Abstract
Endoplasmic reticulum (ER) stress transducers IRE1 (inositol requiring 1), PERK (PKR-like endoplasmic reticulum kinase), and ATF6 (activating transcription factor 6) are well known to transduce signals from the ER to the cytoplasm and nucleus when unfolded proteins accumulate in the ER. Recently, we identified OASIS (old astrocyte specifically induced substance) as a novel ER stress transducer expressed in astrocytes. We report here that BBF2H7 (BBF2 human homolog on chromosome 7), an ER-resident transmembrane protein with the bZIP domain in the cytoplasmic portion and structurally homologous to OASIS, is cleaved at the membrane in response to ER stress. The cleaved fragments of BBF2H7 translocate into the nucleus and can bind directly to cyclic AMP-responsive element sites to activate transcription of target genes. Interestingly, although BBF2H7 protein is not expressed under normal conditions, it is markedly induced at the translational level during ER stress, suggesting that BBF2H7 might contribute to only the late phase of unfolded protein response signaling. In a mouse model of focal brain ischemia, BBF2H7 protein is prominently induced in neurons in the peri-infarction region. Furthermore, in a neuroblastoma cell line, BBF2H7 overexpression suppresses ER stress-induced cell death, while small interfering RNA knockdown of BBF2H7 promotes ER stress-induced cell death. Taken together, our results suggest that BBF2H7 is a novel ER stress transducer and could play important roles in preventing accumulation of unfolded proteins in damaged neurons.
Eukaryotic cells have adapted to deal with unfolded proteins accumulated in the endoplasmic reticulum (ER) via diverse signals from the ER lumen to the cytoplasm and nucleus. This system is termed the unfolded protein response (UPR) (14, 28). The three major transducers of the UPR are IRE1 (inositol requiring 1), PERK (PKR-like endoplasmic reticulum kinase), and ATF6 (activating transcription factor 6), which all sense the presence of unfolded proteins in the ER lumen and transduce signals to the nucleus for transcription of UPR target genes, for translational attenuation of global protein synthesis, and for ER-associated degradation (ERAD).
Recently, we identified OASIS (old astrocyte specifically induced substance) as a novel ER stress transducer in astrocytes (15). OASIS is a basic leucine zipper (bZIP) transcription factor of the cyclic AMP-responsive element-binding protein (CREB)/ATF family, with a transmembrane domain that allows it to associate with the ER. The molecule is cleaved at the membrane in response to ER stress, and its cleaved N-terminal cytoplasmic domain, which contains the bZIP domain, translocates into the nucleus where it activates the transcription of target genes by acting at the ER stress-responsive element (ERSE) and cyclic AMP-responsive element (CRE) (15, 22). Intriguingly, OASIS is induced at the transcriptional level during ER stress in astrocytes of the central nervous system, but not in other types of cells examined.
In humans, more than 55 bZIP transcription factors have been reported (25). By sequence similarity in the coiled-coil region, these bZIP transcription factors can be divided into 16 different families. Among these, OASIS/CREB3L1, CREB-H/CREB3L3, AIbZIP (androgen-induced bZIP)/Tisp40/CREB4/CREB3L4, and Luman/LZIP/CREB3 are identified as members of the OASIS family (25). OASIS family members are type II transmembrane proteins and localize to the ER.
More recently, some OASIS family proteins were reported to act as novel ER stress transducers and to play roles in UPR subpathways. CREB-H is identified as a hepatocyte-specific bZIP transcription factor belonging to the CREB/ATF family (26). In response to ER stress, CREB-H is cleaved by regulated intramembrane proteolysis (RIP) and is required to activate expression of acute phase response (ARP) genes, such as those coding for serum amyloid P-component (SAP) and C-reactive protein (CRP) (37). AIbZIP/Tisp40/CREB4, specifically expressed in prostate/testis, is cleaved by RIP and has the potential to bind directly to the unfolded protein response element (UPRE) (23, 29). Although Luman is also cleaved by RIP, it is never activated in response to ER stressors such as tunicamycin and thapsigargin. However, overexpression of Luman stimulates transcription of EDEM (endoplasmic reticulum degradation-enhancing mannosidase-like protein), a key molecule in ERAD, suggesting that Luman may have a pathway different from that of the common ER stress response (5, 27). More recently, Luman is reported to be activated proteolytically by thapsigargin and to induce transcription of Herp via the ERSE (18).
In the present study, we identified using a homology database search that BBF2H7 (BBF2 human homolog on chromosome 7)/CREB3L2 (cAMP-responsive element binding protein 3-like 2) is a novel member of the OASIS family. BBF2H7 was originally identified as a novel human protein whose C-terminal region is fused to the FUS (fusion) genes in low-grade fibromyxoid sarcoma as a result of chromosomal translocation (30). However, little is known regarding the physiological functions of BBF2H7. The objective of this study is to elucidate the physiological functions of BBF2H7 and the mechanisms of its activation in response to ER stress.
MATERIALS AND METHODS
Cell culture, reagents, and assessment of cell death.
C6 (rat glioma), HEK293T, HeLa, wild-type mouse embryonic fibroblast (MEF), and PERK-deficient MEF cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum. SK-N-SH (human neuroblastoma) cells were maintained in α-modified Eagle's medium (α-MEM; MP Biomedicals, Irvine, CA) containing 10% fetal calf serum. We used tunicamycin, thapsigargin, and brefeldin A (all from Sigma, St. Louis, MO) as ER stress inducers, MG132 (Calbiochem, La Jolla, CA) as a proteasome inhibitor, staurosporine (Sigma) as a classic apoptotic stimulus that acts through mitochondria, cycloheximide (CHX; Sigma) as a translation inhibitor, and actinomycin D (ActD; Sigma) as a transcription inhibitor. To induce amino acid starvation, the medium was exchanged for Hanks' balanced salt solution (HBSS; Invitrogen). For assessment of cell death, cells were treated with thapsigargin for 24 h and the numbers of dead cells were counted based on morphological changes.
Plasmids, transfection, antibodies, and Western blotting.
To prepare pcDNA(3.1+)-Flag-mouse BBF2H7 plasmid, we extracted total RNA from MEF cells, isolated BBF2H7 cDNA by reverse transcription-PCR (RT-PCR), and inserted 1,566 bp of the full-length BBF2H7 cDNA into pcDNA3.1+ between the EcoRI and XhoI sites. Site-directed mutagenesis and a deletion mutation of BBF2H7 were achieved by PCR to create Flag-BBF2H7-376 and Flag-BBF2H7R427A,L430V,R427V/L430V. Expression plasmids for ATF6-373 and the CRE-ERSE reporter [pGL3-short immunoglobulin heavy chain-binding protein (BiP) promoter (−304)-luc] were gifts from Kazutoshi Mori (Kyoto University). OASIS-FL, OASIS-374, and the mutated pGL3-short BiP promoter (−304)-luc were constructed previously (15). C6 glioma, HEK293T, or SK-N-SH cells were transfected with each expression plasmid using Lipofectamine 2000 (Gibco BRL).
Mouse antisera were raised against recombinant BBF2H7 (amino acids 1 to 292) fused to maltose binding protein and recombinant BBF2H7 (amino acids 1 to 292) fused to glutathione S-transferase (GST) and affinity-purified using a HiTrap NHS-activated HP column (Amersham Biosciences). Anti-OASIS antibody was made previously (15). Anti-Flag (Eastman Kodak Company), anti-KDEL (StressGen Biotechnologies), anticalnexin (StressGen Biotechnologies), anti-CHOP (GADD153, Santa Cruz Biotechnology), anti-ATF4 (Santa Cruz Biotechnology), anti-phospho eukaryotic initiation factor 2α (eIF2α) (StressGen Biotechnologies), and antiactin (CHEMICON) antibodies were purchased commercially. Anti-ATF6 antibody was provided by Kazutoshi Mori.
For Western blotting of BBF2H7, cells were lysed in hot sodium dodecyl sulfate (SDS), as described previously (6), prior to electrophoresis in 7.5 to 12.5% SDS-polyacrylamide gels. The protein concentration of each sample was quantified by the Lowry assay (DC protein assay; Bio-Rad), and protein-equivalent samples were subjected to Western blotting.
Northern blotting and RT-PCR.
Total RNA was extracted from tissues using a QIAGEN RNA kit (QIAGEN, Hilden, Germany). Northern blotting and RT-PCR assays were performed according to our published procedures (13). The probe used for Northern blotting of BBF2H7 was a cDNA fragment of 1,020 bp (NM_178661; nucleotides 1441 to 2460), excluding the basic domain and leucine zipper sequences to avoid possible cross-hybridization with known family members. The oligonucleotides used for RT-PCR were as follows: human/mouse/rat-BBF2H7-5′ (5′-CCTTTCCTCTCAGAGAAGAG-3′) and human/mouse/rat-BBF2H7-3′ (5′-TGGCTGTGATGGTCAGAGTGACAG-3′); human-XBP1-5′ (5′-CAGCGCTTGGGGATGGATGC-3′) and human-XBP1-3′ (5′-CCATGGGGAGATGTTCTGGA-3′); mouse-XBP1-5′ (5′-ACACGCTTGGGAATGGACAC-3′), rat-XBP1-5′ (5′-ACACGCTTGGGGATGGATGC-3′), and mouse/rat-XBP1-3′ (5′-CCATGGGAAGATGTTCTGGG-3′); human-BiP-5′ (5′-GTTTGCTGAGGAAGACAAAAAGCTC-3′) and human-BiP-3′ (5′-CACTTCCATAGAGTTTGCTGATAATTG-3′); human-CHOP-5′ (5′-GTCCAGCTGGGAGCTGGAAG-3′) and human-CHOP-3′ (5′-CTGACTGGAATCTGGAGAG-3′); human-PDI-5′ (5′-AAGGGCAAGATCCTGTTCATC-3′) and human-PDI-3′ (5′-GGCTCATCAGGTGGGGCTTGA-3′); and human/mouse/rat-β-actin-5′ (5′-TCCTCCCTGGAGAAGAGCTAC-3′) and human/mouse/rat-β-actin-3′ (5′-TCCTGCTTGCTGATCCACAT-3′).
Immunofluorescence.
HeLa cells were grown on CELLocate coverslips (Eppendorf, Hamburg, Germany) in 60-mm dishes. Cells were fixed with 4% paraformaldehyde for 30 min at 4°C, washed with phosphate-buffered saline for 5 min, and permeabilized in 0.3% Triton X-100 for 5 min at room temperature. Mouse anti-BBF2H7 polyclonal antibody was used at a dilution of 1:50 overnight at 4°C. For overlapping of BBF2H7 with the ER, we stained the cells with rabbit anticalnexin polyclonal antibody diluted to 1:100. Primary antibodies were visualized with fluorescein-conjugated goat anti-rabbit immunoglobulin G (IgG) and Alexa-conjugated goat anti-mouse IgG antibodies for 2 h at room temperature. Stained cells were viewed using a confocal microscope (LSM 510; Carl Zeiss, Jena, Germany).
Luciferase assay.
C6 glioma cells plated onto 24-well plates were transfected with 0.2 μg of a reporter plasmid carrying the firefly luciferase gene and 0.02 μg of the reference plasmid pRL-SV40 carrying the Renilla luciferase gene under the control of the simian virus 40 enhancer and promoter (Promega) with or without 0.1 μg of expression plasmids of BBF2H7-FL, BBF2H7-376, OASIS-FL, or OASIS-374. After 46 h, cells were lysed in 200 μl of Passive lysis buffer (Promega). For induction of ER stress, cells were treated with 1 μM thapsigargin for 16 h prior to harvesting. Firefly luciferase and Renilla luciferase activities were measured in 10 μl of cell lysate using the Promega dual-luciferase reporter assay system (Promega) and a luminometer (Berthold Technologies). Relative activity was defined as the ratio of firefly luciferase activity to Renilla luciferase activity.
EMSA.
The electrophoretic mobility shift assay (EMSA) was performed as follows. In vitro translation of genes of interest was performed using each cDNA and TNT Quick Coupled transcription/translation systems (Promega). Double-stranded synthetic oligonucleotides were radiolabeled using [α-32P]dATP (3,000 Ci/mmol; Amersham) and T4 polynucleotide kinase (Promega). Two microliters of translation products was incubated with each of the oligonucleotide probes (0.1 pmol; ∼9,000 cpm) at 4°C for 1 h in binding buffer [4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol [DTT], 50 mM NaCl, 10 mM Tris-HCl (pH 7.5), and 0.05 mg/ml of poly(dI-dC)-poly(dI-dC)], with or without HeLa cell extract (Promega). Samples were loaded onto nondenaturing polyacrylamide gels and electrophoresed at 200 V at 4°C for 100 min in running buffer (0.5× TBE [Tris, boric acid, EDTA]). The sequences of the oligonucleotides used in the binding were as follows (lowercase letters represent sequences in which the nucleotides were mutated): 5′-CAGCTGGGGGGGCGGAGCAGTGACGTTTATTGCGGAGGGG-3′ (CRE-wt), 5′-CAGCTGGGGGGGCGGAGCctgtcgactcATTGCGGAGGGG-3′ (CRE-mt), 5′-CAGGGCCTTCACCAATCGGCGGCCTCCACGACGGGGCTGG-3′ (ERSE-wt), and 5′-CAGGGCCTTCAgtcgaCGGCGGCCTgagatACGGGGCTGG-3′ (ERSE-mt). For supershift experiments, samples were treated with various antibodies at 4°C for 1 h prior to incubation with a radiolabeled probe.
Permanent focal cerebral ischemia.
Male adult C57BL/6J mice weighing 24 to 34 g (Japan SLC) were used for the experiments and were housed under diurnal lighting conditions. Anesthesia was induced by 2.0% isoflurane and then maintained using 1% isoflurane in 70% N2O and 30% O2 using an animal general anesthesia machine (Soft Lander; Sin-ei Industry Co. Ltd., Saitama, Japan). The body temperature was maintained between 37.0°C and 37.5°C with the aid of a heating pad and heating lamp. A filament occlusion of the left middle cerebral artery (MCA) was carried out as previously described (7, 8). Briefly, the left MCA was occluded with an 8-0 nylon monofilament (Ethicon, Somerville, NJ) coated with a mixture of silicone resin (Xantopren; Bayer Dental, Osaka, Japan), as described (7, 8).
Histology and immunohistochemistry.
At 24 h after the MCA occlusion, mice were anesthetized with sodium pentobarbital and were perfused transcardially with 0.01 M phosphate-buffered saline (PBS; pH 7.4), followed by a fixative containing 4% paraformaldehyde in 0.1 M phosphate buffer. Brains were then immersed for 12 h in 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4), incubated for 24 h in 25% sucrose in 0.1 M phosphate buffer (pH 7.4) at 4°C, embedded in OCT (optimal cutting temperature) medium, and snap-frozen with powdered dry ice. Sections of a 10-μm thickness were cut on a cryostat and stained with hematoxylin-eosin or by immunohistochemistry using both mouse anti-BBF2H7 and rabbit anti-MAP2 (microtubule-associated protein 2) (SIGMA).
Knockdown of BBF2H7.
Annealed double-stranded short interfering RNA (siRNA) for BBF2H7 was obtained from Dharmacon (Lafayette, CO). The sequences of human-BBF2H7 siRNA were 5′-GAGUCUUGUUCAACUGAGAdTdT-3′ (sense) and 5′-UCUCAGUUGAACAAGACUCdTdT-3′ (antisense). BLAST searches (NCBI) confirmed that these sequences were not homologous to any genes other than BBF2H7. A lamin A/C siRNA (QIAGEN, Valencia, CA) was used as a control. SK-N-SH cells at 60% confluence in 24-well plates were transfected with 75 ng of each of the above siRNAs using the HyperFect transfection reagent (QIAGEN) according to the manufacturer's protocol. The transfected cells were incubated at 37°C for 36 h and then stimulated by ER stressors.
RESULTS
BBF2H7, a novel ER-resident transcription factor with a bZIP domain and belonging to the OASIS family, is induced and processed in response to ER stress.
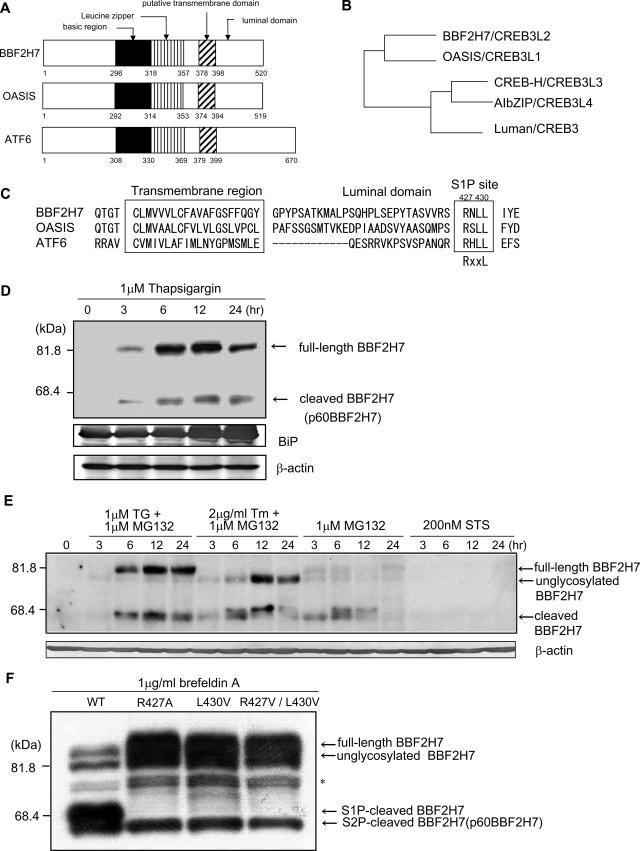
We searched for genes homologous to OASIS using nucleotide databases in order to identify novel ER stress transducers involved in UPR signaling. As a result, we found BBF2H7 (BBF2 human homolog on chromosome 7), a gene originally identified as being homologous to the Drosophila Bbf-2 gene (30). The overall structure of BBF2H7 is similar to those of OASIS and ATF6 (Fig. 1A). BBF2H7 contains a hydrophobic region predicted to be an alpha-helical transmembrane domain and a bZIP motif in the cytoplasmic portion. The bZIP motif of BBF2H7 is highly similar to that in the OASIS family of proteins; OASIS, CREB-H, AIbZIP, and Luman. OASIS is most akin (51% amino acid identity) to BBF2H7 (Fig. 1B). CREB-H (42% identity with BBF2H7), AIbZIP (34% identity), and Luman (37% identity) can be separated into a subgroup of the OASIS family.
FIG. 1.
In response to ER stress, BBF2H7 protein is induced and cleaved at the transmembrane region. (A) Predicted peptide features of human BBF2H7, OASIS, and ATF6. The basic region, leucine zipper, putative transmembrane domain, and luminal domain are indicated. (B) The phylogenetic tree of BBF2H7. The BBF2H7 gene encodes a protein that is akin to OASIS, CREB-H, AIbZIP, and Luman. The relationships between the orthologous proteins are inferred by identity between the proteins. (C) Comparison of amino acid sequences of transmembrane regions of mouse BBF2H7, OASIS, and ATF6. The right box denotes the consensus sequence of the S1P site (RxxL). The left box denotes the position of the transmembrane domains. (D) Expression of endogenous BBF2H7 in C6 glioma cells. Cells were incubated with 1 μM thapsigargin (TG) for the indicated times, and lysates were subjected to Western blotting with anti-BBF2H7 antibody. A strong 85-kDa band (full-length BBF2H7) and weak 60-kDa band (p60BBF2H7) appear in cells treated with ER stress. Anti-KDEL antibody recognizes BiP. Equal amounts of total protein were analyzed in each lane. M.W., molecular mass. (E) Expression of endogenous BBF2H7 in C6 glioma cells treated with ER stress and proteasome inhibitor, proteasome inhibitor only, or protein kinase inhibitor as a classic apoptotic stimulus that acts through mitochondria. Cells were incubated with 1 μM thapsigargin and 1 μM MG132, 2 μg/ml tunicamycin (Tm) (N-glycosylation inhibitor) and 1 μM MG132, 1 μM MG132 only, or 200 nM staurosporine (STS) for the indicated times, and lysates were subjected to Western blotting with anti-BBF2H7 antibody. By inhibiting degradation, a 60-kDa band (p60BBF2H7) appears strongly in cells. The partially N-glycosylated or nonglycosylated forms of BBF2H7 migrate faster than full-length BBF2H7. (F) Mutations of S1P recognition sites in BBF2H7 alter its proteolytic processing in transfected HEK293T cells. HEK293T cells were transfected with plasmids encoding Flag-tagged wild-type BBF2H7 (WT) or BBF2H7 with S1P mutations (R427A, L430V, and R427A/L430V). After 24 h, cells were treated with 1 μg/ml brefeldin A and then harvested after 1 h and subjected to Western blotting with an anti-Flag antibody. Note, the S1P-cleaved BBF2H7 band disappeared when transfection involved S1P mutations. However, S2P-cleaved BBF2H7 (=p60BBF2H7) could be seen via processing independent of S1P cleavage.
OASIS, CREB-H, AIbZIP, and ATF6 are reported to be cleaved at the membrane in response to ER stress (15, 22, 23, 29, 36, 37). Figure 1C compares the sequences of mouse BBF2H7, OASIS, and ATF6 in the transmembrane domain and its immediately surrounding region. In the putative luminal segment, BBF2H7 contains the sequence RNLL (beginning at residue 427, Fig. 1C), which fits the RxxL consensus for S1P, a membrane-anchored serine protease in the Golgi lumen, suggesting that BBF2H7 is cleaved by RIP, similarly to OASIS and ATF6 (22, 36).
To detect BBF2H7 expression at the protein level, Western blotting with an anti-BBF2H7 antibody was performed on C6 glioma cells that had been exposed to the ER stressor thapsigargin (1 μM), an inhibitor of ER Ca2+-ATPase. In the absence of thapsigargin, no BBF2H7 protein was detected (Fig. 1D). Treatment with thapsigargin led to the appearance of an 85-kDa band (full-length BBF2H7) and a 60-kDa band (p60BBF2H7). The expression level of p60BBF2H7 protein was much lower than that of full-length BBF2H7, indicating that cleaved BBF2H7 may be a low-stability protein allowing for easy degradation. Therefore, to determine whether cleaved BBF2H7 is easily degraded, Western blotting was performed on C6 glioma cells exposed to the proteasomal inhibitor MG132 in addition to ER stressors (Fig. 1E). A strong 60-kDa band was observed when proteasomal degradation was inhibited. When C6 glioma cells were exposed to MG132 only, low levels of full-length BBF2H7 and high levels of p60BBF2H7 were observed, because inhibition of proteasome activity also leads to ER stress (3). When C6 glioma cells were exposed to 200 nM staurosporine only, which does not cause ER stress, no BBF2H7 protein was detected. These results suggest that BBF2H7 protein is induced and cleaved in response to ER stress and that the cleaved BBF2H7 is easily degraded by the proteasome.
When cDNA encoding BBF2H7 with mutations at the S1P site was transfected into HEK293T cells, S1P-cleaved BBF2H7 could not be detected after treatment with brefeldin A, which induces ER stress by blocking the pathway from ER to the Golgi apparatus (Fig. 1F). This indicates that BBF2H7 is cleaved by S1P in response to ER stress, similarly to other OASIS family member proteins and ATF6.
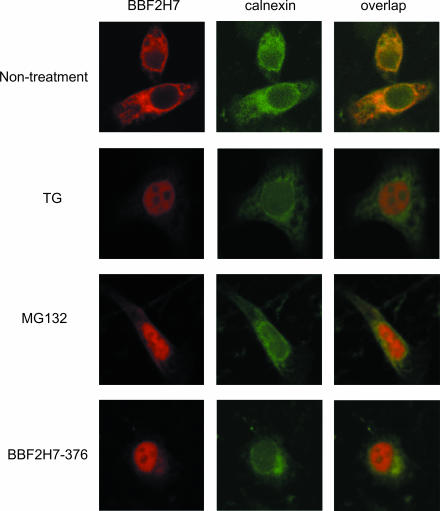
To detect subcellular localization of BBF2H7, immunofluorescence was performed. Immunofluorescence with an anti-BBF2H7 antibody showed fine reticular staining surrounding the nucleus, when BBF2H7 cDNA was transfected into HeLa cells (Fig. 2). The staining pattern was very similar to that obtained with antibody against calnexin, which is known to be a molecule found at the ER membrane. When cells were treated with thapsigargin, BBF2H7 immunoreactivity accumulated in the nucleus and overlap with that of calnexin was dramatically reduced. When cells were treated with MG132, which leads to ER stress by inhibition of proteasome activity (3), BBF2H7 immunoreactivity also accumulated in the nucleus and overlap with that of calnexin was reduced. When the expression plasmid for the BBF2H7 deletion-mutated protein lacking the transmembrane and luminal domains (BBF2H7-376) was transfected into HeLa cells, immunoreactivity also accumulated in the nucleus. We thus concluded that BBF2H7 is induced and cleaved in response to ER stress and that the released p60BBF2H7 is translocated into the nucleus, similarly to other OASIS family member proteins and ATF6. These results suggest that BBF2H7 is a novel ER stress transducer.
FIG. 2.
Subcellular localization of BBF2H7. HeLa cells transfected with BBF2H7 were incubated in the presence or absence of 1 μM thapsigargin (TG) or 10 μM MG132 for 6 h and then costained with anti-BBF2H7 and anticalnexin antibodies. Note that BBF2H7 immunoreactivity completely overlaps that of calnexin under normal conditions, but after ER stress or proteasome inhibition, the BBF2H7 signal accumulates in the nucleus. The lower panel shows that the BBF2H7 deletion-mutated protein lacking the transmembrane and luminal domains (BBF2H7-376) localized in the nucleus under normal conditions.
BBF2H7 protein is markedly induced at the translational level during ER stress.
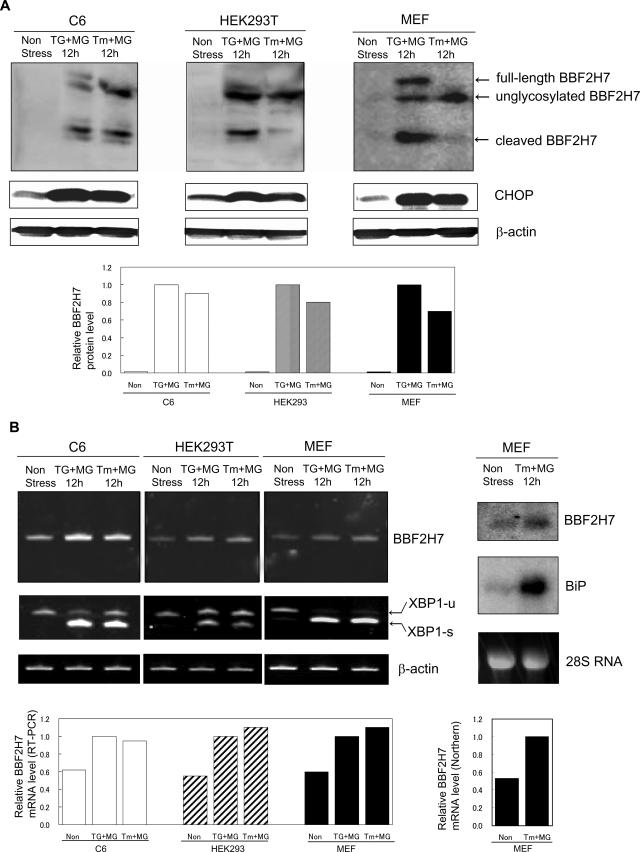
To investigate which type of cells express BBF2H7 at the protein and mRNA levels, Western blotting with an anti-BBF2H7 antibody and RT-PCR analysis were performed on C6 glioma, HEK293T, and MEF cells. In all cell types examined, full-length and cleaved BBFH7 protein appeared in response to ER stress and the amount of total BBF2H7 protein was increased by more than 10-fold (Fig. 3A). On the other hand, while BBF2H7 mRNA levels also increased in response to ER stress, the induction level was only about twofold and therefore was much lower than the magnitude of induction of protein (Fig. 3B). The dissociation between the levels of increase in protein and mRNA for BBF2H7 is similar to that for ATF4 during ER stress. It is well known that stress-induced eIF2α phosphorylation paradoxically increases translation of ATF4, activating the integrated stress response (ISR), a pro-survival gene expression program (9, 19).
FIG. 3.
Up-regulation of BBF2H7 in response to ER stress in various cell types. (A) Expression of endogenous BBF2H7 in C6 glioma cells, HEK293T cells, and MEF cells treated with ER stress and proteasome inhibitor. Cells were incubated with 300 nM thapsigargin (TG) or 1 μg/ml tunicamycin (Tm) with 3 μM MG132 for 12 h, and lysates were subjected to Western blotting with anti-BBF2H7 antibody. (Bottom) Quantitative analysis of BBF2H7 protein expression levels. (B) Weak induction of BBF2H7 mRNA in C6 glioma cells, HEK293T cells, and MEF cells treated with ER stress and proteasome inhibitor. (Left) Cells were incubated with 300 nM thapsigargin or 1 μg/ml tunicamycin with 3 μM MG132 for 12 h, and RT-PCR analysis was performed. (Bottom) Quantitative analysis of BBF2H7 mRNA expression levels. (Right) MEF cells were incubated with 1 μg/ml tunicamycin with 3 μM MG132 for 12 h, and Northern blot analysis was performed. Bottom, quantitative analysis of BBF2H7 mRNA expression levels.
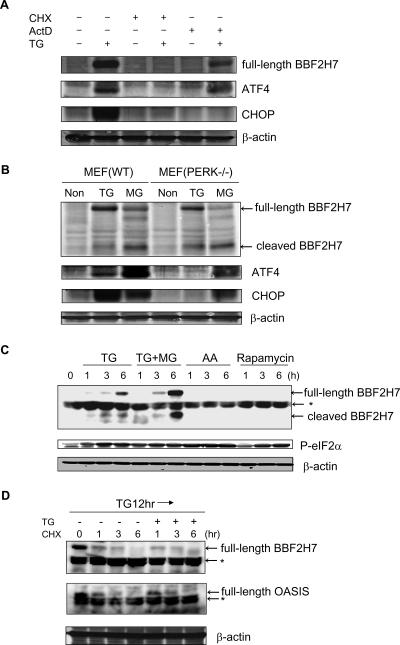
To elucidate whether BBF2H7 protein is up-regulated by posttranscriptional mechanisms similarly to the case of ATF4, we performed Western blotting on MEF cells treated with ER stress for 6 h in the presence or absence of the translation inhibitor CHX or the transcription inhibitor ActD. As expected, BBF2H7 protein, like ATF4, was up-regulated with thapsigargin and ActD (Fig. 4A). On the other hand, as previously reported, CHOP protein was not up-regulated (9). Neither BBF2H7, ATF4, nor CHOP was up-regulated with thapsigargin and CHX. These results suggest that BBF2H7, as in the case of ATF4, is translationally induced after ER stress.
FIG. 4.
BBF2H7 protein is markedly induced at the translational level during ER stress, but not by the PERK-eIF2α pathway. (A) The transcriptional induction step is not essential for up-regulation of BBF2H7 protein. MEF cells were exposed to 1 μM thapsigargin (TG) for 6 h in the presence or absence of 1 μg/ml CHX as a translation inhibitor or 5 μg/ml ActD as a transcription inhibitor. Levels of BBF2H7, ATF4, CHOP, and β-actin were examined by Western blotting. Note that both BBF2H7 protein and ATF4 protein were up-regulated with thapsigargin and ActD, but CHOP protein was not. (B) Wild-type MEF cells or PERK-deficient MEF cells were exposed to 1 μM thapsigargin or 10 μM MG132 (MG) for 6 h or non-stress conditions (Non). Levels of BBF2H7, ATF4, CHOP, and β-actin were measured by Western blotting analysis. Note, in PERK-deficient MEF cells, BBF2H7 protein was up-regulated with thapsigargin, but neither ATF4 protein nor CHOP protein was. (C) C6 glioma cells were exposed to 1 μM thapsigargin, 1 μM thapsigargin with 10 μM MG132 (TG+MG), amino acid starvation (AA), or 10 μM rapamycin for the indicated times. Levels of BBF2H7, phosphorylated eIF2α, and β-actin were examined by Western blotting. An asterisk indicates a nonspecific band. (D) C6 glioma cells were exposed to 1 μM thapsigargin for 12 h and then exposed to 1 μg/ml CHX as a translation inhibitor for the indicated times with or without 1 μM thapsigargin. Levels of BBF2H7, OASIS, and β-actin were examined by Western blotting analysis. An asterisk indicates a nonspecific band. Note that the degradation rate of BBF2H7 protein, as well as that of OASIS protein, is not dependent on ER stress.
Next, to elucidate whether BBF2H7 protein is up-regulated by the PERK-eIF2α pathway in a similar manner to ATF4, we performed Western blotting on wild-type MEF cells and PERK-deficient MEF cells. Unexpectedly, while neither ATF4 nor CHOP was induced in PERK-deficient MEF cells treated with thapsigargin, BBF2H7 protein appeared in PERK-deficient MEF cells as well as in wild-type MEF cells (Fig. 4B). This result suggests that induction of BBF2H7 protein is not dependent on PERK, a major ER stress transducer.
Next, to elucidate whether phosphorylation of eIF2α is necessary for BBF2H7 translation during ER stress, we performed Western blotting on C6 glioma cells exposed to various types of stress (Fig. 4C). Amino acid starvation and rapamycin, which induce eIF2α phosphorylation, did not induce BBF2H7 protein. These results suggest that while BBF2H7 protein is dramatically up-regulated specifically during ER stress, it is not via the PERK-eIF2α pathway.
To exclude the possibility that the increased BBF2H7 protein levels during ER stress were mediated by increased protein stability, we investigated the degradation rate of BBF2H7 protein during ER stress, while inhibiting translation. After treatment with ER stressor for 12 h and then treatment with the translational inhibitor CHX for the indicated times, BBF2H7 protein gradually disappeared either with or without ER stress treatment, in synchronicity with a decrease in OASIS protein (Fig. 4D). This result suggests that the degradation rate of BBF2H7 protein, as well as OASIS protein, does not change during ER stress and that the increase in BBF2H7 protein levels during ER stress is caused by an up-regulation of translation and not by down-regulation of degradation.
BBF2H7 has the potential to activate transcription of target genes via direct binding to the CRE site.
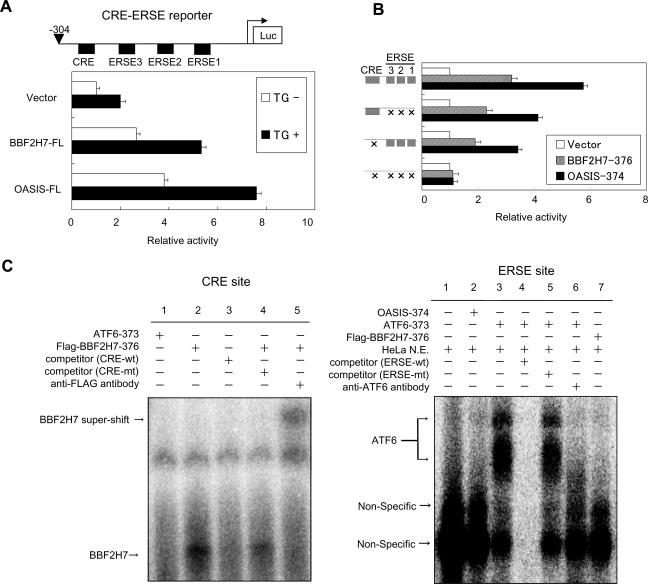
Since the N-terminal portion of BBF2H7 with the bZIP domain translocates into the nucleus in response to ER stress, BBF2H7 could activate the transcription of UPR target genes via direct binding to CRE sites as in the case of OASIS and ATF4, or to ERSE sites, as in the case of ATF6 and XBP1. To investigate whether BBF2H7 can activate the CRE and ERSE sites, we examined the effects of BBF2H7 on the expression of a reporter gene carrying the human BiP promoter, which contains a CRE site and three tandem ERSE sites, immediately upstream of the firefly luciferase gene (CRE-ERSE reporter). Full-length BBF2H7 (BBF2H7-FL) activated expression of luciferase from the CRE-ERSE reporter after ER stress, as in the case of OASIS, although the level of activation by BBF2H7 is somewhat lower (about 30% lower) than that by OASIS (Fig. 5A).
FIG. 5.
Effects of BBF2H7 on CRE and ERSE sites. (A) Reporter assay. C6 glioma cells were transfected with the indicated constructs and the CRE-ERSE reporter plasmid. The relative luciferase activities in the cells incubated in the presence (solid boxes) or absence (open boxes) of 1 μM thapsigargin (TG) for 16 h were determined; the means ± standard deviation of the results from four independent experiments are shown. The upper panel shows the schematic structure of the reporter construct. (B) Reporter assay. CRE and ERSE1-3 of the CRE-ERSE reporter were disrupted by mutating their sequences (15). The left panel represents intact and mutated constructs. The columns show the relative activities of reporter assays after transfection of the indicated constructs in C6 glioma cells. The luciferase (Luc) activity in each sample was normalized and plotted with each nontransfected reporter activity set as 1. (C) EMSA of CRE and ERSE sites. (Left) Direct binding of BBF2H7 to the CRE site. The 32P-labeled CRE sequence of the BiP promoter was incubated with in vitro-translated Flag-BBF2H7-376. Note that competition by a 100-fold excess of the unlabeled CRE sequence (lane 3) and a supershift after incubation with an anti-Flag antibody (lane 5) are detected. (Right) Direct binding of BBF2H7 to the ERSE site. The 32P-labeled ERSE sequence of the BiP promoter was incubated with in vitro-translated Flag-BBF2H7-376. Note that ATF6 can bind to ERSE using nuclear extract (N.E.) from HeLa cells (lane 3). Neither OASIS (lane 2) nor BBF2H7 (lane 7) can bind to ERSE.
Next, to investigate whether the effect of BBF2H7 on the CRE-ERSE reporter was mediated by CRE or ERSE, we examined the CRE-ERSE reporter with mutations in the CRE and/or ERSE sites (Fig. 5B). Mutation of ERSEs alone reduced by about 40% the reporter activity induced by overexpression of BBF2H7-376, compared with the wild-type promoter. Mutation of the CRE site also decreased reporter expression, by more than 60%, and the effects exhibited were therefore more severe than those for mutated ERSE. This indicates that BBF2H7 can potentially target both CRE and ERSE sites, but preferentially acts as a transcription activator at CRE sites, as in the case of OASIS.
To confirm the action at CRE and ERSE sites, EMSA was performed using CRE and ERSE sequences in the BiP promoter (Fig. 5C). EMSA revealed that BBF2H7 bound to the CRE probe but not to the ERSE probe. The reason that no binding of BBF2H7 to ERSE was detected is not known, although it is possible that the binding of BBF2H7 is either very weak or requires other nuclear factors to bind directly to the ERSE. Together, these results suggest that BBF2H7 may modulate transcriptional induction of some ER stress-response genes that contain CRE sites in their promoter regions.
In vivo, BBF2H7 protein is markedly induced in a mouse model of permanent focal brain ischemia.
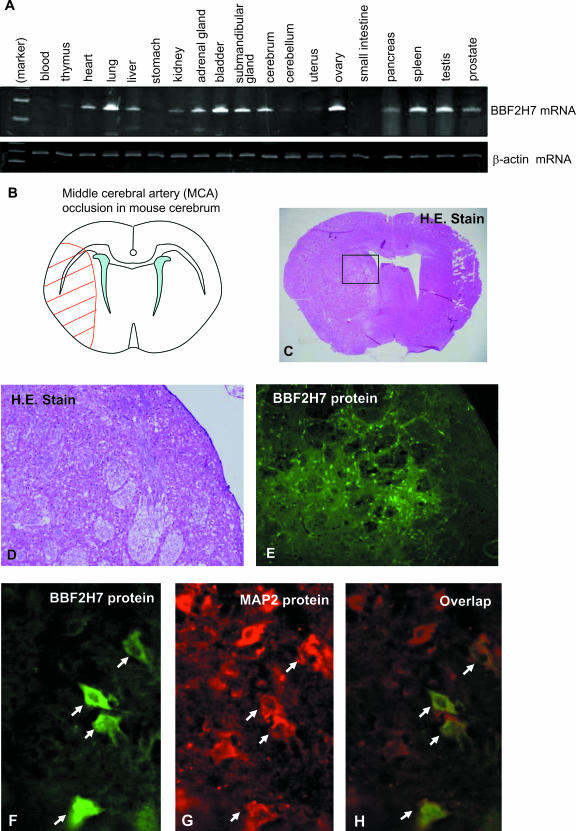
To investigate the expression pattern of BBF2H7 mRNA in mouse adult tissues, we performed RT-PCR analysis using total RNA extracted from 19 organs or tissues. BBF2H7 mRNA was widely detected in many organs or tissues (Fig. 6A). Among these, we focused on the brain, because BBF2H7 is highly expressed in C6 glioma cells (Fig. 1D) and, since reactive astrocytes express OASIS in the cryo-injured brain, BBF2H7 could possibly also be induced in reactive astrocytes. In view of this hypothesis, we decided to assess the expression of BBF2H7 in a mouse model of permanent focal brain ischemia, which induces accumulation of immature proteins in the ER and causes ER stress (4, 12, 21). Moreover, brain ischemia induces hypoxia, which also causes ER stress (16, 31). We therefore performed permanent left MCA occlusion in adult male mice. Twenty-four hours after MCA occlusion, mice developed infarcts affecting both the cortex and striatum (Fig. 6B and C, left side).
FIG. 6.
Expression analysis of the BBF2H7 gene. (A) RT-PCR analysis was performed by using RNA extracted from adult mice. (B) Schematic representation of permanent MCA occlusion in the mouse cerebrum. The red hatched area represents the region of ischemic damage. (C) Hematoxylin-eosin (H.E.) staining of coronal brain section at 24 h after permanent MCA occlusion. Note that only the ischemic side (left side) of the brain is damaged, as shown in panel B. (D) A higher magnification of boxed area in panel C. (E) Immunohistochemistry for BBF2H7 protein in coronal brain section at 24 h after permanent MCA occlusion. Note that BBF2H7-positive cells can be detected only in the peri-infarction region of the striatum. (F to H) Double-labeling immunohistochemistry for BBF2H7 and MAP2 proteins at a higher magnification than in panel E.
Immunohistochemical analysis using the anti-BBF2H7 antibody indicated expression of BBF2H7 protein within areas of the brain subjected to ischemic injury by this procedure, especially in the peri-infarction region (also referred to as ischemic penumbra) in the striatum, but not in the cerebral cortex (Fig. 6E). One possible explanation for this involves the concept of the ischemic core. There should be more damage in the ischemic core (striatum), where there is a constrained blood supply (11). In contrast to the ischemic sides, no BBF2H7 staining was observed in brain tissue from the nonischemic control sides or in brain tissue from mice under normal conditions (data not shown). Double labeling with MAP2, a specific marker for neurons, was utilized for colocalization of BBF2H7. The peri-infarction region in the striatum showed colocalization of BBF2H7 with MAP2, suggesting that some neurons were expressing BBF2H7 (Fig. 6F to H). We did not detect colocalization of BBF2H7 with glial fibrillary acidic protein (GFAP), a specific marker for astrocytes (data not shown). The reason for this is not known, although it is possible that the proliferation of GFAP-positive reactive astrocytes begins more than 2 days after ischemic injury. Therefore, it may be difficult to detect BBF2H7-positive astrocytes.
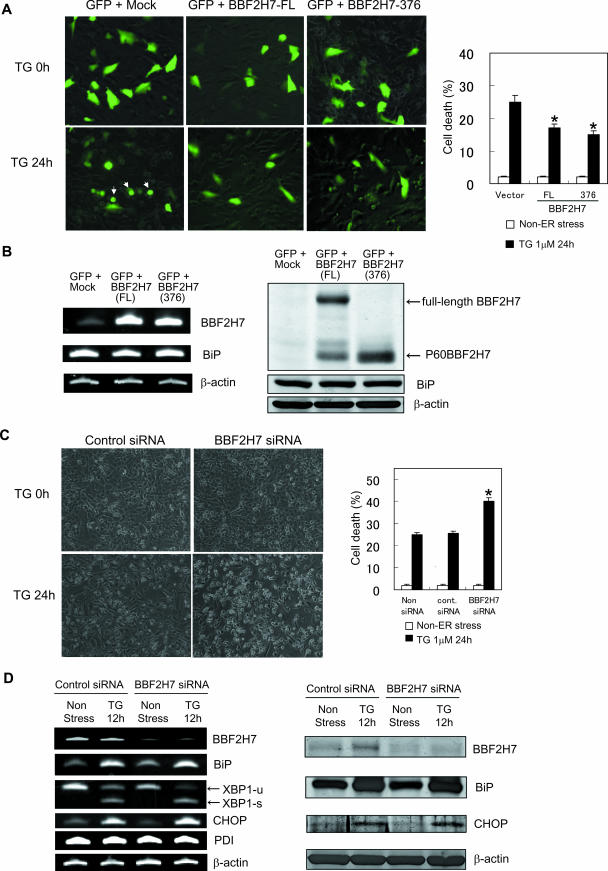
BBF2H7 protects neurons from ER stress-induced cell death.
We have shown that BBF2H7 is induced in damaged neurons in brain ischemia (Fig. 6E to H). We wished, therefore, to explore the physiological significance of this observation. To investigate the physiological function of BBF2H7 during ER stress in neurons, human neuroblastoma cell line SK-N-SH cells were transfected with green fluorescent protein (GFP) with either mock, full-length BBF2H7, or BBF2H7-376, and cell viability was determined 24 h after treatment with thapsigargin. As expected, we observed that overexpression of full-length BBF2H7 or BBF2H7-376 attenuated ER stress-induced cell death (Fig. 7A). To investigate how overexpression of BBF2H7 reduced ER stress-induced cell death in SK-N-SH cells, we tested the expression level of BiP, because BBF2H7 activated expression of luciferase from the CRE-ERSE reporter of BiP (Fig. 5A). Unexpectedly, the expression levels of BiP mRNA and BiP protein were not increased by overexpression of BBF2H7 in SK-N-SH cells (Fig. 7B). The reason for the inconsistency between the significant activity of BBF2H7 on the CRE-ERSE reporter, and no endogenous BiP induction in SK-N-SH cells overexpressing BBF2H7 is not known, although it is possible that some other factors are required to induce endogenous BiP. Another possibility is that BBF2H7 may modulate transcriptional induction of some other genes, which contain CRE site(s) in their promoter regions and have functions in attenuating ER stress-induced cell death.
FIG. 7.
Changes in BBF2H7 levels modulate ER stress-induced cell death in neurons. (A) Overexpression of BBF2H7 protected cells against ER stress-induced cell death. (Left) SK-N-SH cells were transiently transfected with empty, full-length BBF2H7, or BBF2H7-376 with pEGFP vector. After 36 h, cells were treated with 1 μM thapsigargin (TG) for the indicated times. The morphology was observed by fluorescence detection of GFP. The morphological changes such as shrinkage and formation of apoptotic bodies (arrows) were reduced in BBF2H7-transfected cells. (Right) Quantitative analysis of cell death at 24 h after treatment with thapsigargin. Cell death was determined by the morphology (round and shrunken cells were counted as dead cells). Data are the means ± standard deviations of the results of four independent experiments.  , P < 0.05 relative to the control (Student's t test). (B) SK-N-SH cells were transiently transfected with empty, full-length BBF2H7 or BBF2H7-376 with pEGFP vector. After 36 h, total RNA was extracted and subjected to RT-PCR analysis (left) or total protein was extracted and subjected to Western blotting analysis (right). Note that the expression levels of BiP were not changed by overexpression of BBF2H7. (C) BBF2H7 siRNA promoted ER stress-induced cell death. (Left) Representative phase-contrast images. SK-N-SH cells were treated with the indicated siRNAs, incubated for 36 h, and then stimulated with 1 μM thapsigargin for the indicated times. (Right) Quantitative analysis of cell death at 24 h after treatment with thapsigargin. Cell death was determined by the morphology (round and shrunken cells were counted as dead cells). Data are the means ± standard deviation of the results of four independent experiments.
, P < 0.05 relative to the control (Student's t test). (B) SK-N-SH cells were transiently transfected with empty, full-length BBF2H7 or BBF2H7-376 with pEGFP vector. After 36 h, total RNA was extracted and subjected to RT-PCR analysis (left) or total protein was extracted and subjected to Western blotting analysis (right). Note that the expression levels of BiP were not changed by overexpression of BBF2H7. (C) BBF2H7 siRNA promoted ER stress-induced cell death. (Left) Representative phase-contrast images. SK-N-SH cells were treated with the indicated siRNAs, incubated for 36 h, and then stimulated with 1 μM thapsigargin for the indicated times. (Right) Quantitative analysis of cell death at 24 h after treatment with thapsigargin. Cell death was determined by the morphology (round and shrunken cells were counted as dead cells). Data are the means ± standard deviation of the results of four independent experiments.  , P < 0.05 relative to the control (Student's t test). (D) SK-N-SH cells were treated with the indicated siRNAs, incubated for 36 h, and then stimulated with 1 μM thapsigargin for the indicated times. (Left) RT-PCR was performed to examine the levels of the indicated genes. (Right) Western blotting analysis was performed to examine the levels of the indicated proteins. Note that the spliced form of XBP-1 mRNA (XBP1-s) is increased in BBF2H7 siRNA-treated cells compared with the control siRNA-treated cells, when the cells were treated with thapsigargin for 12 h.
, P < 0.05 relative to the control (Student's t test). (D) SK-N-SH cells were treated with the indicated siRNAs, incubated for 36 h, and then stimulated with 1 μM thapsigargin for the indicated times. (Left) RT-PCR was performed to examine the levels of the indicated genes. (Right) Western blotting analysis was performed to examine the levels of the indicated proteins. Note that the spliced form of XBP-1 mRNA (XBP1-s) is increased in BBF2H7 siRNA-treated cells compared with the control siRNA-treated cells, when the cells were treated with thapsigargin for 12 h.
Next, to explore the physiological function of endogenous BBF2H7 in neurons, we silenced the BBF2H7 gene in SK-N-SH cells using siRNA, and cell viability was determined 24 h after treatment with thapsigargin. Conversely, knockdown of BBF2H7 enhanced cell sensitivity to ER stress (Fig. 7C). To investigate how knockdown of BBF2H7 promoted ER stress-induced cell death in SK-N-SH cells, we tested the RNA and/or protein expression levels of BiP, XBP-1, CHOP, and PDI. Although we did not detect significant reductions in the expression of the genes coding for these proteins, the spliced form of XBP-1 mRNA is increased in BBF2H7 siRNA-treated cells compared with the level in control siRNA-treated cells during ER stress (Fig. 7D). The reason for this is not known, although it is possible that knockdown of BBF2H7 decreased transcription of some target genes, enhanced the severity of ER stress, and consequently increased expression of the spliced form of XBP-1 mRNA. Together, these results suggest that BBF2H7 protects neurons from ER stress-induced cell death, by way of modulating transcriptional induction of some unknown ER stress response genes.
DISCUSSION
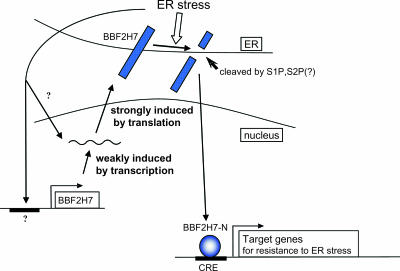
In this study, we have characterized a protein, BBF2H7, that we propose functions as an ER stress transducer in the mammalian UPR. The following facts support the proposed role of BBF2H7 as an ER stress transducer. First, BBF2H7 is an ER-resident transmembrane protein and is cleaved at the membrane in response to ER stress. In the luminal segment, BBF2H7 contains the RxxL consensus for S1P; indicating that BBF2H7 is cleaved by the same mechanisms as are OASIS and ATF6. Second, cleaved fragments of the BBF2H7 N-terminal portion containing the bZIP domain translocate into nuclei. Third, BBF2H7 protein is prominently induced at the translational level during ER stress. Finally, the BBF2H7 N-terminal fragment has the potential to bind directly to the CRE site and activate the transcription of target genes for resistance to ER stress (Fig. 8).
FIG. 8.
Putative mechanisms of BBF2H7 contribution to the UPR. BBF2H7 is weakly induced at the transcriptional level and strongly induced at the translational level during ER stress. Translated BBF2H7 is cleaved at the membrane by S1P and probably S2P in response to ER stress; its cleaved N-terminal cytoplasmic domain then translocates into the nucleus and activates transcription of ER stress resistance genes via direct binding at the CRE site.
In the present study, we showed that BBF2H7 protein is markedly induced at the translational level during ER stress, but not by the PERK-eIF2α pathway (Fig. 4). Generally, when ER stress occurs, activated PERK phosphorylates eIF2α to attenuate the rate of general translation initiation. The rapid and reversible regulation of mRNA translation prevents further synthesis of proteins when the ER lumen is compromised in its protein-folding capacity. Paradoxically, phosphorylation of eIF2α preferentially increases the translation of selective mRNAs that contain inhibitory upstream open-reading frames (uORFs) within their 5′-untranslated region (UTR). The best-studied example in mammalian cells is the eIF2α phosphorylation-dependent translation of ATF4 mRNA (9, 19). Although BBF2H7 in humans, mice, and rats has multiple AUG start codons in the 5′-UTR, and these could be candidates for uORFs, the PERK-eIF2α pathway was not relevant to BBF2H7 translation, as shown in Fig. 4. Although we could not clarify the precise mechanism of up-regulation of BBF2H7 translation, one possibility is that internal ribosome entry site (IRES) may contribute to its translation. BiP has been reported to have IRES activity (20, 33), and translational induction of human inhibitor of apoptosis protein 2 (HIAP2) during ER stress is also mediated via an IRES element (35). It has been reported that IRES-containing 5′-UTRs show a common tendency in terms of the length, number of upstream AUGs, and percent GC content (2). Interestingly, the 5′-UTR of BBF2H7 fits these requirements. In the near future, we would like to analyze whether or not the 5′-UTR of BBF2H7 has IRES activity. Further studies are needed to clarify how BBF2H7 protein is induced at the translational level during ER stress.
Among reported ER stress transducers, BBF2H7 is the first molecule that is up-regulated by translation. Generally, ER stress transducers, such as IRE1, PERK, ATF6, and OASIS, are expressed at the protein level under normal conditions, so as to be prepared to detect ER stress conditions immediately and to transduce signals from the ER to the cytoplasm and nucleus. However, because BBF2H7 protein is not expressed under normal conditions, it cannot function to detect ER stress conditions in the early phase. Only after BBF2H7 protein has been translated in response to ER stress can it function as an ER stress transducer. This unique regulation suggests that BBF2H7 may therefore function in only the late phase of UPR signaling.
In the brains injured by focal ischemia, BBF2H7 protein is dramatically induced only in neurons in the penumbra regions (Fig. 6E to H). However, in vitro experiments showed that BBF2H7 was strongly induced in astrocyte-derived C6 glioma cells. The expression pattern of BBF2H7 is similar to that of ORP150 (oxygen-regulated protein 150 kDa), which was identified as a molecule that is induced in cultured astrocytes exposed to hypoxic stress (17). ORP150, which is an ER-associated chaperone, has been reported to be induced preferentially in neurons after brain ischemia (32). The previous findings as well as those of the present study may indicate that in vivo, neurons exposed to physiological crises rapidly induce both BBF2H7 and ORP150 to protect cells from damage at the early phase of brain injury, at a stage when the reactive astrocytes have not yet proliferated. In contrast, another ER stress transducer, OASIS, was not expressed in neurons at all after brain injury and was induced specifically in reactive astrocytes (15). The clarification of the different mechanisms of regulation of expression for each ER stress transducer in a cell-type-specific manner may help elucidate the detailed mechanisms responsible for acute response to brain damage and for neuronal regeneration.
In the present study, we were unable to obtain clear evidence as to the identity of the target genes of BBF2H7. Our findings indicate that the target genes contain a CRE site or sites in their promoter regions (Fig. 5) and have functions in suppressing cell sensitivity to ER stress (Fig. 7A and C). Although the most likely candidate was BiP, which contains a CRE site and has a function towards suppression of ER stress, we did not observe clear evidence for this in the experiments involving overexpression of BBF2H7 (Fig. 7B) and knockdown of BBF2H7 (Fig. 7D) in SK-N-SH cells. In this case, there are at least three possibilities that could explain the situation. First, although the target of BBF2H7 is BiP, some other factors are required for induction of endogenous BiP in overexpression of BBF2H7. Second, although the target of BBF2H7 is BiP, it is difficult to detect attenuation of BiP induction in BBF2H7 knockdown cells, because BiP is well known to be modulated by many UPR transcription factors, such as XBP1, ATF6, and ATF4 in neurons. Deletion of BBF2H7 and other transcription factors in various combinations would be needed to clarify the requisite role in BiP induction. Third, the target of BBF2H7 is not BiP, but some other genes which contain a CRE site or sites in their promoter regions and have functions in attenuating ER stress-induced cell death. To elucidate the target genes of BBF2H7 more clearly, further studies are needed, such as microarray analysis or those using BBF2H7-deficient mice.
Among the OASIS family, CREB-H knockdown mouse embryos and AIbZIP-deficient mice have already been reported. Although the histopathological analysis of the CREB-H knockdown embryos at E14.5 did not reveal any morphological or developmental defects, CREB-H is cleaved by site 1 and site 2 proteases to liberate an N-terminal fragment that transits to the nucleus to activate transcription of the genes encoding SAP and CRP (37). AIbZIP-deficient mice were born at expected ratios, were healthy, and displayed normal long-term survival rates. Although the seminiferous tubules of AIbZIP-deficient mice contained all of the developmental stages, there was evidence for increased apoptosis of meiotic/postmeiotic germ cells (1). More recently, it has been reported that, in the testis of AIbZIP-deficient mice, epididymal sperm nuclei were abnormally relaxed (24). The phenotypes of mice deficient in members of the OASIS family are milder those of mice deficient in the major ER stress transducers IRE1 and PERK (10, 34). These recent studies might indicate that OASIS family proteins play an auxiliary role in the major ER stress transducer pathway.
In conclusion, this study demonstrates that a novel OASIS family member protein, BBF2H7, acts as a novel ER stress transducer and is markedly induced at the translational level during ER stress. BBF2H7 protein is up-regulated in damaged neurons after ischemia. Furthermore, BBF2H7 protects neurons from ER stress-induced cell death. Our study indicates that future research efforts are merited on the analysis of BBF2H7 and therapy for diseases related to ER stress.
Acknowledgments
We thank Kazutoshi Mori for providing plasmids and antibodies, David Ron for providing the PERK-deficient MEF cells, and Masaya Tohyama and Sadao Shiosaka for helpful discussions and critical reading of the manuscript.
This work was partly supported by the Uehara Memorial Foundation, Japan Health Foundation for the Prevention of Chronic Diseases and the Improvement of QOL of Patients, Suzuken Memorial Foundation, and grants from the JSPS KAKENHI (no. 17200026 and 18700366).
Footnotes
Published ahead of print on 18 December 2006.
REFERENCES
- 1.Adham, I. M., T. J. Eck, K. Mierau, N. Müller, M. A. Sallam, I. Paprotta, S. Schubert, S. Hoyer-Fender, and W. Engel. 2005. Reduction of spermatogenesis but not fertility in Creb3l4-deficient mice. Mol. Cell. Biol. 25:7657-7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baird, S. D., M. Turcotte, R. G. Korneluk, and M. Holcik. 2006. Searching for IRES. RNA 12:1755-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush, K. T., A. L. Goldberg, and S. K. Nigam. 1997. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J. Biol. Chem. 272:9086-9092. [DOI] [PubMed] [Google Scholar]
- 4.DeGracia, D. J., and H. L. Montie. 2004. Cerebral ischemia and the unfolded protein response. J. Neurochem. 91:1-8. [DOI] [PubMed] [Google Scholar]
- 5.DenBoer, L. M., P. W. Hardy-Smith, M. R. Hogan, G. P. Cockram, T. E. Audas, and R. Lu. 2005. Luman is capable of binding and activating transcription from the unfolded protein response element. Biochem. Biophys. Res. Commun. 331:113-119. [DOI] [PubMed] [Google Scholar]
- 6.Erickson, A. H., and G. Blobel. 1979. Early events in the biosynthesis of the lysosomal enzyme Cathepsin D. J. Biol. Chem. 254:11771-11774. [PubMed] [Google Scholar]
- 7.Hara, H., R. M. Friedlander, V. Gagliardini, C. Ayata, K. Fink, Z. Huang, M. Shimizu-Sasamata, J. Yuan, and M. A. Moskowitz. 1997. Inhibition of interleukin 1β converting enzyme family proteases reduces ischemic and excitotoxic neuronal damage. Proc. Natl. Acad. Sci. USA 94:2007-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hara, H., P. L. Huang, N. Panahian, M. C. Fishman, and M. A. Moskowitz. 1996. Reduced brain edema and infarction volume in mice lacking the neuronal isoform of nitric oxide synthase after transient MCA occlusion. J. Cereb. Blood Flow Metab. 16:605-611. [DOI] [PubMed] [Google Scholar]
- 9.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 10.Harding, H. P., H. Zeng, Y. Zhang, R. Jungries, P. Chung, H. Plesken, D. D. Sabatini, and D. Ron. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol. Cell 7:1153-1163. [DOI] [PubMed] [Google Scholar]
- 11.Hossmann, K. A. 1994. Viability thresholds and the penumbra of focal ischemia. Ann. Neurol. 36:557-565. [DOI] [PubMed] [Google Scholar]
- 12.Hu, B. R., S. Janelidze, M. D. Ginsberg, R. Busto, M. Perez-Pinzon, T. J. Sick, B. K. Siesjo, and C. L. Liu. 2001. Protein aggregation after focal brain ischemia and reperfusion. J. Cereb. Blood. Flow Metab. 21:865-875. [DOI] [PubMed] [Google Scholar]
- 13.Imaizumi, K., M. Tsuda, Y. Imai, A. Wanaka, T. Takagi, and M. Tohyama. 1997. Molecular cloning of a novel polypeptide, DP5, induced during programmed neuronal death. J. Biol. Chem. 272:18842-18848. [DOI] [PubMed] [Google Scholar]
- 14.Kaufman, R. J. 2002. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 110:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo, S., T. Murakami, K. Tatsumi, M. Ogata, S. Kanemoto, K. Otori, K. Iseki, A. Wanaka, and K. Imaizumi. 2005. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat. Cell Biol. 7:186-194. [DOI] [PubMed] [Google Scholar]
- 16.Koumenis, C., C. Naczki, M. Koritzinsky, S. Rastani, A. Diehl, N. Sonenberg, A. Koromilas, and B. G. Wouters. 2002. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol. Cell. Biol. 22:7405-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwabara, K., M. Matsumoto, J. Ikeda, O. Hori, S. Ogawa, Y. Maeda, K. Kitagawa, N. Imuta, T. Kinoshita, D. M. Stern, H. Yanagi, and T. Kamada. 1996. Purification and characterization of a novel stress protein, the 150-kDa oxygen-regulated protein (ORP150), from cultured rat astrocytes and its expression in ischemic mouse brain. J. Biol. Chem. 271:5025-5032. [DOI] [PubMed] [Google Scholar]
- 18.Liang, G., T. E. Audas, Y. Li, G. P. Cockram, J. D. Dean, A. C. Martyn, K. Kokame, and R. Lu. 2006. Luman/CREB3 induces transcription of the endoplasmic reticulum (ER) stress response protein Herp through an ER stress response element. Mol. Cell. Biol. 26:7999-8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, P. D., H. P. Harding, and D. Ron. 2004. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macejak, D. G., and P. Sarnow. 1991. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature 353:90-94. [DOI] [PubMed] [Google Scholar]
- 21.Mouw, G., J. L. Zechel, J. Gamboa, W. D. Lust, W. R. Selman, and R. A. Ratcheson. 2003. Activation of caspase-12, an endoplasmic reticulum resident caspase, after permanent focal ischemia in rat. Neuroreport 14:183-186. [DOI] [PubMed] [Google Scholar]
- 22.Murakami, T., S. Kondo, M. Ogata, S. Kanemoto, A. Saito, A. Wanaka, and K. Imaizumi. 2006. Cleavage of the membrane-bound transcription factor OASIS in response to endoplasmic reticulum stress. J. Neurochem. 96:1090-1100. [DOI] [PubMed] [Google Scholar]
- 23.Nagamori, I., N. Yabuta, T. Fujii, H. Tanaka, K. Yomogida, Y. Nishimune, and H. Nojima. 2005. Tisp40, a spermatid specific bZip transcription factor, functions by binding to the unfolded protein response element via the Rip pathway. Genes Cells 10:575-594. [DOI] [PubMed] [Google Scholar]
- 24.Nagamori, I., K. Yomogida, M. Ikawa, M. Okabe, N. Yabuta, and H. Nojima. 2006. The testes-specific bZip type transcription factor Tisp40 plays a role in ER stress responses and chromatin packaging during spermiogenesis. Genes Cells 11:1161-1171. [DOI] [PubMed] [Google Scholar]
- 25.Newman, J. R., and A. E. Keating. 2003. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300:2097-2101. [DOI] [PubMed] [Google Scholar]
- 26.Omori, Y., J. Imai, M. Watanabe, T. Komatsu, Y. Suzuki, K. Kataoka, S. Watanabe, A. Tanigami, and S. Sugano. 2001. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 29:2154-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raggo, C., N. Rapin, J. Stirling, P. Gobeil, E. Smith-Windsor, P. O'Hare, and V. Misra. 2002. Luman, the cellular counterpart of herpes simplex virus VP16, is processed by regulated intramembrane proteolysis. Mol. Cell. Biol. 22:5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 110:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stirling, J., and P. O'Hare. 2006. CREB4, a transmembrane bZip transcription factor and potential new substrate for regulation and cleavage by S1P. Mol. Biol. Cell 17:413-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storlazzi, C. T., F. Mertens, A. Nascimento, M. Isaksson, J. Wejde, O. Brosjo, N. Mandahl, and I. Panagopoulos. 2003. Fusion of the FUS and BBF2H7 genes in low grade fibromyxoid sarcoma. Hum. Mol. Genet. 12:2349-2358. [DOI] [PubMed] [Google Scholar]
- 31.Tajiri, S., S. Oyadomari, S. Yan, M. Morioka, T. Gotoh, J. I. Hamada, Y. Ushio, and M. Mori. 2004. Ischemia-induced neuronal cell death is mediated by the endoplasmic reticulum stress pathway involving CHOP. Cell Death Differ. 11:403-415. [DOI] [PubMed] [Google Scholar]
- 32.Tamatani, M., T. Matsuyama, A. Yamaguchi, N. Mitsuda, Y. Tsukamoto, M. Taniguchi, Y. H. Che, K. Ozawa, O. Hori, H. Nishimura, A. Yamashita, M. Okabe, H. Yanagi, D. M. Stern, S. Ogawa, and M. Tohyama. 2001. ORP150 protects against hypoxia/ischemia-induced neuronal death. Nat. Med. 7:317-323. [DOI] [PubMed] [Google Scholar]
- 33.Thoma, C., G. Bergamini, B. Galy, P. Hundsdoerfer, and M. W. Hentze. 2004. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol. Cell 15:925-935. [DOI] [PubMed] [Google Scholar]
- 34.Urano, F., X. Wang, A. Bertolotti, Y. Zhang, P. Chung, H. P. Harding, and D. Ron. 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287:664-666. [DOI] [PubMed] [Google Scholar]
- 35.Warnakulasuriyarachchi, D., S. Cerquozzi, H. H. Cheung, and M. Holcik. 2004. Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J. Biol. Chem. 279:17148-17157. [DOI] [PubMed] [Google Scholar]
- 36.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355-1364. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, K., X. Shen, J. Wu, K. Sakaki, T. Saunders, D. T. Rutkowski, S. H. Back, and R. J. Kaufman. 2006. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124:587-599. [DOI] [PubMed] [Google Scholar]