these findings indicate that LRIG1 is able to oppose Her2-mediated cell junction breakdown by destabilization of Her2.
We next examined the impact of LRIG1 expression on MDCK invasion in a Matrigel assay. In the absence of HGF, MDCK cells are poorly invasive but their invasion can be modestly stimulated by HGF pretreatment (32). MDCK cells transfected with NeuT are constitutively invasive, and HGF pretreatment of these cells has been found to result in a synergistic increase in their invasive capacity (32). To validate these results using our transduced cells, we examined the invasion of pMX-transduced MDCK cells. As shown in Fig. 7D, control MDCK cells (pMX-3.1) are very poorly invasive, while cells pretreated with HGF are modestly invasive. MDCK cells transfected with NeuT (pMX-NeuT) are constitutively invasive, and pretreatment with HGF results in a synergistic increase in invasion, in agreement with previous results (32). We then examined the effects of LRIG1 on this synergy, as shown in Fig. 7E. LRIG1 expression significantly decreases the invasion of control cells (LRIG1-3.1) and abrogates the synergistic increase observed in NeuT cells (LRIG1-NeuT; compare pMX-NeuT with LRIG1-NeuT). Collectively, the data presented in this paper indicate that LRIG1 opposes Met/Her2 synergy in cellular invasion through a dramatic destabilization of both receptors.
DISCUSSION
Remarkably, the precedent for a leucine-rich repeat protein interacting with the Met receptor comes from the pathogenic bacteria Listeria monocytogenes. Listeria monocytogenes is a food-borne bacterium that gains entry into cells via the action of two bacterial surface proteins, internalin (also known as InlA) and InlB. The host receptor for internalin is E-cadherin (8), while the host receptor for InlB is the Met receptor (8, 50, 57). InlB contains eight leucine-rich repeats followed by an inter-repeat (IR) region with structural homology to Ig-like domains (2), reminiscent of LRIG1 structure. InlB interaction with Met provokes Met ubiquitination in a Cbl-dependent manner and subsequent Met endocytosis, allowing Listeria passage into cells, in essence “hijacking the endocytic machinery to invade cells” (57).
As discussed above, Cbl-mediated degradation of Met receptor plays a critical role in maintaining receptor levels within a range commensurate with normal cellular growth. Uncoupling of Met from Cbl-mediated regulation either by truncation or point mutation results in enhanced Met receptor stability and cellular transformation. Interestingly, a recent report has identified somatic intronic mutations in Met in lung cancer that result in a truncated protein lacking the juxtamembrane domain, analogous to Tpr-Met (33). This truncated protein, ΔEx14, displays decreased interaction with Cbl and heightened stability compared to wild-type receptor. While Cbl clearly plays an important role in Met receptor degradation and is currently the only known means of Met degradation, the Y1003F mutation does not substantially prolong the half life of the Met receptor, suggesting that other mechanisms contribute to Met stability.
Here we uncover a novel mechanism of Met receptor regulation. We demonstrate that the transmembrane leucine-rich repeat protein LRIG1 acts to destabilize the Met receptor, that endogenous LRIG1 and Met interact, and that loss of LRIG1 is sufficient to enhance endogenous Met receptor stability. With respect to EGF receptor degradation, LRIG1 has been reported to function by augmenting the recruitment of Cbl to the EGF receptor (21). However with the Met receptor, LRIG1 functions in a Cbl-independent manner. In addition, while Cbl-mediated regulation of Met receptor is dictated by receptor activation, LRIG1 destabilizes Met receptor regardless of activation status. LRIG1 interacts with Met receptor under basal conditions and destabilizes Met receptor in both the absence and presence of HGF. The mechanism by which LRIG1 destabilizes the Met receptor is currently unknown, but as with the EGF receptor, LRIG1 likely acts to facilitate the interaction of Met with the protein degradation machinery. Receptors that are refractory to Cbl-mediated regulation such as ΔEx14 are unlikely to be resistant to LRIG1-mediated regulation. In agreement with this, Y1003F is as sensitive to LRIG1-induced destabilization as wild-type receptor. Studies are ongoing to identify components of the protein degradation machinery that are essential for LRIG1 action.
LRIG1 destabilizes Met in a variety of cell lines, including human breast cancer cells, and importantly, inhibits their HGF-dependent growth and motility. LRIG1 was previously shown to act as a negative regulator of the ErbB family of receptor tyrosine kinases (21, 34) and has been proposed to function as a tumor suppressor (24). In agreement with this, the LRIG1 gene is located at chromosome 3p14.3, an area frequently deleted in human cancers, and LRIG1 expression is known to be decreased in tumor cell lines and primary tumors of diverse origins (24, 53, 54). Along these lines, we have observed that LRIG1 is underexpressed in the majority of primary human breast tumors (J. Miller and C. Sweeney, unpublished observations).
RTKs are frequently overexpressed and aberrantly active in human tumors. For example, Her2/Neu is overexpressed in ∼25% of breast tumors and correlates with poor patient prognosis. Recently, it has become appreciated that RTKs are not overexpressed in isolation. Rather, RTKs from different families are co-overexpressed in tumors and cross talk among these different families makes a major contribution to tumor growth and therapeutic resistance. For example, in Herceptin-resistant breast tumor cells, IGF-1 receptor cross talk with Her2 contributes to Herceptin resistance (41, 44). There is opportunity for cross talk among Her2 and Met receptor, as approximately 50% of Met receptor-positive breast tumors also overexpress Her2 (37). The consequences of this cross talk are dramatic, as evidenced by Met/Her2 synergy in driving cellular invasion (32).
We show here that LRIG1, by virtue of its ability to interact with and destabilize both Her2 and Met, abolishes Met/Her2 synergy. Loss or down-regulation of LRIG1 in tumors could contribute to overexpression of members of the ErbB family as well as Met receptor, promoting the invasive phenotype. Restoration of LRIG1 to tumors could offer a novel therapeutic strategy for suppression of receptor-positive tumors.
Acknowledgments
This work was supported by NIH grants CA118384 (C.S.) and GM068994 (K.C.) and a UC Davis Health System Award (C.S.). D.S. and J.M. are each recipients of Department of Defense Breast Cancer Research Program predoctoral fellowships.
We thank Andrew G. Manford for assistance in cloning the Met receptor cDNA, Haken Hedman for the gift of LRIG1-151 antibody, Paola Marignani for the gift of the pMX-pie and pJ6Ωpuro vectors and HEK293GPG cells, Hamid Band for the HA-c-Cbl and HA-70Z-Cbl constructs, Andrea Morrione for the HA-tagged ubiquitin construct, Yun Qiu for the pLentiLox 3.7 vector, and Ted M. Dawson for the pRK5-UB-KO construct.
Footnotes
Published ahead of print on 18 December 2006.
REFERENCES
- 1.Abella, J. V., P. Peschard, M. A. Naujokas, T. Lin, C. Saucier, S. Urbé, and M. Park. 2005. Met/hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Mol. Cell. Biol. 25:9632-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bierne, H., and P. Cossart. 2002. InlB, a surface protein of Listeria monocytogenes that behaves as an invasin and a growth factor. J. Cell Sci. 115:3357-3367. [DOI] [PubMed] [Google Scholar]
- 3.Birchmeier, C., W. Birchmeier, E. Gherardi, and G. F. Vande Woude. 2003. Met, metastasis, motility and more. Nat. Rev. Mol. Cell. Biol. 4:915-925. [DOI] [PubMed] [Google Scholar]
- 4.Bladt, F., D. Riethmacher, S. Isenmann, A. Aguzzi, and C. Birchmeier. 1995. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376:768-771. [DOI] [PubMed] [Google Scholar]
- 5.Camp, R. L., E. B. Rimm, and D. L. Rimm. 1999. Met expression is associated with poor outcome in patients with axillary lymph node negative breast carcinoma. Cancer 86:2259-2265. [DOI] [PubMed] [Google Scholar]
- 6.Carter, S., S. Urbe, and M. J. Clague. 2004. The Met receptor degradation pathway. Requirement for Lys48-linked polyubiquitin independent of proteasome activity. J. Biol. Chem. 279:52835-52839. [DOI] [PubMed] [Google Scholar]
- 7.Corso, S., P. M. Comoglio, and S. Giordano. 2005. Cancer therapy: can the challenge be MET? Trends Mol. Med. 11:284-292. [DOI] [PubMed] [Google Scholar]
- 8.Cossart, P. 2001. Met, the HGF-SF receptor: another receptor for Listeria monocytogenes. Trends Microbiol. 9:105-107. [DOI] [PubMed] [Google Scholar]
- 9.Diamonti, A. J., P. M. Guy, C. Ivanoff, K. Wong, C. Sweeney, and K. L. Carraway III. 2002. An RBCC protein implicated in maintenance of steady-state neuregulin receptor levels. Proc. Natl. Acad. Sci. USA 99:2866-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Renzo, M. F., R. P. Narsimhan, M. Olivero, S. Bretti, S. Giordano, E. Medico, P. Gaglia, P. Zara, and P. M. Comoglio. 1991. Expression of the Met/HGF receptor in normal and neoplastic human tissues. Oncogene 6:1997-2003. [PubMed] [Google Scholar]
- 11.Edakuni, G., E. Sasatomi, T. Satoh, O. Tokunaga, and K. Miyazaki. 2001. Expression of the hepatocyte growth factor/c-Met pathway is increased at the cancer front in breast carcinoma. Pathol. Int. 51:172-178. [DOI] [PubMed] [Google Scholar]
- 12.Fournier, T. M., L. Lamorte, C. R. Maroun, M. Lupher, H. Band, W. Langdon, and M. Park. 2000. Cbl-transforming variants trigger a cascade of molecular alterations that lead to epithelial mesenchymal conversion. Mol. Biol. Cell. 11:3397-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallego, M. I., B. Bierie, and L. Hennighausen. 2003. Targeted expression of HGF/SF in mouse mammary epithelium leads to metastatic adenosquamous carcinomas through the activation of multiple signal transduction pathways. Oncogene 22:8498-8508. [DOI] [PubMed] [Google Scholar]
- 14.Gao, C. F., and G. F. Vande Woude. 2005. HGF/SF-Met signaling in tumor progression. Cell Res. 15:49-51. [DOI] [PubMed] [Google Scholar]
- 15.Geetha, T., J. Jiang, and M. W. Wooten. 2005. Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol. Cell 20:301-312. [DOI] [PubMed] [Google Scholar]
- 16.Gentile, A., and P. M. Comoglio. 2004. Invasive growth: a genetic program. Int. J. Dev. Biol. 48:451-456. [DOI] [PubMed] [Google Scholar]
- 17.Gherardi, E., J. Gray, M. Stoker, M. Perryman, and R. Furlong. 1989. Purification of scatter factor, a fibroblast-derived basic protein that modulates epithelial interactions and movement. Proc. Natl. Acad. Sci. USA 86:5844-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghiglione, C., L. Amundadottir, M. Andresdottir, D. Bilder, A. J. Diamonti, S. Noselli, N. Perrimon, and K. L. Carraway III. 2003. Mechanism of inhibition of the Drosophila and mammalian EGF receptors by the transmembrane protein Kekkon 1. Development 130:4483-4493. [DOI] [PubMed] [Google Scholar]
- 19.Ghoussoub, R. A., D. A. Dillon, T. D'Aquila, E. B. Rimm, E. R. Fearon, and D. L. Rimm. 1998. Expression of c-met is a strong independent prognostic factor in breast carcinoma. Cancer 82:1513-1520. [DOI] [PubMed] [Google Scholar]
- 20.Goldoni, S., R. A. Iozzo, P. Kay, S. Campbell, A. McQuillan, C. Agnew, J. X. Zhu, D. R. Keene, C. C. Reed, and R. V. Iozzo. 17. July 2006, posting date. A soluble ectodomain of LRIG1 inhibits cancer cell growth by attenuating basal and ligand-dependent EGFR activity. Oncogene [Online.] doi: 10.1038/sj.onc.1209803. [DOI] [PubMed]
- 21.Gur, G., C. Rubin, M. Katz, I. Amit, A. Citri, J. Nilsson, N. Amariglio, R. Henriksson, G. Rechavi, H. Hedman, R. Wides, and Y. Yarden. 2004. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 23:3270-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond, D. E., S. Urbe, G. F. Vande Woude, and M. J. Clague. 2001. Down-regulation of MET, the receptor for hepatocyte growth factor. Oncogene 20:2761-2770. [DOI] [PubMed] [Google Scholar]
- 23.Hammond, D. E., S. Carter, and M. J. Clague. 2004. Met receptor dynamics and signalling. Curr. Top. Microbiol. Immunol. 286:21-44. [DOI] [PubMed] [Google Scholar]
- 24.Hedman, H., J. Nilsson, D. Guo, and R. Henriksson. 2002. Is LRIG1 a tumour suppressor gene at chromosome 3p14.3? Acta Oncol. 41:352-354. [DOI] [PubMed] [Google Scholar]
- 25.Huang, F., D. Kirkpatrick, X. Jiang, S. Gygi, and A. Sorkin. 2006. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21:737-748. [DOI] [PubMed] [Google Scholar]
- 26.Jeffers, M., L. Schmidt, N. Nakaigawa, C. P. Webb, G. Weirich, T. Kishida, B. Zbar, and G. F. Vande Woude. 1997. Activating mutations for the met tyrosine kinase receptor in human cancer. Proc. Natl. Acad. Sci. USA 94:11445-11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeffers, M., M. Fiscella, C. P. Webb, M. Anver, S. Koochekpour, and G. F. Vande Woude. 1998. The mutationally activated Met receptor mediates motility and metastasis. Proc. Natl. Acad. Sci. USA 95:14417-14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, T., Z. Guo, B. Dai, M. Kang, D. K. Ann, H. J. Kung, and Y. Qiu. 2004. Bi-directional regulation between tyrosine kinase Etk/BMX and tumor suppressor p53 in response to DNA damage. J. Biol. Chem. 279:50181-50189. [DOI] [PubMed] [Google Scholar]
- 29.Jin, L., A. Fuchs, S. J. Schnitt, Y. Yao, A. Joseph, K. Lamszus, M. Park, I. D. Goldberg, and E. M. Rosen. 1997. Expression of scatter factor and c-met receptor in benign and malignant breast tissue. Cancer 79:749-760. [DOI] [PubMed] [Google Scholar]
- 30.Jo, M., D. B. Stolz, J. E. Esplen, K. Dorko, G. K. Michalopoulos, and S. C. Strom. 2000. Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J. Biol. Chem. 275:8806-8811. [DOI] [PubMed] [Google Scholar]
- 31.Kang, J. Y., M. Dolled-Filhart, I. T. Ocal, B. Singh, C. Y. Lin, R. B. Dickson, D. L. Rimm, and R. L. Camp. 2003. Tissue microarray analysis of hepatocyte growth factor/Met pathway components reveals a role for Met, matriptase, and hepatocyte growth factor activator inhibitor 1 in the progression of node-negative breast cancer. Cancer Res. 63:1101-1105. [PubMed] [Google Scholar]
- 32.Khoury, H., M. A. Naujokas, D. Zuo, V. Sangwan, M. M. Frigault, S. Petkiewicz, D. L. Dankort, W. J. Muller, and M. Park. 2005. HGF converts ErbB2/Neu epithelial morphogenesis to cell invasion. Mol. Biol. Cell 16:550-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong-Beltran, M., S. Seshagiri, J. Zha, W. Zhu, K. Bhawe, N. Mendoza, T. Holcomb, K. Pujara, J. Stinson, L. Fu, C. Severin, L. Rangell, R. Schwall, L. Amler, D. Wickramasinghe, and R. Yauch. 2006. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 66:283-289. [DOI] [PubMed] [Google Scholar]
- 34.Laederich, M. B., M. Funes-Duran, L. Yen, E. Ingalla, X. Wu, K. L. Carraway III, and C. Sweeney. 2004. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J. Biol. Chem. 279:47050-47056. [DOI] [PubMed] [Google Scholar]
- 35.Lee, C. C., A. J. Putnam, C. K. Miranti, M. Gustafson, L. M. Wang, G. F. Vande Woude, and C. F. Gao. 2004. Overexpression of sprouty 2 inhibits HGF/SF-mediated cell growth, invasion, migration, and cytokinesis. Oncogene 23:5193-5202. [DOI] [PubMed] [Google Scholar]
- 36.Lee, W. Y., H. H. Chen, N. H. Chow, W. C. Su, P. W. Lin, and H. R. Guo. 2005. Prognostic significance of co-expression of RON and MET receptors in node-negative breast cancer patients. Clin. Cancer Res. 11:2222-2228. [DOI] [PubMed] [Google Scholar]
- 37.Lengyel, E., D. Prechtel, J. H. Resau, K. Gauger, A. Welk, K. Lindemann, G. Salanti, T. Richter, B. Knudsen, G. F. Vande Woude, and N. Harbeck. 2005. C-Met overexpression in node-positive breast cancer identifies patients with poor clinical outcome independent of Her2/neu. Int. J. Cancer 113:678-682. [DOI] [PubMed] [Google Scholar]
- 38.Levkowitz, G., L. N. Klapper, E. Tzahar, A. Freywald, M. Sela, and Y. Yarden. 1996. Coupling of the c-Cbl protooncogene product to ErbB-1/EGF-receptor but not to other ErbB proteins. Oncogene 12:1117-1125. [PubMed] [Google Scholar]
- 39.Liang, T. J., A. E. Reid, R. Xavier, R. D. Cardiff, and T. C. Wang. 1996. Transgenic expression of tpr-met oncogene leads to development of mammary hyperplasia and tumors. J. Clin. Investig. 97:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim, K. L., K. C. Chew, J. M. Tan, C. Wang, K. K. Chung, Y. Zhang, Y. Tanaka, W. Smith, S. Engelender, C. A. Ross, V. L. Dawson, and T. M. Dawson. 2005. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 25:2002-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu, Y., X. Zi, Y. Zhao, D. Mascarenhas, and M. Pollak. 2001. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin). J. Natl. Cancer Inst. 93:1852-1857. [DOI] [PubMed] [Google Scholar]
- 42.Mosesson, Y., and Y. Yarden. 2006. Monoubiquitylation: a recurrent theme in membrane protein transport. Isr. Med. Assoc. J. 8:233-237. [PubMed] [Google Scholar]
- 43.Nagy, J., G. W. Curry, K. J. Hillan, I. C. McKay, E. Mallon, A. D. Purushotham, and W. D. George. 1996. Hepatocyte growth factor/scatter factor expression and c-met in primary breast cancer. Surg. Oncol. 5:15-21. [DOI] [PubMed] [Google Scholar]
- 44.Nahta, R., L. X. Yuan, B. Zhang, R. Kobayashi, and F. J. Esteva. 2005. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 65:11118-11128. [DOI] [PubMed] [Google Scholar]
- 45.Peschard, P., T. M. Fournier, L. Lamorte, M. A. Naujokas, H. Band, W. Y. Langdon, and M. Park. 2001. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol. Cell 8:995-1004. [DOI] [PubMed] [Google Scholar]
- 46.Peschard, P., N. Ishiyama, T. Lin, S. Lipkowitz, and M. Park. 2004. A conserved DpYR motif in the juxtamembrane domain of the Met receptor family forms an atypical c-Cbl/Cbl-b tyrosine kinase binding domain binding site required for suppression of oncogenic activation. J. Biol. Chem. 279:29565-29571. [DOI] [PubMed] [Google Scholar]
- 47.Presnell, S. C., D. B. Stolz, W. M. Mars, M. Jo, G. K. Michalopoulos, and S. C. Strom. 1997. Modifications of the hepatocyte growth factor/c-met pathway by constitutive expression of transforming growth factor-alpha in rat liver epithelial cells. Mol. Carcinog. 18:244-255. [PubMed] [Google Scholar]
- 48.Rubinson, D. A., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, D. L. Rooney, M. M. Ihrig, M. T. McManus, F. B. Gertler, M. L. Scott, and L. Van Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt, L., F. M. Duh, F. Chen, T. Kishida, G. Glenn, P. Choyke, S. W. Scherer, Z. Zhuang, I. Lubensky, M. Dean, R. Allikmets, A. Chidambaram, U. R. Bergerheim, J. T. Feltis, C. Casadevall, A. Zamarron, M. Bernues, S. Richard, C. J. Lips, M. M. Walther, L. C. Tsui, L. Geil, M. L. Orcutt, T. Stackhouse, J. Lipan, L. Slife, H. Brauch, J. Decker, G. Niehans, M. D. Hughson, H. Moch, S. Storkel, M. I. Lerman, W. M. Linehan, and B. Zbar. 1997. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat. Genet. 16:68-73. [DOI] [PubMed] [Google Scholar]
- 50.Shen, Y., M. Naujokas, M. Park, and K. Ireton. 2000. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 103:501-510. [DOI] [PubMed] [Google Scholar]
- 51.Sponsel, H. T., R. Breckon, W. Hammond, and R. J. Anderson. 1994. Mechanisms of recovery from mechanical injury of renal tubular epithelial cells. Am. J. Physiol. 267:F257-F264. [DOI] [PubMed] [Google Scholar]
- 52.Takayama, H., W. J. LaRochelle, R. Sharp, T. Otsuka, P. Kriebel, M. Anver, S. A. Aaronson, and G. Merlino. 1997. Diverse tumorigenesis associated with aberrant development in mice overexpressing hepatocyte growth factor/scatter factor. Proc. Natl. Acad. Sci. USA 94:701-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanemura, A., T. Nagasawa, S. Inui, and S. Itami. 2005. LRIG-1 provides a novel prognostic predictor in squamous cell carcinoma of the skin: immunohistochemical analysis for 38 cases. Dermatol. Surg. 31:423-430. [DOI] [PubMed] [Google Scholar]
- 54.Thomasson, M., H. Hedman, D. Guo, B. Ljungberg, and R. Henriksson. 2003. LRIG1 and epidermal growth factor receptor in renal cell carcinoma: a quantitative RT-PCR and immunohistochemical analysis. Br. J. Cancer 89:1285-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trusolino, L., S. Cavassa, P. Angelini, M. Ando, A. Bertotti, P. M. Comoglio, and C. Boccaccio. 2000. HGF/scatter factor selectively promotes cell invasion by increasing integrin avidity. FASEB J. 14:1629-1640. [DOI] [PubMed] [Google Scholar]
- 56.Uehara, Y., O. Minowa, C. Mori, K. Shiota, J. Kuno, T. Noda, and N. Kitamura. 1995. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature 373:702-705. [DOI] [PubMed] [Google Scholar]
- 57.Veiga, E., and P. Cossart. 2005. Listeria hijacks the clathrin-dependent endocytic machinery to invade mammalian cells. Nat. Cell Biol. 7:894-900. [DOI] [PubMed] [Google Scholar]
- 58.Vigna, E., D. Gramaglia, P. Longati, A. Bardelli, and P. M. Comoglio. 1999. Loss of the exon encoding the juxtamembrane domain is essential for the oncogenic activation of TPR-MET. Oncogene 18:4275-4281. [DOI] [PubMed] [Google Scholar]
- 59.Yamashita, J., M. Ogawa, S. Yamashita, K. Nomura, M. Kuramoto, T. Saishoji, and S. Shin. 1994. Immunoreactive hepatocyte growth factor is a strong and independent predictor of recurrence and survival in human breast cancer. Cancer Res. 54:1630-1633. [PubMed] [Google Scholar]
- 60.Yang, J.-Y., C. S. Zong, W. Xia, Y. Wei, M. Ali-Seyed, Z. Li, K. Broglio, D. A. Berry, and M. C. Hung. 2006. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol. Cell. Biol. 26:7269-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao, Y., L. Jin, A. Fuchs, A. Joseph, H. M. Hastings, I. D. Goldberg, and E. M. Rosen. 1996. Scatter factor protein levels in human breast cancers: clinicopathological and biological correlations. Am. J. Pathol. 149:1707-1717. [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou, P. 2004. Determining protein half lives. Methods Mol. Biol. 284:67-77. [DOI] [PubMed] [Google Scholar]