Abstract
Signaling through receptor protein tyrosine phosphatases (RPTPs) can influence diverse processes, including axon development, lymphocyte activation, and cell motility. The molecular regulation of these enzymes, however, is still poorly understood. In particular, it is not known if, or how, the dimerization state of RPTPs is related to the binding of extracellular ligands. Protein tyrosine phosphatase σ (PTPσ) is an RPTP with major isoforms that differ in their complements of fibronectin type III domains and in their ligand-binding specificities. In this study, we show that PTPσ forms homodimers in the cell, interacting at least in part through the transmembrane region. Using this knowledge, we provide the first evidence that PTPσ ectodomains must be presented as dimers in order to bind heterophilic ligands. We also provide evidence of how alternative use of fibronectin type III domain complements in two major isoforms of PTPσ can alter the ligand binding specificities of PTPσ ectodomains. The data suggest that the alternative domains function largely to change the rotational conformations of the amino-terminal ligand binding sites of the ectodomain dimers, thus imparting novel ligand binding properties. These findings have important implications for our understanding of how heterophilic ligands interact with, and potentially regulate, RPTPs.
Many cell-signaling events are regulated through reversible tyrosine phosphorylation of proteins. This phosphorylation cycle is controlled by the counterbalanced actions of two enzyme families, protein tyrosine kinases and protein tyrosine phosphatases (PTPs), each of which has cytoplasmic and receptor-like members. While the regulation and actions of many receptor protein tyrosine kinases (RTKs) are well characterized, our related understanding of the receptor-type PTPs (RPTPs) remains far from complete.
Twenty-one human RPTPs have been identified, and highly conserved orthologues and homologues exist in vertebrates and invertebrates (3). Several of these RPTPs have particularly well-documented roles in neural development (16, 26, 40), cell adhesion, and motility (7, 51). RPTPs are type I transmembrane proteins with varied extracellular domain structures, and in many cases, it is still unclear what their ligands are. It is also unclear how, and in most cases if, such ligands directly control RPTP activity. Moreover, although RTKs are obligate dimers during ligand activation and signaling, dimerization has been demonstrated for only a small number of RPTPs, and we have only a few clues as to how this dimerization is linked to signaling control. Furthermore, it is not clear whether ligand binding is influenced by, or influences, the RPTP dimerization state.
The RPTPs PTPα, Sap-1, and CD45 form dimers normally in the cell, and this dimerization can block the catalytic action of these enzymes (23, 24, 64). PTPα and CD45 may be inhibited by steric constraints, possibly through an inhibitory wedge structure (8, 29), although this is still controversial (35). With PTPα and Sap-1, there is also evidence that dimerization and the blockade of catalytic function are stabilized, or even initiated, by oxidation and that cis-dimerization can involve cysteine cross-linkage (57, 59). PTPμ can also dimerize, but the influence of this event over enzyme activity is not yet known (12).
Understanding how RPTP dimerization is related to enzyme inhibition is of great importance, given that the monomeric forms of RPTPs appear to be highly active (14, 30, 37, 54). The selective suppression of RPTP activity may indeed be essential in cells. Several studies have shown that negative regulation of PTPσ may enhance nerve growth and even regeneration in adult tissues (32, 46, 56). Similarly, the negative regulation of a Caenorhabditis elegans RPTP, clr-1, is required for the correct netrin-mediated guidance of anterior ventral microtubule axons in the worm (9), and inhibition of CD45 through dimerization is necessary for normal lymphoid homeostasis (31).
Whereas a relationship between RPTP dimerization and enzyme activity is documented, the relationship between RPTP dimerization and ligand interactions is less clear. Several RPTPs interact homophilically, and in one instance, PTPμ, the interaction model places this enzyme in a multimeric interaction interface, with PTPμ binding both in cis and in trans (4, 12). It is not yet clear if cis interactions influence the ability of PTPμ to undergo trans interactions. One of the heterophilic ligands of PTPζ, pleiotrophin, can suppress catalytic activity. While it is conceivable that this occurs by inducing PTPζ dimerization, this has not been directly demonstrated. Recently, an isoform of LAR called LARFN5C, consisting of a single fibronectin type III (FNIII) domain, has been shown to bind homophilically to LAR and may activate it, possibly by favoring the monomer state of LAR (65). If the biological functions of RPTPs are to be better understood, then the relationships between ligand binding, RPTP dimerization, and RPTP signaling activity must be defined more clearly. Similarly, the structural basis for the differential interactions of RPTP protein isoforms with tissue-specific ligands, as seen with LAR (36) and PTPσ (45), for example, must be explained.
In this study, we first investigated the potential dimeric behavior of PTPσ. PTPσ is a large, highly conserved cell adhesion molecule-like RPTP that binds several unrelated extracellular ligands (5, 27, 45). PTPσ functions in the nervous system to control axon development and regeneration (42, 44, 46, 49, 56), the maturation and myelination of nerves (15, 33, 60), and the development of the hypothalamopituitary axis (6, 15, 60). Nevertheless, little is known about either the biochemical action of this enzyme or the mechanism of ligand interactions. Here, we demonstrate for the first time that PTPσ exists in a dimeric state in the cell. Significantly, we have also found that only dimeric, and not monomeric, forms of the receptor are able to bind extracellular ligands. Finally, we propose a structural model that may explain the distinct specificities of ligand binding exhibited by different PTPσ isoforms.
MATERIALS AND METHODS
Epitope tags, mutations, and deletions.
The introduction of cysteines in wild-type chicken PTPσ was carried out using large-template site-directed mutagenesis (62). Epitope tags were introduced either directly, using insertion of oligonucleotides, or by subcloning of chicken PTPσ in the expression vector p3xFLAG-CMV25 (Sigma Aldrich). The FNIII domain deletion series was generated by inserting KpnI sites between each pair of FNIII domains and fusing these sites to a KpnI site in pAPTag2 (10), generating in-frame fusion proteins with placental alkaline phosphatase (PLAP). The fusion points of the sequences were as follows (PTPσ sequences are in boldface): FN0d-AP, VAQ311GTIAIAYVR; FN1d-AP, ESV403GTIAIAYVR; FN2d-AP, IQV504GTIAIAYVR; FN3d-AP, VRE598GTIAIAYVR; FN4d-AP, VIV699GTIAIAYVR; FN5d-AP, PKV802GTIAIAYVR; FN6d-AP, QEL897GTIAIAYVR; FN7d-AP, VQY998GTIAIAYVR; and FN8d-AP, KPE1098GTIAIAYVR. The fusion point of PTPσ1-AP and PTPσ2-AP is VKSSVAVQA1124YVR (19). The single hemagglutinin (HA) tag was inserted at the carboxy-terminal end of PLAP by insertion of the sequence 5′-AGATCGATCTACCCATACGACGTCCCAGACTACGCTGCCTAG-3′ downstream of the BspEI site at the 3′ end of the PLAP gene in the secretable PLAP vector Aptag4 (10). To delete most of the ectodomain, generating protein ICD1, PCR was used to amplify bp 2533 to the stop codon of PTPσ1, and this fragment was ligated between the BglII and XbaI sites of p3XFLAG-myc-CMVTM-25 (Sigma). The site of 5′ fusion at the BglII site has the sequence 5′-AGATCTGATTGATGGAGAGGAA-3′. To delete most of the intracellular domain, generating ECD1, a PCR product containing bp 85 to bp 2650 of PTPσ1 was synthesized using PCR and inserted between the EcoRI and XbaI sites of p3XFLAG-myc-CMVTM-25 (Sigma); this vector supplies a peptide secretion signal upstream of the triple-copy FLAG. The 5′ site of fusion was 5′-GAATTCGGAAAGTCCCC-3′, and the 3′ site was 5′-AACGGAAAGTCTAGA-3′ (the vector sequence is in boldface). The flexible linker was created by annealing the oligonucleotides 5′-GTACGGGCGGAGGAGGATCCGGAGGTGGCGGTTCACCGG-3′ (sense) and 5′-GTACCCGGTGAACCGCCACCTCCGGATCCTCCTCCGCCC-3′ (antisense) and inserting them in frame into an Asp718 (Kpn1) site in FN5d-AP and FN6d-AP.
Transfections, immunoprecipitation, immunofluorescence, and PLAP assays.
DNA vectors were transfected into HEK293T cells using standard calcium phosphate methods. For the cysteine mutations, cells were lysed in 1% Nonidet P-40, 150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 2 mM EDTA, 7 μg/ml pepstatin A, 1.5 μg/ml aprotinin, 2 mM phenylmethylsulfonyl fluoride with or without 20 mM iodoacetamide. For other proteins, cells were lysed in 50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100 plus protease inhibitors (Complete; Roche). For immunoprecipitation, lysates were incubated with antibodies specific for FLAG (Clone M2; Sigma Aldrich, United Kingdom), HA (3F10 [Roche] or HA-7 [Sigma Aldrich, United Kingdom]), or myc (9E10; Sigma Aldrich, United Kingdom) epitopes, and antibody-antigen complexes were purified using protein G Sepharose (Upstate).
For immunofluorescence, cells were fixed 3 days posttransfection in ice-cold 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min, rinsed in PBS, and quenched for 5 min in 2% glycine-PBS. The fixed cells were permeabilized for 5 min in 0.1% Triton X-100, 0.05% sodium dodecyl sulfate (SDS)-PBS. Coverslips were incubated with primary anti-HA and anti-FLAG for 45 min at 37°C. After the PBS washes, secondary anti-rat tetramethyl rhodamine isothiocyanate-conjugated or anti-mouse-fluorescein isothiocyanate-conjugated antibodies (Dako, United Kingdom) were added. The coverslips were washed and mounted with Fluorsave (Calbiochem).
Analysis of disulfide dimers.
Cells were transfected and lysed as described above in the presence of 20 mM iodoacetamide. Protein concentrations were estimated using the Bradford method. After polyacrylamide gel electrophoresis (PAGE), the gels were transferred to polyvinylidene difluoride membranes in 25 mM Tris, 192 mM glycine, 20% methanol. For two-dimensional (2D) gel analysis, protein samples were first subjected to 1D PAGE as described above, except that samples were boiled in 5% SDS, 25% glycerol, 156 mM Tris-HCl, pH 6.8, 50 mM N-ethylmaleimide for 30 s beforehand. Following 1D PAGE, the gels were stained with zinc (Bio-Rad Zinc Stain [18]), and the lanes were excised; destained; incubated in 1× Laemli with or without 5% β-mercaptoethanol plus 50 mM N-ethylmaleimide at 100°C for 5 min; washed twice in 1% SDS, 10% glycerol, 62.5 mM Tris-HCl, pH 6.8, for 5 min to remove reducing agents; and then cast on top of a 4% stack-6% resolving gel.
Immunoblotting.
After the gels were transferred to polyvinylidene difluoride membranes, the membranes were blocked with 10% dry milk powder in Tris-buffered saline overnight before the addition of antibody for 1 h. The primary antibodies used were anti-FLAG (M2; Sigma); anti-HA (HA-7; Sigma); and IG2, anti-PTPσ (52). The filters were washed three times in Tris-buffered saline plus 0.2% Tween 20 before the addition of horseradish peroxidase-conjugated secondary antibodies for 1 h. Horseradish peroxidase was detected with ECL+ (Amersham Biosciences, United Kingdom).
Bis(sulfosuccinimidyl)suberate cross-linking.
A PTPσ1 cDNA was cloned into pTre2 (Clontech) and transfected into PC12 Tet-On cells (Clontech). One selected clone, 12CRYP11, exhibited background levels of PTPσ expression and 5× to 10× inducible expression. Cells were cultured in Dulbecco's modified Eagle's medium, 10% horse serum, 5% fetal calf serum on a collagen IV substrate. The medium was replaced with PBS containing 5 mM bis(sulfosuccinimidyl)suberate (Pearce, Rockford, IL) for 30 min at 4°C, and then Tris-Cl (pH 7.5) was added to a final concentration of 20 mM for 15 min at room temperature. The cells were then washed briefly in ice-cold PBS and lysed in 50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100 plus protease inhibitors (Complete; Roche). Equal amounts of nonreduced lysates were separated by PAGE and immunoblotted using IG2 serum raised against the PTPσ ectodomain (52).
Purification of FN3d-AP and PLAP-HA.
One-milliliter aliquots of anti-PLAP agarose and anti-HA agarose (Sigma Aldrich) were packed into fast protein liquid chromatography columns (Amersham Biosciences). The columns were equilibrated using 5 column volumes of 0.05 M Tris, 0.5 M NaCl (pH 8.0), and 200 ml of precleared, conditioned medium was loaded onto the column. The columns were washed with 5 column volumes of the equilibration buffer, and the bound components were eluted with 0.05 M glycine, 0.5 M NaCl (pH 2.8). Fractions (500 μl) were neutralized with 50 μl of 1.0 M Tris-HCl (pH 9.0), pooled, and desalted using PD10 desalting columns (Amersham Biosciences), and the protein purity was determined by SDS-PAGE on a 6% Tris-glycine gel stained with a PlusOne silverstaining kit (Amersham Biosciences). Immunoblotting was done with the anti-PTPσ antibody IG2 (52). Band intensities were measured using a GS-800 calibrated densitometer and Quantity One software (Bio-Rad).
RAP and solid-phase alkaline phosphatase assays.
Receptor affinity probe (RAP) affinity assays were performed with PLAP fusion proteins as previously described (20). PLAP fusion proteins were first collected in cell culture supernatants, and aliquots were subjected to PAGE before immunoblotting and detection with the anti-PTPσ serum IG2 (52), confirming the fusion protein size and integrity. Solid-phase assays were carried out using plasma polymer-prepared 96-well microplates (Plasso Technology Ltd., Sheffield, United Kingdom; product code P100) designed to bind heparin. Duplicate wells were incubated overnight at 4°C with 200 μl of heparin (20 μg/ml) per well. Bovine serum albumin (BSA)-PBS (2%) was used as a negative control for coating. After the plates were washed, a blocking solution of 5% BSA-PBS-0.05%Tween 20 was added at 37°C for 1 h; 200 μl of fusion protein was then added for 1 h at room temperature. After the plates were washed, 200 μl of secreted alkaline phosphatase buffer containing 12 mM p-nitrophenyl phosphate was added. The PLAP activity was measured for 20 min at 405 nm at room temperature, mean values were taken, and control values from BSA-coated wells were used to normalize all values.
RESULTS
Construction of PTPσ variants with unpaired cysteines.
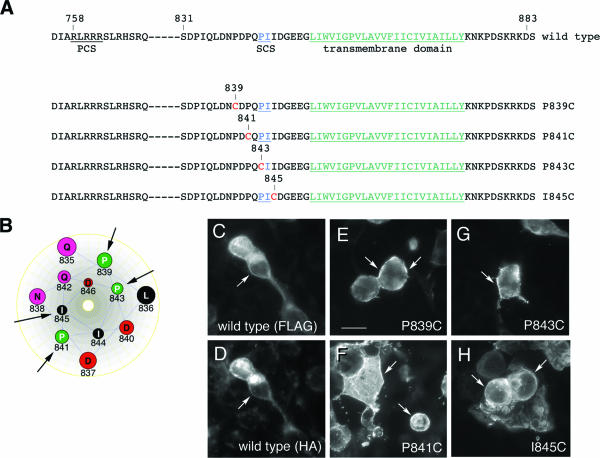
To determine whether PTPσ can exist in a homodimeric state in live cells, unpaired cysteines were introduced in the extracellular juxtamembrane region using site-directed mutagenesis. Cysteines were introduced at four sites, all carboxy terminal to the primary, furin-like protease cleavage site predicted in PTPσ (47) (Fig. 1A). These cysteine positions should also be amino terminal to a putative secondary matrix metalloproteinase cleavage site detected in human PTPσ (amino acids 843 to 844) (47), except for mutation sites 843 and 845, which would be within this site (Fig. 1A). If PTPσ forms dimers, the unpaired cysteines may form covalent linkages, thus “trapping” the protein in the dimeric state. The cysteines were placed 2 amino acids apart to try to ensure that several rotational faces of the peptide structure would be exposed to potential cross-linking (Fig. 1B). The proteins were expressed in HEK293T cells and tested for correct expression on the cell surface. Under the conditions used, the cells were mostly fusiform or rounded in phenotype, and the expression of wild-type (wt) PTPσ and the mutants had no obvious effect on this phenotype. Figure 1 shows that each protein had prominent cell surface localization similar to that seen with wt PTPσ (Fig. 1E to H). The wt PTPσ protein gave similar surface localization when detected with either extracellular or intracellular epitopes (Fig. 1C and D).
FIG. 1.
PTPσ variants containing unpaired cysteine residues. (A) Amino acid sequences are shown for wt PTPσ and the mutated forms P839C, P841C, P843C, and I845C. PCS shows the predicted site of furin-like enzyme cleavage; SCS indicates the site of secondary cleavage by a metalloprotease, occurring in the rat homologues between P843 and I844; the transmembrane domain is underlined in green. (B) A hypothetical helical-wheel representation of the juxtamembrane region of PTPσ showing the sites (arrows) of cysteine mutagenesis (created with Lasergene). (C to H) Indirect-immunofluorescence detection of PTPσ and mutated variants. FLAG-tagged forms of wt PTPσ and the mutated forms P839C, P841C, P843C, and I845C were transfected into 293T cells and detected by indirect immunofluorescence using anti-FLAG primary antibodies and fluorescein isothiocyanate-labeled secondary antibodies. Panel D shows the same field as panel C, but under tetramethyl rhodamine isothiocyanate excitation, detected with anti-HA. Cell surface labeling was detected with each protein (arrows). Scale bar = 10 μm.
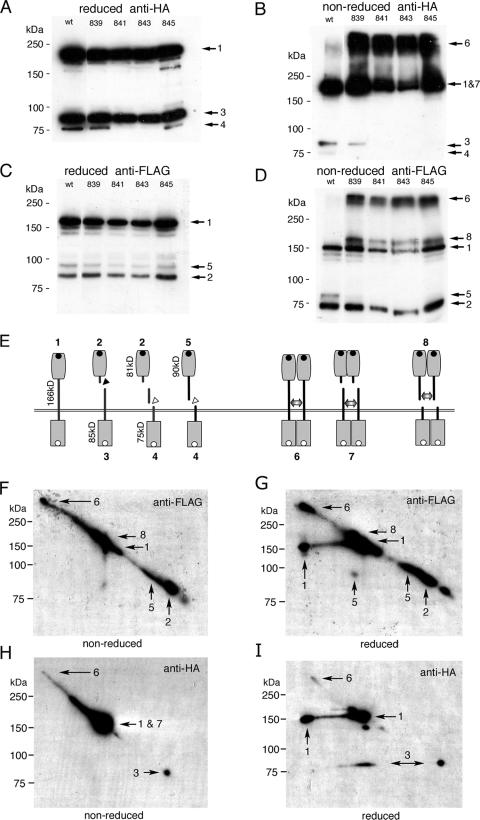
The variant PTPσ proteins were then analyzed by immunoblotting. The pattern of wt PTPσ expression in HEK293T cells was complex, and a schematic of the predicted fragments is shown in Fig. 2E. In tissues, PTPσ1 is thought to be cleaved at a conserved basic region in the ectodomain (Fig. 1A [PCS] and 2E), which is a site found in all LAR-related RPTPs (41). Cleavage produces a membrane-spanning fragment of ∼85 kDa (Fig. 2A, HA band 3, and E, fragment 3) and an 81-kDa ectodomain fragment (Fig. 2C, FLAG band 2, and E, fragment 2). Upon overexpression of PTPσ1, two further proteins were seen. First, a full-length uncleaved protein (theoretical mass, 166 kDa) was generated (Fig. 2A; C, band 1; and E, protein 1), probably due to the inability of cells to process all of the overexpressed protein. A second cleavage event was also observed at various levels between experiments, producing an HA-tagged band of 75 kDa (Fig. 2A, HA band 4, and E, fragment 4). A similar cleavage event has been described for LAR-like RPTPs and usually occurs subsequent to the primary cleavage (1, 47). However, we also observed a FLAG-tagged protein of 90 kDa, which we predict has undergone secondary cleavage without prior primary cleavage (Fig. 2C, FLAG band 5, and E, fragment 5). Although, again, this may be partly an artifact of overexpression, it nonetheless proved useful in analyzing the cysteine dimer data (see below). Under reducing conditions, each mutated protein exhibited a proteolytic pattern similar to that of wt PTPσ (Fig. 2A and C).
FIG. 2.
Cleavage and dimerization of PTPσ with cysteine mutations. Wt PTPσ and its mutated forms P839C, P841C, P843C, and I845C were expressed in HEK293T cells and detected with anti-HA (A and B) or anti-FLAG (C and D). Panels A and C show reduced lysates; B and D show nonreduced lysates. (E) Schematic of PTPσ cleavage and dimer patterns showing protein sizes and numbered fragments (the numbers correspond to those in panels A to D and F to I); primary (black triangle) and secondary (white triangles) protein cleavage sites are indicated; the double-headed arrows indicate cysteine bridges in covalent dimers; the black dots and white dots in panel E represent FLAG and HA tags, respectively. FLAG-tagged PTPσ ectodomains run smaller under nonreducing conditions due to their compact immunoglobulin domain content. Panels F to I show 2D-PAGE analysis of P839C PTPσ proteins. P839C proteins were separated by PAGE and then run in a second, orthogonal dimension under nonreducing (F and H) or reducing (G and I) conditions (reduction is not complete). The protein spots indicated by arrows correspond to the protein numbers in E. Molecular mass markers are indicated to the left of each panel.
Dimers identified by cysteine cross-linking.
The same lysates described above were subjected to PAGE under nonreducing conditions (Fig. 2B and D). The wild-type protein gave a banding pattern very similar to the previous one. In contrast, the mutated forms of PTPσ showed large amounts of reduction-sensitive, high-molecular-mass forms of around 300 to 350 kDa, consistent with their being dimers or multimers of uncleaved PTPσ (Fig. 2B; D, band 6; and E, dimer 6). In nonreduced samples, additional FLAG bands appeared at approximately 180 kDa (Fig. 2D, band 8), and concurrently, the 90-kDa bands (band 5) were greatly reduced in intensity (compare bands 5 in Fig. 2C and D). The 180-kDa bands are therefore predicted to be dimers of the cleaved 90-kDa ectodomain fragment 5 (Fig. 2E, dimer 8). We also predict that under nonreducing conditions, the 85-kDa HA band 3 would dimerize and run at approximately the same size as the uncleaved band 1 (Fig. 2B, bands 1 and 7, and E, dimer 7). In support of this, most of the 85-kDa bands are no longer found under nonreducing conditions (Fig. 2B). To further support these predictions, a 2D gel study was carried out with P839C protein. After P839C was separated under nonreducing conditions, a second dimension was run under reducing and nonreducing conditions (Fig. 2F to I). Upon reduction, the 180-kDa band 8 dropped in size to the predicted 90-kDa spot 5 (Fig. 2G). Moreover, using the HA tag, a proportion of bands “1 and 7” also dropped to the predicted smaller size of band 3, consistent with some of the “1 and 7” band being band 3 dimers (Fig. 2I). Note that this technique does not give quantitative reduction of first-dimension proteins, thus leaving some nonreduced bands present as well. It was also noted that no convincing evidence was seen of bands resembling heterodimers of, for example, band 1 with band 5 or band 1 with band 3.
These data collectively show that when trapped with unpaired cysteines, high levels of dimeric PTPσ can be identified in cells and that these are predominantly homodimers of the same cleavage states.
Dimerization of wild-type PTPσ.
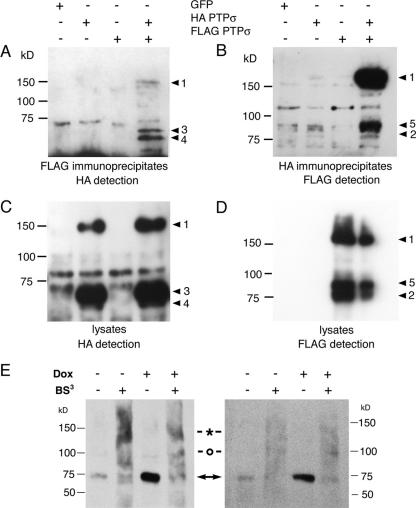
Given that PTPσ dimers could be readily trapped in a cross-linked form by ectopic cysteines, we next wanted to determine whether homodimers of wt PTPσ could be identified in cell lysates. Modified forms of the protein were generated with either an amino-terminal FLAG tag or a carboxy-terminal HA tag. They were cotransfected and immunoprecipitated with one tag, and then the other tag was immunodetected. PTPσ could be coimmunoprecipitated efficiently in both directions, indicating the presence of significant levels of cis homodimers made up of cleaved or uncleaved forms of wt PTPσ (Fig. 3).
FIG. 3.
Wild-type PTPσ homodimer detection. (A to D) Tagged PTPσ proteins were transfected into 293T cells and immunoprecipitated with either anti-FLAG (A) or anti-HA (B). The immunoblots were then detected with either anti-HA (A) or anti-FLAG (B), respectively. The corresponding lysates are shown in panels C and D. The PTPσ protein band numbers to the right of each panel correspond to those described in the legend to Fig. 2E. Panel E shows PTPσ proteins expressed under doxycycline-inducible control in a PC12-derived cell line, 12CRYP11, detected with IG2 serum (52). Two experimental examples are shown in which cells were treated with or without doxycycline (Dox) and with or without the extracellular cross-linker BS3. The native 75-kDa proteins are indicated with arrows, and commonly seen cross-linked 140-kDa and 100-kDa bands are indicated with an asterisk and a circle, respectively. The gels were run under nonreducing conditions because the epitope(s) detected by IG2 antibody is redox sensitive.
To extend this study, we carried out BS3 cross-linking in cells that expressed relatively low levels of PTPσ. A doxycycline-inducible PC12 neuronal cell line derivative was constructed, expressing either low (uninduced) or approximately fivefold-higher (induced) levels of wt PTPσ. Without induction, the cells expressed low levels of the fully cleaved type 2 (nonreduced 75-kDa) protein (Fig. 3E), similar to the situation seen in neuronal cells in vivo (52, 53). BS3 cross-linking of live cells caused the loss of 75-kDa bands and the appearance of bands at approximately 140 kDa, consistent with 75-kDa dimer formation. There was always an extended background smear, with another commonly seen band of unknown origin appearing at 100 kDa, suggesting that PTPσ exhibits variable cross-linking to other cellular proteins as well (Fig. 3E). A similar qualitative pattern was seen in the cells after doxycycline-induced expression of higher PTPσ levels (Fig. 3E). These data suggest that PTPσ dimers can be cross-linked at both low and high expression levels of PTPσ and that much of the protein is exposed at the cell surface.
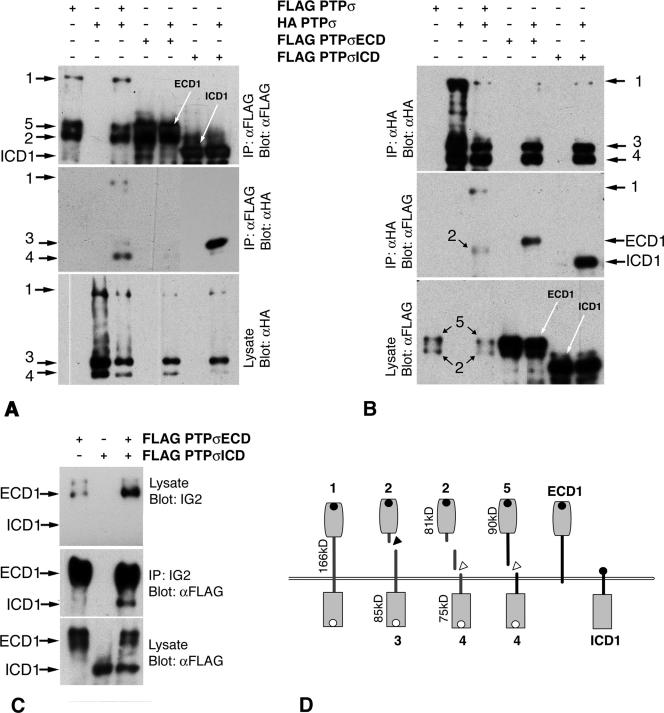
Transmembrane proteins that undergo intramembranous dimerization, including RPTPs, can form interaction interfaces through extracellular, intracellular, and transmembrane regions (11, 23, 48, 61). Two FLAG-tagged forms of PTPσ were constructed to identify which domains of PTPσ facilitate its cis interactions. Protein ICD-1 contains PTPσ1 sequence starting at amino acid 845 in the juxtamembrane region (Fig. 4D). Protein ECD-1 contains all of the ectodomain, stopping at amino acid 883 just inside the cell (Fig. 1 and 4D). These two proteins overlap by 39 amino acids, including the whole transmembrane region.
FIG. 4.
Dimerization of truncated PTPσ proteins. In panels A and B, HA-tagged wild-type protein was cotransfected with FLAG-tagged ECD1 or ICD1, and the proteins were then immunoprecipitated (IP) with either anti-FLAG or anti-HA antibodies. Immunoprecipitates of ICD1 pulled down wt PTPσ fragment 3 very efficiently (A, lane 7 from left; lanes 6 and 7 were exposed for one-fifth of the time of other lanes), whereas immunoprecipitates of ECD1 contained very little wild-type protein (A, lane 5). Immunoprecipitates of wild-type protein pulled down ICD1 and, more weakly, ECD1 (B, lanes 5 and 7). Control coimmunoprecipitations of wt PTPσ are also shown (A and B, lanes 1 to 3). Panel C shows lysates containing ICD1, ECD1, and ICD1 plus ECD1. Immunoprecipitation of ECD1 with extracellular-domain-specific IG2 antiserum also pulled down ICD1 (lane 3, anti-FLAG detection). Panel D shows schematic proteins.
The proteins were coexpressed with wild-type protein, and coimmunoprecipitations were performed. Both ECD-1 and ICD-1 bound to the full-length protein (Fig. 4A and B), with ICD1 consistently binding to wt PTPσ much more efficiently than did ECD-1. For example, no band 3 form of wt PTPσ was observed associating with ECD1, and only very faint bands 1 and 4 were seen (Fig. 4A). Poor accessibility to FLAG is not an explanation, since the coprecipitation of all three wt PTPσ protein bands was seen (Fig. 4A, lane 3, middle). The data currently suggest, therefore, that the transmembrane and catalytic domains together contribute most of the interaction affinity between PTPσ molecules. It was also noted that ICD-1 pulled down the primary cleaved form of PTPσ most efficiently, with little if any uncleaved form being observed even after prolonged filter exposure (Fig. 4A, band 3, lane 7).
Since ECD-1 and ICD1 overlap in the transmembrane region, we cotransfected the two proteins and asked whether they would also associate. When immunoprecipitated with an ectodomain-directed antibody, IG2 (52), ECD1 could efficiently pull down ICD1 (Fig. 4C). Therefore, a significant amount of the interaction interface between PTPσ dimers occurs in the transmembrane region and its immediate surrounding sequences.
PTPσ requires dimerization for ligand binding.
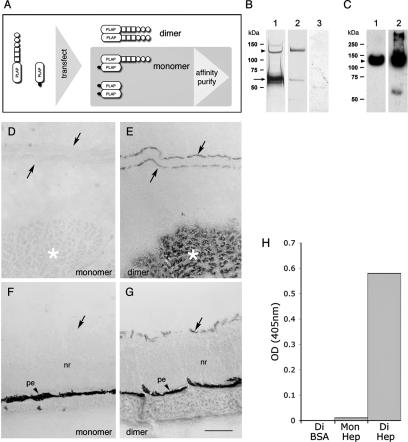
It is not understood whether the extracellular ligands of RPTPs generally bind to monomeric or dimeric forms of the receptors. This information could prove very useful, since it may provide an understanding of the ligand-dependent regulation of RPTPs. Assays of ligand binding to PTPσ in live cells are not currently available, so we have instead developed a novel RAP strategy to begin to address this question. This assay allowed us to examine the stoichiometry of interactions between soluble PTPσ-AP fusion proteins and two PTPσ ligands, the HSPGs (5) and muscle-bound nucleolin (2, 45).
Standard RAP probes consist of a receptor ectodomain fused to PLAP. Since PLAP is active only as a dimer (22), by definition we detect only oligomers of paired ectodomains using the PLAP readout. To test instead whether RPTP ectodomain monomers have any ligand binding function, a method was established using heterodimers of the ectodomain-PLAP with PLAP. An HA-tagged form of secreted PLAP was constructed (PLAP-HA) and cotransfected into 293T cells, along with the PTPσ-AP probe (Fig. 5A). This allowed cells to secrete three protein complexes: (i) homodimers of full-length PTPσ-AP, (ii) homodimers of PLAP-HA, and (iii) heterodimers of PLAP-HA-PTPσ-AP (Fig. 5A). Cell supernatants were passed through an anti-HA affinity column to purify the PLAP-HA proteins. The PLAP homodimer has no binding activity in RAP assays; therefore, any binding activity in the affinity-purified mixture should come only from PLAP-HA-PTPσ-AP heterodimers. During affinity purification, the flowthrough (containing PTPσ-AP homodimers) was shown to have RAP activity (data not shown), confirming that the purification did not affect its binding competence.
FIG. 5.
Ligand binding by dimeric PTPσ. PLAP probes containing monomeric PTPσ ectodomains were generated as shown in panel A (see the text for further details). (B) Silver stained proteins after affinity purification. Lane 1, anti-HA affinity-purified FN3d-AP/PLAP-HA, showing PLAP-HA (arrow) and FN3d-AP (arrowhead). Lane 2, anti-PLAP affinity eluate from Fn3d-AP-conditioned medium. Lane 3, anti-HA affinity eluate from FN3d-AP-conditioned medium. (C) Eluates from affinity matrices immunodetected with IG2 (anti-PTPσ) antibody. Lanes 1 and 2 are the same as in panel B. (D to G) E10 chicken embryo head cryosections after RAP assay and nitroblue tetrazolium/BCIP (5-bromo-4-chloro-3-indolylphosphate) reactions, probed with monomeric (D and F) and dimeric (E and G) forms of FN3d-AP. Panels D and E show the nasal neuroepithelium (arrows, basement membranes) and nearby skeletal muscle (asterisks). Panels F and G show retinal tissue with neural retina (nr), inner limiting basement membrane (arrows), and pigmented epithelium (pe). Scale bar = 0.1 mm. Panel H shows data from a representative solid-phase binding assay. Wells coated with either heparin (Hep) or BSA (shown on the x axis) were treated with either dimer (Di) or monomer (Mon) forms of FN3d-AP. The amounts of AP probe bound were then quantified.
Two proteins were then tested in the RAP assay, one an affinity-purified positive control of PTPσ-AP homodimers (purified using anti-PLAP antibody; see Materials and Methods) and the other the purified heterodimer (PLAP-HA-PTPσ-AP). Fractions of these protein solutions were first immunoblotted using anti-PTPσ antibodies, to allow us to normalize for PTPσ ectodomain content. They were then applied to tissue sections for RAP assays (Fig. 5D to G). This revealed that the monomeric PTPσ-AP had no detectable ligand binding activity (Fig. 5D and F), even when RAP reactions were allowed to run overnight. A second experiment was carried out with the monomeric form of PTPσ-AP, to address directly whether the monomer could bind to purified heparin, a polysaccharide known to bind to this protein. This assay was carried out in solid phase, with heparin-coated 96-well plates (supplied by PLASSO Ltd., United Kingdom). Normalized amounts of the PLAP fusion proteins were added to the plates in solution, and their binding capacities were measured. Again, the monomeric form of the ectodomain had no significant affinity for heparin (Fig. 5H). These RAP and solid-phase affinity data therefore support the conclusion that the PTPσ ectodomain must be in a dimeric structure in order to bind ligands efficiently.
Dimerization influences isoform specificity.
The two main protein isoforms of PTPσ have either four (PTPσ1) or eight (PTPσ2) FNIII-type domains, and these isoforms have distinct ligand binding capacities (20, 45, 50). As demonstrated above, ligand interactions require a dimerized PTPσ structure, at least in vitro. This led us to investigate whether the ligand binding specificities of PTPσ1 and PTPσ2 are controlled purely by the FNIII subdomain content or by additional supramolecular influences brought about through dimer coupling.
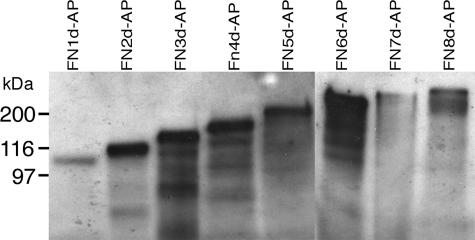
A protein deletion series was constructed from PTPσ2 in which FNIII subdomains were deleted progressively from the carboxy terminus. Our previously reported PTPσ-AP probes were fused to PLAP at alanine residue 1124 in PTPσ, approximately 27 amino acids downstream of FNIII domain 8 (taken as ending in the sequence KPE) (20). Both PTPσ1-AP and PTPσ2-AP thus have 119 amino acids in common at their carboxy-terminal ends. The current deletion series started with construct FN8d-AP, which had the non-FNIII stretch of 27 amino acids deleted. This was followed by progressive deletion of FNIII domains (FN7d-AP to FN1d-AP). Each fusion protein was expressed in 293T cells and identified in the supernatant by immunoblotting (Fig. 6). This enabled us to confirm their predicted sizes and to normalize for the amounts of PLAP fusion proteins present. These normalized probes were then screened for their ligand binding properties (Fig. 7).
FIG. 6.
FNIII domain deletion series. CDNAs encoding a series of FNIII deletions in the PTPσ ectodomain were fused in frame to PLAP and expressed by 293T cells. Supernatants containing similar levels of these fusion proteins, FN1d-AP to FN8d-AP, were immunoblotted and detected with anti-PTPσ serum IG2. Proteins of the correct sizes were detected in each case. Molecular mass markers are indicated to the left.
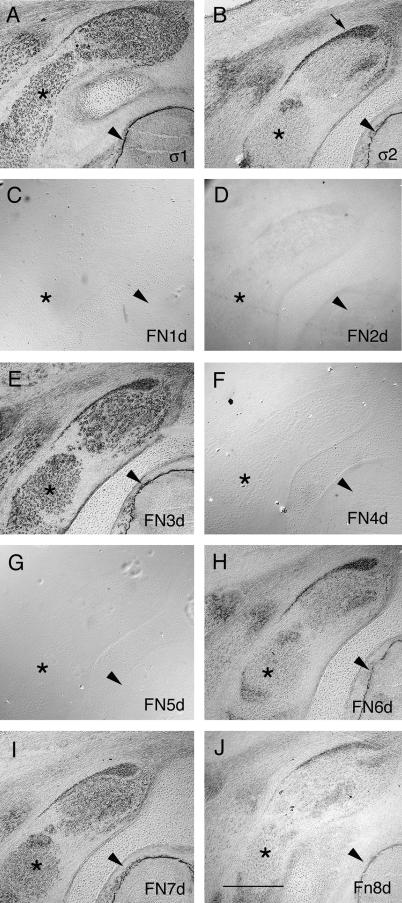
FIG. 7.
RAP assays with PTPσ FNIII deletion series. (A to J) RAP assays with PTPσ1-AP (A), PTPσ2-AP (B), FN1d-AP (C), FN2d-AP (D), FN3d-AP (E), FN4d-AP (F), FN5d-AP (G), FN6d-AP (H), FN7d-AP (I), and FN8d-AP (J) in the brachial spinal cord region of E10 chicken embryos. The arrowheads indicate basement membranes surrounding the spinal cord, the asterisks indicate skeletal muscle, and the arrow in panel B indicates associated tendon tissue. Scale bar = 1 mm.
We monitored the binding of probes to two different ligand types: (i) HSPG ligands in basement membranes (BMs) (5) and (ii) a non-HSPG ligand in skeletal muscle, a component of which is nucleolin (2, 45). Proteins FN1d-AP, FN4d-AP, and FN5d-AP had no detectable binding capacity, and FN2d-AP had extremely low activity (Fig. 7). Proteins with fewer domains than FN1d-AP also had no activity (data not shown). In contrast, FN3d-AP and FN7d-AP had strong binding capacities, similar qualitatively to that of PTPσ1-AP (Fig. 7A, E, and I). These proteins bound to both BMs and muscles. FN8d-AP had some weak RAP activity (Fig. 7J), and Fn6d-AP had stronger activity (Fig. 7H), but both of these probes bound in similar fashions to PTPσ2-AP, with little if any muscle binding (Fig. 7B). The binding specificities of these deletion forms are summarized in Fig. 8A.
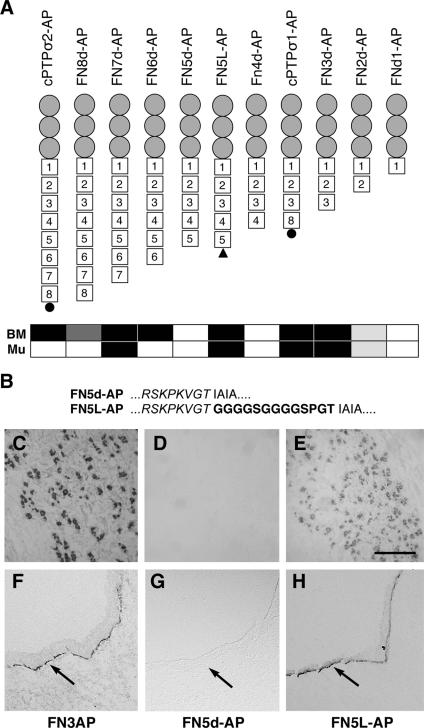
FIG. 8.
Recovery of ligand binding function after addition of peptide linker. (A) Schematic diagram of the FNIII deletion series shown in Fig. 6, together with FN5L-AP containing a peptide linker (black triangle). Gray circles, IG domains; squares, FNIII domains. The black circles in PTPσ2-AP and PTPσ1-AP represent an additional 29-amino-acid segment (see the text). The binding pattern of each fusion protein is summarized as either nonbinding (white rectangle), strongly binding (black rectangle), or weakly binding (two gray shades) to BMs or muscle (Mu). (B) Amino acid sequence of Fnd5L-AP at the junction between FN5d (italic) and PLAP, with the added linker in boldface. (C to H) FN3d-AP (C and F), FN5d-AP (D and G), and FN5L-AP (E and H) RAP assays in E10 chicken embryo head sections, showing relative binding to muscle (C to E) or basement membrane ligands (F to H, arrows). Scale bar = 50 μm.
The data indicate that the amino-terminal segment consisting of the immunoglobulin domains and the first three FNIII domains, is a region that is sufficient for ligand binding. With ectodomains larger than FN3d, there was no simple correlation between the presence of specific FNIII domains and the relative ability to bind specific ligands. This suggests that some factor other than simply FNIII domain content is influencing the specificity of ligand binding. It is possible that some of these artificial deletion forms are simply incapable of ligand binding due to gross protein misfolding. Nevertheless, given that these constructs are all anchored to dimerized PLAP, it is more likely that the ligand binding capacities of these ectodomains are being restricted by rotational constraints. If so, this might indicate that there are only a limited number of energetically favorable rotational conformations in dimers of the PTPσ ectodomain, resulting in either a PTPσ1- or PTPσ2-like ligand binding pattern (Fig. 7A and B). To test whether ligand binding potential can be recovered by simply providing greater rotational freedom, we reengineered FN5d-AP by inserting a flexible linker, GGGGSGGGGSPGT, between the PTP ectodomain and PLAP in FNd5-AP (Fig. 8B). The resulting protein, FN5L-AP (Fig. 8A), was then tested for its RAP activity in comparison to FN5d-AP. The addition of the linker to FN5d-AP caused a striking recovery of RAP binding activity, so that FN5L-AP now quite closely resembled FN3d-AP (Fig. 8C-H). These data suggest that PTPσ dimers can fall broadly into one of two different conformations with the distinct ligand binding specificities of PTPσ1 or PTPσ2. We suggest that the alternative FN4 to -7 region found in PTPσ2 affects the ligand binding specificity largely by forcing the amino-terminal ligand binding region into a novel rotational state.
DISCUSSION
This study has shown that the CAM-like RPTP, PTPσ, is homodimerized in the cell and that dimerization is not greatly influenced by the posttranslational proteolytic state of the protein. Our data further reveal that PTPσ requires dimerization to bind to its ligands, since monomeric ectodomains have no ligand binding capacity, at least under in vitro conditions. Lastly, our data also now indicate that the distinct ligand binding capacities of two major PTPσ isoforms are brought about through their alternative use of FNIII domains, which in turn imposes distinct rotational or flexional constraints on their ectodomain dimers.
PTPσ can dimerize efficiently in the cell, and we found that the transmembrane and juxtamembrane regions are sufficient for at least some of this interaction. Nevertheless, the data suggest that the intracellular and, to a lesser extent, the extracellular domain of the protein also contribute. This is similar to the situation with several other receptors, including PTPα and RTKs (11, 23, 48, 61). Dimers are found between uncleaved PTPσ proteins and between fully cleaved forms of PTPσ. It therefore seems unlikely that cleavage per se is necessary for dimerization. This study used four cysteine-containing mutations in the juxtamembrane domain region, all of which trapped PTPσ in its dimerized state. All of the mutated proteins underwent a juxtamembrane cleavage carboxy terminal to their cysteine bridges. Given that the secondary cleavage site (*) in rat PTPσ is in the amino acid sequence DPQP*IVD846 (1), our data indicate that, at least for I845C, the avian PTPσ cleavage is more carboxy terminal. This suggests either that there is little sequence specificity or that the replacement of mammalian valine 845 in PTPσ with isoleucine in the chicken may alter the cleavage specificity.
RPTPs, such as PTPα, CD45, and SAP-1, exist as homodimers, and this is believed to negatively regulate catalytic activity (11, 55, 58, 59, 64). It is not known, however, if such dimerization can affect the potential ligand binding properties of their ectodomains. In comparison, many RTKs and cytokine receptors are normally activated only when they can dimerize and are bound to their ligands (17, 21, 38, 63). In some cases, RTKs are monomeric, and ligand binding then induces dimerization. Ligands can either form a cross-link in a receptor dimer (63) or allosterically trigger a receptor-only dimerization event (17). In other receptors, such as those for insulin (38) or erythropoietin (43), the receptors are already dimerized and ligand binding induces conformational changes and receptor activation. With RPTPs, the relationships between ligand binding, dimerization, and changes in RPTP activity have proved difficult to assess. RPTP dimers are potentially inactive, while monomers are likely to be highly active (13, 14, 29, 54, 58, 59). The binding of ligands, such as pleiotrophin, to PTPζ inactivates the enzyme (34, 39), but it is not known if this occurs through enforced dimerization. The recent identification of a homophilic binding domain in LAR revealed that this domain can positively regulate LAR, possibly by binding to LAR monomers and preventing inactive LAR dimer formation (65). A similar situation is seen with CD45-associated protein, which associates with CD45 in cis and reduces dimer formation, consequently activating CD45 (54). The general picture concerning the influence of ectodomain ligands over RPTP function has therefore yet to be formed.
PTPσ can bind to at least three extracellular ligands (5, 27, 45). Although the role of ligand interactions is currently unclear, it is likely that they influence the biological functions of this phosphatase (6, 15, 33, 42, 44, 46, 49, 56, 60). Because it has not proven technically possible to test the behavior of ligand binding to PTPσ in live cells, we chose a complementary approach instead. This examined how soluble PTPσ ectodomains bind to heterotypic ligands in vitro. The most significant finding is that PTPσ binds to these ligands only when it is presented as a dimer. The ligands we have tested include HSPGs and a muscle site that includes nucleolin (2). HSPGs have been shown to modulate PTPσ interactions with retinal axon receptors (44), and HSPGs have been shown to interact functionally with DLAR, a Drosophila ancestor of PTPσ, during motor axon guidance and synaptogenesis (19, 25). Such HSPGs may therefore be binding to preexisting PTPσ dimers during these events. To understand whether such ligand binding alters the catalytic function of the dimer, we must await assays that can directly quantify the activation state of PTPσ in the cell.
PTPσ is expressed as two major protein isoforms (50). The longer isoform, PTPσ2, is distinct from PTPσ1 in being unable to bind to skeletal-muscle ligands (20, 45). It has been a puzzle why the increased number of FNIII domains in PTPσ2 should apparently result in different or possibly fewer ligand partners. The current study demonstrates that the ligand binding specificity does not correlate straightforwardly with the FNIII domain content. For example, FN7d has all of the unique domains of PTPσ2 (FN4 to -7), but unlike PTPσ2, it does bind to muscle. On the other hand, addition of the final FN8 domain to FNd7-AP causes loss of muscle interaction: however, FN8 is found in PTPσ1, which does bind muscle. We have also ruled out a simple effect of increasing ectodomain length on ligand binding specificity, since FN6d-AP behaves like PTPσ2, whereas the longer FNd7-AP again behaves like PTPσ1. The lack of a straightforward relationship between the ligand binding specificity and the FNIII content, together with the sensitivity of ligand binding to small increments in protein length, suggests that ligand binding is defined by additional molecular constraints. We propose a parsimonious model in which the binding patterns of PTPσ1 and PTPσ2 reflect alternative rotational conformers of the dimerized ectodomains. FNIII domains form relatively inflexible, rod-like structures (28). One effect of adding four FNIII domains to PTPσ1 to produce PTPσ2 (5), therefore, might be to induce a new conformational or rotational change in the region amino terminal to FNIII domain 4, having the effect of masking the muscle binding site while retaining the HSPG binding site. From our deletion series, it appears that all ligand binding is effected by this amino-terminal region contained within FN3d-AP. If this model of regulation using rotational conformation holds true, then we predict that some of the alternatively encoded exons found in the related RPTPs LAR and PTPδ (41, 66) could also be imposing novel rotational conformations on ectodomain dimers. Multiple ligand types might therefore be able to bind to different RPTP isoforms through two distinct mechanisms, first, through the intrinsic binding properties of alternative peptide inserts (36), and second, through larger-scale conformational changes in the molecules.
One important question to be answered is whether rotational conformation is an intrinsic property of ectodomains or whether it can be imposed in an “inside out” manner by intracellular domains. Data using PTPα suggest that for type IV RPTPs, this is possible (57). Similarly, we need to address the possibility that ligand binding “locks” ectodomain dimers into distinct rotational conformations that in turn differentially influence intracellular signaling. Our current data indicate that the addition of soluble heparin to PTPσ-expressing cells does not affect the overall degree of dimerization that we can detect in the cells (C. Faux and S. Lee, unpublished data).
One problem that has dogged the RPTP field is the relatively poor success in identifying ligands using ectodomain fusion assays. We have now demonstrated that, with PTPσ at least, the rotational freedom of the ectodomain with respect to PLAP plays a highly critical role in determining whether ligand binding capacity is retained. This could now prove useful in the future application of this RAP assay in ligand characterization.
In summary, we have shown for the first time that PTPσ is dimerized in the cell and that ectodomain dimerization is necessary for heterophilic ligand binding. We also provide new evidence of how alternative use of FNIII domain complements in two major isoforms of PTPσ can alter the ligand binding specificities of the ectodomains. The data suggest a model in which the inclusion of alternative FNIII domains functions largely to change the rotational conformations of the amino-terminal ligand binding sites of the PTPσ ectodomain dimers, thus imparting novel ligand binding properties. Thus, by alternative encoding of ectodomain exons, PTPσ has a sensitive mechanism available to it to control its ligand binding potential. These findings could have important implications for our understanding of how heterophilic ligands interact with, and potentially regulate, RPTPs.
Acknowledgments
The research was funded by the Wellcome Trust (D.A. and M.H.; 071418), the Child Health Research Appeal Trust (S.L.), the BBSRC (United Kingdom) (C.F. and J.N.; 31/C17480), and EC Network HPRN-CT-2000-00085.
We thank Radu Aricescu for critical reading of the manuscript and Plasso Technology Ltd. for their generous gift of heparin binding plates.
Footnotes
Published ahead of print on 18 December 2006.
REFERENCES
- 1.Aicher, B., M. M. Lerch, T. Muller, J. Schilling, and A. Ullrich. 1997. Cellular redistribution of protein tyrosine phosphatases LAR and PTPσ by inducible proteolytic processing. J. Cell Biol. 138:681-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alete, D. E., M. E. Weeks, A. G. Hovanession, M. Hawadle, and A. W. Stoker. 2006. Cell surface nucleolin on developing muscle is a potential ligand for the axonal receptor protein tyrosine phosphatase-sigma. FEBS J. 273:4668-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, A., J. Sasin, N. Bottini, I. Friedberg, A. Osterman, A. Godzik, T. Hunter, J. Dixon, and T. Mustelin. 2004. Protein tyrosine phosphatases in the human genome. Cell 117:699-711. [DOI] [PubMed] [Google Scholar]
- 4.Aricescu, A. R., W. C. Hon, C. Siebold, W. Lu, P. A. van der Merwe, and E. Y. Jones. 2006. Molecular analysis of receptor protein tyrosine phosphatase mu-mediated cell adhesion. EMBO J. 25:701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aricescu, A. R., I. W. McKinnell, W. Halfter, and A. W. Stoker. 2002. Heparan sulfate proteoglycans are ligands for receptor protein tyrosine phosphatase sigma. Mol. Cell. Biol. 22:1881-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batt, J., S. Asa, C. Fladd, and D. Rotin. 2002. Pituitary, pancreatic and gut neuroendocrine defects in protein tyrosine phosphatase- sigma-deficient mice. Mol. Endocrinol. 16:155-169. [DOI] [PubMed] [Google Scholar]
- 7.Beltran, P. J., and J. L. Bixby. 2003. Receptor protein tyrosine phosphatases as mediators of cellular adhesion. Front Biosci. 8:D87-D99. [DOI] [PubMed] [Google Scholar]
- 8.Bilwes, A. M., J. Den Hertog, T. Hunter, and J. P. Noel. 1996. Structural basis for inhibition of receptor protein-tyrosine phosphatase-alpha by dimerization. Nature 382:555-559. [DOI] [PubMed] [Google Scholar]
- 9.Chang, C., T. W. Yu, C. I. Bargmann, and M. Tessier-Lavigne. 2004. Inhibition of netrin-mediated axon attraction by a receptor protein tyrosine phosphatase. Science 305:103-106. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, H. J., and J. G. Flanagan. 1994. Identification and cloning of ELF-1, a developmentally expressed ligand for the Mek4 and Sek receptor tyrosine kinases. Cell 79:157-168. [DOI] [PubMed] [Google Scholar]
- 11.Chin, C. N., J. N. Sachs, and D. M. Engelman. 2005. Transmembrane homodimerization of receptor-like protein tyrosine phosphatases. FEBS Lett. 579:3855-3858. [DOI] [PubMed] [Google Scholar]
- 12.Cismasiu, V. B., S. A. Denes, H. Reilander, H. Michel, and S. E. Szedlacsek. 2004. The MAM (Meprin/A5-protein/PTPmu) domain is a homophilic binding site promoting the lateral dimerization of receptor-like protein-tyrosine phosphatase micro. J. Biol. Chem. 279:26922-26931. [DOI] [PubMed] [Google Scholar]
- 13.den Hertog, J., A. Groen, and T. van der Wijk. 2005. Redox regulation of protein-tyrosine phosphatases. Arch. Biochem. Biophys. 434:11-15. [DOI] [PubMed] [Google Scholar]
- 14.den Hertog, J., T. van der Wijk, L. G. Tertoolen, and C. Blanchetot. 2003. Receptor protein-tyrosine phosphatase dimerization. Methods Enzymol. 366:224-240. [DOI] [PubMed] [Google Scholar]
- 15.Elchebly, M., J. Wagner, T. E. Kennedy, C. Lanctot, E. Michaliszyn, A. Itie, J. Drouin, and M. L. Tremblay. 1999. Neuroendocrine dysplasia in mice lacking protein tyrosine phosphatase sigma. Nat. Genet. 21:330-333. [DOI] [PubMed] [Google Scholar]
- 16.Ensslen-Craig, S. E., and S. M. Brady-Kalnay. 2004. Receptor protein tyrosine phosphatases regulate neural development and axon guidance. Dev. Biol. 275:12-22. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson, K. M. 2004. Active and inactive conformations of the epidermal growth factor receptor. Biochem. Soc. Trans. 32:742-745. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Patron, C., L. Castellanos-Serra, and P. Rodriguez. 1992. Reverse staining of sodium dodecyl sulfate polyacrylamide gels by imidazole-zinc salts: sensitive detection of unmodified proteins. BioTechniques 12:564-573. [PubMed] [Google Scholar]
- 19.Fox, A. N., and K. Zinn. 2005. The heparan sulfate proteoglycan syndecan is an in vivo ligand for the Drosophila LAR receptor tyrosine phosphatase. Curr. Biol. 15:1701-1711. [DOI] [PubMed] [Google Scholar]
- 20.Haj, F., I. McKinnell, and A. Stoker. 1999. Retinotectal ligands for the receptor tyrosine phosphatase CRYPα. Mol. Cell Neurosci. 14:225-240. [DOI] [PubMed] [Google Scholar]
- 21.Hibi, M., and T. Hirano. 1998. Signal transduction through cytokine receptors. Int. Rev. Immunol. 17:75-102. [DOI] [PubMed] [Google Scholar]
- 22.Hoylaerts, M. F., T. Manes, and J. L. Millan. 1997. Mammalian alkaline phosphatases are allosteric enzymes. J. Biol. Chem. 272:22781-22787. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, G., J. den Hertog, and T. Hunter. 2000. Receptor-like protein tyrosine phosphatase alpha homodimerizes on the cell surface. Mol. Cell. Biol. 20:5917-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, G., J. den Hertog, J. Su, J. Noel, J. Sap, and T. Hunter. 1999. Dimerization inhibits the activity of receptor-like protein-tyrosine phosphatase-alpha. Nature 401:606-610. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, K. G., A. P. Tenney, A. Ghose, A. M. Duckworth, M. E. Higashi, K. Parfitt, O. Marcu, T. R. Heslip, J. L. Marsh, T. L. Schwarz, J. G. Flanagan, and D. Van Vactor. 2006. The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron 49:517-531. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, K. G., and D. Van Vactor. 2003. Receptor protein tyrosine phosphatases in nervous system development. Physiol. Rev. 83:1-24. [DOI] [PubMed] [Google Scholar]
- 27.Krasnoperov, V., M. A. Bittner, W. Mo, L. Buryanovsky, T. A. Neubert, R. W. Holz, K. Ichtchenko, and A. G. Petrenko. 2002. Protein-tyrosine phosphatase-sigma is a novel member of the functional family of alpha-latrotoxin receptors. J. Biol. Chem. 277:35887-35895. [DOI] [PubMed] [Google Scholar]
- 28.Leahy, D. J., I. Aukhil, and H. P. Erickson. 1996. 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 84:155-164. [DOI] [PubMed] [Google Scholar]
- 29.Majeti, R., A. M. Bilwes, J. P. Noel, T. Hunter, and A. Weiss. 1998. Dimerization-induced inhibition of receptor protein tyrosine phosphatase function through an inhibitory wedge. Science 279:88-91. [DOI] [PubMed] [Google Scholar]
- 30.Majeti, R., and A. Weiss. 2001. Regulatory mechanisms for receptor protein tyrosine phosphatases. Chem. Rev. 101:2441-2448. [DOI] [PubMed] [Google Scholar]
- 31.Majeti, R., Z. Xu, T. G. Parslow, J. L. Olson, D. I. Daikh, N. Killeen, and A. Weiss. 2000. An inactivating point mutation in the inhibitory wedge of CD45 causes lymphoproliferation and autoimmunity. Cell 103:1059-1070. [DOI] [PubMed] [Google Scholar]
- 32.McLean, J., J. Batt, L. C. Doering, D. Rotin, and J. R. Bain. 2002. Enhanced rate of nerve regeneration and directional errors after sciatic nerve injury in receptor protein tyrosine phosphatase sigma knock-out mice. J. Neurosci. 22:5481-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meathrel, K., T. Adamek, J. Batt, D. Rotin, and L. C. Doering. 2002. Protein tyrosine phosphatase sigma-deficient mice show aberrant cytoarchitecture and structural abnormalities in the central nervous system. J. Neurosci. Res. 70:24-35. [DOI] [PubMed] [Google Scholar]
- 34.Meng, K., A. Rodriguez-Pena, T. Dimitrov, W. Chen, M. Yamin, M. Noda, and T. F. Deuel. 2000. Pleiotrophin signals increased tyrosine phosphorylation of beta-catenin through inactivation of the intrinsic catalytic activity of the receptor-type protein tyrosine phosphatase beta/zeta. Proc. Natl. Acad. Sci. USA 97:2603-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nam, H. J., F. Poy, H. Saito, and C. A. Frederick. 2005. Structural basis for the function and regulation of the receptor protein tyrosine phosphatase CD45. J. Exp. Med. 201:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Grady, P., T. C. Thai, and H. Saito. 1998. The laminin-nidogen complex is a ligand for a specific splice isoform of the transmembrane protein tyrosine phosphatase LAR. J. Cell Biol. 141:1675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostman, A., and F. D. Bohmer. 2001. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatases. Trends Cell Biol. 11:258-266. [DOI] [PubMed] [Google Scholar]
- 38.Ottensmeyer, F. P., D. R. Beniac, R. Z. Luo, and C. C. Yip. 2000. Mechanism of transmembrane signaling: insulin binding and the insulin receptor. Biochemistry 39:12103-12112. [DOI] [PubMed] [Google Scholar]
- 39.Pariser, H., P. Perez-Pinera, L. Ezquerra, G. Herradon, and T. F. Deuel. 2005. Pleiotrophin stimulates tyrosine phosphorylation of beta-adducin through inactivation of the transmembrane receptor protein tyrosine phosphatase beta/zeta. Biochem. Biophys. Res. Commun. 335:232-239. [DOI] [PubMed] [Google Scholar]
- 40.Paul, S., and P. J. Lombroso. 2003. Receptor and nonreceptor protein tyrosine phosphatases in the nervous system. Cell Mol. Life Sci. 60:2465-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pulido, R., N. X. Krueger, C. Serra Pages, H. Saito, and M. Streuli. 1995. Molecular characterization of the human transmembrane protein-tyrosine phosphatase delta. Evidence for tissue-specific expression of alternative human transmembrane protein-tyrosine phosphatase delta isoforms. J. Biol. Chem. 270:6722-6728. [DOI] [PubMed] [Google Scholar]
- 42.Rashid-Doubell, F., I McKinnell, A. R. Aricescu, G. Sajnani, and A. W. Stoker. 2002. Chick PTPσ regulates the targeting of retinal axons within the optic tectum. J. Neurosci. 22:5024-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Remy, I., I. A. Wilson, and S. W. Michnick. 1999. Erythropoietin receptor activation by a ligand-induced conformation change. Science 283:990-993. [DOI] [PubMed] [Google Scholar]
- 44.Sajnani, G., A. R. Aricescu, E. Y. Jones, J. Gallagher, D. Alete, and A. Stoker. 2005. PTPσ promotes retinal neurite outgrowth non-cell-autonomously. J. Neurobiol. 65:59-71. [DOI] [PubMed] [Google Scholar]
- 45.Sajnani-Perez, G., J. K. Chilton, A. R. Aricescu, F. Haj, and A. W. Stoker. 2003. Isoform-specific binding of the tyrosine phosphatase PTPσ to a ligand in developing muscle. Mol. Cell Neurosci. 22:37-48. [DOI] [PubMed] [Google Scholar]
- 46.Sapieha, P. S., L. Duplan, N. Uetani, S. Joly, M. L. Tremblay, T. E. Kennedy, and A. Di Polo. 2005. Receptor protein tyrosine phosphatase sigma inhibits axon regrowth in the adult injured CNS. Mol. Cell Neurosci. 28:625-635. [DOI] [PubMed] [Google Scholar]
- 47.SerraPages, C., H. Saito, and M. Streuli. 1994. Mutational analysis of proprotein processing, subunit association, and shedding of the LAR transmembrane protein tyrosine phosphatase. J. Biol. Chem. 269:23632-23641. [PubMed] [Google Scholar]
- 48.Shiang, R., L. M. Thompson, Y. Z. Zhu, D. M. Church, T. J. Fielder, M. Bocian, S. T. Winokur, and J. J. Wasmuth. 1994. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell 78:335-342. [DOI] [PubMed] [Google Scholar]
- 49.Stepanek, L., A. W. Stoker, E. Stoeckli, and J. L. Bixby. 2005. Receptor tyrosine phosphatases guide vertebrate motor axons during development. J. Neurosci. 25:3813-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoker, A. W. 1994. Isoforms of a novel cell adhesion molecule-like protein tyrosine phosphatase are implicated in neural development. Mech. Dev. 46:201-217. [DOI] [PubMed] [Google Scholar]
- 51.Stoker, A. W. 2005. Protein tyrosine phosphatases and signalling. J. Endocrinol. 185:19-33. [DOI] [PubMed] [Google Scholar]
- 52.Stoker, A. W., B. Gehrig, F. Haj, and B. H. Bay. 1995. Axonal localisation of the CAM-like tyrosine phosphatase CRYP alpha: a signalling molecule of embryonic growth cones. Development 121:1833-1844. [DOI] [PubMed] [Google Scholar]
- 53.Stoker, A. W., B. Gehrig, M. R. Newton, and B. H. Bay. 1995. Comparative localisation of CRYP-alpha, a CAM-like tyrosine phosphatase, and NgCAM in the developing chick visual system. Dev. Brain Res. 90:129-140. [DOI] [PubMed] [Google Scholar]
- 54.Takeda, A., A. Matsuda, R. M. Paul, and N. R. Yaseen. 2004. CD45-associated protein inhibits CD45 dimerization and upregulates its protein tyrosine phosphatase activity. Blood 103:3440-3447. [DOI] [PubMed] [Google Scholar]
- 55.Tertoolen, L. G., C. Blanchetot, G. Jiang, J. Overvoorde, T. W. Gadella, Jr., T. Hunter, and J. den Hertog. 2001. Dimerization of receptor protein-tyrosine phosphatase alpha in living cells. BMC Cell Biol 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, K. M., N. Uetani, C. Manitt, M. Elchebly, M. L. Tremblay, and T. E. Kennedy. 2003. Receptor protein tyrosine phosphatase sigma inhibits axonal regeneration and the rate of axon extension. Mol. Cell Neurosci. 23:681-692. [DOI] [PubMed] [Google Scholar]
- 57.van der Wijk, T., C. Blanchetot, J. Overvoorde, and J. den Hertog. 2003. Redox-regulated rotational coupling of receptor protein-tyrosine phosphatase alpha dimers. J. Biol. Chem. 278:13968-13974. [DOI] [PubMed] [Google Scholar]
- 58.van der Wijk, T., J. Overvoorde, and J. den Hertog. 2004. H2O2-induced intermolecular disulfide bond formation between receptor protein-tyrosine phosphatases. J. Biol. Chem. 279:44355-44361. [DOI] [PubMed] [Google Scholar]
- 59.Walchli, S., X. Espanel, and R. H. van Huijsduijnen. 2005. Sap-1/PTPRH activity is regulated by reversible dimerization. Biochem. Biophys. Res. Commun. 331:497-502. [DOI] [PubMed] [Google Scholar]
- 60.Wallace, M. J., J. Batt, C. A. Fladd, J. T. Henderson, W. Skarnes, and D. Rotin. 1999. Neuronal defects and posterior pituitary hypoplasia in mice lacking the receptor tyrosine phosphatase PTPσ. Nat. Genet. 21:334-338. [DOI] [PubMed] [Google Scholar]
- 61.Weiner, D. B., J. Liu, J. A. Cohen, W. V. Williams, and M. I. Greene. 1989. A point mutation in the neu oncogene mimics ligand induction of receptor aggregation. Nature 339:230-231. [DOI] [PubMed] [Google Scholar]
- 62.West, M., and V. G. Wilson. 2002. Method for transferring mutations between plasmids. BioTechniques 32:44, 46. [DOI] [PubMed] [Google Scholar]
- 63.Westermark, B., L. Claesson-Welsh, and C. H. Heldin. 1989. Structural and functional aspects of the receptors for platelet-derived growth factor. Prog. Growth Factor Res. 1:253-266. [DOI] [PubMed] [Google Scholar]
- 64.Xu, Z., and A. Weiss. 2002. Negative regulation of CD45 by differential homodimerization of the alternatively spliced isoforms. Nat. Immunol. 3:764-771. [DOI] [PubMed] [Google Scholar]
- 65.Yang, T., W. Yin, V. D. Derevyanny, L. A. Moore, and F. M. Longo. 2005. Identification of an ectodomain within the LAR protein tyrosine phosphatase receptor that binds homophilically and activates signalling pathways promoting neurite outgrowth. Eur. J. Neurosci. 22:2159-2170. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, J. S., and F. M. Longo. 1995. LAR tyrosine phosphatase receptor: alternative splicing is preferential to the nervous system, coordinated with cell growth and generates novel isoforms containing extensive CAG repeats. J. Cell Biol. 128:415-431. [DOI] [PMC free article] [PubMed] [Google Scholar]