Abstract
Cleavage and polyadenylation is an essential processing reaction required for the maturation of pre-mRNAs into stable, export- and translation-competent mature mRNA molecules. This reaction requires the assembly of a multimeric protein complex onto a bipartite core sequence element consisting of an AAUAAA hexamer and a GU/U-rich downstream sequence element. In this study we have analyzed 3′ end processing of the human melanocortin 1 receptor gene (MC1R). The MC1R gene is an intron-free transcription unit, and its poly(A) site lacks a defined U/GU-rich element. We describe two G-rich sequence elements that are critical for efficient cleavage at the MC1R poly(A) site. The first element is located 30 nucleotides downstream of the cleavage site and acts as an essential closely positioned enhancer. The second G-rich region is positioned more than 440 nucleotides downstream of the MC1R processing site and is instrumental for optimal processing efficiency. Both G-rich sequences contain clusters of heterogeneous nuclear ribonucleoprotein binding motifs and act together to enhance cleavage at the MC1R poly(A) site.
The expression of eukaryotic protein-encoding genes requires that the primary RNA transcripts undergo three major processing reactions. The very 5′ end of each emerging pre-mRNA is cotranscriptionally capped, intron sequences are excised via a complex splicing mechanism, and 3′ ends of almost all genes are generated by a two-step mechanism involving an endonucleolytic cleavage reaction followed by the polymerization of a poly(A) tail.
In mammals, the cleavage and polyadenylation reaction requires the cotranscriptional assembly of a large complex consisting of five multimeric factors on a core cis element embedded in the 3′ end of the pre-mRNA (41). The core poly(A) recognition site is a bipartite sequence encompassing a conserved hexamer, AAUAAA or AUUAAA, and a lesser conserved U- and/or GU-rich region of variable length located up to 60 nucleotides further downstream (59). Both of these cis elements directly interact with subunits of the two major cleavage and polyadenylation factors, the cleavage and polyadenylation specificity factor (CPSF) and the cleavage stimulatory factor (CstF), in a cooperative manner (13, 59). After the assembly of additional essential factors, including cleavage factors I (CFI) and II (CFII) and in most cases the poly(A) polymerase, cleavage occurs cotranscriptionally, probably catalyzed by the CPSF-73 subunit (45).
The cleavage efficiency at canonical poly(A) sites is determined by the sequence composition of both the hexamer and the U/GU-rich downstream sequence element (DSE). Mutation of each individual nucleotide of the optimal AAUAAA sequence severely reduces the efficiency of cleavage at the mutant sites (24, 46, 54). The strength of the DSE is dependent on its U and GU composition but also on how far it is positioned from the cleavage site (20, 35). The excision of introns has also been associated with the stimulation of the poly(A) cleavage reaction in spliced genes, and the artificial removal of the intron sequences in expression constructs generally results in a dramatic reduction in cleavage efficiency at poly(A) sites (31, 37, 38).
Several examples have been reported where the efficiency of a poly(A) site is significantly enhanced or reduced by additional auxiliary sequence elements positioned upstream or downstream of the core processing signal.
Upstream sequence elements (USEs) are generally uridine rich (25, 59) and are found in several viral transcripts, including adenovirus, adeno-associated virus (42), human immunodeficiency virus type 1 (49, 50), simian virus 40 (SV40) (59), and human papillomavirus (60). Both the human lamin B2 and C2 complement genes are examples of cellular transcripts that contain critical USEs which modulate processing efficiency by interacting with the polypyrimidine rich binding protein (PTB) and CstF-64, respectively (7, 8, 36). Furthermore, the cyclooxygenase 2 gene is alternatively polyadenylated, and the use of the proximal poly(A) site is controlled by three short uridine-rich USEs (24). In addition, a novel upstream sequence element with several copies of the consensus UGUAN sequence has been shown to alter cleavage efficiency of both canonical and noncanonical poly(A) sites via the direct interaction with the essential cleavage factor I (CFI) (9, 51).
Importantly, the presence of USEs is often associated with the enhanced expression of intronless transcripts. The herpes simplex virus type 1 intronless thymidine kinase pre-mRNA contains a specific pre-mRNA processing enhancer which stimulates 3′ end formation. The molecular mechanism appears to be dependent on the interaction of the cellular heterogeneous nuclear ribonucleoproteins with the pre-mRNA processing enhancer (22, 32). Functionally similar posttranscriptional regulatory elements (PREs) in cis have been described in hepatitis B viral transcripts (30, 57) and the woodchuck hepatitis RNAs (16). An analogous element within the polyadenylated but non-protein-encoding nuclear-retained RNA from the Kaposi's sarcoma-associated herpesvirus is able to confer efficient expression of an intron-free globin mRNA by enhancing 3′ end processing (14). A similar cellular functional homologue of the above-described viral sequences was found in the polyadenylated intron-free mouse histone H2a gene (26, 29), where 3′ end processing and nuclear cytoplasmic export is stimulated by the interaction of this sequence element with the splicing factors 9G8 and SRp20 (28). An element in the intronless human c-jun gene has been identified which may enhance 3′ end formation and cytoplasmic accumulation by an as-yet unknown mechanism (23).
In contrast to the many examples where USEs play a critical role in enhancing cleavage at the poly(A) site, elements that are located downstream of the core processing site are less frequently reported. The best characterized such element is a 14-nucleotide-long G-rich sequence located immediately downstream of the SV40 late poly(A) signal (3, 4, 12). Similar stimulatory G-rich sequences positioned immediately adjacent to the core poly(A) sites are predicted at the 3′ ends of several cellular genes (3, 25, 58). In addition, a regulatory sequence element consisting of six GGGU motifs positioned 174 nucleotides downstream of the human papillomavirus L1 early poly(A) site has recently been shown to regulate tissue- and stage-specific expression of viral early and late genes (39).
Furthermore, another G-rich sequence, the artificial MAZ4 site, enhances the efficiency of poly(A) cleavage and transcription termination in vitro and in vivo (21, 55, 56) by a so-far unknown mechanism which involves pausing of RNA polymerase II. In addition, non-G-rich sequences have also been implicated in influencing cleavage activity at poly(A) sites. A pyrimidine-rich sequence in the calcitonin/calcitonin gene-related peptide pre-mRNA stimulates poly(A) site use via its interaction with PTB (33).
In this study, we have investigated how the intronless melanocortin 1 receptor pre-mRNA (MC1R) is efficiently processed at the 3′ end. The MC1R gene encodes a 317-amino-acid seven-transmembrane receptor expressed on the cell surface of melanocytes. MC1R plays a key role in the formation of skin and hair pigmentation by regulating the relative ratio of the two major classes of melanin, eumelanin (brown/black) and pheomelanin (red/yellow) (43).
The MC1R gene is intron free and lacks a defined U- or GU-rich downstream poly(A) site sequence element. This led us to explore if additional alternative poly(A) enhancer elements are present within the MC1R transcription unit that compensate both for the lack of a defined DSE and the lack of stimulation by terminal intron removal that directs efficient cleavage in spliced genes. Our investigation has revealed two G-rich sequence elements that are both critical for efficient cleavage at the MC1R poly(A) site. The first G-rich element is located 35 nucleotides downstream of the poly(A) processing site and acts as an essential closely positioned enhancer. The second G-rich element is positioned 440 nucleotides downstream of the MC1R poly(A) site and is instrumental for optimal processing efficiency. Our data suggest that both G-rich sequences act jointly to stimulate cleavage at the MC1R 3′ end processing site. Both G-rich elements consist of several copies of defined binding motifs for RNA binding proteins of the hnRNPH family, and their interaction with the G-rich elements may play a role in stimulation of the cleavage step. In addition, it appears that the MC1R DSE constitutes a single essential diuridine which, in conjunction with the downstream-located G boxes, directs cleavage at the poly(A) site.
MATERIALS AND METHODS
Plasmid construction. (i) CMV-driven MC1R reporter gene (MC1R-GFP).
The cytomegalovirus (CMV) promoter was amplified by PCR from a CMV-driven human immunodeficiency virus minigene (19) and cloned into the AflIII and EcoRI restriction sites in the pUC18 plasmid. The MC1R 5′-untranslated region (UTR) was amplified by PCR using genomic DNA isolated from HeLa cells and cloned downstream of the CMV promoter as an EcoRI-BamHI fragment. The coding sequence of the green fluorescent protein (GFP) was amplified and ligated downstream of to the 5′UTR sequence using the BamHI and XbaI restriction sites. The MC1R 3′UTR sequences, including 1 kb of 3′-flanking regions, was amplified from genomic HeLa DNA and inserted into the above construct using XbaI and SphI restriction sites.
(ii) 5′UTR deletion and MC1R coding region replacement clones.
Using the MC1R-GFP plasmid, the 394-bp 5′UTR was progressively shortened from the 5′ end using restriction sites EcoRI-XmnI, EcoRI-BglII, and EcoRI-BamHI, resulting in deletions of 180, 243, and 394 nucleotides, respectively.
Using the MC1R-GFP plasmid, the entire GFP cDNA was removed and replaced by one of two Pfu-amplified halves of the MC1R open reading frame using the restriction sites BamHI and XbaI, resulting in clones MCIR-5′C and MC1R3′C.
(iii) 3′UTR and 3′ flank deletion clones.
The 3′UTR deletion clones ΔNH and ΔXH were made by Pfu amplification of the wild-type (wt) MC1R 3′UTR using the primer pairs mdN1-mdH3R and mdX1-mdH3R, respectively (mdN1, 5′-GTTGTGCGGCCGCAGGCTC-3′; mdH3R, 5′-GTGCCAAGCTTGCATGCCTC-3′; mdX1, 5′-CCTCTAGAGGTATGGGGCTG-3′). Truncated NotI (XbaI)-HindIII-amplified fragments were religated into NotI (XbaI)-HindIII-digested MC1R-GFP followed by ligation of the wt HindIII fragment, screened for orientation using AflII. The 3′UTR deletion clone Δh34 was amplified using mdh34 (5′-CAGTGCCTGTAAGCTTGGGG-3′) and mdH3R, and the resulting HindIII fragment was ligated into HindIII-digested MC1R-GFP vector and screened for orientation using AflII.
The 3′ flank clones Δh36 through Δh46 were made by Pfu amplification using forward primer mdH3F (5′-GACCAGAAAGCTTCATCCAG-3′) paired with one of the reverse primers mdh36 (5′-ATCCCTCAAGCTTCAAACTTC-3′), mdh37 (5′-ACCCCTGCCAAGCTTCCCCT-3′), mdh38 (5′-CTGGGGCTGAAGCTTCCTCA-3′), mdh39 (5′-GGTCAGCGCAAGCTTGGTTG-3′), mdh40 (5′-CTAGGAAAGCTTCAAGAGTG-3′), mdh41 (5′-CACAGGGAAGCTTGGGAACC-3′), mdh45 (5′-ATCTTCAGAAGCTTCACATC-3′), and mdh46 (5′-GACGCCCAAGCTTCAGCAAGA-3′), respectively. Truncated HindIII-HindIII-amplified fragments were religated into HindIII-digested MC1R-GFP and subsequently screened for orientation using AflII.
(iv) Internal deletion clones.
Three deletion clones were made covering GB1 and/or GB2, named ΔG1, ΔG2, and ΔG1/2. All clones were made by Pfu amplification of fragments juxtaposed immediately 5′ and 3′ of the area to be deleted using phosphorylated internal primers (i.e., the primers located adjacent to the targeted sequence). The resulting two fragments were then ligated together and subsequently reamplified using the external primer pair containing HindIII restriction sites. The primers used were the following: for ΔG1, mdH3F-mdGR3 (5′-CGCAAACTTCAAGCACTTTC-3′) and mdGR2 (5′-GAAGGTGGGCAGGGGTGAGA-3′)-mdH3R; for ΔG2, mdH3F-mdG21 (5′-CTGGGAACCCGCCTCTTCTG-3′) and mdG22 (CTTCTGAAGATGGCAGTGAG-3′)-mdH3R; for ΔG1/2 the primers were the same as those for ΔG2, save starting with ΔG1 as the template.
The uridine-to-cytidine conversion clones were constructed in a similar manner, save that the phosphorylated internal primers were placed immediately next to each other, resulting in no loss of sequence, save the U-to-C changes encoded by the primers themselves. The primers were the following: for Mut1, mdH3F-mdGU1 (5′-CGGGCGCTTTCTGGAAGGGG-3′) and mdGU2 (5′-AAGCTCGCGGGCGGAGGGAT-3′)-mdH3R; for Mut2, mdH3F-mdGU5 (5′-CAAGCACTTTCTGGAAGGGG-3′) and mdGU6 (5′-AAGCTCGCGGGTGGAGGGAT-3′)-mdH3R; for Mut3, mdH3F-mdGU7 (5′-CGGGCACTTTCTGGAAGGGG-3′) and mdGU3 (5′-AAGTTTGCGGGTGGAGGGAT-3′)-mdH3R.
(v) GB1 and control templates for band shift.
The plasmid used to in vitro transcribe RNA containing GB1 was created by inserting a PCR-amplified fragment using the primers mdG1F (5′-CTTGAAGCTTGCGGGTGGAG-3′) and mdG1R (5′-GCCACGAATTCCGGGACTCC-3′), resulting in a 74-nucleotide fragment which was cloned into HindII-EcoRI-digested pGEM-4. The control RNA was a 77-nucleotide fragment corresponding to the extreme 3′ end of the MC1R 3′ flank which was obtained by digesting the MC1R-GFP construct with ApaI and HindIII and cloned into SmaI-HindIII-digested pGEM-4.
The G1 template was linearized with EcoRI and subsequently transcribed by T7 RNA polymerase, and the control RNA was linearized with HindIII and transcribed by Sp6 RNA polymerase. The resulting radiolabeled transcripts were 84 and 86 nucleotides long, respectively.
Tissue culture.
Human SK23 (a kind gift from Angelo Scibetta, Cancer Research United Kingdom, London), embryonic kidney 293 (HEK293), and HeLa cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 1% l-glutamine, and 1% penicillin-streptomycin at 37°C with 5% CO2.
Cells of 50 to 70% confluence were transfected using Polyfect transfection reagent from QIAGEN. For the transient transfections, 3 μg MC1R plasmids and 0.05 μg VA plasmid (a cotransfectional control of pUC-18 vector containing the adenovirus VA I gene that is constitutively expressed by RNA polymerase III) were added to 150 μl DMEM with 25 μl Polyfect solution and incubated for 20 min at room temperature. This mix was then added to the cells in 5 ml DMEM-10% FCS and incubated at 37°C in 5% CO2. After 5 h the medium was replaced with 10 ml of fresh DMEM-10% FCS.
RNA preparation.
Total RNA was isolated approximately 16 h after transfection as described previously (18). Hypotonic lysis was used to collect nuclear and cytoplasmic fractionated RNA. Cell pellets were resuspended in 400 μl of lysis buffer (0.14 M NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl, pH 7.5) and then underlayed with a 400-μl 40% sucrose cushion and centrifuged to precipitate nuclei. Four-hundred microliters of supernatant was collected, phenol-chloroform extracted, and ethanol precipitated. Pellets were resuspended in 100 μl DNase I buffer supplemented with 5 μl DNase I, incubated at 37°C for 30 min, phenol extracted, and precipitated. The final pellets were resuspended in R-loop buffer (18) and stored at −20°C. For the isolation of nuclear RNA, the collected nuclei were directly subjected to hot phenol treatment (18).
RNA analysis.
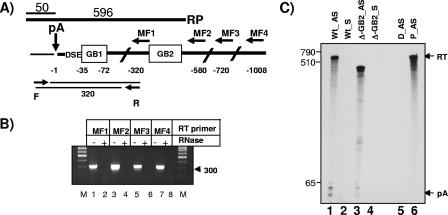
EcoRI-linearized pGEM-4 plasmids (0.5 μg) containing a 410-nucleotide-long PstI MC1R fragment encompassing the poly(A) site or the VA gene, respectively, were transcribed by T7 RNA polymerase. The in vitro-transcribed antisense RNA probes were then gel purified on a 6% polyacrylamide gel and stored in 100 μl R-loop buffer. The long RNase protection probes used in Fig. 7C were transcribed either by Sp6 for the antisense or T7 for the sense probe from a pGEM-4 plasmid that contained MC1R sequences from the region 50 nucleotides upstream of the major poly(A) cleavage site to 5 nucleotides downstream of GB2.
FIG. 7.
GB2 is part of the uncleaved MC1R pre-mRNA. A) The MC1R poly(A) site (pA) DSE and the 3′-flanking region (boldface black line) is shown, and GB1 and GB2 are indicated in open boxes. Sequences of the 3′UTR are indicated by the thin black line. The position of the reverse transcription primers MF1 to MF4 are shown by horizontal arrows, and their distance relative to the site of cleavage (vertical arrow with pA above) is indicated in negative values below. F and R represent forward and reverse primers used for the PCR amplification of the cDNA products obtained with the MF primers. The resulting PCR product (two thin black lines) and its predicted length are shown. The RNase protection probe is indicated above the graph, and the resulting protected fragments are indicated. B) Reverse transcription-PCR of total RNA isolated from cells transfected with the MC1R wt reporter gene. Reverse transcription primers used are shown above the gel. −/+ RNase indicates whether or not total RNA was treated with RNase prior to reverse transcription. M, size markers; the 300-nucleotide band is indicated. C) RNase protection using total RNA from HEK293 cells transfected with either the wt-MC1R construct or the Δ-GB2 plasmid using universally labeled protection probes complementary to either the sense MC1R pre-mRNA sequence (wt_AS and Δ-GB2_AS) or complementary to the antisense pre-mRNA sequence (wt_S and Δ-GB2_S). D_AS represents full digestion of the antisense protection probe in the absence of cellular RNA, and P_AS stands for undigested full-length probe. pA and RT are the same as described in the legend to Fig. 2B.
RNase protection analysis.
Three to 15 μl of RNA sample (depending of transfection efficiencies) was added to 2 to 3 μl of 300- to 500-cps/μl [α-32P]UTP radiolabeled riboprobe in a total volume of 30 μl R-loop buffer. This hybridization mix was then denatured at 94°C for 20 min and incubated at 56°C for 15 to 18 h. RNase digestion was carried out by adding 300 μl RNase mix (1 ml RNase buffer: 300 mM NaCl, 10 mM Tris-HCl, pH 7.4, 5 mM EDTA supplemented with 4 μl RNase A [10 mg/ml] and 2 μl RNase T1 [550 U/ml]) to the hybridized samples and subsequent incubation at 30°C for 45 min. RNase digestion was followed by Proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation. RNA pellets were resuspended in 15 μl RNA loading buffer, and the protected RNA fragments were fractionated on a 6% polyacrylamide gel. Undigested control probes were resuspended in 60 μl loading buffer, and 12 μl was loaded onto a gel. Quantitations were carried out using software provided with the Fuji phosphorimager, and unless otherwise stated poly(A) cleavage efficiency is depicted as the ratio between cleaved and uncleaved RNA.
RNA band shift.
In vitro-transcribed [α-32P]UTP-labeled RNA containing the GB1 sequences was incubated with 0.25 μl of HEK293 nuclear extract (purchased from Jena Bioscience GBH, Germany) on ice for 30 min. Subsequently, 4 μl (0.8 μg) of hnRNPH-specific antibody [(N-16):sc-10042; Santa Cruz Biotechnology] was added, and the reaction mixture was incubated for a further 10 min on ice. If indicated, 0.6 μg secondary antibody (anti-goat immunoglobulin G [IgG]) was subsequently added and incubated for a further 10 min on ice. The RNA protein mix was then separated using a 4% glycine gel as described previously (53).
Reverse transcription-PCR analysis.
One microliter of total RNA isolated from cells which were transiently transfected with the MC1R wild-type expression plasmid was subjected to reverse transcription using superscript III reverse transcriptase (Invitrogen) and various reverse primers. Resulting cDNAs were amplified in a PCR with two universal primers (mdMF1, mdMF2, mdMF3, and mdMF4). Resulting cDNAs were amplified in a PCR designed to quantitate both processed and pre-mRNA by amplifying a fragment dependent on transcribed sequences upstream of the poly(A) site (mdOX6) and downstream of the known cleavage sites (mdGR3).
siRNA treatment.
Small interfering RNAs (siRNAs) against hnRNPH (sc-35579) and control siRNAs (sc-37007) where purchased from Santa Cruz Biotechnology. HEK293 cells grown in 6-well plates were twice transfected at 24-h intervals with 60 pmol of either siRNA targeting hnRNPH or the control siRNA prior to MC1R plasmid transfection (0.5 μg/well).
Western blotting was performed with ECL on HEK293 cell protein extract using a 1:300 dilution of goat anti-human hnRNPH/H′ or rabbit anti-human cyclin H (Santa Cruz Biotechnology).
RESULTS
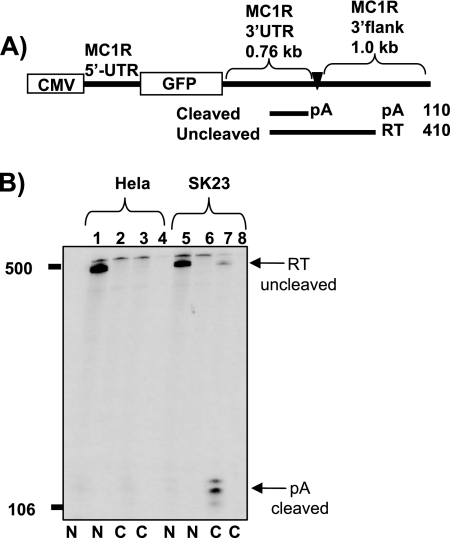
The MC1R reporter gene is efficiently expressed in SK23 melanoma and HEK293 cells.
To investigate how the human intronless MC1R gene is efficiently cleaved and polyadenylated, we designed a reporter construct based on genomic MC1R sequences. We fused the MC1R 5′-untranslated region (UTR) to the open reading frame of the green fluorescence protein (GFP) and added the 3′UTR, including 1 kb 3′-flanking sequences from the MC1R gene. A cytomegalovirus (CMV) promoter was introduced to drive efficient ligand- and tissue-independent transcription from this construct (Fig. 1A). The reporter plasmid was transfected into HeLa cells together with a cotransfection control plasmid containing the RNA polymerase III-transcribed adenovirus VA I gene. The expression of nuclear and cytoplasmic RNA fractions was subsequently analyzed by RNase protection using a detection probe encompassing the MC1R cleavage site. This RNase protection probe allowed us to discriminate between cleaved and uncleaved MC1R RNA species (Fig. 1A). A VA-specific probe was used to control for transfection efficiencies, RNA quality, and equal loading. As expected, the plasmid was efficiently transcribed by the CMV promoter in HeLa cells. However, MC1R RNA could only be detected in the nuclear fraction and was undetectable in the cytoplasmic preparation (Fig. 1B, lanes 1 and 3). Most importantly, the protected MC1R RNA species represented unprocessed readthrough pre-mRNAs (RT) but clearly lacked a 110-nucleotide-long protected band corresponding to mature cleaved and polyadenylated mRNA (pA) in either fraction (Fig. 1B, compare RT and pA in lanes 1 and 3). This surprising result suggested that cell-specific factors may be required to allow efficient cleavage at the poly(A) site or that a specific sequence may destabilize the MC1R mRNA in the HeLa cytoplasm. In order to find a more suitable cell line for the analysis of MC1R gene expression, we next employed the melanoma-derived cell line SK23. RNase protection of RNA isolated from transiently transfected SK23 cells showed that the reporter gene was efficiently expressed, and uncleaved MC1R RNA could be detected in the nuclear fraction (Fig. 1B, lane 5). However, in contrast to the analysis using HeLa cells, short protected bands representing cleaved mature MC1R mRNA (confirmed by 3′ rapid amplification of cDNA ends; data not shown) could now clearly be identified in the SK23 cytoplasmic RNA preparation (Fig. 1B, compare lanes 3 and 7). Note that control transfections with an empty pUC-18 vector (Fig. 1B, lanes 2, 4, 6, and 8) confirmed that the endogenous MC1R gene does not produce detectable signals in either cell line.
FIG. 1.
The MC1R reporter gene shows cell type-specific expression patterns. A) Diagram depicting the MC1R reporter gene. The position and the lengths of the MC1R-derived UTRs are indicated above the diagram. The promoter (CMV) and the GFP open reading frame (GFP) are represented by open boxes. The MC1R poly(A) site is pointed out by a filled triangle and pA. RNase protection fragments representing uncleaved (RT) and cleaved (pA) RNA species are shown as black lines under the diagram, and the expected lengths of the protected bands are given on the right side. B) RNase protection analysis of nuclear (N) and cytoplasmic (C) RNA isolated from HeLa cells (lanes 1 to 4) and melanoma-derived SK23 cells (lanes 5 to 8), which were transiently transfected with the MC1R wt reporter construct. Arrows with pA and RT indicate RNase protection bands representing cleaved and uncleaved MC1R RNA species. Even-numbered lanes are control transfections where the VA plasmid is cotransfected with an empty pUC18 vector to control for potential stimulation of endogenous MC1R expression. Note that RNA was normalized to the cotransfectional VA control.
We conclude that the MC1R 5′UTR and/or the 3′UTR, including flanking sequences, prevent efficient expression of the reporter gene in HeLa cells either at the level of cell type-specific cleavage of the pre-mRNA or by destabilizing the MC1R mRNA.
Searching for cis elements in the 3′UTR which affect MC1R gene expression.
To address how the intronless MC1R gene is efficiently processed at the 3′ end, we first focused on identifying regulatory sequence elements within the untranslated regions of the MC1R pre-mRNA. This was justified, since previously published work on the regulation of both viral and cellular intronless polyadenylated transcripts suggested that efficient cleavage of such transcripts depends on the presence of enhancer sequences in the 3′UTR (14, 16, 22, 23, 26, 28-30, 32, 57).
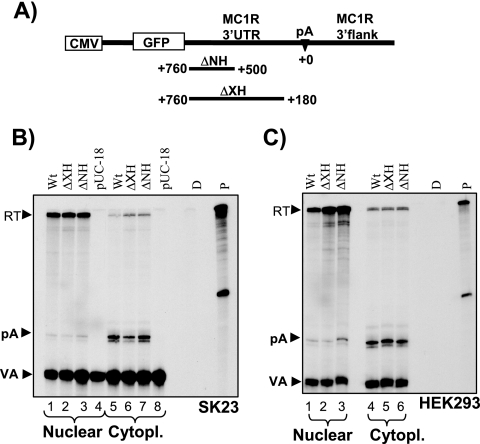
We created a series of plasmids in which 260 (ΔNH) or 580 (ΔXH) bases of the 3′UTR were deleted in a 5′ to 3′ direction and a clone where the region between the 3′UTR nucleotides 580 and 726 was removed. To our surprise, RNase protection analysis using the above-described 3′ protection probe revealed that none of these deletions had any effect on unprocessed nuclear or processed cytoplasmic MC1R RNA levels in SK23 cells (Fig. 2B, compare lanes 1 to 3 and lanes 5 to 7). The deletion clone where the sequences located between nucleotides 580 and 726 were removed was analyzed using an RNase protection probe covering 5′UTR sequences and had no significant effect on reporter mRNA levels in either fraction (data not shown). Note that a pUC18 control transfection again confirmed that the detected MC1R RNA species in the transfected cells originated from the transfected plasmids and were not the results of stimulating endogenous gene expression (Fig. 2B, lanes 4 and 8). We subsequently established that our MC1R reporter gene was also efficiently expressed in transiently transfected 293 human embryonic kidney cells (HEK293), and as can be seen in Fig. 2C, identical results were obtained using nuclear (lanes 1 to 3) and cytoplasmic (lanes 4 to 6) RNA from this cell line. For all subsequent analyses, we employed the HEK293 cells because of ease of handling.
FIG. 2.
The MC1R 3′UTR lacks processing enhancer sequences. A) CMV-MC1R reporter construct with the promoter and GFP open reading frame shown as open boxes. 3′UTR and 3′-flanking sequences are pointed out above the construct. The poly(A) site is depicted as described in the legend to Fig. 1A. The nucleotides in the 3′UTR are numbered in a 3′ to 5′ direction as positive values, with the adenosine of the primary cleavage site (CA) set as 0. The region and lengths of the introduced 3′UTR deletions in the two mutant constructs ΔNH and ΔXH are indicated below with a black line. B) RNase protection analysis of nuclear (lanes 1 to 4) and cytoplasmic (Cytopl.; lanes 5 to 8) steady-state RNA isolated from SK23 cells transiently transfected with wt, ΔNH, and ΔXH MC1R constructs and the cotransfection control VA plasmid. The same 3′ RNase protection probe used for Fig. 1A was used. Protected bands representing readthrough (RT) MC1R pre-mRNA, cleaved MC1R mRNA (pA), and RNA from the cotransfection control VA (VA) are shown. pUC18 control transfections of empty pUC-18 vector controlling for endogenous MC1R expression are shown in lanes 4 and 8. D and P are controls representing RNase protection probes incubated with or without RNase and in the absence of cellular RNA. C) RNase protection analysis of nuclear (lanes 1 to 3) and cytoplasmic (lanes 4 to 6) RNA from transfections with the wt, ΔNH, and ΔXH plasmids using the human embryonic kidney cell line HEK293.
Since a previously reported cleavage and transport enhancer element in the histone H2a gene was located within the coding region (26-29), we also replaced the GFP open reading frame with MC1R coding sequences. Transfection and subsequent RNA analysis of these clones again had no effect on RNA levels (data not shown). Interestingly, removal of either the 5′ or the 3′UTR sequences in the GFP MC1R construct had little effect on overall expression levels of cleaved mRNA in HeLa cells (data not shown). These results suggest that the 3′UTR of the MC1R mRNA plays no significant role in 3′ end processing of the MC1R pre-mRNA and subsequent nuclear cytoplasmic export of the cleaved and polyadenylated transcripts. In addition, the lack of MC1R reporter expression observed in our initial HeLa experiments (see Fig. 1) is likely to be the consequence of cell-specific regulation at the level of poly(A) cleavage rather than specific destabilization of the MC1R reporter RNA by sequences located in the UTRs.
The role of the MC1R 3′-flanking region in directing efficient 3′ end processing.
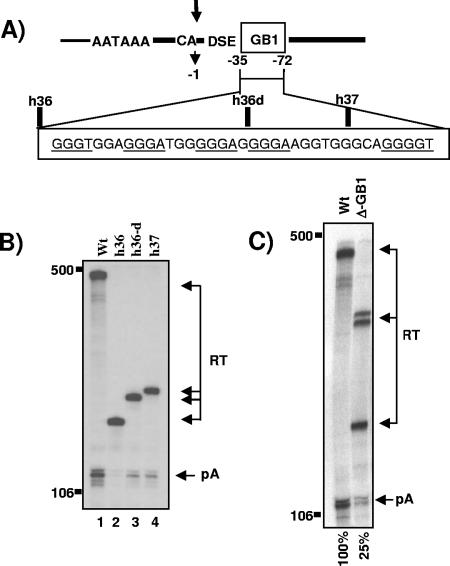
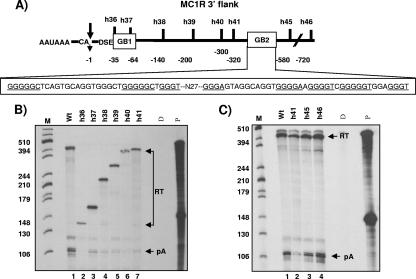
The lack of a processing enhancer element in the 3′UTR and the finding that cell-specific expression may be regulated at the level of 3′ end processing prompted us to explore whether regulatory sequences could be found further downstream of the core poly(A) site in the 3′-flanking region of the MC1R gene. Such sequences which would not be part of the mRNA could still compensate for a weak DSE and the lack of terminal intron removal by conferring efficient and tissue-specific poly(A) cleavage at the processing site. To identify such potential elements, we removed large parts of the MC1R 3′-flanking regions in the MC1R reporter plasmid without affecting the core poly(A) sequences (Fig. 3A). The resulting clones h36, h36d, and h37 retained 35, 55, and 65 nucleotides, respectively, of the 3′-flanking region (counted from the site of cleavage). These deletion constructs were transiently transfected into HEK293 cells, and cleavage was analyzed by RNase protection of total RNA using the above-described 3′ protection probe. The use of the 3′ protection probe in the analysis of the deletion clones resulted in protected readthrough fragments (RT) that are significantly shorter in length compared to uncleaved wild-type transcripts, because the probe overlaps the introduced deletions (Fig. 3B, RT in lanes 2 to 4). To our surprise, however, the deletion of flanking sequences downstream of the h36 position (Fig. 3A) almost completely abolished cleavage at the poly(A) site despite the presence of the complete core poly(A) sequence (Fig. 3B, compare pA in lanes 1 and 2). Cleavage was partially recovered with two clones (h36d and h37) that retained at least half of a 37-nucleotide-long G-rich element, and we hereby name this sequence GB1 (Fig. 3A and B, lanes 3 and 4). From this RNA protection analysis we conclude that a sequence which constitutes more than 70% G residues and is located adjacent to the DSE may be essential to direct poly(A) cleavage in the MC1R pre-mRNA. GB1 holds five binding motifs for the hnRNPH binding proteins and is therefore similar to an enhancer sequence found downstream of the core SV40 late poly(A) site (3, 4, 12). However, it is important to note that the h36 construct was obtained by deleting 1 kb of the downstream 3′-flanking region. By introducing such a large deletion, we could not rule out that the observed drop in levels of cleaved transcripts was not due to the absence of the G-rich sequence in this plasmid but rather caused by positioning a potentially negative element (located in plasmid sequences) closer to the processing site. To address this possibility, we designed a new plasmid in which the 37-nucleotide-long G-rich sequence is excised without affecting the core poly(A) signal or the remaining downstream-positioned 1-kb flanking region. As can be seen in Fig. 3C, lane ΔGB1, the precise excision of the G-rich box reduced the efficiency of poly(A) site cleavage to an average of 25% of wild-type levels, calculated as the ratio between cleaved RNA and readthrough RNA levels (pA/RT). It is notable that several protected RT bands can be detected in the ΔGB1 lane, because the 3′ probe overlaps with the introduced deletion in this clone. Also, the transfection of clones that contained GB1 deletions, ΔGB1 (h36, 36d, and h37) and ΔGB1/2 (see below), repeatedly resulted in a significant accumulation of readthrough transcripts, hinting at a possible role of this element in transcription termination.
FIG. 3.
G-rich sequence adjacent to the core poly(A) site is critical for basal cleavage activity. A) The structure of the MC1R poly(A) site with the adjacent G-rich sequence is shown. CA and the arrow indicate the primary site of cleavage, DSE represents the downstream sequence element, and the G-rich sequence is shown as an open box named GB1. The position of the G-rich sequence is indicated with numbers, where the site of cleavage is set to 0 and nucleotides in the 3′ flank are numbered in negative values in a 5′ to 3′ direction. GB1 is enlarged below, the hnRNPH binding protein motifs are underlined, and the deletion clones are indicated above the enlarged sequence. B) RNase protection analysis of total RNA isolated from HEK293 cells transiently transfected with the wt, h36, h36-d, and h37 mutant MC1R reporter plasmids (lanes 1 to 4). pA and RT are the same as described in the legend to Fig. 2B. Note that the length of the protected readthrough bands (RT) differs in each lane, because the introduced deletions overlap with the protection probe, which contains wild-type 3′ flanking sequences. C) Representative RNase protection analysis of transfections with wt and the Δ-GB1 deletion construct. Phosphorimager quantitations of cleavage efficiency calculated as the ratio between cleaved and uncleaved transcripts (pA/RT) are indicated at the bottom of the gel, and the numbers are the mean values of three independent experiments with a standard deviation of ±8.6%. pA and RT are the same as described in the legend to Fig. 2B. Note that the protected fragments representing readthrough transcripts are split into several bands due to the deletion of GB1 overlapping with the protection probe.
The above-described results show that a 37-nucleotide-long G-rich sequence (called GB1) located immediately 3′ of the core poly(A) sequences plays a critical role in directing a basal level of cleavage activity in the MC1R poly(A) site.
A second processing enhancer located deep in the 3′ flank is critical for optimal poly(A) site use.
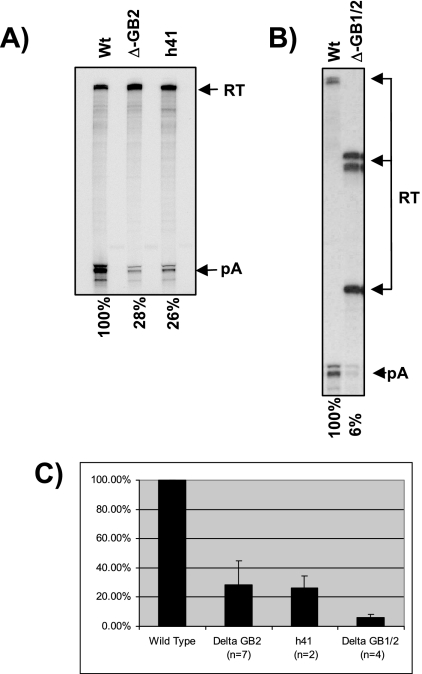
The above-described analysis implied that the GB1 sequence alone is not sufficient to direct optimal cleavage activity at the MC1R poly(A) site. Extra sequences must be present in the 3′ flank which, in conjunction with the core poly(A) signal and GB1, can direct cleavage to levels observed with the wild-type clone. Further support for the presence of such an additional stimulatory region was found when we transfected a number of clones (h36 to h41) that contained the core poly(A) signal, GB1 plus additional 3′-flanking sequences, which gradually increased in length up to 720 nucleotides counted from the site of cleavage (Fig. 4A). RNase protection analysis of total RNA from these transfected clones revealed that despite the presence of GB1 and up to 320 nucleotides of 3′-flanking sequences, the levels of cleaved MC1R RNA did not increase significantly (Fig. 4B, compare the pA band in lane 1 and lanes 2 to 7). In contrast, wild-type cleavage levels were reached with two clones, h45 and h46, that retained more than 450 nucleotides of the MC1R flanking region (Fig. 4C, compare lanes 1 and 2 to lanes 3 and 4). This finding indicated that a second enhancer region located more than 320 nucleotides downstream of the poly(A) cleavage site must be present in the MC1R 3′ flank. Notably, the constructs (h45 and h46) that regain wild-type cleavage levels both contain a second 100-nucleotide-long G-rich element, named GB2 (Fig. 4A), harboring two G-rich clusters. Interestingly, this second G-rich element, like GB1, also contains several hnRNPH binding motifs. To establish that this second G-rich element is a critical processing enhancer, we designed a construct in which sequences located between the h41 and h45 position, including the 100-nucleotide-long second G-rich region, were removed (see Fig. 4A). This clone, named Δ-G2, was then transfected into HEK293 cells together with the h41 deletion clone, and cleavage efficiency relative to that of the wild-type clone was monitored. The results from these transfections are presented in Fig. 5A and C and demonstrate that the removal of the second G-rich box results in a substantial decrease of MC1R mRNA levels (pA/RT) to an average of 28% compared to wild-type mRNA levels. This reduction in cleavage efficiency is similar to that obtained with the h41 deletion clone (Fig. 5C, compare lanes wt, ΔGB2, and h41). In addition, the drop in cleavage activity upon deletion of the second G-rich sequence was similar to the effects observed with GB1 deletion clones (Fig. 3B), suggesting that the two elements act together. Furthermore, note that unlike with GB1 deletion constructs, only a marginal increase in overall readthrough transcripts was observed in the ΔGB2 construct, suggesting that GB2, unlike GB1, may not be a critical element for transcription termination.
FIG. 4.
Second G-rich enhancer is critical for optimal MC1R poly(A) site use. A) Diagram of the MC1R core poly(A) site (AAUAAA and DSE) and the 3′-flanking region. CA and the arrow represent the primary site of cleavage, and the two G-rich regions are indicated in the open boxes named GB1 and GB2. Individual deletion clones (h36 to h46) are depicted above the black line representing the 3′ flank. The numbers of 3′ flank nucleotides that are retained in each individual clone are indicated as negative values relative to the site of cleavage. The sequence of GB2 is enlarged below, and hnRNPH binding sites are underlined. N27 represents 27 nucleotides which separate the two clusters of hnRNPH binding sites in GB2. B) RNase protection analysis of total RNA from transfections of the h36 to h41 deletion clones. The lengths of the RT bands from the individual deletion plasmids differ in size because the introduced deletions overlap with the RNase protection probe. A size marker, M, is present in lane 1, and the length of each fragment is shown on the left. D, P, RT, and pA are the same as described in the legend to Fig. 2B. C) RNase protection of total RNA isolated from transfections with MC1R deletion clones h41, h45, and h46.
FIG. 5.
Deletion of sequences encompassing GB2 result in a dramatic reduction of cleavage efficiency. A) RNase protection analysis of HEK293 cells transiently transfected with a ΔGB2 clone containing an internal deletion spanning the second G-rich element. Quantitations were performed from seven (Δ-GB2) and three (h41) independent experiments, and the mean values are indicated below the gel. Cleavage efficiency was calculated as the pA/RT ratio. B) Representative RNase protection of transfections with a ΔGB1/2 double deletion clone where both GB1 and GB2 are removed. Quantitations (pA/RT) are the mean values of four independent experiments. C) Plot summarizing quantitation with standard deviation of cleavage efficiency of clones that lack GB2 sequences. The number of independent experiments is indicated in brackets below the clone names. Quantitation of RNase protection was carried out using a Fuji phosphorimager.
As mentioned above, GB2 contains two obvious clusters of G-rich sequence elements, and both clusters contain three and five hnRNPH binding motifs, respectively (Fig. 4A). This prompted us to test whether both G clusters located in the second enhancer region are critical for cleavage or if the presence of one group is sufficient to direct optimal cleavage activity. Deletion of either of the GB2 subclusters only had a marginal effect on cleavage and reduced activity by about twofold (data not shown).
The above-described analysis of the deletion clones shows that the removal of either GB1 or GB2 still allows a basal cleavage activity at the MC1R poly(A) site. This is in contrast to the results observed with the h36 deletion clone, which lacks all downstream flanking sequences, including GB1 and GB2, and where cleaved MC1R RNA species were almost undetectable (Fig. 3B). It appears that either of the two G boxes in conjunction with the core poly(A) sequences can direct low levels of cleavage at the 3′ end processing site independent of its position. If this were the case, then a double deletion of GB1 and GB2 should reduce cleavage activity even further. We therefore analyzed the effects on cleavage efficiency from a construct in which both GB1 and GB2 have been removed. The removal of both G-rich sequences had a similar effect on the h36 clone and reduced cleavage efficiency at the MC1R poly(A) site more than 10-fold compared to wt levels (Fig. 5C, compare wt with ΔGB1/2). The further reduction of cleaved mRNA from the double-deletion clone indicates that the presence of a single G-rich enhancer at either of the two locations in the 3′ flank can stimulate minimal cleavage activity at the processing site.
In summary, we have identified a second G-rich cluster positioned far into the 3′-flanking region that is essential for optimal cleavage activity. We show that both G-rich elements act together to enhance poly(A) cleavage efficiency at the MC1R pre-mRNA.
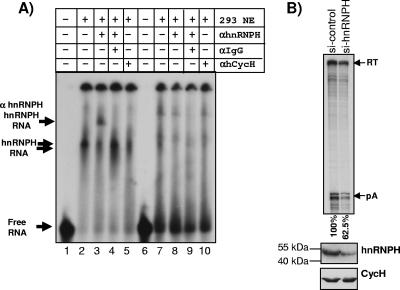
The hnRNPH binding motifs located in GB1 can interact with hnRNPH.
As mentioned earlier, optimal cleavage efficiency at the SV40 late poly(A) site depends on the interaction between hnRNPH and the G-rich element located downstream of the core poly(A) sequences. This prompted us to clarify whether the hnRNPH binding sites that are present in both GB1 and GB2 can interact with hnRNPH/H′ proteins by employing native band shift experiments. We in vitro transcribed the GB1 sequences and incubated the gel-purified RNA in HEK293 nuclear extract (NE). As can be seen in Fig. 6, the GB1 RNA exposed to nuclear extract resulted in a specific shift of the RNA (compare lane 1 with 2, 4, and 5) which is not present in the control RNA of equivalent length consisting of sequences originating from the extreme end of the MC1R 3′-flanking region of the wt reporter plasmid (lanes 6 to 10). In order to verify whether the observed shift is caused by the interaction of hnRNPH/H′ with the radiolabeled G1 RNA and not by any other protein present in the extract, we added a hnRNPH/H′-specific antibody to the reaction mix. Strikingly, the addition of this antibody resulted in a supershift of the RNA-hnRNPH/H′ complex (lane 3) which is clearly absent when an unrelated anti-human cyclin H antibody (lane 5) or control RNA (lane 8) is used instead. Since the observed supershift could be the result of an unspecific interaction between the antibody and the RNA, we incubated αhnRNPH/H′ with the RNA in the absence of nuclear extract. This incubation had no effect on the migration of the RNA (data not shown). Furthermore, the incubation of the GB1 RNA with NE, anti-hnRNPH/H′ antibody, and an additional anti-IgG antibody resulted in the abolition of the RNA-hnRNPH-αhnRNPH complex, which is most likely due to the formation of an additional complex consisting of RNA-hnRNPH-αhnRNPH-αIgG, which would be retained in the slot of the gel (lane 4).
FIG. 6.
A) hnRNPH can interact with in vitro-transcribed GB1 sequences. Shown is an RNA band shift using in vitro-transcribed radiolabeled GB1 RNA (lanes 1 to 5) or control RNA (lanes 6 to 10) and HEK293 nuclear extract (NE). The table above the gel indicates addition (+) or omission (−) of components to the incubation mix: lane 1, no NE added; lanes 2 to 5 and 7 to 10, 0.25 μl nuclear extract added; lanes 3, 4, 8, and 9, 2 μl anti-hnRNPH antibody (αhnRNPH) added; lanes 4 and 9, anti-IgG antibody (αIgG) added; and lanes 5 and 10, 2 μl control anti-human cyclin H antibody (αhCycH) added. Free RNA and the formed complexes RNA-hnRNPH and RNA-hnRNPH-αhnRNPH are indicated by arrows on the left side. B) Upper panel, RNase protection of siRNA-treated HEK293 cells transiently transfected with the MC1R wild-type plasmid. si-control indicates HEK293 cells transfected with nontargeting siRNAs. si-hnRNPH represents HEK293 cells transfected with siRNAs targeting hnRNPH. Quantitation was performed using a Fuji phosphorimager, and the cleavage efficiency was calculated as the pA/RT ratio. Relative values of the processing activity are indicated below the gel and represent mean values of two independent experiments (62% and 63%). Lower panel, Western blot analysis of siRNA-treated HEK293 cells. hnRNPH, Western blot using an anti-hnRNPH-specific antibody; CycH, Western blot using a cyclin H-specific antibody.
The above-described results show that hnRNPH can interact with GB1. However, in order to verify if hnRNPH plays a critical role in the actual processing of the MC1R pre-mRNA, we employed siRNA to knock down hnRNPH protein levels in HEK293 cells. Although we successfully used this approach to knock down hnRNPH to near background levels in control HeLa cells (data not shown), siRNA-treated HEK293 cells always retained some hnRNPH protein (Fig. 6B, 293 hnRNPH). Nevertheless, RNase protection analysis of two independent experiments shows that when hnRNPH protein levels are significantly reduced in HEK293 cells compared to cells transfected with scrambled control siRNAs, cleavage efficiency dropped by 1.6-fold in both experiments. This is comparable to the results obtained in a recent study which showed a similar reduction in the expression of a transiently transfected reporter gene containing the 14-nucleotide SV40 G-rich element in HEK293 cells that were exposed to hnRNPH siRNAs (2).
We conclude from these results that the hnRNPH binding motifs which are present in both GB1 and GB2 are able to interact with hnRNPH/H′ proteins in HEK293 nuclear extract. This suggests that the interaction between hnRNPH/H′ proteins and the GB1/GB2 sequences plays an important role in poly(A) cleavage at the MC1R poly(A) site.
Contiguous uncleaved pre-mRNAs containing the GB2 element can be detected.
As described above, the second enhancer element is located more than 440 nucleotides downstream of the core poly(A) site. This element can only function as an enhancer of cleavage at the processing site if it is transcribed by RNA polymerase II (pol II) as part of a contiguous MC1R transcript. We therefore used a reverse transcription-PCR-based approach to test if uncleaved MC1R pre-mRNAs containing the second G-rich element can be detected. Total RNA isolated from HEK293 cells transfected with the wt MC1R reporter gene was subjected to reverse transcription using four primers (MF1 to MF4) corresponding to sequences located in the 3′ flank as indicated in Fig. 7A. The resulting cDNA products were subsequently amplified using a forward primer (F) positioned in the 3′UTR upstream of the poly(A) cleavage site and a universal reverse primer (R) corresponding to sequences immediately 5′ of GB1. The use of these primers results in PCR products of equal length regardless of the employed reverse transcription primer. As can be seen in Fig. 7B, PCR products of similar intensity are obtained with all used reverse primers, including reverse transcription primers that are complementary to sequences located downstream of GB2 (Fig. 7B, lanes 1, 3, 5, and 7). In contrast, reverse transcription-PCR of RNase-treated samples did not result in any detectable products (Fig. 7B, lanes 2, 4, 6, and 8).
Although the above-described reverse transcription-PCR analysis evidently demonstrates that some MC1R pre-mRNAs contain the poly(A) site and 3′-flanking sequences located downstream of GB2, this experiment does not give a clear indication as to how much pre-mRNA the GB2 sequences contain before cleavage occurs. To address this question, we employed an RNase protection analysis with a 619-nucleotide-long protection probe that contains 596 nucleotides complementary to sequences between 50 nucleotides upstream of the major cleavage site and 5 nucleotides downstream of GB2. As can be seen (Fig. 7C, lane 1), RNase protection with an antisense protection probe (AS) on total RNA isolated from HEK293 cells transiently transfected with the wt-MC1R plasmid results in a single major 596-nucleotide-long readthrough band. The protected band represents pre-mRNAs that are uncleaved at the poly(A) site and contain the 3′-flanking region including GB2. Importantly, there are no additional shorter bands of significant intensity that would correspond to uncleaved pre-mRNAs that lack GB2.
To confirm that the full-length protected band is not the result of a contamination of the total RNA preparation with plasmid DNA, we also performed an RNase protection with a protection probe that is complementary to the same region but is in the sense orientation and hence would only result in a signal if plasmid DNA is present. As can be seen in Fig. 7C, lanes 2 and 4, no bands can be detected in these lanes, confirming that the RNA samples are free of plasmid DNA. We next wanted to clarify if this unusually long protection probe can be efficiently digested using our standard RNase protection protocol. We therefore included the analysis of total RNA from cells transfected with the Δ-GB2 deletion construct (Fig. 7C, lanes 3 and 4). RNase protection of the Δ-GB2 pre-mRNA should give a shorter 368-nucleotide-long protected fragment, because the deletion overlaps with the protection probe. In addition, we also performed a full RNase digestion control without any cellular RNA (Fig. 7C, lane 5). Both controls confirm that the long RNase protection probes can be efficiently digested into smaller fragments or indeed be fully digested if no cRNA is present.
These experiments verify that the MC1R reporter gene can be transcribed at least up to 550 nucleotides past the poly(A) site before cleavage at the processing site occurs. Furthermore, our RNase protection analysis suggests that indeed the vast majority of detectable pre-mRNAs are uncleaved at the poly(A) site and do contain the GB2 sequence element. We therefore conclude that the MC1R gene is transcribed into a contiguous pre-mRNA that contains the GB2 region, which is then available to contribute to cleavage efficiency at the poly(A) site.
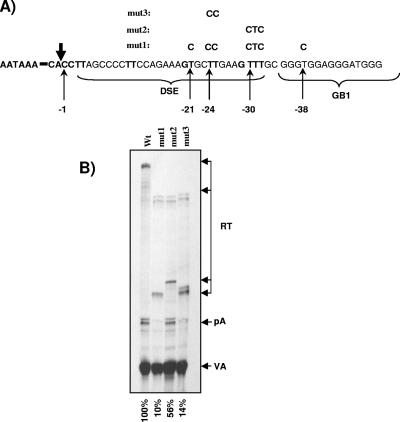
Two consecutive uridine residues located in the DSE are crucial for the recognition of the MC1R poly(A) site.
We demonstrate above that efficient cleavage at the MC1R poly(A) signal requires, in addition to the core poly(A) signal, two G-rich sequences located in the 3′-flanking region. The results obtained with the h36 and the GB1/2 deletion clones suggested that the sequences that constitute the classic core poly(A) signal (AAUAAA and the GU-rich element) are unable to direct significant cleavage activity at the processing site without the G-rich enhancers. This is likely due to the fact that the MC1R DSE is suboptimal and consists of only four pairs of consecutive uridine residues, which could potentially serve as CstF binding sites (15). We therefore wanted to clarify whether the uridine residues located in the DSE play any role in the recognition of the MC1R processing site or if this process is solely dependent on the AAUAAA GB1 and GB2. For this purpose, we designed a set of plasmids which contained intact GB1 and GB2 sequences but had specific U-to-C substitutions in the DSE (Fig. 8A, mut1, mut2, and mut3). Substitution of the uridines at positions −21, −24, −25, −30, −31, −32, and −38 relative to the poly(A) cleavage site reduced cleavage activity at the MC1R processing site to a degree similar to that previously observed with clones that lacked both G-rich sequences (Fig. 8B, mut1). This result confirms that the uridines positioned immediately downstream of the MC1R processing signal, together with the AAUAAA hexamers GB1 and GB2, are absolutely required for poly(A) cleavage at this site.
FIG. 8.
DSE sequences are essential to direct cleavage at the MC1R poly(A) site. A) Detailed picture of the wt MC1R poly(A) site, including the AAUAAA hexamer, the DSE, and GB1. The site of cleavage is indicated by the boldface vertical arrow, and the T-to-C substitutions introduced into the mutant constructs mut1, mut2, and mut3 are indicated by the thin arrows. The positions of the substituted nucleotides relative to the site of cleavage are shown below in negative numbers. B) RNase protection analysis of HEK293 cells transiently transfected with wt, mut1, mut2, and mut3 MC1R plasmids. RT, pA, and VA are the same as indicated in previous figure legends. RT bands from transfections with mutant clones are split into multiple bands due to the overlap of the mutations with the protection probe. Quantitation of RNase protection was carried out using a Fuji phosphorimager. The cleavage efficiency was calculated as the pA/RT ratio. Relative values are indicated below each lane.
We next investigated if all the uridines located in the DSE are required for cleavage or if this degenerate poly(A) sequence still retained some degree of redundancy, which is a characteristic feature of DSEs (59). We therefore designed two more constructs containing U-to-C substitutions of the diuridines at position −24/−25 and the triple uridines at positions −30 to −32, respectively. Interestingly, replacing the GUUU sequence with GCUC only had a marginal effect on cleavage (less than twofold) (Fig. 8B, mut2). In contrast, substitution of the UU at positions −24 and −25 reduced cleavage efficiency by sevenfold (Fig. 8B mut3), highlighting the importance of these two nucleotides for the cleavage activity.
These results demonstrate that the two consecutive uridine residues positioned −24 and −25 nucleotides downstream of the cleavage site are critical for the recognition and cleavage at the MC1R poly(A) site. Surprisingly, the degenerate DSE still retains some degree of redundancy, in that the longest uridine stretch present in the GUUU sequence at positions −30 to −32 appears to play a minor role in the poly(A) site recognition.
DISCUSSION
Cleavage and polyadenylation is a critical processing reaction which is essential for the maturation of pre-mRNA molecules into fully functional mRNA transcripts. Here we investigated how the naturally intronless melanocortin 1 receptor gene is efficiently expressed in spite of the lack of a stimulatory effect splicing exhibits on poly(A) cleavage and despite the presence of a suboptimal DSE in the core processing signal. Our investigation revealed that the MC1R poly(A) site consists of at least three critical elements: the core poly(A) signal constituting the AAUAAA hexamer plus at least one critical diuridine in the DSE and finally two related essential enhancer elements that contain clusters of hnRNPH binding motifs, GB1, immediately adjacent to the core signal, and GB2, located 440 nucleotides downstream of the core site.
A G-rich sequence immediately downstream of the core poly(A) site is critical for poly(A) cleavage.
Little is known about how naturally intronless genes are efficiently cleaved and polyadenylated at their 3′ end without the stimulatory effect of splicing. Most of our knowledge of how such uninterrupted genes are properly processed comes from studies on the expression of unspliced viral transcripts. Many of these viral transcripts contain processing enhancer elements in their 3′UTRs, which have been shown to affect both cleavage efficiency at the poly(A) sites and stimulate nuclear cytoplasmic export (16, 22, 30, 32, 57). Similar functionally homologous sequences have been identified in a few intronless cellular genes (14, 23, 26, 28, 29). In contrast, our results presented in Fig. 2 suggest that the MC1R gene has no similarly positioned enhancer element. Removal of 3′UTR or coding sequences had no effect on cleavage and cytoplasmic accumulation of MC1R reporter mRNA. On the contrary, it appears that efficient cleavage at the MC1R site is solely dependent on the core poly(A) site sequences and two G-rich enhancer sequences located in the 3′-flanking region.
The first G-rich sequence located immediately downstream of the core poly(A) site proved to be essential for efficient cleavage. Similar G-rich enhancer sequences located adjacent to poly(A) sites have previously been reported in viral and cellular genes. The best known example is the GGGGGAGGUGUGGG sequence located immediately downstream of the SV40 late poly(A) signal (3, 4, 12). This enhancer element contains a single GGGA tetranucleotide representing a minimal protein recognition site for several members of the hnRNPH protein family, including hnRNPH/H′/F and 2H9 (11). Importantly, in vitro experiments have shown that hnRNPH can stimulate cleavage of the SV40 poly(A) site by interacting with this 14-bp sequence element if it is positioned immediately downstream of the core poly(A) site (3).
Interestingly, the GB1 processing enhancer in the MC1R pre-mRNA also contains recognition sites for members of the hnRNPH family, and our results indicate that hnRNPH can indeed interact with these binding motifs (Fig. 6). However, MC1R GB1 differs considerably from the SV40 element by harboring clusters of binding sites that are specific for certain hnRNPH family members. GB1 is a 37-nucleotide-long sequence, and it contains three repeats of the minimal sequence (GGGA) sufficient to interact with all members of the hnRNPH family flanked by two single GGGU sequences which can only bind hnRNPH and H′ (10). It is therefore possible that these sequences interact with several members of the hnRNPH protein family, and it is reasonable to suggest that such an interaction plays a role in enhancing cleavage activity at the MC1R poly(A) site (Fig. 3 to 6). This is further strengthened by our observation that cleavage efficiency at the MC1R poly(A) site is significantly reduced in HEK293 cells when hnRNPH protein levels are decreased by siRNA (Fig. 6B).
A second G-rich element located deep in the 3′ flank is required for optimal cleavage efficiency.
It is clear from our results that the presence of GB1 is not sufficient to direct optimal use of the MC1R cleavage and polyadenylation site. The finding that a second distant G-rich element is critical for optimal poly(A) site use is supported by two sets of experiments.
First, the deletion clones that contain 3′ flanks of up to 320 nucleotides but lack the second G-rich sequence clearly fail to direct wild-type cleavage activity. This is in sharp contrast to the deletion clones that still retain this region (Fig. 4).
Second, an internal deletion encompassing an additional G-rich element results in a similar reduction in cleavage activity, as has been observed with the deletion clones h37 to h41. This second G-rich element GB2 has a sequence composition analogous to that of GB1, in that it constitutes several hnRNPH binding motifs. GB2 has two G-rich clusters which are separated by 27 nucleotides. The first stretch consists of two GGGGGC hexanucleotides and one GGGU sequence which are selective for hnRNPH/H′ binding (34). The second cluster consists of 32 nucleotides containing three GGGA elements followed by two GGGU sequences. Interestingly, the deletion of either half of GB2, which effectively removes three and five hnRNPH sites, respectively, from the reporter construct resulted in a modest reduction of cleavage efficiency (data not shown). This indicates that the GB2 sequence has some degree of functional redundancy. It is therefore plausible that GB2 is interacting with multiple hnRNPH proteins and that the binding of multiple hnRNPH molecules has a cumulative effect on cleavage efficiency.
The architecture of the MC1R poly(A) site with an enhancer element located far downstream of the actual site of cleavage is similar to an arrangement found in the human papillomavirus genome. In this virus, six GGGU hnRNPH H-specific binding motifs located 174 nucleotides downstream of the L1 early poly(A) site have recently been shown to regulate tissue- and stage-specific expression of early and late genes. The interaction between these GGGU repeats and hnRNPH is essential for the efficient use of the early processing site (39). This clearly demonstrates that a sequence similar to GB2 located deep in the 3′ flank can act as an enhancer at an upstream poly(A) site via its interaction with hnRNPH proteins.
However, the distant location of GB2 requires that it is transcribed as part of a contiguous uncleaved pre-mRNA in which both the core poly(A) site and GB2 are present. Our reverse transcription-PCR and RNase protection experiments prove that with uncleaved pre-mRNAs containing the core poly(A) signal, GB1 and GB2 can readily be detected within a contiguous pre-mRNA transcript (Fig. 7). Further, these data reveal that RNA pol II transcribes at least 1 kb past the poly(A) site, which is consistent with previous studies showing that it is not unusual for RNA pol II to transcribe far into the downstream 3′-flanking regions before cleavage at the poly(A) site occurs (6, 17). Interestingly, transcription of 3′-flanking regions may be necessary for poly(A) sites to function. A recent investigation on the cotranscriptional nature of the cleavage event at poly(A) sites suggests that the stable assembly of a functional poly(A) complex may be dependent on the tether between the downstream-positioned 3′ flank RNA and RNA pol II. Maintenance of this tether would allow the transition of poly(A) factors associated with the C-terminal domain of the large subunit of RNA pol II to the poly(A) signal at a time when the polymerase is transcribing the 3′ flank (44). This tether-based model could explain how the second MC1R element can affect the cleavage efficiency from such a distant position. GB2 may perhaps act by promoting the offloading of poly(A) factors from the C-terminal domain onto the pre-mRNA, possibly with the help of hnRNPH proteins. In addition, it is plausible that GB2 is positioned in close proximity to the actual processing site by means of folding the pre-mRNA sequences between GB1 and GB2. A similar situation occurs for the human T-cell leukemia virus I, where folding of the 255-nucleotide-long REX-responsive element juxtaposes the AAUAAA hexamer with a distant downstream sequence element (1).
The role of the uridine residues in the MC1R DSE.
Our investigation revealed that the two uridine residues located 24 and 25 nucleotides downstream of the poly(A) site are central for efficient cleavage at the processing site. The substitution of these two nucleotides significantly reduced cleavage efficiency. Recently published work suggests that CstF64 interacts with the DSE by recognizing UU dinucleotides (15, 40) within a GU-rich environment. We therefore speculate that uridines 24 and 25 are the major contact point between the MC1R DSE sequence and the CstF64 subunit. We further suggest that CstF64 is unable to efficiently recognize the MC1R DSE because it lacks a distinct GU or U rich sequence. Instead, the CstF subunit may be recruited to the pre-mRNA via the two G-rich sequence elements, possibly as a consequence of hnRNPH binding. Subsequently, the cleavage and stimulation factor would then be anchored onto the U24U25 dinucleotide in the DSE.
GB1 may affect transcription termination in the MC1R transcription unit.
We suspect that GB1, unlike GB2, not only affects MC1R expression at the level of 3′ end processing but also controls transcription termination. This hypothesis is supported by the observation that readthrough RNA levels sharply increase with constructs that lack GB1 (Fig. 3 and 5) due to read-around transcription. We believe that the deletion of GB1, similar to the recently described G-rich MAZ4 pause sites (21), can also affect transcription termination at the end of the MC1R transcription unit. It has been suggested that the MAZ4 sites positioned close to a poly(A) site causes RNA polymerase to pause and thus promotes more efficient transcription termination facilitated by the 5′- to 3′-specific exoribonuclease Xrn2 (21). However, it is unlikely that GB1 acts as a pause site that facilitates 5′ to 3′ exonuclease digestion of the polymerase-associated cleaved transcript, as the second G-rich box located 400 nucleotides downstream has to be transcribed prior to cleavage at the poly(A) site.
We speculate that GB1 facilitates termination by promoting the recruitment of the 5′ to 3′ exonuclease Xrn2 to the cleaved RNA. This hypothesis is plausible when one takes into consideration that GB1 has the potential to form a G-quadruplex structure (for a review, see reference 47). The formation of this structure may induce efficient termination by presenting an optimal substrate to the exoribonuclease. Interestingly, the mouse Xrn1 exonuclease, the cytoplasmic close relative of the nuclear exonuclease Xrn2, has been shown to have a preference for G4 tetraplex RNA structures (5).
The presence of a sequence that can both enhance cleavage efficiency and transcription termination at the end of the MC1R gene could indeed be important to prevent transcription interference at the downstream-positioned gene. In this context it is worth noting that the MC1R gene is positioned unusually close to its downstream neighboring tubulin beta III gene (TUBB3). The MC1R poly(A) site and the ATG start codon of the TUBB3 open reading frame are separated by only 2,500 nucleotides. It is therefore highly likely that TUBB3 expression would be seriously affected by transcriptional interference without efficient termination soon after cleavage at the MC1R poly(A) site.
Cell type-specific expression of MC1R reporter mRNA.
We initially intended to analyze the control of MC1R gene expression in HeLa cells, but to our surprise we found that the MC1R poly(A) site in a transiently transfected CMV-driven reporter gene is not efficiently used in this cell line. In contrast, in both SK23 melanoma-derived cells and HEK293 cells, processed mature reporter transcripts are readily detectable (Fig. 1). Our data suggest that the lack of MC1R mRNA in this cell type is a direct consequence of reduced cleavage efficiency. This is comparable to findings presented in a recent study that analyzed the effects of three processing enhancer elements located in the mRNAs of viral and an intronless cellular transcript in different cell lines. All three tested elements enhanced poly(A) cleavage in a β-globin cDNA reporter gene only in selected cell lines (23). It appears that 3′ processing enhancers may frequently be used to contribute to the control of cell- or tissue-specific gene expression, and it is likely that the cell type-specific use of the melanocortin poly(A) site in the reporter plasmid reflects an additional mechanism to control tissue-specific expression of the MC1R gene. Two possible molecular mechanisms could account for this specific poly(A) site use.
First, the cell-specific processing of the MC1R poly(A) site may simply be due to different levels of poly(A) factors in the three tested cell types. Indeed, differential levels of CstF64 have long been known to affect alternative poly(A) site usage in the immunoglobulin M (IgM) pre-mRNA processing (48). Likewise, it may be that increased CstF64 levels would allow a more efficient recognition of the suboptimal MC1R poly(A) signal.
Second, a recent study showed that hnRNPF bound to pre-mRNA can inhibit the cleavage and polyadenylation reaction at the promoter-proximal poly(A) site in the alternatively polyadenylated mouse IgM pre-mRNA. It was further demonstrated that hnRNPF and hnRNPH protein levels differ between plasma and mature B cells. Based on these observations, a model was proposed in which hnRNPF acts as an inhibitor of the promoter-proximal poly(A) site in mature B cells and hnRNPH/H′ as an activator of processing at the more distal poly(A) in plasma cells (52). Analogous to this model, it is plausible that the balance between the binding of the positive processing enhancer hnRNPH/H′ and the negative processing factor hnRNPF to their corresponding motifs in GB1 and GB2 regulates the use of the MC1R processing site in a cell type-specific manner.
Weak core poly(A) sites may frequently contain enhancer sequences located deep in the flanking regions which significantly strengthen their use.
The presence of regulatory elements located 440 nucleotides downstream of the 3′ processing site highlights the importance of these flanking regions within transcription units. It is possible that poly(A) sites which so far have mostly been ignored containing weak core processing sequences might regularly be strengthened by sequences located far downstream in the 3′ flank. In addition, the presence of distant enhancer elements may be a general mechanism to enhance weak poly(A) sites in a tissue-specific manner, and it may represent a possible mechanism that allows intronless genes to efficiently cleave and polyadenylate pre-mRNAs independent of splicing.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Ahmed, Y. F., G. M. Gilmartin, S. M. Hanly, J. R. Nevins, and W. C. Greene. 1991. The HTLV-I Rex response element mediates a novel form of mRNA polyadenylation. Cell 64:727-737. [DOI] [PubMed] [Google Scholar]
- 2.Alkan, S. A., K. Martincic, and C. Milcarek. 2006. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem. J. 393:361-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arhin, G. K., M. Boots, P. S. Bagga, C. Milcarek, and J. Wilusz. 2002. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 30:1842-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagga, P. S., L. P. Ford, F. Chen, and J. Wilusz. 1995. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 23:1625-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashkirov, V. I., H. Scherthan, J. A. Solinger, J. M. Buerstedde, and W. D. Heyer. 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136:761-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauren, G., S. Belikov, and L. Wieslander. 1998. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 12:2759-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brackenridge, S., H. L. Ashe, M. Giacca, and N. J. Proudfoot. 1997. Transcription and polyadenylation in a short human intergenic region. Nucleic Acids Res. 25:2326-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brackenridge, S., and N. J. Proudfoot. 2000. Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol. Cell. Biol. 20:2660-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, K. M., and G. M. Gilmartin. 2003. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol. Cell 12:1467-1476. [DOI] [PubMed] [Google Scholar]
- 10.Buratti, E., M. Baralle, L. De Conti, D. Baralle, M. Romano, Y. M. Ayala, and F. E. Baralle. 2004. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHbeta genes. Nucleic Acids Res. 32:4224-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caputi, M., and A. M. Zahler. 2001. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem. 276:43850-43859. [DOI] [PubMed] [Google Scholar]
- 12.Chen, F., and J. Wilusz. 1998. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 26:2891-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 14.Conrad, N. K., and J. A. Steitz. 2005. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 24:1831-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deka, P., P. K. Rajan, J. M. Perez-Canadillas, and G. Varani. 2005. Protein and RNA dynamics play key roles in determining the specific recognition of GU-rich polyadenylation regulatory elements by human Cstf-64 protein. J. Mol. Biol. 347:719-733. [DOI] [PubMed] [Google Scholar]
- 16.Donello, J. E., J. E. Loeb, and T. J. Hope. 1998. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J. Virol. 72:5085-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dye, M. J., and N. J. Proudfoot. 2001. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105:669-681. [DOI] [PubMed] [Google Scholar]
- 18.Furger, A., J. Monks, and N. J. Proudfoot. 2001. The retroviruses human immunodeficiency virus type 1 and Moloney murine leukemia virus adopt radically different strategies to regulate promoter-proximal polyadenylation. J. Virol. 75:11735-11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furger, A., J. M. O'Sullivan, A. Binnie, B. A. Lee, and N. J. Proudfoot. 2002. Promoter proximal splice sites enhance transcription. Genes Dev. 16:2792-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil, A., and N. J. Proudfoot. 1987. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′ end formation. Cell 49:399-406. [DOI] [PubMed] [Google Scholar]
- 21.Gromak, N., S. West, and N. J. Proudfoot. 2006. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 26:3986-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guang, S., A. M. Felthauser, and J. E. Mertz. 2005. Binding of hnRNP L to the pre-mRNA processing enhancer of the herpes simplex virus thymidine kinase gene enhances both polyadenylation and nucleocytoplasmic export of intronless mRNAs. Mol. Cell. Biol. 25:6303-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guang, S., and J. E. Mertz. 2005. Pre-mRNA processing enhancer (PPE) elements from intronless genes play additional roles in mRNA biogenesis than do ones from intron-containing genes. Nucleic Acids Res. 33:2215-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall-Pogar, T., H. Zhang, B. Tian, and C. S. Lutz. 2005. Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res. 33:2565-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, J. U. N., C. S. Lutz, J. Wilusz, and B. I. N. Tian. 2005. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11:1485-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, Y., and G. G. Carmichael. 1997. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1-related mRNAs. Proc. Natl. Acad. Sci. USA 94:10104-10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, Y., R. Gattoni, J. Stevenin, and J. A. Steitz. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11:837-843. [DOI] [PubMed] [Google Scholar]
- 28.Huang, Y., and J. A. Steitz. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7:899-905. [DOI] [PubMed] [Google Scholar]
- 29.Huang, Y., K. M. Wimler, and G. G. Carmichael. 1999. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 18:1642-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, Z. M., and T. S. Yen. 1995. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol. Cell. Biol. 15:3864-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyburz, A., A. Friedlein, H. Langen, and W. Keller. 2006. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol. Cell 23:195-205. [DOI] [PubMed] [Google Scholar]
- 32.Liu, X., and J. E. Mertz. 1995. HnRNP L binds a cis-acting RNA sequence element that enables intron-dependent gene expression. Genes Dev. 9:1766-1780. [DOI] [PubMed] [Google Scholar]
- 33.Lou, H., D. M. Helfman, R. F. Gagel, and S. M. Berget. 1999. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 19:78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markovtsov, V., J. M. Nikolic, J. A. Goldman, C. W. Turck, M. Y. Chou, and D. L. Black. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDevitt, M. A., R. P. Hart, W. W. Wong, and J. R. Nevins. 1986. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 5:2907-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreira, A., Y. Takagaki, S. Brackenridge, M. Wollerton, J. L. Manley, and N. J. Proudfoot. 1998. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 12:2522-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niwa, M., C. C. MacDonald, and S. M. Berget. 1992. Are vertebrate exons scanned during splice-site selection? Nature 360:277-280. [DOI] [PubMed] [Google Scholar]
- 38.Niwa, M., S. D. Rose, and S. M. Berget. 1990. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4:1552-1559. [DOI] [PubMed] [Google Scholar]
- 39.Oberg, D., J. Fay, H. Lambkin, and S. Schwartz. 2005. A downstream polyadenylation element in human papillomavirus type 16 L2 encodes multiple GGG motifs and interacts with hnRNP H. J. Virol. 79:9254-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez Canadillas, J. M., and G. Varani. 2003. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 22:2821-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Proudfoot, N. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16:272-278. [DOI] [PubMed] [Google Scholar]
- 42.Qiu, J., R. Nayak, and D. J. Pintel. 2004. Alternative polyadenylation of adeno-associated virus type 5 RNA within an internal intron is governed by both a downstream element within the intron 3′ splice acceptor and an element upstream of the P41 initiation site. J. Virol. 78:83-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rees, J. L. 2004. The genetics of sun sensitivity in humans. Am. J. Hum. Genet. 75:739-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rigo, F., A. Kazerouninia, A. Nag, and H. G. Martinson. 2005. The RNA tether from the poly(A) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol. Cell 20:733-745. [DOI] [PubMed] [Google Scholar]
- 45.Ryan, K., O. Calvo, and J. L. Manley. 2004. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10:565-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheets, M. D., S. C. Ogg, and M. P. Wickens. 1990. Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 18:5799-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simonsson, T. 2001. G-quadruplex DNA structures-variations on a theme. Biol. Chem. 382:621-628. [DOI] [PubMed] [Google Scholar]
- 48.Takagaki, Y., and J. L. Manley. 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell 2:761-771. [DOI] [PubMed] [Google Scholar]
- 49.Valsamakis, A., N. Schek, and J. C. Alwine. 1992. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol. Cell. Biol. 12:3699-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valsamakis, A., S. Zeichner, S. Carswell, and J. C. Alwine. 1991. The human immunodeficiency virus type 1 polyadenylylation signal: a 3′ long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc. Natl. Acad. Sci. USA 88:2108-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venkataraman, K., K. M. Brown, and G. M. Gilmartin. 2005. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 19:1315-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veraldi, K. L., G. K. Arhin, K. Martincic, L. H. Chung-Ganster, J. Wilusz, and C. Milcarek. 2001. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 21:1228-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 54.Wickens, M., and P. Stephenson. 1984. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3′ end formation. Science 226:1045-1051. [DOI] [PubMed] [Google Scholar]
- 55.Yonaha, M., and N. J. Proudfoot. 1999. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell 3:593-600. [DOI] [PubMed] [Google Scholar]
- 56.Yonaha, M., and N. J. Proudfoot. 2000. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 19:3770-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zang, W. Q., and T. S. Yen. 1999. Distinct export pathway utilized by the hepatitis B virus posttranscriptional regulatory element. Virology 259:299-304. [DOI] [PubMed] [Google Scholar]
- 58.Zarudnaya, M. I., I. M. Kolomiets, A. L. Potyahaylo, and D. M. Hovorun. 2003. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 31:1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao, X., D. Oberg, M. Rush, J. Fay, H. Lambkin, and S. Schwartz. 2005. A 57-nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2, and polypyrimidine tract binding protein. J. Virol. 79:4270-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]