Abstract
Repair of DNA double-strand breaks (DSBs) protects cells and organisms, as well as their genome integrity. Since DSB repair occurs in the context of chromatin, chromatin must be modified to prevent it from inhibiting DSB repair. Evidence supports the role of histone modifications and ATP-dependent chromatin remodeling in repair and signaling of chromosome DSBs. The key questions are, then, what the nature of chromatin altered by DSBs is and how remodeling of chromatin facilitates DSB repair. Here we report a chromatin alteration caused by a single HO endonuclease-generated DSB at the Saccharomyces cerevisiae MAT locus. The break induces rapid nucleosome migration to form histone-free DNA of a few hundred base pairs immediately adjacent to the break. The DSB-induced nucleosome repositioning appears independent of end processing, since it still occurs when the 5′-to-3′ degradation of the DNA end is markedly reduced. The tetracycline-controlled depletion of Sth1, the ATPase of RSC, or deletion of RSC2 severely reduces chromatin remodeling and loading of Mre11 and Yku proteins at the DSB. Depletion of Sth1 also reduces phosphorylation of H2A, processing, and joining of DSBs. We propose that RSC-mediated chromatin remodeling at the DSB prepares chromatin to allow repair machinery to access the break and is vital for efficient DSB repair.
In eukaryotic cells, the genome is packaged into chromatin containing nucleosomes (12). By limiting the enzymes' access to the target DNA sequence, chromatin poses a unique challenge to a variety of DNA transactions, including gene expression, replication, recombination, and DNA repair (36, 40, 47). To overcome this inhibitory effect of chromatin, two classes of chromatin modifications have been developed. One involves posttranslational modification of histones (18, 44), and the other is ATP-dependent chromatin remodeling, which mobilizes nucleosomes by weakening the contacts between histone octamers and DNA (4, 13, 28, 55).
The repair of DNA double-strand breaks (DSBs) is fundamental to the survival of cells and organisms (20) because inaccurate repair can lead to genome instability (38). Several highly efficient mechanisms have been developed to eliminate DSBs, and many gene products are dedicated to these processes (22). Because DSB repair occurs in the context of the chromatin, chromatin modification is a prerequisite for this process. A few recent findings, listed below, support the role of chromatin in DSB repair (36, 37, 46, 51). First, several unique histone modifications occur in response to DSB damage at the chromatin domain closely associated with the damage (1, 3, 7, 9, 17, 43). In Saccharomyces cerevisiae, phosphorylation of the carboxy terminus of H2A is induced by genotoxic agents over a very large domain (∼50 kb) surrounding the DSB. Cohesin and the Ino80 chromatin remodeling complex are then recruited to phosphorylated H2A to promote efficient DSB repair by homologous recombination (HR) and nonhomologous end joining (NHEJ) (10). In mammals, a conserved motif in the C-terminal tail of the histone variant H2AX is rapidly phosphorylated in response to DNA damage. It helps signal and recruit DNA repair machinery to DSBs (2, 5, 35). A phosphatase complex (HTP-C) that dephosphorylates γH2AX was recently identified (8, 19). Surprisingly, dephosphorylation of γH2AX by HTP-C is not required for DSB repair but rather is required for recovery from a DNA damage checkpoint (19).
Histones near DSBs are further modified by acetylation (36). Histone H4 molecules are acetylated at DSBs by Esa1 for efficient NHEJ of DSBs (3). Localized acetylation of histones H3 and H4 is triggered by HR through the activity of Gcn5 and Esa1 and subsequently removed by Rpd3, Sir2, and Hst2 histone deacetylases (45). HAT1, a type II histone acetyltransferase, and Sin3 histone deacetylase are recruited to the break and are implicated in reestablishing the chromatin after DSB repair (17, 39).
Increasing evidence also supports that at least four distinct ATP-dependent chromatin remodeling complexes—Ino80, RSC, Swi/Snf, and Swr1—are directly involved in DSB repair (14). The components of these complexes are all recruited to the persistent DSB induced by HO endonuclease (6, 9, 31, 42, 48, 50). Functional interaction by the components of Ino80 or Swr1 with phosphorylated H2A or by Rsc1/Rsc2 with Ku and Mre11 proteins further supports the role of these remodeling complexes in DSB repair (9, 31, 42, 50). Interestingly, Ino80, RSC, and Swi/Snf may participate in distinct steps of yeast mating type gene conversion. Ino80 displaces nucleosomes from 3′ single-stranded DNA for the Rad51-coated presynaptic filament formation (48), and Swi/Snf is needed for invasion of homologous duplex DNA (6). RSC has been thought to play a role in the late stage of HR, following synapsis between donor and recipient DNA molecules (6).
Biochemical and genetic evidence is accumulating about the factors involved in chromatin modification during DSB repair. However, little is known about how chromatin is altered by the break and the way chromatin remodeling facilitates repair. Here, we determined the chromatin structure at the MAT locus of yeast chromosome III, before and after inducing a DSB, by measuring its susceptibility to micrococcal or restriction nuclease cleavage. We also evaluated the role of the RSC chromatin remodeling complex in DSB-induced nucleosome repositioning, processing, and joining of a chromosomal DSB. Our studies support the idea that chromatin prevents the repair machinery from reaching the damaged chromatin, as well as an early role for the RSC chromatin remodeling complex in efficient DSB repair.
MATERIALS AND METHODS
Strains.
All strains are derivatives of JKM179, which has the genotype hoΔ MATα hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG′ ura3-52 ade3::GAL::HO (see Table S1 in the supplemental material). The tet-STH1 strain is a segregant from a cross between JKM179 and the Hughes tet promoter strain (YSC1180-9491319; Open Biosystems).
Microccocal nuclease sensitivity assay.
Cultures were grown to a density between 1 × 107 and 2 × 107 cells/ml in preinduction medium (yeast extract-peptone [YEP]-glycerol) in the absence or presence of 10 μg/ml doxycycline for 12 h. HO endonuclease activity was induced by adding 2% galactose, and the yeast was lysed with glass beads in buffer Z after cross-linking with 2% formaldehyde. MNase (45 U) was added to 4.5 × 108 cells at 37°C for 14 min with 20 mM MgCl2 and 2 mM CaCl2. Reactions were stopped by the addition of 1% sodium dodecyl sulfate and 5 mM EDTA and cleared by centrifugation for 15 min. The samples were reverse cross-linked, and DNA was purified by deproteinization and ethanol precipitation. The amount of DNA enriched by MNase treatment was determined with real-time quantitative PCR (qPCR) using the ABI Prism 7900 (PE Applied Biosystems). The threshold was set to cross a point at which PCR amplification was linear, and the number of cycles required to reach the threshold was analyzed using Microsoft Excel. The relative enrichment was calculated by dividing the amount of a given target sequence protected from MNase treatment with the amount of the control sequence (PRE1) and was further normalized by determining the efficiency of the PCR using each primer set compared to that of the control primer set (PRE1). A list of primers and their sequences are available upon request.
Indirect end labeling and primer extension assay.
Cultures in preinduction medium (YEP-glycerol) were induced for HO expression by addition of 2% galactose for 1 h. Permeabilized spheroplasts were isolated as described previously (11), with the following modifications. Zymolyase was used at 10 mg/ml, and the spheroplasting step was extended to 3 min, 40 s. For indirect end-labeling experiments, 0.05 and 0.1 unit of MNase were used per 200 μl spheroplasts. For primer extension analysis, 0.5 to 1.5 unit was used for 200 μl spheroplasts. In all cases, the reactions were stopped by the addition of sodium dodecyl sulfate and EDTA to the digests. After RNase A and proteinase K treatment, DNA was purified by phenol-chloroform extraction and ethanol precipitation. Approximately half of the sample was digested with NdeI, BamHI, and EcoRI and then subjected to Southern blot hybridization for indirect end labeling using a radiolabeled probe that anneals to the region +337 to +1211 (875 bp) or +1680 to +2313 (634 bp) distal to the HO cut or −2606 to −2034 (573 bp) proximal to the HO cut. For a primer extension analysis, primers corresponding to either bp +120 to +144 (for the Crick strand) or +696 to +725 (for the Watson strand) with respect to the HO cut site were used for primer extension, as described by Fazzio and Tsukiyama (11). Densitometry of the bands in each lane was performed using a phosphorimager.
Restriction endonuclease cleavage.
Spheroplasts were digested with several different concentrations of enzymes for 20 min at 37°C, deproteinized, and subsequently digested fully with the following restriction enzymes: HincII (+44) with HindIII and NdeI; HaeIII (+106, +913) with EcoRV and Sty1 or StyI and HindIII; Hinf1 (+302 bp) with HaeIII and StyI; NlaIV (−97 bp) with HaeIII; EcoRV (−268 bp) with HaeIII and StyI; and NdeI (+232 bp, −514 bp) with DraI. Purified DNA was then subjected to Southern blot hybridization with a radiolabeled probe specific to the MAT sequence. The fraction of total chromatin cut with each of the restriction enzymes was quantified by using a phosphorimager.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (42). For histone H3 ChIP, samples were sonicated six times for 20 s instead of the three times for 20 sec used in all other ChIP assays. In the modified ChIP assay with MNase digestion, chromatin was digested with 45 U of MNase prior to immunoprecipitation in place of sonication. The antibodies for YKu and Mre11 were generous gifts from A. Tomkinson and P. Sung, respectively. The anti-H3 antibody was purchased from Abcam (ab1791).
Nonhomologous end-joining assay.
Exponentially growing cultures were incubated with 2% galactose to induce HO for 1 h and then switched to a glucose-containing medium to shut off HO expression. Cells were harvested at several times before (t = 0) and after glucose addition, and DSB efficiency and NHEJ efficiency were measured by qPCR with the primers that anneal to either side of the DSB.
RESULTS
A DSB rapidly realigns nucleosomes at the site of damage.
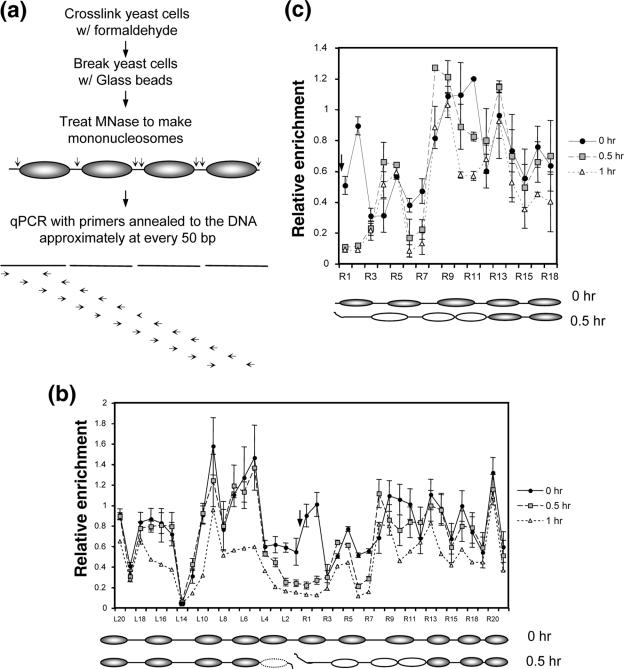
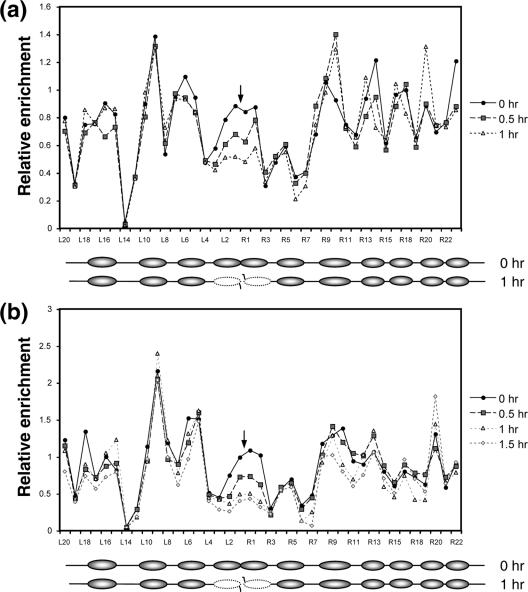
To evaluate the role of chromatin and chromatin modifications in DSB repair, we need to know the nature of the chromatin altered by DSBs and how chromatin modification enzymes catalyze this reaction. We thus used the galactose-controlled HO endonuclease (24) to introduce a single DSB into a donorless (i.e., no HML and HMR) strain of yeast, JKM179, at the MATα locus of chromosome III. We then determined the DSB-induced chromatin changes at this genomic locus. Persistent HO expression and the lack of homologous templates confined the DSB repair to inefficient NHEJ with small insertions/deletions (24, 30). Almost all cells eventually die from DSB damage. Nuclei were isolated at 0 h, 0.5 h, or 1 h after HO expression, cross-linked with formaldehyde, and digested with micrococcal nuclease (MNase). The MNase sensitivity of the chromatin was analyzed using qPCR with primers designed to anneal to the DNA approximately every 50 bp for up to ∼1 kb on either side of the DSB (Fig. 1a). Expression of HO for 1 h led to highly efficient (>90%) DSB formation (see Fig. S1 in the supplemental material). We found that without DSB induction, the DNA is sensitive to MNase about every 200 bp (Fig. 1b, 0 h). The HO recognition site is among those segments susceptible to MNase digestion, likely due to its location in linker DNA between nucleosomes. The nucleosomes at the MAT locus are thus positioned nonrandomly before DSB induction (Fig. 1b, 0 h) (53). More importantly, HO expression realigned the nucleosomes in such a way that at the distal end three nucleosomes were positioned away from the break, and ∼100 bp immediately adjacent to the DSB were exposed to nuclease digestion (Fig. 1b, 0.5 h). About 200 bp immediately next to the proximal end of the DSB also became vulnerable to MNase digestion, likely from the eviction of a nucleosome in response to the DSB (Fig. 1b, 0.5 h).
FIG. 1.
The MNase cleavage profile at the yeast MAT locus before and after a DSB. MNase sensitivity was measured by digesting chromatin with 45 U of MNase and quantifying the amount of undigested DNA with qPCR (a). The relative resistance of a given target sequence to MNase was measured by normalizing the PCR efficiency of each of a series of primers that anneal every 50 bp proximal (L series) or distal (R series) to the DNA surrounding the DSB with that of control primers (PRE1). The MNase cleavage profiles of JKM179 (b) and the G1-arrested JKM139 (MATa derivative of JKM179) (c) are shown at 0, 0.5, and 1.0 h following the addition of galactose to the culture medium. Shown below each graph are the hypothetical positions of nucleosomes inferred from this assay as well as indirect end-labeling and primer extension assays (Fig. 2; see Fig. S3 in the supplemental material). Each oval represents an individual nucleosome; blank ovals indicate mobilized nucleosomes, and the dotted ovals are the nucleosomes lost upon DSB induction. The arrow indicates the location of the HO-induced break. The average for three independent experiments is shown, along with the standard deviation.
Prolonged expression of HO (>1 h) makes the entire region surrounding the DSB twice as vulnerable to MNase digestion (Fig. 1b, 1 h). This broad sensitivity to MNase digestion after 1 to 2 h of HO expression is likely caused by the exonucleolytic degradation of the 5′ DNA ends (48). The deletion of the MRE11 gene or HO expression at G1 considerably slows down resection of the 5′ strand (16) but does not prevent the rapid repositioning of nucleosomes at the distal end of the DSB or the heightened MNase cleavage adjacent to the DSB, indicating that nucleosome movement is independent of end processing (see Fig. 1c).
The unique nucleosome movement induced by a DSB was further confirmed by indirect end-labeling and primer extension analysis of the MNase-treated chromatin at the MAT locus following 1 h of HO expression (Fig. 2) (see Fig. S2 in the supplemental material). New MNase-hypersensitive sites emerged at locations slightly more distal to the DSB, while the MNase-hypersensitive sites existing before DSB formation simultaneously disappeared. A yeast strain lacking the HO gene, SLY558, did not undergo DSB-induced nucleosome repositioning (data not shown). Thus, the DSB-induced shift in the nucleosome position at the MAT locus is a direct consequence of the DSB and not of the metabolic change caused by adding galactose to the culture medium. We also noted that the single unrepaired DSB did not alter the pattern of MNase digestion of bulk chromatin (see Fig. S3 in the supplemental material).
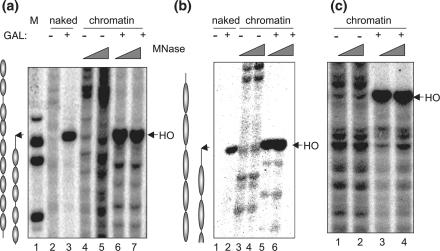
FIG. 2.
Nucleosomes are repositioned to sites more distal from the DSB. Chromatin DNA isolated from cells induced with 1.0 h of HO expression (+) or with no induction (−) was partially digested with MNase and then deproteinized and fully digested with BamHI (a), HindIII (b), or EcoRI (c) to yield common end fragments. Southern blot hybridization was carried out using a radiolabeled probe that anneals 2 kb distal to the break (a) (+1680 to +2313 with respect to the HO cut), 1 kb distal (b) (+337 to +1211 relative to the HO cut), or 2 kb proximal (c) (−2606 to −2034 relative to the HO cut) to map the nucleosome positions adjacent to a DSB at the MAT locus. Molecular weight markers (M) and the MNase digest of naked DNA (naked) are shown for comparison. The arrows indicate the locations of the HO breaks. The inferred nucleosome positions are shown next to the panels of indirect end labeling.
DSB allows nearby chromatin to be more easily digested by nuclease.
The elevated MNase sensitivity of chromatin adjacent to a DSB may indicate that these regions are more accessible to other DNA processing enzymes, including DSB repair factors. We therefore examined the accessibility of multiple sites surrounding the HO recognition sequence at the MAT locus by digesting nuclei with several restriction enzymes before and after 1 h of HO expression. We found a >5-fold increase in the cleavage at the HincII site (+44 bp relative to the HO cleavage site) and the HaeIII (+106 bp) site within an hour of DSB induction (Fig. 3) (see Fig. S4 in the supplemental material). The DSB also increased the cutting at the NlaIV (−97 bp) and EcoRV (−268 bp) sites located proximal to the break (Fig. 3) (see Fig. S4 in the supplemental material). In contrast, the NdeI (+232 bp) and HinfI (+302 bp) sites and a second HaeIII site (+913 bp) were all resistant (∼10% cutting) to enzyme cleavage after the DSB. Cleavage at the second NdeI site, 514 bp proximal to the break, was efficient regardless of break formation, suggesting that it may be located in the linker DNA (Fig. 3a and b). The result shows that the region immediately adjacent to the break becomes highly susceptible to MNase and restriction enzyme cleavage, supporting its enhanced accessibility.
FIG. 3.
The DNA is more accessible to restriction enzymes adjacent to the DSB. (a) Location of restriction enzyme recognition sequences surrounding a DSB at the MAT locus. (b) Graph summarizing the restriction enzyme digests of chromatin before and after the DSB. Chromatin DNA was digested by the enzymes listed in panel a, deproteinized, and then digested by one or two additional restriction enzymes. The DNA was separated by gel electrophoresis, and any changes in the accessibility of restriction endonucleases were analyzed quantitatively by Southern blot hybridization using a probe specific for the MAT locus. Percent cleavage was calculated by dividing the amount of signal of a cut fragment by the combined intensity of cut and uncut fragments (see Fig. S4 in the supplemental material), as quantified with a phosphorimager.
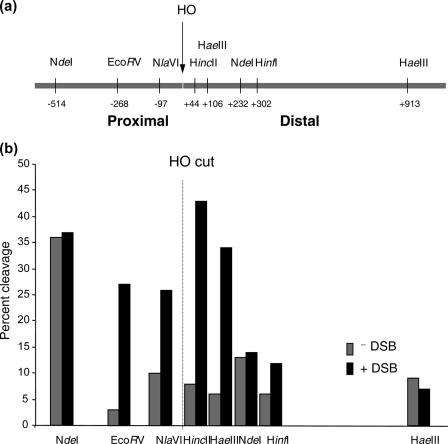
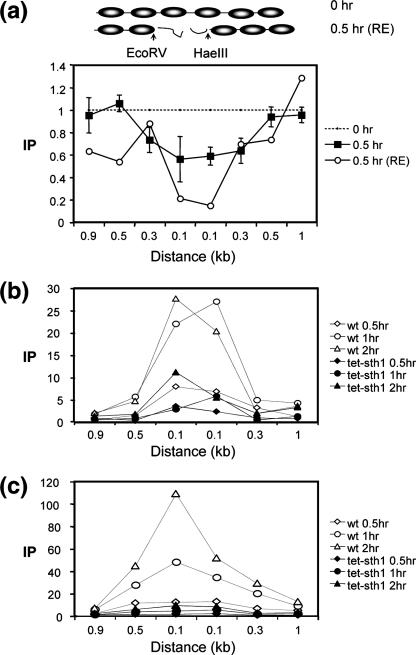
We postulated that the increased accessibility of the chromatin associated with the DSB may stem from the loss of nucleosomes in this region. To test this possibility, we examined the association of histone H3 with DNA surrounding the break using a ChIP assay with an anti-histone H3 antibody. Using a series of primer sets that anneal to DNA on either side of the break, we found that the chromatin immediately next to the break lost ∼50% of its histone H3 (Fig. 4a). In contrast, the MAT sequences positioned at 0.4 or 0.9 kb from the break were fully associated with histone H3 after 0.5 h of HO expression. The remaining histone H3 signal next to the break did not come from recurrent end-joining, because it persisted even in the end-joining-defective dnl4Δ cells (data not shown). Rather, it likely came from long DNA molecules that contain the histone H3-free DNA adjacent to the DSB and other DNA still associated with histone H3 due to incomplete shearing of chromatin during sonication. To test this possibility, we digested chromatin with EcoRV and HaeIII enzymes that cleave sequences located between proximal (−268) and distal (+106 bp) borders of both nucleosome-free and nucleosomal DNA (Fig. 4a), and we examined the association of H3 with DNA on either side of enzyme cleavage using a ChIP assay. We found that DSB formation displaced almost all histone H3 (>80%) from DNA proximal to the HaeIII site but not from DNA distal to the HaeIII site (Fig. 4a). Likewise, histone H3 was absent from DNA distal to the EcoRV site following DSB induction (Fig. 4a). Thus, the results support the idea that a few hundred base pairs from DNA ends are free of nucleosomes. Histone H3 disappears from the sequence immediately adjacent to the DSB at G1 when end processing is slowed, excluding an effect of end processing on this reaction (data not shown).
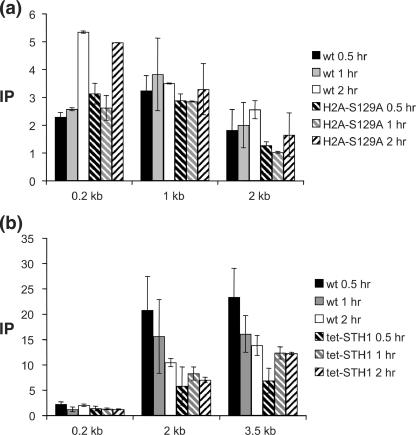
FIG. 4.
DNA in the region of the DSB becomes histone free and is a preferential target for Yku or Mre11 binding. ChIP assays were used to assess the levels of histone H3 (a) or Yku70 (b) and Mre11 (c) in the presence or absence of Sth1 at the DSB using an anti-H3 antibody (Abcam), an anti-Yku antibody (a gift from A. Tomkinson), and an anti-Mre11 antibody (a gift from P. Sung). Yeast cells grown in the preinduction medium were treated with 10 μg/ml doxycycline for 12 h or left untreated. Chromatin was isolated 0 and 0.5 h (a) or 0, 0.5, 1.0, and 1.5 h (b and c) after galactose addition, cross-linked with formaldehyde, and fragmented by extensive sonication or by restriction enzyme cleavage. After immunoprecipitation and reverse cross-linking, purified DNA was analyzed by qPCR using multiple sets of primers that anneal 0.1 kb to 0.9 kb proximal (L) or distal (R) to the DSB, as well as primers specific for the PRE1 gene situated on chromosome V as a control. PCR signals from each primer set at different durations of HO expression were quantified and plotted as a graph. IP represents the ratio of the histone H3, Yku70, or Mre11 PCR signal before and after HO induction, normalized by the PCR signal of the PRE1 control. Above the graph (a), the positions of recognition sites for EcoRV and HaeIII, two restriction enzymes used to fragment chromatin prior to immunoprecipitation by anti-histone H3 antibody, are shown. Each point is the average for at least two separate experiments. The standard deviation is shown.
The yeast Ku and Mre11 complexes bind to nucleosome-free sequences at the DSB.
Formation of nucleosome-free DNA that is highly accessible to MNase and restriction endonucleases may be crucial to recruit repair proteins to the broken chromatin. We tested this idea using a ChIP assay, carefully mapping the binding sites of Yku70 and the Mre11 proteins at or near the DSB. The Yku70 and Mre11 proteins were chosen for this study because these two repair proteins likely arrive at the DSB early (26). As shown in Fig. 4b and c, both Yku70 and Mre11 associate primarily with the DNA immediately adjacent to the DSB. The binding of these proteins beyond the nucleosome-free DNA is reduced significantly as the distance from the break becomes greater (Fig. 4b and c). The result is thus consistent with the premise that Yku70 and Mre11 bind primarily to the nucleosome-free DNA formed immediately after the DSB induction.
RSC remodels damaged chromatin.
Recently, multiple histone modification enzymes and ATP-dependent chromatin remodeling complexes have been implicated in DSB repair (51). Among them, RSC seems uniquely suited for a role in early chromatin remodeling at the DSB, because it is rapidly recruited to the DSB and functions in both end-joining and homologous recombination (6, 42). In addition, two subunits of RSC, known as Rsc1 and Rsc2, physically interact with the key repair proteins Mre11 and Yku80. These interactions are critical for cell survival after genotoxic damage (42). To examine whether RSC is responsible for the DSB-induced chromatin change, we analyzed MNase cleavage of rsc mutant nuclei after a DSB occurred at the MAT locus.
RSC is essential for cell viability (54), and thus, the available rsc mutants to date possess rsc activity that is limited but sufficient to sustain cell survival. Instead of using these rsc mutants, we established a donorless yeast strain with the key ATPase subunit (Sth1) of the RSC complex under the control of the Tet-off system and the galactose-inducible HO integrated at the ade3 locus (29). We then monitored the MNase cleavage of chromatin at the MAT locus after adding galactose and doxycycline to the medium, simultaneously inducing HO expression but shutting off Sth1 expression. The conditional repression of STH1 by doxycycline was confirmed by Western blotting, using an anti-Sth1 antibody (see Fig. S5a in the supplemental material). The induction of a break by HO in the Sth1-depleted strain or the strain with a gene deletion of RSC2, encoding a nonessential subunit of RSC, was moderately reduced to approximately 70% of that in control cells at 1 h (see Fig. S1 in the supplemental material). As shown in Fig. 5a, doxycycline-induced repression of Sth1 dramatically inhibited the nucleosome movement at the distal side of the break, and the chromatin failed to show the broad MNase sensitivity after 1 h or more of HO expression, although the nucleosome-free and MNase-hypersensitive zone next to the DSB still formed, albeit with a delay. This suggests that RSC plays a critical role in remodeling chromatin at the DSB. Deletion of RSC2 also delayed the nucleosome positioning and formation of the MNase-hypersensitive sites surrounding the DSB in a similar but less severe fashion than doxycycline-mediated repression of STH1 expression (compare Fig. 5a and b). The results indicate that chromatin remodeling at the DSB requires an RSC complex containing Sth1 and Rsc2.
FIG. 5.
Lack of RSC reduces the chromatin remodeling at the DSB. The MNase cleavage profiles of the tet-Sth1 (SLY579) with 10 μg/ml doxycycline (A) or SLY403 (rsc2Δ) (B) are shown at 0, 0.5, and 1.0 h after galactose was added to the culture medium as described in Fig. 1. Inferred nucleosome positions before and after the DSB are shown below each graph. Each oval represents an individual nucleosome; the dotted ovals are the nucleosomes lost upon DSB induction. The arrow indicates the location of the HO-induced break. The average for three independent experiments is shown.
RSC facilitates binding of Ku and Mre11 and efficient phosphorylation of H2AX at the damaged chromatin.
Since Yku70 and Mre11 bind preferentially to MNase-sensitive, nucleosome-free DNA and RSC is required to form this DNA next to the DSB, we reasoned that the absence of functional RSC may impair the binding of these two proteins at the DSB. To test this idea, we performed ChIP assays with anti-Yku70 or anti-Mre11 antibody to examine the level of Yku70 or Mre11 binding at the DSB when STH1 expression was repressed.
We found that repression of Sth1 markedly reduced the binding of both Yku70 and Mre11 to the DSB (Fig. 4b and c). No change in the expression of Yku70 or Mre11 was detected using immunoblot analysis of yeast extract with anti-Yku70 or anti-Mre11 antibody (see Fig. S5b and S5c in the supplemental material). The results suggest that RSC facilitates the binding of Yku70 and Mre11 to the DSB by creating nucleosome-free DNA suitable for the binding of these proteins.
Damage-induced phosphorylation of H2AX is another early chromatin modification that occurs in every eukaryote from yeast to humans (10, 35). The available evidence suggests that phosphorylated H2AX marks the targeting of the Ino80 chromatin-remodeling complex and recruitment of cohesin to the DSBs (48, 49). Therefore, we tested whether phosphorylated H2AX targets RSC to the DSB by ChIP assays with anti-FLAG antibody with yeast strains carrying the FLAG-tagged Rsc1 and the hta1-S129A hta2-S129A mutation, which blocks damage-induced H2AX phosphorylation (10, 43). We found that lack of H2AX phosphorylation did not interfere in the recruitment of Rsc1-FLAG to the DSB (Fig. 6a). In contrast, the level of H2AX phosphorylation is significantly decreased when Sth1 is depleted (Fig. 6b). The reduced level of H2AX phosphorylation at a later time may come from end processing that induces nucleosome eviction from broken DNA ends (48). The result supports the early involvement of RSC in DSB repair, one purpose of which is to facilitate targeting of the repair enzymes to the DSB.
FIG. 6.
Depletion of RSC reduces phosphorylation of H2A. The level of Rsc1-FLAG at the DSB in the wild-type SLY1103 or SLY1101 (hta1-S129A hta2-S129A) (a), where damage-induced phosphorylation of carboxy-terminal H2A is blocked, or the level of phosphorylated H2A in cells that were replete or deficient in Sth1 (b) was determined using ChIP assays. Chromatin isolated from yeast cells grown in the preinduction medium with 10 μg/ml doxycycline for 12 h and HO expression induced by the addition of 2% galactose were cross-linked and immunoprecipitated with an anti-FLAG antibody (a) or a H2AX antibody (b). Purified DNA was analyzed by qPCR using multiple sets of primers. See the legend to Fig. 4 for the definition of IP.
Depletion of Sth1 severely reduced processing and joining of a DSB at the MAT locus.
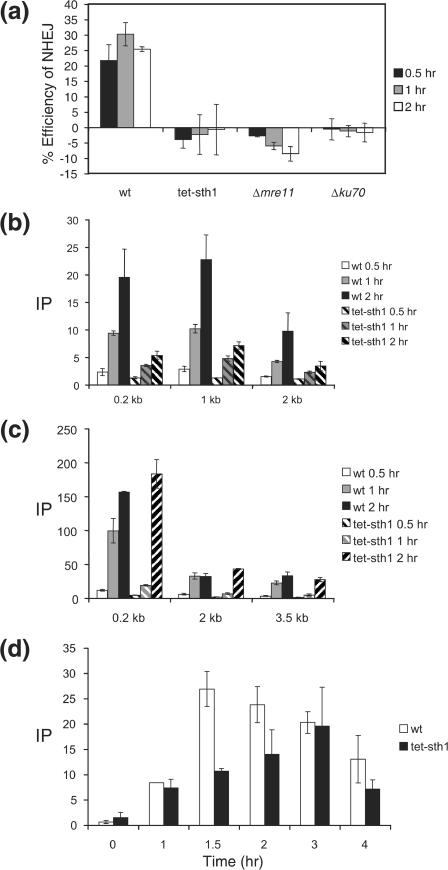
Reduced binding of the Yku and Mre11 proteins at a DSB prompted us to examine whether depletion of Sth1 decreases end joining of a DSB at the MAT locus. To test this idea, we induced HO expression for 1 h and then quickly shut it off by adding glucose to the culture medium. We then measured the rate of end joining by qPCR using a set of primers annealed to either side of the break and genomic DNA isolated at several times during recovery (Fig. 7a). As controls, we measured the rate of end joining for the YKU-deleted or MRE11-deleted strain using the same qPCR assay. We found that depletion of Sth1 dramatically reduced end joining at the MAT locus, with its end-joining defect being as severe as that for strains lacking the Yku or Mre11 protein.
FIG. 7.
Depletion of Sth1 severely reduced joining and processing of DSB at the MAT locus. (A) NHEJ proficiency was assayed by using qPCR with oligonucleotides that anneal to each side of DSB. Cultures were induced to express HO for 1 h and switched to the YEP-glucose medium that shuts off HO expression. The percent NHEJ efficiency was calculated by the equation (Tx − T0)/(Tun− T0) × 100, using the amount of PCR product prior to HO expression (Tun), immediately after 1 h of HO expression (T0), and during the recovery following HO expression (TX), all of which were normalized by the control (PRE-1) PCR. In this assay, almost every surviving cell religated and restored the complete MATα locus by precise end joining (25). Each point represents the average for three or more separate experiments. The standard deviation is shown. ChIP assays were used to assess the levels of RPA (b) or Rad51 (c) in the presence or absence of Sth1 at the DSB using an anti-RPA antibody (a gift from S. Brill) or anti-Rad51-antibody (a gift from P. Sung) and the level of Rad51 at the HML donor (d). Purified DNA was analyzed by qPCR using multiple sets of primers. See the legend to Fig. 4 for the definition of IP.
Processing of DSBs by 5′-to-3′ nuclease(s) and the resulting formation of recombinogenic single-stranded DNA depend on a functional Mre11 complex and are required for efficient homologous recombination (33). Reduction of Mre11 binding at a DSB in Sth1 depletion led us to test whether the lack of RSC also affects the DNA end-processing step. We determined the rate of resection at the DSB by measuring the level of binding of RPA, a trimeric single-strand binding protein, to single-stranded DNA formed at or near the DSB with a ChIP assay followed by qPCR using primer sets annealed to multiple regions at the MAT locus (23). We found that depletion of Sth1 markedly reduced binding of RPA to the DSB and thus decreased the formation of single-stranded DNA (Fig. 7b). Deficient nucleolytic end processing and the observed reduction in single-stranded DNA formation are consistent with the lack of broad sensitivity to MNase after 1 to 2 h of HO expression in rsc mutants (Fig. 5). We also measured the binding of Rad51 to the DSB for yeast strains depleted of Sth1 using a ChIP assay with anti-Rad51 antibody (a gift from P. Sung). Association of Rad51 to the DSB was significantly delayed for the first hour for the Sth1-depleted strain (Fig. 7c) but was fully recovered to a level identical to that of the wild type after 2 h of HO expression. In addition, Rad51 binds well to the HML donors in the absence of Sth1, albeit with a delay (Fig. 7d). Consistent with the previous reports, repression of Sth1 resulted in a moderate reduction (∼2-fold) of mating type switch recombination (6; data not shown).
The results suggest that the slower end processing in rsc mutants delayed the initial binding of Rad51 but failed to block its subsequent binding at the DSB.
DISCUSSION
We have shown here that creation of a barely repairable DSB causes a rapid reorganization of the surrounding nucleosomes by the internal migration of two to four nucleosomes at the distal end and the displacement of a nucleosome at the proximal end (Fig. 1b). These lead to the generation of histone H3-free DNA of several hundred base pairs near the DSB that serves as a binding site for the Yku and Mre11 repair enzymes (Fig. 1 and 4) and facilitates processing and joining of chromosome breaks (Fig. 7). Nucleosome alignment occurring within 0.5 h of HO expression is likely independent from the end processing that normally begins at 1 h or later (Fig. 1b). Consistent with this, nucleosomes still realign at G1, when resection is severely limited (Fig. 1c). The increased MNase cleavage of the entire MAT locus at 1 h after adding galactose coincides with the 5′ exonucleolytic degradation of the break; this supports the role of end processing at this time and that of the Ino80 complex in nucleosome displacement and the subsequent recruitment of homologous recombination proteins (48).
Chromatin remodeling at DSB requires the RSC complex. When STH1 expression was repressed, the nucleosome at the distal end failed to reposition, and the MNase cleavage of the entire region surrounding the DSB was reduced after 1 h of HO expression. A very similar, albeit less severe, chromatin defect in rsc2Δ mutants further supports the role of RSC in this process. Recent biochemical studies showed that RSC disassembles and removes nucleosomes from the DNA end in a stepwise manner (27). RSC is also capable of moving nucleosomes by directional translocation of the DNA (41). Therefore, RSC is well suited for the observed nucleosome repositioning after a DSB and is directly responsible for these changes. Interestingly, RSC remodels chromatin differently at two DNA ends. It displaces a nucleosome at the proximal end, but pushes two to four nucleosomes further inward at the distal end. We do not yet know the reason for such asymmetry and the way RSC remodels nucleosomes at either end of the break. Nevertheless, the difference between the two sides of the break has been proposed for some time, since one end participates more preferentially in strand invasion than the other for recombination (15, 33). Furthermore, Ino80, another chromatin remodeler required for DSB repair, asymmetrically binds proximal to the MAT locus and migrates to the distal end upon DSB induction (48). We speculate that this asymmetry is caused by asymmetric end processing of HO cleavage or by the preexisting chromatin architecture before DSB formation.
We have shown that RSC participates in an early step of DSB repair by preparing the binding site for the Mre11 and Yku complexes. RSC arrives at the DSB very early (in fact, almost at the same time as Yku and Mre11) and physically and functionally interacts with DSB repair enzymes that act at the earliest stage of DSB repair (6, 42). RSC is also needed for the high-level H2AX phosphorylation at DSBs, an early marker of DNA DSB sites (35, 49). Early involvement in DSB repair explains why RSC is required for both the Yku-dependent end joining and homologous recombination (6, 42). By controlling the binding of the Mre11 complex at the DSB, it also regulates end processing and timely execution of the downstream HR steps. RSC may also mobilize nucleosomes in front of nuclease(s) to facilitate end processing of chromatin. Previously, characterization of rsc2Δ demonstrated additional deficiency in the postsynaptic step of gene conversion using assays that measure intermediates from distinct steps of yeast mating-type gene conversion (6). Based on these observations, we propose that RSC plays a role in at least two distinct steps of DSB repair: targeting repair enzymes to the DSB and following strand invasion and gap synthesis, in resolution and/or ligation of recombination intermediates.
We noted that enhanced MNase cleavage adjacent to the DSB still occurs in rsc mutants, but with a delay. This suggests that other mechanism(s) exist for nucleosome displacement at the DSB and can promote the binding of the Yku70/80 or Mre11 complex even when RSC is not available. Other chromatin remodelers, like Ino80, are clearly good candidates for such a role. Indeed, we found that inactivation of both RSC and Ino80 caused a synergistic reduction in survival after genotoxic damage (Y.Y. and S.E.L., unpublished observations). Alternatively, the nucleosomes immediately adjacent to the DSB may be inherently unstable and may slowly fall from the end. The alternative mechanism(s) of nucleosome displacement then allow few Yku70 and Mre11 proteins to associate with the DSB in rsc mutants; that is why the available rsc mutations are not as defective in DSB repair as conventional repair mutants, such as the yku70Δ mutant (42). However, we would also like to point out that all available rsc mutants tested for their role in DSB repair still retain enough activity to support its essential cellular functions and minimal repair functions, but a true null mutation would not be able to catalyze DSB repair so competently. Indeed, we showed that depletion of Sth1 caused an end-joining defect as severe as those associated with deletion of the YKU70 or MRE11 gene.
Like yeast, both humans and mice have two distinct Swi/Snf ATP-dependent remodeling complexes, one of which is essential for cell viability (52). Inactivation of mammalian swi/snf complexes also causes inefficient DSB repair, heightened damage sensitivity, and a deficit in damage-induced H2AX phosphorylation (34). Conditional inactivation of murine Snf5, a subunit shared between two Swi/Snf-like chromatin remodelers, is highly sensitive to DSB-causing agents (21). Given the remarkable conservation of DSB repair mechanisms from yeast to humans, it will be interesting to test whether chromatin remodelers catalyze equivalent reactions in DSB repair of higher organisms.
Supplementary Material
Acknowledgments
We are thankful to Steven Brill, Jim Haber, Brehon Laurent, Kyungjae Myung, Patrick Sung, Alan Tomkinson, Toshio Tsukiyama, Jerry Workman, and the members of the S.E.L. laboratory for the helpful comments and discussion on the manuscript.
This work was supported by the Sydney Kimmel Cancer Research Foundation and NIH grant ES012244 to S.E.L.
Footnotes
Published ahead of print on 18 December 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ai, X., and M. R. Parthun. 2004. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell 14:195-205. [DOI] [PubMed] [Google Scholar]
- 2.Bassing, C. H., K. F. Chua, J. Sekiguchi, H. Suh, S. R. Whitlow, J. C. Fleming, B. C. Monroe, D. N. Ciccone, C. Yan, K. Vlasakova, D. M. Livingston, D. O. Ferguson, R. Scully, and F. W. Alt. 2002. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. USA 99:8173-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. [DOI] [PubMed] [Google Scholar]
- 4.Cairns, B. R. 2005. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15:185-190. [DOI] [PubMed] [Google Scholar]
- 5.Celeste, A., S. Petersen, P. J. Romanienko, O. Fernandez-Capetillo, H. T. Chen, O. A. Sedelnikova, B. Reina-San-Martin, V. Coppola, E. Meffre, M. J. Difilippantonio, C. Redon, D. R. Pilch, A. Olaru, M. Eckhaus, R. D. Camerini-Otero, L. Tessarollo, F. Livak, K. Manova, W. M. Bonner, M. C. Nussenzweig, and A. Nussenzweig. 2002. Genomic instability in mice lacking histone H2AX. Science 296:922-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chai, B., J. Huang, B. R. Cairns, and B. C. Laurent. 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19:1656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, W. L., F. B. Turner, T. Krishnamoorthy, B. Wolner, S. H. Ahn, M. Foley, J. A. Dorsey, C. L. Peterson, S. L. Berger, and C. D. Allis. 2005. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15:656-660. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury, D., M. C. Keogh, H. Ishii, C. L. Peterson, S. Buratowski, and J. Lieberman. 28. November 2005, posting date. Gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20:801-809. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 9.Downs, J. A., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger, N. Bouchard, S. J. Kron, S. P. Jackson, and J. Cote. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16:979-990. [DOI] [PubMed] [Google Scholar]
- 10.Downs, J. A., N. F. Lowndes, and S. P. Jackson. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408:1001-1004. [DOI] [PubMed] [Google Scholar]
- 11.Fazzio, T. G., and T. Tsukiyama. 2003. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol. Cell 12:1333-1340. [DOI] [PubMed] [Google Scholar]
- 12.Felsenfeld, G., and M. Groudine. 2003. Controlling the double helix. Nature 421:448-453. [DOI] [PubMed] [Google Scholar]
- 13.Fyodorov, D. V., and J. T. Kadonaga. 2001. The many faces of chromatin remodeling: SWItching beyond transcription. Cell 106:523-525. [DOI] [PubMed] [Google Scholar]
- 14.Huang, J., B. Liang, J. Qiu, and B. C. Laurent. 5. December 2005, posting date. ATP-dependent chromatin-remodeling complexes in DNA double-strand break repair: remodeling, pairing and (re)pairing. Cell Cycle 4:1713-1715. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 15.Hunter, N., and N. Kleckner. 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106:59-70. [DOI] [PubMed] [Google Scholar]
- 16.Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani, W. Carotenuto, G. Liberi, D. Bressan, L. Wan, N. M. Hollingsworth, J. E. Haber, and M. Foiani. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431:1011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jazayeri, A., A. D. McAinsh, and S. P. Jackson. 7. January 2004, posting date. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 101:1644-1649. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 19.Keogh, M. C., J. A. Kim, M. Downey, J. Fillingham, D. Chowdhury, J. C. Harrison, M. Onishi, N. Datta, S. Galicia, A. Emili, J. Lieberman, X. Shen, S. Buratowski, J. E. Haber, D. Durocher, J. F. Greenblatt, and N. J. Krogan. 20. November 2006, posting date. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 439:497-501. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 20.Khanna, K. K., and S. P. Jackson. 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27:247-254. [DOI] [PubMed] [Google Scholar]
- 21.Klochendler-Yeivin, A., E. Picarsky, and M. Yaniv. 2006. Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Mol. Cell. Biol. 26:2661-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krejci, L., L. Chen, S. Van Komen, P. Sung, and A. Tomkinson. 2003. Mending the break: two DNA double-strand break repair machines in eukaryotes. Prog. Nucleic Acid Res. Mol. Biol. 74:159-201. [DOI] [PubMed] [Google Scholar]
- 23.Lee, S. E., D. A. Bressan, J. H. J. Petrini, and J. E. Haber. 2002. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair 1:27-40. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. E., F. Paques, J. Sylvan, and J. E. Haber. 1999. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 9:767-770. [DOI] [PubMed] [Google Scholar]
- 26.Lisby, M., J. H. Barlow, R. C. Burgess, and R. Rothstein. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699-713. [DOI] [PubMed] [Google Scholar]
- 27.Lorch, Y., B. Maier-Davis, and R. D. Kornberg. 21. February 2006, posting date. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. USA 103:3090-3093. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 29.Mnaimneh, S., A. P. Davierwala, J. Haynes, J. Moffat, W. T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, M. Trochesset, D. Morse, N. J. Krogan, S. L. Hiley, Z. Li, Q. Morris, J. Grigull, N. Mitsakakis, C. J. Roberts, J. F. Greenblatt, C. Boone, C. A. Kaiser, B. J. Andrews, and T. R. Hughes. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118:31-44. [DOI] [PubMed] [Google Scholar]
- 30.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison, A. J., J. Highland, N. J. Krogan, A. Arbel-Eden, J. F. Greenblatt, J. E. Haber, and X. Shen. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767-775. [DOI] [PubMed] [Google Scholar]
- 32.Reference deleted.
- 33.Pâques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, J. H., E. J. Park, H. S. Lee, S. J. Kim, S. K. Hur, A. N. Imbalzano, and J. Kwon. 24. August 2006, posting date. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 25:3986-3997. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, C. L., and J. Cote. 2004. Cellular machineries for chromosomal DNA repair. Genes Dev. 18:602-616. [DOI] [PubMed] [Google Scholar]
- 37.Petrini, J. H. 2005. At the end, remodeling leads to eviction. Nat. Struct. Mol. Biol. 12:1028-1029. [DOI] [PubMed] [Google Scholar]
- 38.Pierce, A. J., J. M. Stark, F. D. Araujo, M. E. Moynahan, M. Berwick, and M. Jasin. 2001. Double-strand breaks and tumorigenesis. Trends Cell Biol. 11:S52-S59. [DOI] [PubMed] [Google Scholar]
- 39.Qin, S., and M. R. Parthun. 2006. Recruitment of the type B histone acetyltransferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol. Cell. Biol. 26:3649-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 41.Saha, A., J. Wittmeyer, and B. R. Cairns. 2002. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16:2120-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shim, E. Y., J. L. Ma, J. H. Oum, Y. Yanez, and S. E. Lee. 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell. Biol. 25:3934-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shroff, R., A. Arbel-Eden, D. Pilch, G. Ira, W. M. Bonner, J. H. Petrini, J. E. Haber, and M. Lichten. 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14:1703-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamburini, B. A., and J. K. Tyler. 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 25:4903-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiriet, C., and J. J. Hayes. 2005. Chromatin in need of a fix: phosphorylation of H2AX connects chromatin to DNA repair. Mol. Cell 18:617-622. [DOI] [PubMed] [Google Scholar]
- 47.Tsukiyama, T., and C. Wu. 1997. Chromatin remodeling and transcription. Curr. Opin. Genet. Dev. 7:182-191. [DOI] [PubMed] [Google Scholar]
- 48.Tsukuda, T., A. B. Fleming, J. A. Nickoloff, and M. A. Osley. 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438:379-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unal, E., A. Arbel-Eden, U. Sattler, R. Shroff, M. Lichten, J. E. Haber, and D. Koshland. 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16:991-1002. [DOI] [PubMed] [Google Scholar]
- 50.van Attikum, H., O. Fritsch, B. Hohn, and S. M. Gasser. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777-788. [DOI] [PubMed] [Google Scholar]
- 51.van Attikum, H., and S. M. Gasser. 2005. The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell. Biol. 6:757-765. [DOI] [PubMed] [Google Scholar]
- 52.Wang, W. 2003. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr. Top. Microbiol. Immunol. 274:143-169. [DOI] [PubMed] [Google Scholar]
- 53.Weiss, K., and R. T. Simpson. 1998. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα. Mol. Cell. Biol. 18:5392-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson, B., H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 3. October 2006, posting date. The RSC chromatin remodeling complex bears an essential fungal-specific protein module with broad functional roles. Genetics 172:795-809. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, C., T. Tsukiyama, D. Gdula, P. Georgel, M. Martinez-Balbas, G. Mizuguchi, V. Ossipow, R. Sandaltzopoulos, and H. M. Wang. 1998. ATP-dependent remodeling of chromatin. Cold Spring Harbor Symp. Quant. Biol. 63:525-534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.