Abstract
Tetrahymena thermophila macronuclear histone H1 is phosphorylated by a cdc2 kinase, and H1 phosphorylation regulates CDC2 expression by a positive feedback mechanism. In starved wild-type cells, decreased expression of the CDC2 gene is correlated with a global reduction in the phosphorylation of H1 and reduced phosphorylation of H1 in the region upstream of the CDC2 gene. To determine whether the reduced H1 phosphorylation upstream of the CDC2 gene is merely a reflection of global dephosphorylation or is due to specific targeting of dephosphorylation of H1 to the CDC2 promoter during starvation, the CDC2 promoter was mapped, and the distributions of phosphorylated and unphosphorylated H1 across the CDC2 gene were determined using chromatin immunoprecipitation. Unphosphorylated H1 is specifically enriched in a region of the CDC2 promoter when the gene's expression is reduced during starvation but not when CDC2 is highly active in growing cells. The region of unphosphorylated H1 coincides with a region that is essential for CDC2 expression. These studies are the first in vivo demonstration that the phosphorylation of H1 is being regulated at a fine level and that unphosphorylated H1 can be specifically targeted to a promoter, where it likely regulates transcription in a gene-specific manner.
Linker histones (H1s) are associated with the linker DNA between nucleosome core particles in chromatin. They are less conserved than core histones and often exhibit multiple isotypes and cell type-specific variants (11). Metazoan H1s have three domains: a more conserved central globular domain and less structured N- and C-terminal tails (29). In the protozoan Tetrahymena thermophila, the transcriptionally active macronucleus contains a single H1 whose linker location and solubility properties are typical of other H1s. The amino acid composition of Tetrahymena macronuclear H1 is similar to that of the lysine-rich C-terminal domain of metazoan H1 (25, 30, 36), which controls the binding of H1 to chromatin in vivo (32, 56) and plays a major role in stabilizing chromatin secondary structure in vitro (42). However, as in some other protists, Tetrahymena H1 lacks a globular domain.
In vitro studies suggested that H1 is required for formation and stabilization of higher order chromatin structure (49, 60) and acts as a general repressor of transcription (21, 38, 46, 47). However, in vivo, H1 is not essential for chromosome compaction per se but is required for complete condensation of interphase and mitotic chromosomes (43, 53), and the in vivo function of linker histones on transcription is gene specific and can be either positive or negative. In Xenopus laevis embryos, increased amounts of an H1 variant repressed oocyte but not somatic 5S rRNA genes or other Pol III transcripts (7, 35). Also in Xenopus, somatic, but not maternal, H1 selectively silences the transcription of regulatory genes required for mesodermal differentiation (55). In Tetrahymena, knockout of the HHO1 gene encoding macronuclear H1 had no detectable effect on the expression of most genes tested but induced the basal expression of the repressed NgoA gene. Surprisingly, H1 was required for the induced transcription of the CyP1 gene (54). Overexpression of two H1 variants in mouse cultured cells increased both basal and induced expression from the mouse mammary tumor virus (MMTV) promoter (27). In yeast, disruption of the gene encoding H1 also resulted in decreased expression of specific genes (31). In Neurospora crassa, elimination of H1 expression resulted in specific derepression of the pyruvate decarboxylase gene (20). In mice, deletion of genes encoding specific H1 subtypes can attenuate or enhance genes subject to position effect (1). In mouse embryonic stem cells, when half of the total H1 was depleted by knocking out three H1 genes, only about 0.56% of over 4,500 genes examined showed significant differences in expression (19). Thus, the function of linker histones in vivo is usually not essential, and effects on gene expression can be positive or negative. The apparent exception to this, embryonic lethality (but not general cell-lethality) caused by reducing H1 in mice (18), could be explained by effects on the expression of specific genes critical to specific developmental stages.
In vivo studies of the effects of H1 on transcription have generally involved elimination or overexpression of H1. As a result, the specific cellular mechanism(s) by which H1 affects transcription in vivo is not known. Recent studies indicate H1 may regulate transcription by competing for nucleosome binding with site-specific factors or with more abundant, less specific high-mobility-group chromatin proteins (10), by interacting with chromosome remodeling complexes (33, 48), by effects on DNA methylation (19, 61), or by phosphorylation (4, 6, 8, 16, 34, 37, 58).
Numerous studies have suggested that global changes in H1 phosphorylation, like posttranslational modifications of core histones, can be regulated to modify histone-chromatin interactions and are correlated with changes of transcription in vivo (reviewed in reference 63). Phosphorylation of H1 also has been implicated in chromatin decondensation that occurs at sites where DNA is being replicated (2) and in reduced binding of Hp1α and disassembly of higher order chromatin structures during interphase in mammalian cells (12, 28). We have been studying H1 and its phosphorylation in the ciliated protozoan Tetrahymena thermophila. In Tetrahymena, macronuclear H1 was previously found by Edman sequencing to be phosphorylated at three major, canonical cdc2 sites and, in a hierarchical fashion, at two minor, non-cdc2 sites whose phosphorylation depends on prior phosphorylation of the three cdc2 sites. Mutation of these five sites in combination abolished all detectable 32P signal from radiolabeled H1 (44), suggesting that they represented all of the phosphorylation sites on Tetrahymena H1 (44). However, recent mass spectroscopic analyses identified two additional minor, non-cdc2 phosphorylation sites whose phosphorylation requires prior phosphorylation on at least one of the three cdc2 sites. Thus, elimination of the five sites that were originally described did indeed completely eliminate phosphorylation of Tetrahymena H1 (22).
Tetrahymena H1 phosphorylation levels change dramatically in response to various physiological conditions. H1 is highly phosphorylated in growing cells (25) and is hyperphosphorylated upon heat shock (23) and during prezygotic stages of conjugation (50). H1 phosphorylation decreases in stationary phase and decreases still further during prolonged starvation (22, 50), and H1 is rapidly and almost completely dephosphorylated during late conjugation when the parental macronuclei cease transcription and become highly condensed prior to their elimination from the cell (40). These changes in H1 phosphorylation in physiological and developmental conditions that alter gene expression patterns suggest that H1 phosphorylation functions in transcription in Tetrahymena. Indeed, phosphorylated and dephosphorylated macronuclear H1 are enriched in transcriptionally active and inactive subdomains of macronuclear chromatin, respectively (41).
In vitro mutagenesis coupled with gene replacement studies showed that global phosphorylation of Tetrahymena macronuclear H1 regulates transcription of specific genes both positively and negatively in vivo and that phosphorylation of H1 mimicked the effects of H1 knockout (16, 54). More detailed mutagenic analyses indicated that phosphorylation of H1 creates a negative-charge patch (15) and causes a small increase in the rapid rate of association-dissociation of H1 on chromatin (14).
Genes whose expression is affected by H1 phosphorylation were cloned by suppression-subtractive hybridization based on their differential expression in starved A5 cells (in which the five phosphorylation sites identified by Edman sequencing were mutated to alanines, eliminating all phosphorylation) and starved E5 cells (in which these five phosphorylation sites were mutated to glutamic acid to mimic hyperphosphorylation [17]). A number of Tetrahymena genes were shown to be regulated by the change in H1 phosphorylation levels that occurs during starvation, including the CDC2 gene encoding a cdc2 kinase, which was expressed more strongly in starved E5 cells than in A5 cells. In wild-type (WT) growing cells, when H1 is highly phosphorylated, the CDC2 gene is highly expressed. During starvation of WT cells, when H1 is extensively but incompletely dephosphorylated, the CDC2 gene is weakly expressed, but if the E5 mutant H1 is overexpressed, endogenous H1 becomes hyperphosphorylated and CDC2 expression increases in starved cells. When H1 dephosphorylation during starvation was inhibited by okadaic acid (OA), CDC2 expression also increased. These studies argue that there is a positive feedback between CDC2 expression and phosphorylation of H1. Chromatin immunoprecipitation (ChIP) demonstrated that phosphorylation of H1 upstream of the CDC2 gene was decreased in starved cells (17). These studies indicated that the phosphorylation level of H1 associated with the CDC2 upstream region decreased in starved cells but did not determine whether this change was simply a reflection of the global dephosphorylation of H1 in the entire macronucleus, was due to specific dephosphorylation of H1 or binding of unphosphorylated H1, or reflected loss of H1 from the promoter. To distinguish among these possibilities, here we performed more extensive ChIP analyses, coupled with promoter mapping, demonstrating that unphosphorylated H1 is specifically enriched in a restricted region of the CDC2 promoter in starved but not in growing cells. We also show that this region is essential for CDC2 expression. These results argue that unphosphorylated H1 can be localized to a specific promoter region where it likely regulates transcription in a gene-specific manner. Coupled with studies demonstrating that Cdc45 can target Cdk2 and H1 phosphorylation to replication foci in mammalian cells (2), these studies indicate that, like posttranslational modifications of core histones, targeted regulation of the H1 phosphorylation state is a general mechanism for regulating chromatin function.
MATERIALS AND METHODS
Strains and culture conditions.
WT strains CU428 and B2086 were provided by P. J. Bruns (Cornell University). A5 and E5 strains were created by Y. Dou (16). Macronuclear H1 knockout strain ΔH1 was created by X. Shen (53, 54). Cells were grown in super proteose peptone (SPP) medium containing 1% proteose peptone (1× SPP) at 30°C (26) with vigorous shaking. Cell number was determined using a Coulter counter (Coulter Electronics, Inc.). For starvation, cells were washed and resuspended in 10 mM Tris (pH 7.5) at 30°C without shaking for 24 h.
Genomic walking.
Genomic walking libraries with different restriction fragments ligated to an adaptor at both ends (13) were provided by K. Mochizuki and were used as templates for nested PCR of genomic fragments using two primers in the adaptor and two CDC2 gene primers obtained from the cDNA sequence. The PCR products were cloned and sequenced.
5′ RACE and 3′ RACE.
cDNA synthesized from log-phase total RNA by NotI-adaptor and oligo(dT) primers through “template switched” reverse transcription (62) by SuperScript II RNaseH− RTase (Life Technologies) was provided by C. Tsao. NotI-adaptor primer and two CDC2 primers were used for nested PCR of this cDNA. PCR products were cloned and sequenced, revealing at least four clustered transcription start sites. The most frequent start site, found in 7 of 11 clones, occurred 52 bp upstream, another two clones started 56 bp upstream, and the remaining two started at 36 and 44 bp upstream of the initiator ATG. Short 5′-untranslated regions (UTRs) and multiple start sites are typical of Tetrahymena genes. A similar strategy, using 3′ rapid amplification of cDNA ends (RACE), identified the 3′ UTR at a single site 226 bases downstream from the TGA.
Gene replacement.
To analyze the function of the promoter in its endogenous location, upstream deletions of the CDC2 5′-flanking region were used to drive GFP expression. The Cd2+-inducible neo3 drug selection cassette (52) was inserted downstream of the 3′ UTR to enable selection of cells containing the transformed genes. Transformation of Tetrahymena macronuclei occurs by homologous integration. The macronucleus is polycopy (∼45 ploid) and divides amitotically: chromosomes assort randomly without segregation of sister chromatids. As a result, initial replacement of 1 of the 45 copies of the CDC2 gene by the transforming GFP can be selected at increasing concentrations of paramomycin sulfate (pm; Sigma) to partially replace the endogenous CDC2 gene while cells remain viable, even if the gene has an essential phenotype.
To create the deletions, the Tetrahymena macronuclear genome sequence (http://www.tigr.org/tdb/e2k1/ttg/index.shtml) was used to construct primers to obtain the 5′ upstream regions of the deletions (CDC2 far 5′) and the distal 3′ downstream region, enabling homologous recombination with the endogenous CDC2 gene during somatic transformations (Fig. 1). CDC2 knockout and promoter-mapping constructs were digested with KpnI and SacI and transformed into 15- to 17-h conjugating CU428 and B2086 cells using the Biolistic PDS-1000/He particle delivery system (Bio-Rad) as described previously (9). Transformants were selected initially at 70 μg/ml pm and 1 μg/ml Cd2+ and were serially transferred every 2 to 3 days to fresh media containing 0.5 μg/ml Cd2+ with increasing concentrations of pm.
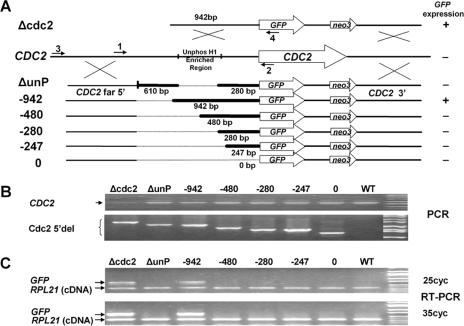
FIG. 1.
Mapping the CDC2 promoter at the CDC2 locus using GFP fusions. A. Serial deletion constructs were made using GFP as a reporter and a neo3 selectable marker inserted downstream of the 3′ UTR. Homologous recombination occurs in the 5′ region flanking the deletion (CDC2 far 5′) and the distal 3′-flanking region of the CDC2 gene to insert the reporter constructs into the endogenous CDC2 gene during somatic transformations. The CDC2 knockout construct, Δcdc2, lacking any 5′ deletion is shown at the top of the diagram. Below the endogenous CDC2 locus are the deletion constructs, with the heavy lines indicating the bases immediately 5′ of the initial ATG that are retained in the constructs. The numbered arrows indicate the primers used in panels B and C. The GFP expression status determined by RT-PCR shown in panel C is listed to the right of the deletion constructs. B. Genomic PCR using primers 1 and 2 showed the presence of the endogenous CDC2 gene (CDC2), and genomic PCR using primers 3 and 4 demonstrated that the various deletion constructs (Cdc2 5′del) localized to the endogenous CDC2 locus in the transformed cells. Shown are the ethidium bromide (EB)-stained PCR products separated on agarose gels. C. GFP expression was analyzed by RT-PCR using total RNA extracted from the above-mentioned transformants in log phase. Shown are the EB-stained RT-PCR products from 25 or 35 cycles of PCRs separated on agarose gels. The two primers for the ribosomal protein RPL21 gene span an intron, so true RT-PCR products are distinguishable from contaminating genomic DNA. Unphos H1, unphosphorylated H1.
RT-PCR.
Total RNA (3 μg) isolated with Trizol (Life Technologies), oligo(dT) primer, and Stratascript RTase (Stratagene) were used for reverse transcription (RT) according to the manufacturer's instructions. The cDNA was amplified by gene-specific primers GFP-N and GFP-C, together with L21 FW and L21 RV. The L21 primers span an intron, and the PCR product is 850 bp from genomic DNA and 360 bp from cDNA.
H1 extraction and gel electrophoresis.
H1 was extracted from whole cells using 5% perchloric acid (PCA [25]). PCA-soluble proteins were analyzed on both sodium dodecyl sulfate (SDS)-12% polyacrylamide and 15% acid-urea polyacrylamide gels (23). To dephosphorylate H1, 25 μg was treated with λ-protein phosphatase (10 U/μl for 5 h at 30°C; New England Biolabs, Inc.) and precipitated with 20% trichloroacetic acid.
Purification of anti-unphosphorylated H1 antibody.
Anti-unphosphorylated H1 was affinity purified by a modification of the method of Olmsted (45). About 40 μg of highly phosphorylated H1 extracted from heat-shocked (40°C for 10 min) CU428 cells was separated by SDS-12% polyacrylamide gel electrophoresis (PAGE). The gel was electroblotted to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore) with transfer buffer (48 mM Tris, 39 mM glycine, 18% methanol, 0.1% SDS) at 15 V for 1.5 h. The portion of the membrane containing phosphorylated H1 was cut into small pieces, blocked with 5% nonfat dry milk in TBST (100 mM Tris, 0.9% NaCl, 0.1% Tween 20, pH 7.5) for 1 h, and washed with Buffer 1 as described previously (45). The blot fragments were then mixed with 10 μl unphosphorylated H1 antiserum (originally called dephosphorylated H1 antiserum in reference 41) and 490 μl 1% bovine serum albumin in TBST and incubated at 4°C overnight with gentle shaking, and the supernatant was saved. Adsorption was repeated two more times by treating the blot fragments with 0.2 M glycine-HCl (pH 2.8) to remove the antibodies and using them to retreat the supernatant. The purified anti-unphosphorylated H1 antibody was assayed by Western blotting and used for ChIP.
Chromatin immunoprecipitation.
ChIP was performed as described previously (17). Primer pairs were designed to amplify overlapping fragments with similar GC contents of different sizes along the CDC2 gene 5′-flanking and coding regions to enable multiplex PCR.
Isolation of macronuclei and micrococcal nuclease digestion.
Macronuclei were isolated as described previously (26), washed three times with RSB buffer (10 mM Tris, pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.2 mM phenylmethylsulfonyl fluoride), and resuspended with 600 μl RSB buffer with 0.1 mM CaCl2 at 5 × 107 macronuclei/ml. Micrococcal nuclease (MNase; Worthington) was added to 50 U/ml, and the nuclei were digested for 0, 1, 2, 3, 4, and 5 min at 37°C. Digestion was stopped by addition of 3.5 volumes of stop buffer (1% SDS, 0.5 M EDTA, 18 mM Tris, pH 9.5; prewarmed to 65°C), followed by incubation at 65°C for at least 20 min. Proteinase K was then added to 1 mg/ml, and incubation continued at 55°C for 4 h. An equal volume of water was added to the digestion, and the DNA was extracted with phenol-chloroform, precipitated with ethanol in the presence of 0.3 M sodium acetate, and resuspended in double-distilled water. As a control, an equal amount of genomic DNA was resuspended in RSB and treated with 5 U/ml MNase for 0, 0.5, 1, and 2 min at 37°C, and DNA was reisolated as described above for macronculei. The purified MNase-digested DNA was then digested with TaqI, and digests were separated on a 1.2% agarose gel, blotted, and hybridized (3). Probe DNA was PCR products amplified from Tetrahymena genomic DNA by primer sets CDC2 840 up and CDC2 1105 down, labeled with [α-32P]dATP by random priming.
Primers used in this study.
The CDC2 primers used for nested PCR and sequencing in genomic walking were the following: CDK1, 5′-AGATTTCTCCGATTGCGGTAGAGGG-3′; CDK5, 5′-CGCTCAAGCTTGCTATTTTTCATAACC-3′; CDK10, 5′-TGAATAAGCTCGAAGGTAGC-3′. The adaptor primers were the following: L1, 5′-GTTCATCTTTACAAGCTAGCG-3′; L2, 5′-TCCTGAACAATGCTGTGGAC-3′. The NotI-adaptor primer used in 5′ RACE was 5′-GAGGCGGCCGCAATGAACACTGCGTTTGCGGG-3′. The CDC2 primers used in 5′ RACE were CDK1 and CDK5 (see above). Gene-specific primers used in RT-PCR were the following: GFP-N, 5′-ATGAGTAAAGGAGAAGAACTTTTC-3′; GFP-C, 5′-TTATTTGTATAGTTCATCCATGCC-3′; L21 FW, 5′-AAGTTGGTTATCAACTGTTGCGTT-3′; L21 RV, 5′-CCCAGAAAGTTCCTGCTGCAT-3′. Primers used to amplify the different fragments in ChIP were the following: 181-bp fragment, CDK 65 down (5′-ATAAATACTTATCAAAAAGCAAATG-3′) and CDK 245 up (5′-TACCTCCTAATTTTAGTTTATCTAC-3′); 310-bp fragment, CDK 243 down (5′-GTAAATGAAGAACATTAAGAATGC-3′) and CDK 552 up (5′-ATATACTTGGATAACAAAAAGAGG-3′); 222-bp fragment, CDK 524 down (5′-TATAGCCTCTTTTTGTTATCCAAG-3′) and CDK 745 up (5′-TAAAAATTTCCTATCATTCTAGTAC-3′); 199-bp fragment, CDK 727 E down (5′-AGTCCGGAATTCGAATGATAGGAAATT TTTACTAAGC-3′) and CDK 913 up (5′-TTGCTTAAAGGTACTTTAACCTTC-3′); 242-bp fragment, CDK 901 down (5′-TACCTTTAAGCAATTAAGAAAGTTG-3′) and CDK 1142 up (5′-AAATATATTTGATTAAAAACCAACC-3′); 260-bp fragment, CDK 1130 down (5′-ATCAAATATATTTATTAGTAACCAC-3′) and CDK 8 (5′-TTGATAATCAATAAAGAGAGACAC-3′); 308-bp fragment, CDK 1389 down (5′-AATAGTTCAAGAAGTTCTATTAGTC-3′) and CDK 5 (5′-CGCTCAAGCTTGCTATTTTTCATAACC-3′); 343-bp fragment, CDC 1646 coding (5′-AATAAATAATCTGACAGTAAAAATGG-3′) and CDK 1 (5′-AGATTTCTCCGATTGCGGTAGAGGG-3′); 472-bp fragment, CDC 1989 coding (5′-TCTCTCTTAAAGGAGTTATAACATC-3′) and CDC 2225 coding (5′-TTGAGGCTTCAAATCTCTATGAAG-3′); 539-bp fragment, CDK 1130 down (5′-ATCAAATATATTTATTAGTAACCAC-3′) and CDK 5′f EcoRI (5′-AGTCCGGAATTCTTTTACTGTCAGATTATTTATTTGC 3′); GTU1 280-bp fragment, GTU C13AL (5′-CTTGGCCGACTTAAAGTGTAATGATAT CTCTAGGC-3′) and GTU 521 (5′-AATGTGAGGAGTGAGTGAG-3′. Primers used to amplify the DNA fragment used for the probe template in MNase mapping were CDC2 840 down (5′-CGATCAGAAAGCAAATTAAAAATATA-3′) and CDC2 1105 up (5′-TGAAACACTCAAAAAAGCCTCTC-3′).
Nucleotide sequence accession number. The GenBank accession number for CDC2 is AY706655.
RESULTS
Mapping the promoter of the CDC2 gene using GFP fusions.
Prior to analyzing H1 phosphorylation at the CDC2 gene, we characterized the CDC2 mRNA and mapped the promoter at its endogenous locus using GFP fusions. A total of 1.6 kb of CDC2 5′-flanking sequence was cloned by genomic walking, and the transcription start sites and 3′ UTR were determined by 5′ and 3′ RACE. A series of upstream deletions of the CDC2 5′ region attached to the GFP coding region were introduced into WT cells (Fig. 1A). Since we showed previously that CDC2 expression is linked to H1 phosphorylation (17), we reasoned that some endogenous cdc2p would be necessary for H1-regulated expression of GFP in the CDC2 locus, so the promoter mapping was done in transformants whose endogenous CDC2 gene was only partially replaced (Fig. 1B, upper panel). The introduced genes containing the 5′ deletions and the GFP reporter were easily detected by PCR of genomic DNA in these transformants (Fig. 1B, lower panel), and RT-PCR was done to detect GFP mRNA (Fig. 1C). Note that we have restricted our analyses to log-phase cells where the CDC2 gene is normally highly expressed and have used a sensitive RT-PCR assay to determine whether any mRNA was detectable rather than measuring the precise amount of GFP mRNA. Note also that mRNA from ribosomal gene RPL21 serves as a positive control for each PCR. A strain with an intact promoter in which most copies of the CDC2 coding regions have been replaced with GFP (Δcdc2) is the positive control for the GFP message. A region required to drive GFP expression in log-phase cells was mapped to between −480 and −942 bp upstream from the initial ATG (about 430 to 890 bp upstream of transcription start sites). A region (−280 to −942) that was found to be enriched in unphosphorylated H1 in starved cells (see below) was also shown to be required for GFP expression (Fig. 1).
Specificity of the anti-phosphorylated and anti-unphosphorylated H1 antibodies.
We showed previously that a 539-bp region upstream of the CDC2 coding sequence was enriched when immunoprecipitated by anti-phosphorylated H1 antibody in log-phase cells when CDC2 is highly expressed but not in starved cells when CDC2 is weakly expressed (17). There are three possible explanations for this finding: (i) H1 associated with this region could simply mirror the behavior of global H1, which is highly phosphorylated in growing cells and largely dephosphorylated in starved cells (25, 50); (ii) unphosphorylated H1 is specifically localized in this region or becomes inaccessible to the antibody (possibly due to interference from other proteins) in starved cells; and (iii) H1 is present in this region in growing cells but absent in starved cells. To distinguish among these possibilities, we wished to perform ChIP analyses with an antibody that preferentially reacted with unphosphorylated H1, in addition to the antibody against phosphorylated H1 used previously. These antibodies (kindly provided by C. David Allis) were raised against phosphorylated or unphosphorylated H1 peptides (41).
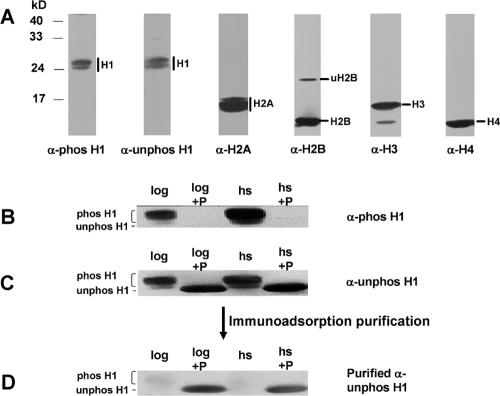
We checked the specificity of the antibodies by Western blots using both SDS-PAGE (Fig. 2) and acid-urea-PAGE (data not shown), which showed that both antisera recognized histone H1 but not the core histones (Fig. 2A). Anti-phosphorylated H1 antiserum was highly specific for phosphorylated H1 (Fig. 2B), and its reactivity was not detectable if H1 was treated with phosphatase (Fig. 2B). However, we found that this antibody could not distinguish between phosphorylated H1 isoforms differing in the number of phosphates they contained (X. Song and M. A. Gorovsky, unpublished observations). The anti-unphosphorylated H1 antiserum recognized both phosphorylated and unphosphorylated H1s (Fig. 2C). We purified this serum (see Materials and Methods) by adsorption to phosphorylated H1 immobilized on a PVDF membrane. The antibodies remaining in the supernatant were again tested using Western blotting and now showed strong preference for unphosphorylated H1 (Fig. 2D). We used these purified anti-unphosphorylated H1 antibodies as well as the anti-phosphorylated antibodies in the subsequent ChIP assays.
FIG. 2.
Specificity and purification of anti-H1 antibodies. A. Macronuclei were isolated as described previously (26) from heat-shocked (40°C for 1 h) cells. A total of 3 × 106 macronuclei/lane were subjected to SDS-12% PAGE, transferred to PVDF membranes, and probed with antisera that were produced against phosphorylated H1 (phos-H1), unphosphorylated H1 (unphos-H1), or H2A. Blots were stripped and reprobed with anti-H2B, -H3, or -H4 antisera. B, C, and D. Histone H1 was extracted with 5% PCA from WT cells in log phase (log) or cells heat shocked at 40°C for 10 min (hs). Aliquots of H1 were treated with λ-protein phosphatase (+P). H1 samples were subjected to SDS-PAGE, transferred to a PVDF membrane, and then probed with antisera produced against phosphorylated or unphosphorylated H1 peptides. The blot was stripped and reprobed for each successive analysis. Phosphorylated H1 and unphosphorylated H1 isoforms are labeled. B. Phosphatase treatment abolishes the signal recognized by anti-phosphorylated H1 antiserum. C. Unpurified anti-unphosphorylated H1 antiserum recognizes both phosphorylated and unphosphorylated H1s. D. Anti-unphosphorylated H1 antibodies purified by immunoadsorption demonstrate high specificity for unphosphorylated H1.
Unphosphorylated H1 is enriched in the CDC2 promoter in starved cells.
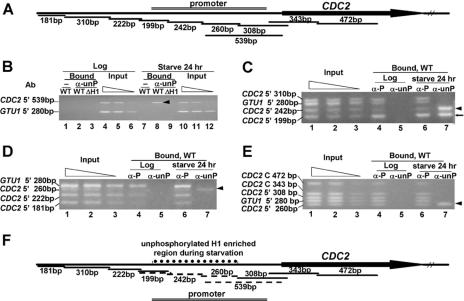
Our previous studies had shown that the 539- bp region upstream of the CDC2 gene was enriched when chromatin from log-phase cells was immunoprecipitated with anti-phosphorylated H1 but not when chromatin was immunoprecipitated from starved cells (17). In contrast, using anti-unphosphorylated H1 antibody (Fig. 3B, lane 8, arrowhead), this 539-bp PCR fragment within the CDC2 promoter (Fig. 3A) was greatly enriched in DNA immunoprecipitated from 24-h-starved WT cells but was not detected in chromatin immunoprecipitated from log-phase cells (Fig. 3B, lane 2). This result demonstrates that H1 is still associated with the CDC2 promoter in starved cells when the gene is relatively inactive, but it is unphosphorylated. No PCR product was detected with DNA isolated from chromatin immunoprecipitated with anti-unphosphorylated H1 antibody from cells lacking H1 (Fig. 3B, lanes 3 and 9) (54) or in the absence of antibody (Fig. 3B, lanes 1 and 7), further demonstrating the specificity of the antibody. The 539-bp CDC2 fragment and a 280-bp fragment upstream of the control GTU1 coding region were amplified to similar extents and in proportion to the amount of the template when DNA isolated from the input chromatin prior to immunoprecipitation was used (Fig. 3B, lanes 4, 5, and 6 as well as 10, 11, and 12), validating the conditions used for the PCR.
FIG. 3.
Unphosphorylated H1 is specifically localized to a restricted region of the CDC2 promoter during starvation when CDC2 is weakly expressed. A. Diagram of the CDC2 PCR fragments analyzed in the ChIP assay. Primer sets that amplify adjacent fragments differing in length by 19 to 130 bp were designed across the 5′ and coding regions of the CDC2 gene. The 539-bp fragment overlaps two fragments (260 bp and 308 bp) that were also analyzed. Different fragments were amplified in multiplex PCR of input or immunoprecipitated (Bound) DNA together with a 280-bp control fragment immediately upstream of the GTU1 coding region. GTU1 gene expression is not affected by H1 phosphorylation. B. Chromatin in the CDC2 promoter is enriched in unphosphorylated H1 in starved cells. ChIP was done on log-phase or 24-h-starved WT CU428 cells or H1 knockout cells (ΔH1) using purified anti-unphosphorylated H1 antibody (α-unP) or no antibody (−). Input DNA and immunoprecipitated DNA were purified and used in multiplex PCR to amplify a 539-bp fragment immediately upstream of the CDC2 coding region and the GTU1 280-bp fragment. The input sample was diluted in a 2.5-fold series to serve as a quantitation control. Shown are the PCR products from the bound or input DNA analyzed on a 3% agarose gel and stained with ethidium bromide. The arrowhead shows the CDC2 539-bp fragment that was enriched by immunoprecipitation with purified anti-unphosphorylated H1 antibody only in starved-cell chromatin. No PCR fragments were detectable in the absence of antibody or in chromatin from cells lacking H1. C, D, and E. Mapping the region enriched in unphosphorylated H1 in starved cells. ChIP was done on log-phase or starved cells, and the immunoprecipitated DNA was analyzed as described above. The input results are for the starved cells, where the critical observations have been obtained. The inputs from the growing cells are indistinguishable from those of starved cells (data not shown). Arrowheads indicate the fragments that were enriched by purified anti-unphosphorylated H1 antibody in starved cells. The arrow (←) in panel C points to a fragment that was enriched by immunoprecipitation with both anti-phosphorylated H1 (α-P) and purified anti-unphosphorylated H1 (α-unP) antibodies. F. Summary of the ChIP results. Fragments reproducibly enriched by immunoprecipitation with anti-unphosphorylated H1 antibody in 24-h-starved cells are indicated by dashed lines. Fragments that were not enriched were labeled by solid lines. The left boundary of the region containing unphosphorylated H1 resides within the 199-bp fragment, since it was enriched by both anti-phosphorylated and anti-unphosphorylated H1 antibodies. The 539-bp fragment contains the right-hand boundary, indicated by the fact that it can be split into one (260-bp) fragment that is enriched in unphosphorylated H1 and another (308-bp) fragment that is not.
The 280-bp GTU1 fragment was not detectable in DNA precipitated by anti-unphosphorylated H1 from either log-phase (Fig. 3B, lane 2) or starved cells (Fig. 3B, lane 8). However, it could be immunoprecipitated from both growing and starved cells using the anti-phosphorylated H1 antibody (17). Thus, the upstream region of the GTU1 gene probably contains little or no unphosphorylated H1 in either physiological state. In contrast, the finding that the 539-bp CDC2 fragment is specifically immunoprecipitated with the anti-unphosphorylated H1 only in starved cells indicates that some or all of this region is more enriched in unphosphorylated H1 than the GTU1 5′-flanking region.
Unphosphorylated H1 is specifically enriched in a restricted region of the CDC2 promoter during starvation when CDC2 is down-regulated.
The experiments described above do not allow a determination of whether the presence of unphosphorylated H1 is specific to the promoter of CDC2 or the absence of unphosphorylated H1 is specific to the GTU1 5′-flanking region. To distinguish between these alternatives, we mapped the location of unphosphorylated H1 along the 5′-flanking region as well as the coding region of the CDC2 gene using primer sets that amplify overlapping DNA fragments (Fig. 3A). In each experiment, multiplex PCR was used to amplify three to four different CDC2 fragments together with the control GTU1 5′ fragment (Fig. 3C, D, and E).
As seen in one example of the multiplex PCR, three CDC2 5′-flanking fragments as well as the GTU1 5′-flanking fragment were similarly amplified in input DNA, and their products were proportional to the amount of template used (Fig. 3C, lanes 1, 2, and 3). Thus, relative changes in the intensities of these bands in bound DNA precipitated with anti-phosphorylated or anti-unphosphorylated H1 antibodies compared to input DNA indicate enrichment or depletion of phosphorylated or unphosphorylated H1 in all or part of the affected fragment(s). ChIP results from 24-h-starved cells show that the CDC2 5′ 242-bp fragment was greatly enriched by immunoprecipitation with anti-unphosphorylated H1 (Fig. 3C, lane 7, arrowhead), while it was less efficiently amplified after immunoprecipitation with anti-phosphorylated H1 (Fig. 3C, lane 6). The CDC2 5′ 199-bp fragment is enriched with both anti-unphosphorylated H1 (Fig. 3C, lane 7, arrow) and anti-phosphorylated H1 (Fig. 3C, lane 6) in 24-h-starved cells. Because these ChIP experiments determine whether any part of a chromatin fragment has relatively more or less affinity for the antibodies than fragments to which it is being compared, this result suggests that the 199-bp fragment contains both a region where H1 is phosphorylated and one that is unphosphorylated. Similar ChIP experiments were done on other fragments (Fig. 3D and E).
All of the ChIP results are summarized in Fig. 3F. These studies indicate that unphosphorylated H1 is specifically enriched in a region of the CDC2 promoter during starvation when CDC2 expression is reduced but not in growing cells when CDC2 is highly active. The 5′ boundary of the region preferentially associated with unphosphorylated H1 resides within the 199-bp fragment, since it was enriched by both anti-phosphorylated and anti-unphosphorylated H1 (Fig. 3C). Fragments immediately upstream of it are enriched by anti-phosphorylated H1, and those immediately downstream of it are enriched by anti-unphosphorylated H1. Enrichment of the 539-bp fragment (Fig. 3B) can be explained by the fact that it contains the right-hand boundary of the region containing unphosphorylated H1, as indicated by the observations that this fragment can be split into a 260-bp fragment that is enriched in chromatin immunoprecipitated by anti-unphosphorylated H1 (Fig. 3E) and a 308-bp overlapping fragment that is not. Thus, the region enriched in unphosphorylated H1 in starved cells is restricted to a domain of 500 to 600 bp in the CDC2 promoter (dotted line in Fig. 3F) and does not include either the downstream coding region or regions further upstream of the CDC2 gene. Importantly, this region is upstream of the transcription start sites that occur 36 to 52 bp upstream of the initial ATG (see Materials and Methods).
After we obtained the results localizing the region preferentially associated with unphosphorylated H1, we analyzed expression of a GFP fusion lacking this region (Fig. 1, ΔunP) and found it is required for CDC2 promoter function. Thus, in starved cells, a highly specific region containing unphosphorylated H1 is localized to nontranscribed sequences upstream of the CDC2 gene that are required for promoter activity.
Enrichment of unphosphorylated H1 occurs without major alteration of chromatin structure.
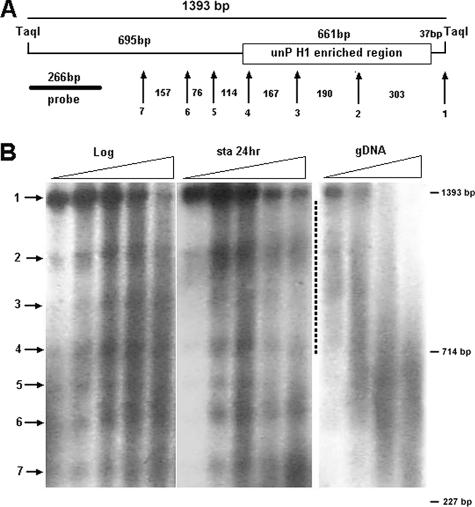
To examine whether the chromatin structure in the CDC2 promoter region enriched in unphosphorylated H1 differs between log-phase and starved cells, we mapped that region using MNase digestion (Fig. 4). No MNase-hypersensitive sites were detected in this region. MNase digestion of chromatin in Tetrahymena macronuclei has been reported to produce fuzzy bands with a spacing of ∼200 bp, probably owing to the extreme AT richness of Tetrahymena DNA and the high level of transcription in macronuclei (24). In both growing and starved cells, a series of fuzzy bands is observed. Except for the spacing between bands 4 and 5 and bands 5 and 6, these spacings are compatible with the presence of nonrandomly positioned nucleosomes. The region between bands 1 and 2 could accommodate two tightly packed nucleosomes. No reproducible differences in the spacing of these MNase-sensitive sites were detected between growing and starved cells (Fig. 4B, region indicated by a dotted line). Thus, the differences in chromatin structure associated with the localization of unphosphorylated H1, if any, are slight.
FIG. 4.
Localization of unphosphorylated H1 occurs without major alteration of chromatin structure. Macronuclei from log-phase and 24-h-starved WT CU428 cells as well as genomic DNA from CU428 cells were digested with MNase, followed by TaqI digestion, and analyzed by Southern blotting. A. A diagram of the mapped region. The probe used in panel B is indicated. The numbered arrows correspond to the bands shown in panel B. The numbers in between the arrows are the spacing (in base pairs) between the bands. B. Southern blot showing DNA isolated from macronuclei from log-phase and starved cells as well as genomic DNA. All samples were analyzed on the same gel from which the last, overdigested lane of each sample set and empty lanes separating the different sets of samples were removed to facilitate visual comparisons. The dotted line indicates the position of the part of the blot corresponding to the region enriched in unphosphorylated H1.
DISCUSSION
Links between gene expression and targeted posttranslational modifications of core histones have been extensively documented (for recent global analyses of acetylation and methylation and for references, see references 5 and 51). There is less information concerning the relationship between H1 phosphorylation and transcription. In vitro studies have shown that H1 facilitates binding of steroid hormone receptor to MMTV promoter-containing minichromosomes, which in turn results in promoter-specific phosphorylation of H1 (37, 58), and that phosphorylated H1 inhibits the activity of ATP-dependent chromatin-remodeling enzymes (34). Lee and Archer (39) demonstrated that, in tissue culture cells, histone H1 on the MMTV promoter adjacent to the hormone response element was dephosphorylated upon prolonged hormone treatment and that specific H1 isoforms were dephosphorylated (4). However, the loss of histone H1 phosphorylation under these conditions was global, and there was no direct demonstration of the localization of differentially phosphorylated H1 to specific sites on chromatin. Interestingly, the expression of different genes responds differently to altered levels H1 phosphorylation in both mammalian cells (6) and in Tetrahymena (16), indicating that there is no simple relationship between H1 phosphorylation state and the transcriptional state of genes.
We showed previously that the 539-bp fragment derived from the CDC2 5′ region is phosphorylated (precipitatable with anti-phosphorylated H1 antibody) in growing cells (17) and that OA prevents the starvation-induced repression of CDC2, as does E5, a constitutively phosphorylated H1 (17). Together with the studies described here, these observations argue that H1 in this regulatory region of CDC2 was phosphorylated in growing cells and becomes dephosphorylated upon starvation. We also showed that there was a small but reproducible reduction in the precipitation of this fragment with anti-phosphorylated H1 antibody in starved cells. Consistent with this observation, this fragment can be divided into two regions, one of which is enriched in starved-cell chromatin immunoprecipitated with anti-unphosphorylated H1 antibody and one which is not. However, no other fragment that was enriched in unphosphorylated H1 in starved cells showed a similar reduction in phosphorylated H1. We believe that this reflects the fact that the anti-phosphorylated H1 antibody cannot differentiate different isoforms of phosphorylated H1 and thus cannot distinguish, for example, when a 5-phosphate isoform changes to a 2-phosphate isoform. As a result, the lack of a reduction in signal for the anti-phosphorylated precipitated material after starvation seen in this work does not preclude that there was reduced phosphorylation; it only means that there was not enough complete dephosphorylation to make a detectable difference with the anti-phosphorylated H1 antibody. What is being measured by the multiplex PCR approach utilized here is relative enrichment (or depletion, although that is more difficult to demonstrate). If part of a fragment has more unphosphorylated H1 than other regions and part of it has more phosphorylated H1 than other regions, it will be enriched in both precipitations, as is the case for the 199-bp fragment. We wish to emphasize that it is the positive result with the anti-unphosphorylated antibody that is critical and that is easily detected.
The detailed mechanism whereby unphosphorylated H1 replaces phosphorylated H1 in this region is not known. There is little evidence that phosphorylated or unphosphorylated H1 can bind to specific DNA sequences, although some preferential binding to four-way junctions and preferential binding based on GC content have been reported (57, 59). We have examined the CDC2 upstream sequence that is associated with unphosphorylated H1 in starved cells for these features and found it lacks inverted repeats and binding sites for known transcription factors and that it is similar to adjacent sequences in its GC content. Another possible mechanism is that alteration of chromatin composition or structure causes unphosphorylated H1 to bind to this region more strongly than phosphorylated H1. However, as described above, the changes in chromatin structure in this region, if any, are small. In addition, fluorescence recovery after photobleaching analyses indicate that while unphosphorylated H1 exchanges more slowly than phosphorylated H1, the rate of difference is not large (14), making it unlikely that differential binding of one over the other could result in the striking localization of the unphosphorylated isoform that we have observed here. An alternative mechanism for regulating localization of unphosphorylated H1 upstream of the CDC2 gene during starvation is the targeting of the catalytic and/or a regulatory subunit of an H1 phosphatase (14). Consistent with this, a temporally regulated, large-scale, but incomplete dephosphorylation of H1 in Tetrahymena cells has been shown to occur between 6 and 12 h of starvation (16), and treatment of starved cells with a phosphatase inhibitor prevents both H1 dephosphorylation and reduction of CDC2 expression during starvation (17). Finally, we cannot rule out the possibilities that the region enriched in unphosphorylated H1 is produced by recruitment of new, unphosphorylated H1 to this region either to replace phosphorylated H1 that has been displaced by transcription initiation as has been proposed for the MMTV promoter (see Fig. 6 in reference 58) or to occupy regions that are devoid of H1 in growing cells.
An important question is whether the alteration in H1 phosphorylation upstream of the CDC2 gene is the cause or the consequence of transcription. The unphosphorylated region is upstream of the region encoding the 5′ end of the CDC2 mRNA and is unlikely to be transcribed. Thus, the change in H1 phosphorylation state is unlikely to be produced by transcription per se, and its position suggests it is more likely to play a role in transcription initiation than elongation. Also, replacement of WT H1 with a mutant H1 that mimics constitutive phosphorylation and treatment of starved cells with a phosphatase inhibitor both enhance CDC2 gene expression (17). These observations argue that the targeting of unphosphorylated H1 plays an important role in regulating transcription from the CDC2 promoter rather than vice versa. Thus, the studies described here provide evidence indicating that the phosphorylation of H1 is being regulated at a very fine level and that unphosphorylated H1 can be specifically targeted to a promoter in vivo, and they suggest it likely regulates transcription. These results, coupled with recent studies showing that Cdk2 and H1 phosphorylation are targeted to replication foci in mammals (2), where they result in chromatin decondensation and probably facilitate the progression of replication forks, argue that the targeted regulation of the phosphorylation state of H1 is a general mechanism for regulating chromatin function.
Acknowledgments
We are grateful to Josephine Bowen for technical assistance and critically reading the manuscript, to C. David Allis for providing the anti-phosphorylated and anti-unphosphorylated antibodies, and to Q. Yu, X. Bi, and B. Cui for suggestions on MNase digestion.
This work was supported by National Institutes of Health grant GM21793.
Footnotes
Published ahead of print on 28 December 2006.
REFERENCES
- 1.Alami, R., Y. Fan, S. Pack, T. M. Sonbuchner, A. Besse, Q. Lin, J. M. Greally, A. I. Skoultchi, and E. E. Bouhassira. 2003. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc. Natl. Acad. Sci. USA 100:5920-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrow, M. G., and J. L. Hamlin. 2005. Chromatin decondensation in S-phase involves recruitment of Cdk2 by Cdc45 and histone H1 phosphorylation. J. Cell Biol. 168:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1988. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 4.Banks, G. C., L. J. Deterding, K. B. Tomer, and T. K. Archer. 2001. Hormone-mediated dephosphorylation of specific histone H1 isoforms. J. Biol. Chem. 276:36467-36473. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman, J. S. Liu, T. Kouzarides, and S. L. Schreiber. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99:8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharjee, R. N., G. C. Banks, K. W. Trotter, H. L. Lee, and T. K. Archer. 2001. Histone H1 phosphorylation by Cdk2 selectively modulates mouse mammary tumor virus transcription through chromatin remodeling. Mol. Cell. Biol. 21:5417-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouvet, P., S. Dimitrov, and A. P. Wolffe. 1994. Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev. 8:1147-1159. [DOI] [PubMed] [Google Scholar]
- 8.Bruhat, A., and J.-P. Jost. 1996. Phosphorylation/dephosphorylation of the repressor MDBP-2-H1 selectively affects the level of transcription from a methylated promoter in vitro. Nucleic Acids Res. 24:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassidy-Hanley, D., J. Bowen, J. Lee, E. S. Cole, L. A. VerPlank, J. Gaertig, M. A. Gorovsky, and P. J. Bruns. 1997. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146:135-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catez, F., T. Ueda, and M. Bustin. 2006. Determinants of histone H1 mobility and chromatin binding in living cells. Nat. Struct. Mol. Biol. 13:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, R. D. 1984. A minireview of microheterogeneity in H1 histone and its possible significance. Anal. Biochem. 136:24-30. [DOI] [PubMed] [Google Scholar]
- 12.Daujat, S., U. Zeissler, T. Waldmann, N. Happel, and R. Schneider. 2005. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J. Biol. Chem. 280:38090-38095. [DOI] [PubMed] [Google Scholar]
- 13.Devon, R. S., D. J. Porteous, and A. J. Brookes. 1995. Splinkerettes-improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 23:1644-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dou, Y., J. Bowen, Y. Liu, and M. A. Gorovsky. 2002. Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. J. Cell Biol. 158:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dou, Y., and M. A. Gorovsky. 2000. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol. Cell 6:225-231. [DOI] [PubMed] [Google Scholar]
- 16.Dou, Y., C. A. Mizzen, M. Abrams, C. D. Allis, and M. A. Gorovsky. 1999. Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol. Cell 4:641-647. [DOI] [PubMed] [Google Scholar]
- 17.Dou, Y., X. Song, Y. Liu, and M. A. Gorovsky. 2005. The H1 phosphorylation state regulates expression of CDC2 and other genes in response to starvation in Tetrahymena thermophila. Mol. Cell. Biol. 25:3914-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan, Y., T. Nikitina, E. M. Morin-Kensicki, J. Zhao, T. R. Magnuson, C. L. Woodcock, and A. I. Skoultchi. 2003. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 23:4559-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan, Y., T. Nikitina, J. Zhao, T. J. Fleury, R. Bhattacharyya, E. E. Bouhassira, A. Stein, C. L. Woodcock, A. I. Skoultchi, E. M. Morin-Kensicki, T. R. Magnuson, and S. Dimitrov. 2005. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123:1199-1212. [DOI] [PubMed] [Google Scholar]
- 20.Folco, H. D., M. Freitag, A. Ramon, E. D. Temporini, M. E. Alvarez, I. Garcia, C. Scazzocchio, E. U. Selker, and A. L. Rosa. 2003. Histone H1 is required for proper regulation of pyruvate decarboxylase gene expression in Neurospora crassa. Eukaryot. Cell. 2:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, B., H. Jaffe, and G. Kunos. 1998. Histone H1 isoforms purified from rat liver bind nonspecifically to the nuclear factor 1 recognition sequence and serve as generalized transcriptional repressors. Mol. Cell. Biochem. 178:187-196. [DOI] [PubMed] [Google Scholar]
- 22.Garcia, B. A., S. Joshi, C. E. Thomas, R. K. Chitta, R. L. Diaz, S. A. Busby, P. C. Andrews, R. R. Ogorzalek Loo, J. Shabanowitz, N. L. Kelleher, C. A. Mizzen, C. D. Allis, and D. F. Hunt. 2006. Comprehensive phosphoprotein analysis of linker histone H1 from Tetrahymena thermophila. Mol. Cell Proteomics 5:1593-1609. [DOI] [PubMed] [Google Scholar]
- 23.Glover, C. V. C., K. J. Vavra, S. D. Guttman, and M. A. Gorovsky. 1981. Heat shock and deciliation induce phosphorylation of histone H1 in Tetrahymena pyriformis. Cell 23:73-77. [DOI] [PubMed] [Google Scholar]
- 24.Gorovsky, M. A. 1986. Ciliate chromatin and histones, p. 227-261. In J. G. Gall (ed.), The molecular biology of ciliated protozoa. Academic Press, Inc., Orlando, FL.
- 25.Gorovsky, M. A., J. B. Keevert, and G. L. Pleger. 1974. Histone F1 of Tetrahymena macronuclei. Unique electrophoretic properties and phosphorylation of F1 in an amitotic nucleus. J. Cell Biol. 61:134-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorovsky, M. A., M.-C. Yao, J. B. Keevert, and G. L. Pleger. 1975. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. IX:311-327. [DOI] [PubMed] [Google Scholar]
- 27.Gunjan, A., and D. T. Brown. 1999. Overproduction of histone H1 variants in vivo increases basal and induced activity of the mouse mammary tumor virus promoter. Nucleic Acids Res. 27:3355-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hale, T. K., A. Contreras, A. J. Morrison, and R. E. Herrera. 2006. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1alpha. Mol. Cell 22:693-699. [DOI] [PubMed] [Google Scholar]
- 29.Hartman, P. G., G. E. Chapman, T. Moss, and E. M. Bradbury. 1977. Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The three structural regions of the histone H1 molecule. Eur. J. Biochem. 77:45-51. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi, T., H. Hayashi, and K. Iwai. 1987. Tetrahymena histone H1. Isolation and amino acid sequence lacking the central hydrophobic domain conserved in other H1 histones. J. Biochem. 102:369-376. [DOI] [PubMed] [Google Scholar]
- 31.Hellauer, K., E. Sirard, and B. Turcotte. 2001. Decreased expression of specific genes in yeast cells lacking histone H1. J. Biol. Chem. 276:13587-13592. [DOI] [PubMed] [Google Scholar]
- 32.Hendzel, M. J., M. A. Lever, E. Crawford, and J. P. Th'ng. 2004. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 279:20028-20034. [DOI] [PubMed] [Google Scholar]
- 33.Hill, D. A., and A. N. Imbalzano. 2000. Human SWI/SNF nucleosome remodeling activity is partially inhibited by linker histone H1. Biochemistry 39:11649-11656. [DOI] [PubMed] [Google Scholar]
- 34.Horn, P. J., L. M. Carruthers, C. Logie, D. A. Hill, M. J. Solomon, P. A. Wade, A. N. Imbalzano, J. C. Hansen, and C. L. Peterson. 2002. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol. 9:263-267. [DOI] [PubMed] [Google Scholar]
- 35.Kandolf, H. 1994. The H1A histone variant is an in vivo repressor of oocyte-type 5S gene transcription in Xenopus laevis embryos. Proc. Natl. Acad. Sci. USA 91:7257-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasinsky, H. E., J. D. Lewis, J. B. Dacks, and J. Ausió. 2001. Origin of H1 linker histones. FASEB J. 15:34-42. [DOI] [PubMed] [Google Scholar]
- 37.Koop, R., L. Di Croce, and M. Beato. 2003. Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. EMBO J. 22:588-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laybourn, P. J., and J. T. Kadonaga. 1991. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science 254:238-245. [DOI] [PubMed] [Google Scholar]
- 39.Lee, H. L., and T. K. Archer. 1998. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 17:1454-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, R., R. G. Cook, and C. D. Allis. 1991. Proteolytic removal of core histone amino termini and dephosphorylation of histone H1 correlate with the formation of condensed chromatin and transcriptional silencing during Tetrahymena macronuclear development. Genes Dev. 5:1601-1610. [DOI] [PubMed] [Google Scholar]
- 41.Lu, M. J., S. S. Mpoke, C. A. Dadd, and C. D. Allis. 1995. Phosphorylated and dephosphorylated linker histone H1 reside in distinct chromatin domains in Tetrahymena macronuclei. Mol. Biol. Cell. 6:1077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu, X., and J. C. Hansen. 2004. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J. Biol. Chem. 279:8701-8707. [DOI] [PubMed] [Google Scholar]
- 43.Maresca, T. J., B. S. Freedman, and R. Heald. 2005. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J. Cell Biol. 169:859-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizzen, C. A., Y. Dou, Y. Liu, R. G. Cook, M. A. Gorovsky, and C. D. Allis. 1999. Identification and mutation of phosphorylation sites in a linker histone. Phosphorylation of macronuclear H1 is not essential for viability in Tetrahymena. J. Biol. Chem. 274:14533-14536. [DOI] [PubMed] [Google Scholar]
- 45.Olmsted, J. B. 1981. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J. Biol. Chem. 256:11955-11957. [PubMed] [Google Scholar]
- 46.Owen-Hughes, T., and J. L. Workman. 1994. Experimental analysis of chromatin function in transcription control. Crit. Rev. Eukaryot. Gene Expr. 4:403-441. [PubMed] [Google Scholar]
- 47.Paranjape, S. M., R. T. Kamakaka, and J. T. Kadonaga. 1994. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu. Rev. Biochem. 63:265-297. [DOI] [PubMed] [Google Scholar]
- 48.Ramachandran, A., M. Omar, P. Cheslock, and G. R. Schnitzler. 2003. Linker histone H1 modulates nucleosome remodeling by human SWI/SNF. J. Biol. Chem. 278:48590-48601. [DOI] [PubMed] [Google Scholar]
- 49.Ramakrishnan, V. 1997. Histone H1 and chromatin higher-order structure. Crit. Rev. Eukaryot. Gene Expr. 7:215-230. [DOI] [PubMed] [Google Scholar]
- 50.Roth, S. Y., I. G. Schulman, R. Richman, R. G. Cook, and C. D. Allis. 1988. Characterization of phosphorylation sites in histone H1 in the amitotic macronucleus of Tetrahymena during different physiological states. J. Cell Biol. 107:2473-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schubeler, D., D. M. MacAlpine, D. Scalzo, C. Wirbelauer, C. Kooperberg, F. van Leeuwen, D. E. Gottschling, L. P. O'Neill, B. M. Turner, J. Delrow, S. P. Bell, and M. Groudine. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18:1263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shang, Y., X. Song, J. Bowen, R. Corstanje, Y. Gao, J. Gaertig, and M. A. Gorovsky. 2002. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 99:3734-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen, X., L. Yu, J. W. Weir, and M. A. Gorovsky. 1995. Linker histones are not essential and affect chromatin condensation in vivo. Cell 82:47-56. [DOI] [PubMed] [Google Scholar]
- 54.Shen, X. T., and M. A. Gorovsky. 1996. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86:475-483. [DOI] [PubMed] [Google Scholar]
- 55.Steinbach, O. C., A. P. Wolffe, and R. A. Rupp. 1997. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature 389:395-399. [DOI] [PubMed] [Google Scholar]
- 56.Th'ng, J. P., R. Sung, M. Ye, and M. J. Hendzel. 2005. H1 family histones in the nucleus: control of binding and localization by the C-terminal domain. J. Biol. Chem. 280:27809-27814. [DOI] [PubMed] [Google Scholar]
- 57.Varga-Weisz, P., K. Van Holde, and J. Zlatanova. 1993. Preferential binding of histone H1 to four-way helical junction DNA. J. Biol. Chem. 268:20699-20700. [PubMed] [Google Scholar]
- 58.Vicent, G. P., R. Koop, and M. Beato. 2002. Complex role of histone H1 in transactivation of MMTV promoter chromatin by progesterone receptor. J. Steroid Biochem. Mol. Biol. 83:15-23. [DOI] [PubMed] [Google Scholar]
- 59.Wellman, S. E. 1996. Carboxyl-terminal peptides from histone H1 variants: DNA binding characteristics and solution conformation. Biopolymers 39:491-501. [DOI] [PubMed] [Google Scholar]
- 60.Widom, J. 1998. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 27:285-327. [DOI] [PubMed] [Google Scholar]
- 61.Wierzbicki, A. T., and A. Jerzmanowski. 2005. Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation. Genetics 169:997-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, Y. Y., E. M. Machleder, A. Chenchik, R. Li, and P. D. Siebert. 2001. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques 30:892-897. [DOI] [PubMed] [Google Scholar]
- 63.Zlatanova, J., P. Caiafa, and K. Van Holde. 2000. Linker histone binding and displacement: versatile mechanism for transcriptional regulation. FASEB J. 14:1697-1704. [DOI] [PubMed] [Google Scholar]