Abstract
Thyroid hormone (T3) is critical for growth, differentiation, and maintenance of metabolic homeostasis. Mice with a knock-in mutation in the thyroid hormone receptor α gene (TRα1PV) were created previously to explore the roles of mutated TRα1 in vivo. TRα1PV is a dominant negative mutant with a frameshift mutation in the carboxyl-terminal 14 amino acids that results in the loss of T3 binding and transcription capacity. Homozygous knock-in TRα1PV/PV mice are embryonic lethal, and heterozygous TRα1PV/+ mice display the striking phenotype of dwarfism. These mutant mice provide a valuable tool for identifying the defects that contribute to dwarfism. Here we show that white adipose tissue (WAT) mass was markedly reduced in TRα1PV/+ mice. The expression of peroxisome proliferator-activated receptor γ (PPARγ), the key regulator of adipogenesis, was repressed at both mRNA and protein levels in WAT of TRα1PV/+ mice. Moreover, TRα1PV acted to inhibit the transcription activity of PPARγ by competition with PPARγ for binding to PPARγ response elements and for heterodimerization with the retinoid X receptors. The expression of TRα1PV blocked the T3-dependent adipogenesis of 3T3-L1 cells and repressed the expression of PPARγ. Thus, mutations of TRα1 severely affect adipogenesis via cross talk with PPARγ signaling. The present study suggests that defects in adipogenesis could contribute to the phenotypic manifestation of reduced body weight in TRα1PV/+ mice.
Thyroid hormone (T3) is critical for growth, differentiation, development, and maintenance of metabolic homeostasis. Its action is initiated by interaction with thyroid hormone receptors (TRs) that are members of the steroid hormone/retinoic acid receptor superfamily. Two TR genes located on two different chromosomes encode four T3 binding isoforms: α1, β1, β2, and β3. TRs bind to specific DNA sequences (thyroid hormone response elements) on promoters to regulate target gene transcription (3). TR transcription is regulated at multiple levels (10). In addition to that by T3 and types of thyroid hormone response elements, TR transcription is modulated by tissue- and development-dependent TR isoform expression (3) and by a host of corepressors and coactivators (7, 30).
Given the important biological functions of TRs, it is reasonable to expect that mutations of TRs could have deleterious effects. Indeed, mutations of the TRβ gene are known to cause the genetic syndrome of resistance to thyroid hormone (RTH) (45). TRβ mutants identified in patients with RTH lose T3 binding activity and transcription capacity and act in a dominant negative manner to cause clinical phenotypes (43, 45). Patients with RTH are usually heterozygotes with only one mutated TRβ gene (43). Some of the reported clinical features include goiter, short stature, decreased weight, tachycardia, hearing loss, attention deficit hyperactivity disorder, decreased IQ, and dyslexia (5, 43). One patient homozygous for a mutant TRβ who displayed an extraordinary and complex phenotype of extreme RTH, with very high levels of thyroid hormone and thyroid-stimulating hormone (TSH), has been reported (31).
A mouse model that faithfully recapitulates human RTH has been created (TRβPV mouse [23]). This knock-in mutant mouse harbors a potent dominant negative TRβ mutation found in an RTH patient known as PV (29, 33). This mouse model not only allows the elucidation of the molecular basis of RTH in vivo (9) but also enables the discovery of other diseases caused by the mutations of both TRβ alleles. Indeed, homozygous TRβPV/PV mice spontaneously develop follicular thyroid carcinoma (39, 47) and pituitary tumors (16), indicating the severe consequences of mutations of both TRβ alleles (8, 11, 12).
One central issue in understanding the biology of TR is whether TRα1 and TRβ serve redundant or specific roles. Studies of mice deficient for either of the two TR genes or for both TR genes indicate that TR isoforms have both redundant roles and specific functions (14). To ascertain whether mutations of the TRα gene cause common or distinct abnormalities compared with mutations of the TRβ gene, we have created another knock-in mouse (TRα1PV mouse [22]) by targeting the same PV mutation to the corresponding locus of the TRα gene. Strikingly, the TRα1PV mouse exhibits a phenotype distinct from that of the TRβPV mouse. In contrast to the case with TRβPV mice, TRα1PV mice do not display the RTH phenotype. This lack of an RTH phenotype is consistent with the observation that no mutations of the TRα gene have ever been detected in patients with RTH. Homozygous mutations of the TRα gene are more deleterious than are homozygous mutations of the TRβ gene in that TRα1PV/PV mice die either near the end of the gestation period or very shortly after birth (22). Moreover, TRα1PV/+ mice are dwarfs with reduced body lengths and weights, have reduced fertility and high mortality rates, and display distinct abnormal T3 target gene expression profiles (22). These observations indicate that mutations of TRα1 and TRβ genes result in different abnormalities, suggesting that the actions of these two TR mutant isoforms are distinct in vivo.
We have previously shown that the reduced body lengths of TRα1PV/+ mice are due to delayed bone development and shorter long bones (32). To further understand the molecular action of TRα1PV in vivo, the purpose of this study was to identify the defects contributing to severe weight reduction of TRα1PV/+ mice. We found that TRα1PV/+ mice had impairments in the adipogenesis of white adipose tissue (WAT) but not brown adipose tissue (BAT). The impaired adipogenesis of WAT was, at least in part, due to the repression of the expression and transcription activity of the master regulator of adipogenesis, peroxisome proliferator-activated receptor γ (PPARγ). Thus, TRα1PV mediates the reduction of WAT mass via the repression of critical genes in adipogenesis, contributing to the phenotypic expression of severe reduced body weight observed in dwarfism.
MATERIALS AND METHODS
Animals.
This animal study was carried out according to the protocol approved by the National Cancer Institute Animal Care and Use Committee. Genotyping of TRα1PV mice was performed using specific primers for TRα1 (22). Wild-type littermates were used for the comparison of phenotypes.
To determine the effect of T3 on target gene expression in vivo, wild-type male mice (age, 2 to 3 months) were divided into three groups (n = 4 to 5). One group of mice was rendered into a hypothyroid state by feeding with a low-iodine diet supplemented with 0.15% propylthiouracil (PTU) (Harlan Teklad, Madison, WI) for 35 days. Another group of mice was fed the same diet (low-iodine with PTU) for 35 days, but was rendered into a hyperthyroid state by daily T3 intraperitoneal injections (5 μg per 20 g of body weight) during the last 5 days. They were dissected 24 h after the last T3 injection. Control groups received no treatment. The hypothyroid or hyperthyroid state of mice was confirmed by the determination of serum thyroid hormone levels (48).
Construction of pcDNA3.1-TRα1 and pcDNA3.1-TRα1PV.
The plasmid pCLC61 (26) was used as the PCR template to clone human TRα1PV into pFLAG-CMV2 (Sigma). To replace the human TRα1 C-terminal region (amino acids 394 to 410) with the human PV mutation (16 amino acids derived from TRβ PV), two primers (TRα1-EcoRI, GCGAATTCAGAACAGAAGCCAAGCAAGGTG, and TRα1PV-BamHI, AATGGATCCTCAGTCTAATCCTCGAACACTTCCAAGAACAAAGGG [underlined portions of sequences indicate restriction sites]) and an adaptor (TRα1PV adaptor, CTTCCAAGAACAAAGGGGGGAAGAGTTCTGTGGGGGCACTCGACTTTCATG) were used in the PCR. This cloned plasmid was designated Flag-TRα1PV. To generate pcDNA3.1-TRα1 and pcDNA3.1-TRα1PV expression plasmids, pCLC61 and Flag-TRα1PV were used as templates, respectively, and primers hTRα1-BamHI-5′ (CGCGGATCCATGGAACAGAAGCCAAGC), hTRα1-EcoRI-3′ (CCGGAATTCTTAGACTTCCTGATCCTC), and hTRβ PV-EcoRI-3′ (CCGGAATTCTCAGTCTAATCCTCGAAC) were designed. Underlined portions of sequences indicate the restriction sites. All of the plasmid constructs were confirmed by DNA sequencing.
Determination of serum hormones and glucose.
Serum levels of total T4 and T3 were determined by using a GammaCoat T4 or T3 radioimmunoassay kit (DiaSorin, Stillwater, MN) according to the manufacturer's instructions. Serum glucose (nonfasting) was determined by using the method of glucose oxidation (Accu-Chek glucose monitor; Roche Diagnostics Co., Indianapolis, IN). Triglycerides and free fatty acids were measured using assay kits (catalog no. TR22421 [Thermo Trace Ltd., Melbourne, Australia] and catalog no. 1383175 [Roche Diagnostics GmbH, Mannheim, Germany], respectively) according to the manufacturer's instructions. Serum adiponectin, insulin, and leptin were measured by radioimmunoassays (catalog no. MADP-60HK, SRI-13K, and ML-82K, respectively; Linco Research, Inc., St. Charles, MO).
Glucose tolerance test.
Fasted mice were given 1.5 g glucose per kilogram of body weight intraperitoneally. One drop of whole blood was obtained from the mouse tails and placed on a LifeScan glucometer glucose test strip immediately. Blood was obtained 0, 15, 30, 45, 60, and 120 min after the glucose injection.
Insulin tolerance test.
Fasted mice were given 1 mU of regular insulin per gram body weight intraperitoneally. Blood glucose levels were measured 0, 15, 30, 45, 60, and 120 min after the insulin injection, by a method similar to that described above for the glucose tolerance test.
Determination of G6PDH activity.
Inguinal fat was dissected and processed, and glucose-6-phosphate dehydrogenase (G6PDH) activity was measured as described previously (6), with modifications. Briefly, inguinal fat was minced, homogenized in 5 volumes of ice-cold 10 mM Tris buffer (pH 7.4) containing 0.32 M sucrose, 2 mM EDTA, and 5 mM 2-mercaptoethanol, and filtered through two layers of gauze. After centrifugation at 10,000 × g for 10 min and skimming off of the top fat layer, the supernatant was centrifuged for 1 h at 100,000 × g to obtain the cytosolic fraction. G6PDH activity was determined in 100 mM Tris buffer (pH 8.0) containing 1 mM glucose 6-phosphate, 1 mM NADP+, and a suitable amount of diluted cytosolic protein (∼50 to 100 μg). The formation of NADPH at room temperature (absorbance, A340) was measured for 10 min at 30-s intervals. The enzyme activity was expressed as the change in optical density per minute per milligram of protein.
Lipolysis assay.
Epididymal adipose tissue was removed and washed in Krebs-Ringer bicarbonate buffer (pH 7.4). Fat cells were isolated as previously described (27, 35). The lipolysis assays were carried out similarly as previously described (13, 27, 42). Briefly, fat cells (500 cells/100 μl) were incubated in a 96-well microtiter plate for 2 h at 37°C in Krebs-Ringer bicarbonate buffer containing 40 mg/ml bovine serum albumin, 3 μmol of glucose, and 0.1 mg/ml ascorbic acid in the absence or presence of an increasing concentration of norepinephrine, forskolin, or dibutyryl cyclic AMP (cAMP). Lipolytic activity was determined by glycerol release using a standard kit assay (Sigma-Aldrich Co.). Each assay was run in triplicate.
For the comparison of adipocytes in WAT between TRα1PV/PV mice and wild-type mice, inguinal fat pads were removed and tissues with same weight (150 mg) were minced and digested using 1 ml type 2 collagenase (0.2% in Hanks' balanced salt solution containing 1% bovine serum albumin, 3 mM CaCl2, and 50 ng/ml gentamicin). The digestion was carried out in a 37°C shaker for about 20 to 30 min to complete the dissociation of cells. After centrifugation at 1,000 rpm for 10 min, the mature adipocytes on the top white layer and the stromal vascular fraction containing preadipocytes in the pellet were resuspended in 10% fetal bovine serum-Dulbecco's modified Eagle's medium (DMEM) and counted using a Beckman Coulter Z1.
Cell cultures.
The 3T3-L1 cells (ATCC CL-173) were maintained in DMEM with 4.5 g/liter glucose, 10% calf serum, and penicillin-streptomycin (Gibco) in a humidified incubator at 5% CO2. Cells were subcultured at a split ratio of 1:4. Adipocyte differentiation was induced as described previously (38, 44), with modifications. After cells reached confluence, they were incubated with DMEM containing 10% resin-stripped calf serum with or without 2 nM T3 for 36 h, at which time (day 0) the cells were treated with 1 μM dexamethasone (Sigma, MO) and 0.5 mM 3-isobutyl-1-methyl xanthine (IBMX) (Sigma) in the presence or absence of 2 nM T3 for 60 h. Thereafter, the cells were fed every other day with DMEM containing 10% resin-stripped fetal bovine serum with or without 2 nM T3 until being stained by Oil Red O or harvested for Western blot analysis at day 9.
For Oil Red O staining of cells, 35-mm dishes or six-well plates were washed once in phosphate-buffered saline, and cells were fixed in 10% formalin solution (Sigma) for 50 min, followed by staining with Oil Red O for 1 h. Oil Red O was prepared by diluting a stock solution (0.12 g of Oil Red O [Sigma] in 24 ml of isopropanol) with water (6:4), followed by filtration. After staining, dishes or plates were washed three times in water and scanned by the Astra 6450 scanner (UMAX Technologies, Dallas, TX) or photographed with an Eclipse TE2000 (Nikon Instruments, Inc., NY) inverted microscope system at ×100 magnification or an Eclipse TS100 with a Coolpix (Nikon) digital camera at ×40 magnification.
To determine the effect of TRα1PV on the adipogenesis of 3T3-L1 cells, cells were transfected with 4 or 6 μg of the expression vector for TRα1PV (FLAG-TRα1PV) or pFLAG-CMV2, as a control, using the Nucleofector II device with cell line Nucleofector solution V according to the manufacturer's protocol (Amaxa, Inc.). After transfection, cells were plated into 35-mm dishes or six-well plates and cultured for 24 h and induced to differentiate by the protocol described above. Eleven days after transfection, cells were stained with Oil Red O. Cells were also lysed for Western blot analysis by using anti-Flag and C4 antibodies to detect Flag-TRα1PV and TRα1, respectively.
For the generation of the cell lines stably expressing Flag-TRα1PV, human TRα1PV cDNA was cloned into pFH-IRESneo (a generous gift of Robert G. Roeder, Rockefeller University, New York, NY) to obtain pFH-TRα1PV as described previously (28, 46). 3T3-L1 cells were transfected with pFH-TRα1PV or with pFH-IRESneo as a control by using Lipofectamine 2000 (Invitrogen) as a transfection reagent according to the manufacturer's protocol and selected with 350 μg/ml of G418 (Gibco) as previously described (28, 46). Pooled G418-resistant clones were expanded in selection medium. The expression of the stably transfected gene was confirmed by immunoblotting with anti-FLAG (Sigma) and TRα1PV antibodies (C3 or 302).
Quantitative real-time RT-PCR.
RNA from fat tissues was extracted using an RNeasy lipid tissue mini kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions. The determination of mRNA by real-time reverse transcription-PCR (RT-PCR) was carried out as described previously by Ying et al. (47) by using total fat RNA (100 ng) and the following primers and sequences: mTRα1 forward (nucleotides [nt] 1286 to 1304), CAGCTCAAGAATGGTGGCT, and reverse (nt 1660 to 1639), GACTTCCTGATCCTCAAAGACC; mTRβ1 forward (nt 889 to 908), ACAGCAAGAGGCTAGCCAAG, and reverse (nt 1154 to 1135), ACTGAAGGCTTCCAGGTCAA; mAdipsin forward (nt 273 to 292), TCCGCCCCTGAACCCTACAA, and reverse (nt 591 to 572), TAATGGTGACTACCCCGTCA; maP2 forward (nt 219 to 241), CTGGACTTCAGAGGCTCATAGCA, and reverse (nt 106 to 82), TACTCTCTGACCGGATGGTGACCAA; mACC forward (nt 6036 to 6056), GAGACGCTGGTTTGTAGAAGT, and reverse (nt 6285 to 6267), TCGCTGGGTGGGTGAGATG; mFAS forward (nt 65 to 84), CGGTATGTCGGGGAAGTTGC, and reverse (nt 346 to 326), CGGAGTGAGGCTGGGTTGATA; mG6PD forward (nt 642 to 662), GAGGAGTTCTTTGCCCGTAAT, and reverse (nt 968 to 948) CATCTCTTTGCCCAGGTAGTG; and m18S forward, ACCGCAGCTAGGAATAATGGA, and reverse CAAATGCTTTCGCTCTGGTC.
The primer sequences for lipoprotein lipase (LpL), PPARγ, and the control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) have been described previously by Ying et al. (47).
Western blot analysis.
The liver and fat tissues were dissected, cut into small pieces, and homogenized with Dounce homogenizer in 0.32 M sucrose, 2 mM EDTA, and 5 mM 2-mercaptoethanol in 10 mM Tris buffer (pH 7.4). This homogenate was centrifuged at 10,000 × g for 10 min. The top fat layer of white fat homogenate was discarded, and the pellets were resuspended for the preparation of nuclear or cytosolic extract using a NE-PER kit (Pierce; catalog no. 78833) according to the manufacturer's protocols. The protein concentration was determined by the Bradford method (Pierce Chemical Co., Rockford, IL) with bovine serum albumin (Pierce Chemical Co.) as the standard. Western blot analysis was carried out as described previously (16). For the detection of PPARγ, nuclear fractions (25 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The primary antibody used in the Western blot analysis was monoclonal anti-PPARγ antibody (1:100 dilution; catalog no. sc-7273; Santa Cruz, Inc.). For the control of protein loading of nuclear extracts, poly(ADP-ribose) polymerase (PARP) was used (anti-PARP antibody [1:100 dilution; catalog no. sc-7150; Santa Cruz, Inc.]).
Transient transfection.
Transient transfection experiments were carried out in CV1 cells as described previously by Araki et al. (2). Cells (1.5 × 105 cells/well) in six-well plates were transfected with pPPRE-TK-Luc (0.5 μg) and PPARγ1 expression vector (1 μg of pSG5/stop-mPPARγ) in the absence of TRα1 or PV expression plasmids (pcDNA3.1-TRα1 and pcDNA3.1-TRα1PV, respectively). Twenty-four hours after transfection, 20 μM troglitazone (TZD) or 100 nM T3 was added and incubated for an additional 24 h before harvesting of the cells to determine the luciferase activity. All experiments were performed in triplicate and repeated three times. The results shown are the means ± standard errors of the means (SEM).
Electrophoretic motility shift assay (EMSA).
The double-stranded oligonucleotide containing the peroxisome proliferator response element (PPRE) (PPRE-5′, GAACGTGACCTTTGTCCTGGTCCCCTTTGCT, and PPRE-3′, GGGACCAGGACAAAGGTCACGTTCGGGAAAGG; underlined is the PPRE for acyl coenzyme A (acyl-CoA) oxidase, a target gene for PPARγ [19]) was labeled with [32P]dCTP as described previously by Araki et al. (2). About 0.2 ng of probe (3 × 104 to 5 × 104 cpm) was incubated with in vitro-translated PPARγ1, TRα1, or TRα1PV with or without RXRβ (2 μl) in the binding buffer. DNA-bound proteins were detected by autoradiography.
Preparation of nuclei and ChIP.
Nuclei from thyroid tissues were isolated as described above for the Western blot analysis. Nuclei pellets were suspended in 0.74 ml of buffer, and the cross-link and subsequent steps were carried out as previously described (1). The chromatin immunoprecipitation (ChIP) assay was performed using a ChIP assay kit (Upstate, Inc.) according to the manufacturer's instructions. Chromatin solution (1 ml) was immunoprecipitated with 5 μl of anti-TRb1 antibody C4 (4) or anti-PV (T1 [49] or 302 [4]), anti-PPARγ (1 μg; Santa Cruz; catalog no. sc-7196), or anti-NCoR antiserum (5 μl; a generous gift of J. Wong, Baylor College of Medicine); immunoglobulin G (IgG) (2 μg) and MOPC (2 μg) were used as negative controls. The recovered DNA was used as a template for amplification using real-time PCR. Two percent of the chromatin solution (20 μl) was used for the control of input DNA. The primer sequences for the lipoprotein lipase PPRE were 5′-CCTCCCGGTAGGCAAACTG-3′ (forward) and 5′-AACGGTGCCAGCGAGAAG-3′ (reverse).
The amplified DNA was analyzed on a 2% agarose gel with ethidium bromide staining.
Statistical analysis.
All data are expressed as means ± SEM. Statistical analysis was performed with the use of analysis of variance, and a P value of <0.05 was considered significant.
RESULTS
Reduced white adipose tissue mass in TRα1PV/+ mice.
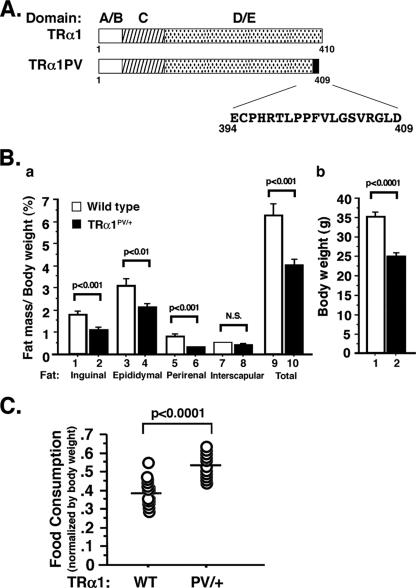
TRα1PV/+ mice were created by targeted mutation of TRα1PV into the TRα gene locus (22). The mutated TRα1PV sequence is shown in Fig. 1A. The severe reduced body weight exhibited by TRα1PV/+ mice prompted us to ascertain whether a reduction in fat tissues is one of the impairments contributing to dwarfism. Compared with that in the wild-type siblings, among TRα1PV/+ mice there was a reduction in the mass of inguinal, epididymal, and perirenal fat tissues (WAT). Figure 1B, panel a, shows the percentage ratios of fat mass versus body weight. Figure 1B, panel b, shows that compared with wild-type siblings, TRα1PV/+ mice had a significant reduction in body weight consistent with the previous observation (22). As shown in Fig. 1B, panel a, compared with those of the wild-type siblings, significant 38, 32, and 60% reductions in ratios of fat mass per body weight for inguinal (bar 1, versus bar 2), epididymal (bars 3 versus 4), and perirenal fat (bars 5 versus 6), respectively, were observed in male mice at ages 3 to 6 months (n = 15 to 16; P < 0.01). However, no significant differences in interscapular fat (BAT) mass between TRα1PV/+ mice and wild-type siblings (Fig. 1B, panel a, bar 7 versus bar 8) were observed. Thus, the decrease in WAT mass accounts for the 40% reduction in the total fat mass of TRα1PV/+ mice (Fig. 1B, panel a, bar 9 versus bar 10). The reduction in WAT mass persisted up to the age of more than 1 year. A similar reduction in WAT mass was also observed for female mice (data not shown).
FIG. 1.
(A) Schematic representation of the structure of wild-type TRα1 and TRα1PV. The C-terminal, mutated sequence of TRα1PV is indicated. (B) Reduced WAT mass in TRα1PV/+ mice. Inguinal, epididymal, perirenal, and interscapular fat tissues were weighed after dissection from mice at ages of 3 to 6 months. Total fat represents the sum of these four isolated fat tissues. Ratios of fat mass versus body weight were determined and are shown in panel a. Mouse body weights are shown in panel b. The data are expressed as means ± SEM (error bars) (n = 15 to 16). (C) Increased food consumption in TRα1PV/+ mice. Food consumed by mice in 2 days was measured, and the food intake was normalized to the body weight of each mouse and expressed as g/g body weight−0.75 (24, 50). Each circle represents an individual mouse, and the horizontal bars represent the mean values. The differences are statistically significant (P < 0.0001). WT, wild type; N.S., no significant difference.
The decrease in WAT was not due to a reduction in food uptake by TRα1PV/+ mice. On the contrary, TRα1PV/+ mice consumed significantly more food (∼34%) than did their wild-type siblings (P < 0.001; n = 15 to 16) (Fig. 1C). Table 1 shows the changes in lipid-related serum hormones and factors accompanied by reductions in adipose tissues in TRα1PV/+ mice compared to those in their wild-type siblings. Among males, there was a significant reduction in the serum levels of free fatty acids (36% [P < 0.01]), total triglycerides (31% [P < 0.05]), and leptin levels (70% [P < 0.0001]) in TRα1PV/+ mice but a significant increase in serum levels of adiponectin (22% [P < 0.05]). In contrast, no significant changes in glucose or insulin were detected in TRα1PV/+ mice relative to those in their wild-type siblings (Table 1). No significant differences in glucose tolerance and insulin tolerance tests were observed between TRα1PV/+ and wild-type mice (data not shown). Similar serum hormonal changes were observed in female TRα1PV/+ mice except that a smaller reduction in leptin was detected in female mice than in male mice (36% [P < 0.05]) (Table 1). The reduction in serum leptin levels is consistent with an increase in food consumption by TRα1PV/+ mice (Fig. 1C). The patterns in changes of serum adipokine levels in TRα1PV/+ mice (i.e., a decrease in leptin and an increase in adiponectin levels) are consistent with the reduced adipose mass (17, 25).
TABLE 1.
Serum hormone levels in wild-type and TRα1PV/+ micea
| Mouse characteristic or ratio | Age (mo) | BW (g) | Serum hormone levels
|
|||||
|---|---|---|---|---|---|---|---|---|
| Glucose (mg/dl) | FFA (mM) | TG (mg/dl) | Insulin (ng/ml) | Adiponectin (μg/ml) | Leptin (ng/ml) | |||
| Male | ||||||||
| Wild type (n = 22) | 4.30 ± 0.22 | 34.0 ± 0.87 | 121.5 ± 6.64 | 0.36 ± 0.04 | 99.0 ± 11.2 | 0.41 ± 0.06 | 10.53 ± 0.44 | 7.01 ± 0.88 |
| TRα1PV/+ (n = 23) | 3.82 ± 0.19 | 23.73 ± 0.51 | 129.9 ± 8.48 | 0.23 ± 0.02 | 68.5 ± 5.36 | 0.29 ± 0.04 | 12.84 ± 0.96 | 2.09 ± 0.43 |
| W/M ratio | 1.43**** | 0.94 (NS) | 1.57** | 1.45* | 1.41 (NS) | 0.82* | 3.4**** | |
| Female | ||||||||
| Wild type (n = 9) | 2.7 ± 0.14 | 24.41 ± 0.45 | 125.40 ± 8.23 | 0.54 ± 0.06 | 16.10 ± 10.96 | 0.68 ± 0.12 | 15.68 ± 2.49 | 7.01 ± 1.3 |
| TRα1PV/+ (n = 15) | 3.3 ± 0.22 | 19.40 ± 0.42 | 130.50 ± 7.64 | 0.34 ± 0.03 | 13.36 ± 7.18 | 0.83 ± 0.14 | 21.53 ± 1.53 | 4.47 ± 0.35 |
| W/M ratio | 1.26*** | 0.96 (NS) | 1.56** | 1.21* | 0.82 (NS) | 0.73* | 1.57* | |
*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant; W, wild type; M, TRα1PV/+ mutant; BW, body weight; FFA, free fatty acid; TG, total triglycerides. Data are means ± standard errors.
Differential expression of TR isoforms in mature adipocytes and preadipocytes of WAT.
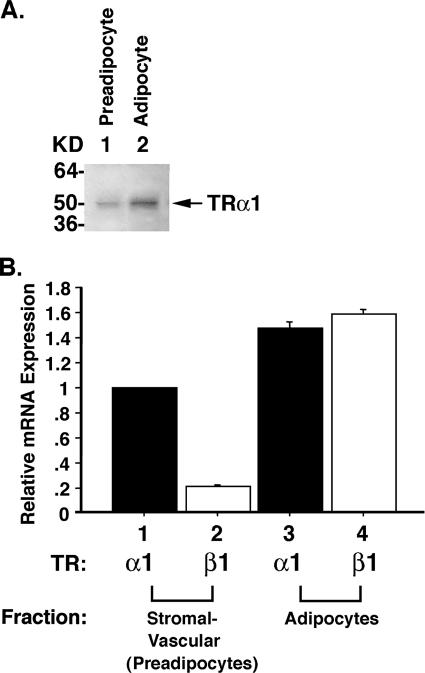
That WAT but not BAT exhibits fat mass reduction prompted us to focus our efforts on understanding the molecular basis underlying the reduced WAT masses of TRα1PV/+ mice. We hypothesized that TRα1PV could interfere with the critical functions of wild-type TRα1 during the differentiation of preadipocytes to adipocytes. Indeed, recent findings indicate that during the adipogenesis of 3T3-L1 cells, TRα1 mRNA is constitutively expressed in preadipocytes (15). Its expression continues to increase during adipogenesis, concurrent with the appearance of lipid droplets (15, 21). In contrast, very little, if any, TRβ1 mRNA is detectable in either preadipocytes or adipocytes (15, 21). These findings suggest a critical role of TRα1 during adipogenesis of 3T3-L1 cells (21). Because discordance in the expression of TR isoforms at the mRNA and protein levels has been reported (37), we evaluated the protein abundance of TR isoforms in 3T3-L1 preadipocytes and mature adipocytes. Consistent with the mRNA expression, Fig. 2A shows that, indeed, TRα1 protein was detected in preadipocytes (Fig. 2A, lane 1), and its abundance was increased in mature adipocytes (Fig. 2A, lane 2). However, no TRβ1 proteins were detectable in either preadipocytes or adipocytes.
FIG. 2.
Expression of TRs in 3T3-L1 cells (A) and WAT of wild-type mice (B). (A) Cellular extracts were prepared from 3T3-L1 preadipocytes (lane 1) and mature adipoctyes (lane 2), and Western blot analysis was carried out using monoclonal anti-TR antibody C4 that recognized TRα1 and TRβ1 as described in Materials and Methods. Only TRα1 was detected. (B) WAT of wild-type mice was fractionated into mature adipocytes and stromal vascular fraction enriched with preadipocytes as described in Materials and Methods. A QuantiTect SYBR green RT-PCR kit (QIAGEN) and LightCycler software were used for the absolute quantification of TRα1 and TRβ1 mRNA levels in mature adipocytes and stromal vascular fraction, respectively. mRNA abundance of TRα1 and TRβ1 was normalized by 18S and shown as mRNA expression relative to the expression of TRα1 in the stromal vascular fraction, defined as 1. Error bars indicate standard errors.
The differential expression of TR isoforms in 3T3-L1 preadipocytes and adipocytes prompted us to examine whether the expression of TR isoforms is similarly developmentally regulated in the WAT of wild-type mice. We therefore fractionated the WAT of adult wild-type mice into mature adipocytes and the stromal vascular fraction enriched with preadipocytes and determined the expression of TRα1 and TRβ1 in these two fractions. Figure 2B shows that TRα1 was the major TR isoform expressed in preadipocytes (bar 1 versus bar 2), whereas TRβ1 was more abundantly expressed in mature adipocytes than in preadipocytes (bar 2 versus bar 4). The differential TR isoform expression patterns are consistent with those reported by Fu et al. (15). Taken together, these differential expression patterns of TR isoforms suggest that TRα1 could play a critical role in the adipogenesis of WAT.
TRα1PV-mediated impairment in adipogenesis via repression of PPARγ functions.
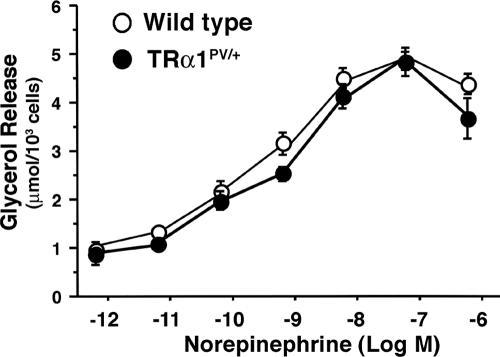
The reduction in WAT prompted us to determine first whether it was due to an increase in catecholamine-induced lipolysis. We therefore compared catecholamine-induced lipolysis levels in the WAT of wild-type and TRα1PV/+ mice. Figure 3 shows that the dose-dependent, norepinephrine-induced lipolysis levels measured by glycerol release did not differ significantly between the white fat cells of wild-type and TRα1PV/+ mice. Similar results were found when the release of glycerol was induced by forskolin and dibutyl cAMP (data not shown), indicating that the reduced fat accumulation in the adipocytes of TRα1PV/+ mice was not due to an increase of catecholamine-induced lipolysis.
FIG. 3.
Comparison of in vitro norepinephrine-mediated lipolysis in isolated white fat cells. White fat cells were isolated from the epididymal pad, and the amount of glycerol released in the presence of increasing concentrations of norepinephrine was determined as described in Materials and Methods. Closed circles and open circles represent data from TRα1PV/+ mice and wild-type mice, respectively. Data are expressed as means ± SEM (error bars) (n = 9).
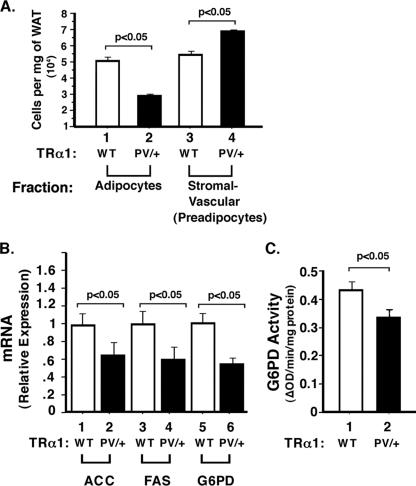
The above results suggest that impaired adipogenesis could underlie the reduced fat mass. This would predict that TRα1PV/+ mice have reduced mature adipocytes. We therefore fractioned WAT and separated mature adipocytes from the stromal vascular fraction enriched with preadipocytes. Figure 4A shows that in TRα1PV/+ mice, the number of mature adipocytes was reduced by ∼50% (compare bars 1 and 2), and the number of preadipocytes was increased by ∼20% (compare bars 3 and 4). These results show that there were fewer mature adipocytes and more preadipocytes in TRα1PV/+ mice, indicative of impaired adipogenesis in TRα1PV/+ mice.
FIG. 4.
Impaired adipogenesis in WAT of TRα1PV/+ mice. (A) There was a reduced number of mature adipocytes in TRα1PV/+ mice compared with that in wild-type (WT) mice. Mature adipocytes were separated from the stromal vascular fraction enriched with preadipocytes as described in Materials and Methods. (B) Reduced expression of key lipogenic enzymes in TRα1PV/+ mice. Total RNAs were prepared from mice aged 4 to 5 months, and quantitative real-time RT-PCR was performed using 0.1 μg of total RNA as described in Materials and Methods. Relative changes (n-fold) of the expression of mRNA are shown. The differences are significant (P < 0.05). The activity of G6PD was reduced in TRα1PV/+ mice compared with that in wild-type mice (C). The enzyme activity was determined as described in Materials and Methods. The differences are significant (P < 0.05). ΔOD, change in optical density. Error bars indicate standard errors.
We further tested whether the expression of key lipogenic enzymes was affected in WAT. Indeed, Fig. 4B shows that the expression of acetyl-CoA-carboxylase, fatty acid synthase, and glucose-6P-dehydrogenase (G6PD) was significantly reduced in WAT of TRα1PV/+ mice (compare bars 2, 4, and 6 with 1, 3, and 5, respectively). Furthermore, the G6PD enzymatic activity was also significantly reduced (Fig. 4C). The reduced expression of these key lipogenic enzymes further supports the notion that the reduced WAT mass of TRα1PV/+ mice is due to impairment in adipogenesis.
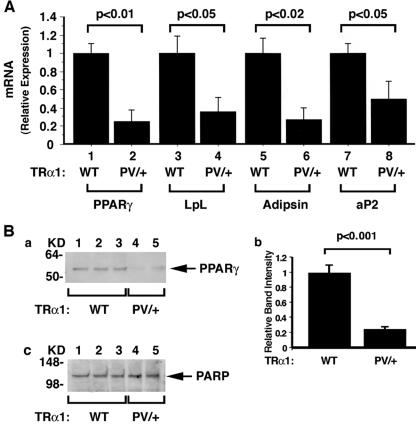
To understand how TRα1PV mediates impaired adipogenesis, we focused on the study of PPARγ, the key regulator of adipogenesis in adipocytes. Fig. 5A, bar 2, shows that the expression levels of PPARγ mRNA were reduced by 80% in the WAT of TRα1PV/+ mice compared with those in their wild-type siblings (bar 1). We further determined PPARγ protein levels by Western blot analysis. Figure 5B further shows that the PPARγ protein abundance was reduced in WAT of TRα1PV/+ mice compared with that in their wild-type siblings (panel a, lanes 4 to 5 versus lanes 1 to 3). Quantitative analysis showed an 80% reduction in TRα1PV/+ mice compared with that in their wild-type siblings (Fig. 5B, panel b). Figure 5B, panel c, shows the protein loading control using the nuclear marker PARP.
FIG. 5.
Comparison of the expression levels of PPARγ and its downstream target genes at the mRNA levels in TRα1PV/+ mice and wild-type (WT) siblings (A) and PPARγ protein abundance (B). (A) mRNA expression of PPARγ and its downstream target genes in the WAT of TRα1PV/+ mice and wild-type siblings. Total RNAs were prepared from mice aged 4 to 5 months, and quantitative real-time RT-PCR was performed using 0.1 μg of total RNA as described in Materials and Methods. Relative changes (n-fold) of the expression of mRNA are shown. The differences are significant (P < 0.05). (B) Panel a represents nuclear extracts (25 μg) isolated from WAT (inguinal fat) of TRα1PV/+ mice (n = 2) and wild-type siblings (n = 3). Western blot analysis was carried as described in Materials and Methods. The primary antibody was the monoclonal anti-PPARγ antibody (Santa Cruz; catalog no. sc-7273; 1:100 dilution). Panel b shows the quantification of band intensities from panel a, and relative differences were plotted. The differences are significant (P < 0.001). For panel c, PARP was used for the control of protein loading (anti-PARP antibody [Santa Cruz; catalog no. sc-7150]; 1:100 dilution). Error bars indicate standard errors.
Consistent with the reduced expression of PPARγ, the expression levels of LpL, adipsin, and aP2, for the PPARγ downstream target genes involved in adipogenesis, were all significantly reduced (Fig. 5A, compare bars 4, 6, and 8 with bars 3, 5, and 7, respectively). These results suggest that mutations of TRα1 led to the reduced expression of PPARγ and the attenuation of its signaling to reduce the expression of several downstream target genes to impair adipogenesis in WAT.
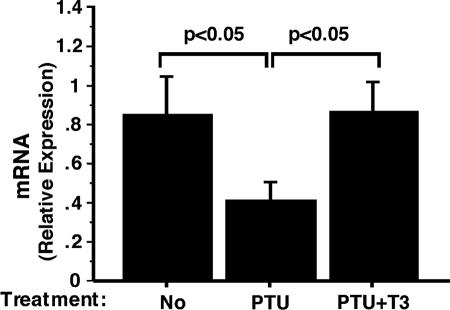
The above findings suggest that the PPARγ gene could be a T3-regulated gene. To test this possibility, we rendered wild-type mice into a hypothyroid state by treating them with PTU and subsequently treated these mice with T3. The PTU-induced hypothyroid state and the subsequent T3-induced hyperthyroid state were confirmed by thyroid function tests (Table 2). As expected, hypothyroid mice induced by PTU had significantly reduced total T4 and highly elevated TSH levels (Table 2) and, upon injection of T3, total T3 was elevated 15-fold accompanied by a lowering of TSH (Table 2). We then compared the expression levels of the PPARγ gene in PTU-induced hypothyroid mice and T3-treated hyperthyroid mice. As shown by quantitative RT-PCR analysis, PPARγ mRNA was reduced by 50% in hypothyroid mice but T3 treatment restored its expression to a level similar to that in the euthyroid state (Fig. 6). We also isolated primary adipocytes and showed that T3 activated the expression of PPARγ mRNA (data not shown). These data indicate that the expression of the PPARγ gene is regulated by T3. These findings are consistent with the repression of the PPARγ gene expression detected in WAT in TRα1PV/+ mice in which TRα1 is mutated (Fig. 5).
TABLE 2.
Thyroid function test results of PTU-treated wild-type mice
| Treatment | Total T4 (μg/dl) | Total T3 (ng/ml) | TSH (ng/ml) |
|---|---|---|---|
| Control | 2.91 ± 0.29 | 1.06 ± 0.09 | 32.7 ± 6.9 |
| PTU | 1.07 ± 0.04a | 0.37 ± 0.01a,b | 63,669 ± 8501a,b |
| PTU + T3 | 0.87 ± 0.06a | 5.65 ± 0.43a,b | 61.0 ± 9.2a,b |
Compared with that for the control, P < 0.001 (mean ± SE; n=4 to 6).
The effect of T3 compared with only PTU; P was <0.001 (mean ± SE; n = 4 to 6).
FIG. 6.
Effect of T3 on the mRNA expression of PPARγ in WAT of wild-type mice. Hypothyroid mice were induced by PTU treatment, and hyperthyroid mice were induced from injection of T3 as described in Materials and Methods. mRNA expression was determined by real-time RT-PCR. The data are expressed as means ± SEM (error bars) (n = 4 to 5).
Repression of the PPARγ transcription activity by TRα1PV.
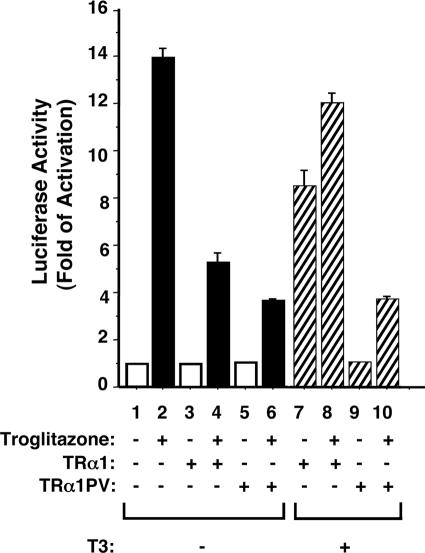
In addition to the repression of the PPARγ gene at both the mRNA and protein levels, we postulated that TRα1PV could also act to interfere with the transcription activity of PPARγ. We therefore evaluated the effect of TRα1PV on TZD-dependent, PPARγ-mediated transcription activity (Fig. 7). Compared with bar 1, Fig. 7, bar 2, shows the PPARγ-mediated, TZD-dependent activation of transcription (14-fold). In the absence of T3, the unliganded TRα1 (Fig. 7, bar 4) as well as TRα1PV (bar 6) repressed the TZD-dependent, PPARγ-mediated transcription activity (compare bars 4 and 6 with bar 2). In the presence of T3, the repression effect on the TZD-dependent, PPARγ-mediated transcription activity was derepressed by the liganded TRα1 (Fig. 7, bar 8 versus bar 4). However, no such derepression was observed for TRα1PV (Fig. 7, bar 10 versus bar 6). Thus, the reduced activity of PPARγ induced by TRα1PV was mediated two ways: by the repression of expression and by the inhibition of the transcription activity. These data suggest that the impairment in adipogenesis in WAT of TRα1PV/+ mice was due, at least in part, to the TRα1-mediated, reduced activity of PPARγ.
FIG. 7.
TRα1PV represses the ligand-dependent transactivation of PPARγ in CV-1 cells. CV-1 cells were cotransfected with 0.5 μg of the reporter plasmid (pPPRE-TK-Luc), 0.1 μg of PPARγ expression vector (pSG5-mPPARγ 1), and 0.1 μg of TRα1 or TRα1PV expression vector (pcDNA3.1-TRα1 or pcDNA3.1-TRα1PV, respectively). Cells were treated with either dimethyl sulfoxide as vehicle or troglitazone (20 μM) in the absence (−) or presence (+) of T3 (100 nM), as marked. Data were normalized against the protein concentration in the lysates. Relative luciferase activity was calculated and shown as induction relative to the luciferase activity of PPRE in the cells treated with dimethyl sulfoxide in the absence of T3, defined as 1. The data are expressed as means ± SEM (error bars) (n = 3).
TRα1PV competes with PPARγ for binding to PPRE.
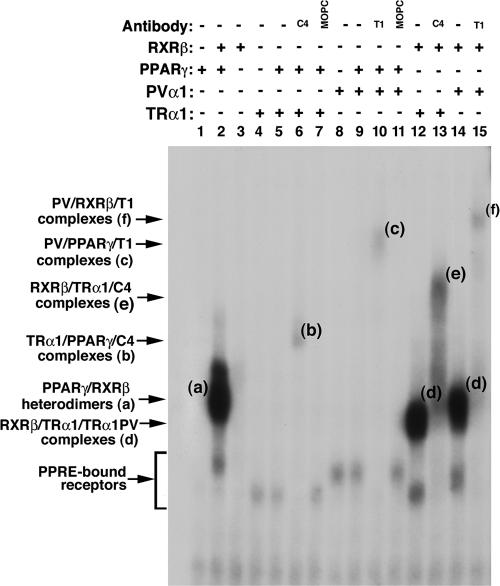
To understand how TRα1PV inhibited TZD-dependent PPARγ transcription activity, we considered the possibility that the repression could be due to the competition of TRα1PV with PPARγ for binding to PPRE. We therefore evaluated the binding of TRα1PV to PPRE by EMSA (Fig. 8). Fig. 8, lane 2, shows that PPARγ binds to PPRE strongly as a heterodimer with RXRβ (band a), but binding to PPRE as a homodimer was not detectable under the experimental conditions (lane 1). Although no binding of RXR homodimers to PPRE was observed (Fig. 8, lane 3), the binding of TRα1 to PPRE as a homodimer (lane 4) or as a heterodimer with PPARγ was detected (lane 5). The latter was confirmed by using supershift experiments in which anti-TRα1 antibody C4 (Fig. 8, lane 6) specifically shifted the PPRE-bound PPARγ/TRα1 heterodimers to a more retarded position by EMSA (band b), but not by an irrelevant antibody (MOPC) (Fig. 8, lane 7). TRα1PV also bound to PPRE as a homodimer (Fig. 8, lane 8) or as a heterodimer with PPARγ (lane 9). The latter was confirmed by supershifting the PPRE-bound PPARγ/PV to a more retarded position with anti-PV antibody T1 (Fig. 8, lane 10, band c). However, an irrelevant antibody failed to do so (Fig. 8, lane 11), indicating the specific interaction of PPRE-bound TRα1PV with anti-PV antibody T1.
FIG. 8.
Binding of PPARα, TRα1, or TRα1PV to PPRE by EMSA. Lysate containing in vitro-translated PPARγ, TRα1, or TRα1PV proteins (5 μl) in the presence (+) or absence (−) of RXRβ or anti-TRβ1 antibody (C4; 2 μg), anti-PV antibody (T1; 2 μg), or an irrelevant antibody, MOPC (2 μg); antibodies were incubated with 32P-labeled PPRE and analyzed by gel retardation as described in Materials and Methods. Amounts of lysate were kept constant by the addition of unprogrammed lysate as needed.
TRα1 or TRα1PV also bound to PPRE as a heterodimer with RXRβ (band d in Fig. 8, lanes 12 and 14, respectively). This binding was confirmed by supershifting the PPRE-bound TRα1/RXRβ or TRα1PV/RXRβ with anti-TRα1 antibody C4 or anti-PV antibody T1 to a more retarded position (Fig. 8, lane 13, band e, and lane 15, band f, respectively). These results indicate that TRα1 and the mutant TRα1PV can bind to PPRE as homodimers and as heterodimers with PPARγ or RXRβ.
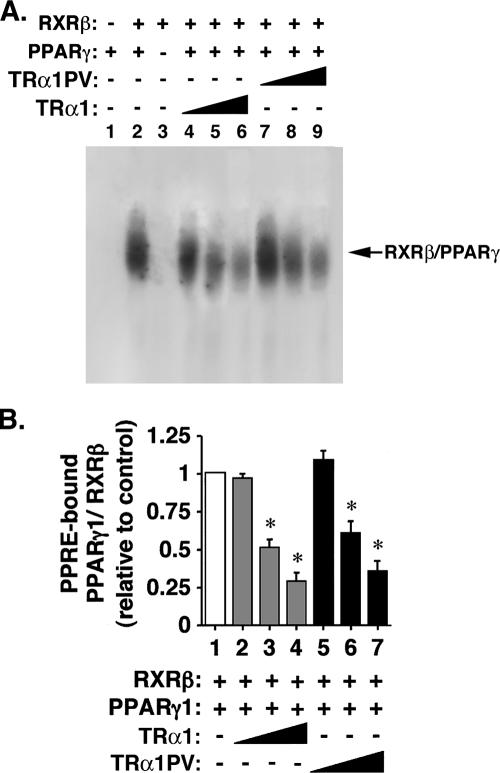
Since both PPARγ and TRs heterodimerize with RXR on their cognate hormone response elements, the finding that both TRα1 and TRα1PV bound to PPRE as heterodimers with RXRβ or PPARγ suggests that TRα1 or TRα1PV could compete with PPARγ for binding to PPRE as a heterodimer with RXR or PPARγ. Figure 9 shows that, indeed, compared with PPRE-bound PPARγ/RXR in the absence of TRα1 (Fig. 9A, lane 2), binding of PPARγ/RXRβ to PPRE was decreased in the presence of increasing concentrations of TRα1 (Fig. 9A, lanes 4 to 6) or TRα1PV (Fig. 9A, lanes 7 to 9). This concentration-dependent decrease in the binding by TRα1 or TRα1PV can be seen more readily in Fig. 9B, in which the band intensities in Fig. 9A are quantified and graphed. Taken together, these findings indicate that TRα1PV could act to repress the transcription activity of PPARγ by competition with PPARγ for PPRE and for heterodimerization with RXR. Thus, the TRα1PV-mediated repression of the PPARγ transcription leads to the repression of expression of downstream target genes, thereby contributing to impairment in the adipogenesis of WAT.
FIG. 9.
Inhibition of the PPARγ/RXR heterodimer binding to PPRE by increasing amounts of TRα1 or TRα1PV. (A) Lysates containing PPARγ and RXRβ proteins (1 μl) were incubated in the absence (+) or presence (−) of TRα1 (1, 5, and 10 μl for lanes 4, 5, and 6, respectively) or TRα1PV (1, 5, and 10 μl for lanes 7, 8, and 9, respectively) with 32P-labeled PPRE and analyzed by EMSA as described in Materials and Methods. (B) Band intensities of lanes 2 and 4 through 9 from panel A were quantified by using UMAX Astra 6450 (UMAX Technologies, Inc., Dallas, TX), and the data were analyzed by using NIH Image 1.61. The data are expressed as means ± SEM (error bars) (n = 3). The asterisks indicate that the differences are significant (P < 0.01).
Recruitment of TRα1PV to the promoter of a PPARγ target gene, the lipoprotein lipase, in WAT of TRα1PV/+ mice.
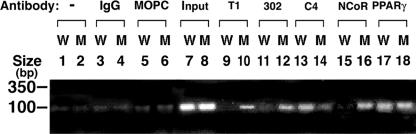
To further support the notion that the constitutive association of TRα1PV with corepressors, such as nuclear receptor corepressor (NCoR), could lead to the repression of PPARγ transcription activity, we carried out ChIP assays using WAT nuclear extracts of TRα1PV/+ mice to determine whether TRα1PV and NCoR were recruited to the promoter of the LpL gene in vivo. The LpL gene is a direct target of PPARγ that contains a PPRE in its promoter between positions −169 and −157 (36). Its mRNA expression is repressed in the WAT of TRα1PV/+ mice (Fig. 5A). Figure 10 shows the recruitment of TRα1PV, NCoR, and PPARγ to the LpL promoter by ChIP assays. TRα1PV was clearly recruited to the LpL promoter since an 81-bp PCR product was detected when DNA-protein complexes were immunoprecipitated by either polyclonal anti-PV-specific antibody T1 (Fig. 10, lane 10) or the monoclonal antibody 302 (Fig. 10, lane 12). As expected, no positive signals were detected in wild-type mice (Fig. 10, lanes 9 and 11). A clear signal for wild-type TRs was detected in wild-type mice (Fig. 10, lane 13) when anti-TR antibody (C4) was used in the assay. As shown in Fig. 10, lane 14, a weak signal was also apparent in TRα1PV/+ mice. That NCoR was also recruited to the LpL promoter was demonstrated with TRα1PV/+ mice (by the positive signal shown in Fig. 10, lane 16) but not with wild-type mice (lane 15). When anti-PPARγ antibody was used to immunoprecipitate the DNA-protein complexes, clear signals were detected to indicate the recruitment of PPARγ to the LpL promoter in TRα1PV/+ mice (Fig. 10, lane 18) as well as in wild-type mice (lane 17). No antibody (Fig. 10, lanes 1 and 2), rabbit IgG (lanes 3 and 4), and MOPC (lanes 5 and 6), an irrelevant mouse monoclonal antibody, were the negative controls for immunoprecipitation, and Fig. 10, lanes 7 and 8, shows the input.
FIG. 10.
Recruitment of NCoR to PPRE-bound PPARγ and TRα1PV in the promoter of the lipoprotein lipase gene in WAT of TRα1PV/+ mice by ChIP assay. Nuclear extracts from wild-type (W) or TRα1PV/+ (M) mice were processed for the ChIP assay as described in Materials and Methods. Anti-PPARγ, anti-NCoR, anti-PV (polyclonal antibody T1 or monoclonal antibody 302) (4, 49) antibodies and IgG (for negative controls) were used for immunoprecipitation. The precipitated DNA was amplified by PCR with primers specific for PPRE in the lipoprotein lipase promoter, and the products were analyzed. −, no antibody.
Inhibition of 3T3-L1 adipogenesis by TRα1PV.
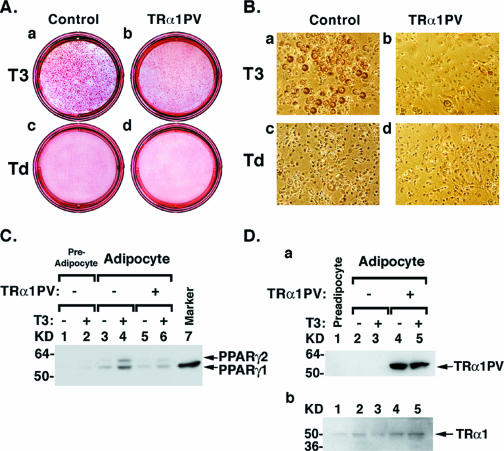
The studies of TRα1PV/+ mice support the notion that TRα1PV impairs the adipogenesis of WAT by the repression of PPARγ expression and also by the inhibition of PPARγ transcription activity. To demonstrate directly the inhibitory effect of adipogenesis by TRα1PV, we adopted the approach of using a well-characterized 3T3-L1 adipogenesis model (15). Consistent with findings by Jiang et al. (21), we found that T3 increased the adipogenesis of 3T3-L1 cells as indicated by an increased number of adipocytes with lipid droplets (Fig. 11A and B, compare panels a and c). This T3-induced adipogenesis was concurrently accompanied by an increased abundance of PPARγ1 and PPARγ2 proteins in adipocytes (Fig. 11C, compare lanes 3 and 4), while no PPARγ1 and PPARγ2 proteins were visible in preadipocytes (Fig. 11C, lanes 1 and 2). The expression of the transfected TRα1PV (Fig. 11D, panel a, lanes 4 and 5) significantly blocked the T3-induced adipogenesis as evidenced by the reduction of mature adipocytes (Fig. 11A and B, compare panels a and b). The reduction of lipid droplet accumulation in TRα1PV-expressed cells was accompanied by a concurrent reduction of T3-activated expression of the PPARγ1 and PPARγ2 proteins (Fig. 11C, compare lanes 4 and 6). Figure 11D, panel b, shows that TRα1 protein was detected in the preadipocytes (Fig. 11D, panel b, lane 1) as well as in the adipocytes (lanes 2 to 5). Taken together, these cell-based findings further support the notion that TRα1PV interferes with the functions of TRα1 in adipogenesis via the repression of expression and the transcription activity of PPARγ.
FIG. 11.
TRα1PV impairs T3-dependent 3T3-L1 adipogenesis. 3T3-L1 cells were transfected with 6 μg of the expression vector for TRα1PV (FLAG-TRα1PV) or pFLAG-CMV2 as a control, and the transfected cells were induced to differentiate into mature adipocytes as indicated in Materials and Methods. Mature adipocytes with lipid droplets were visualized by staining with Oil Red-O (A) and shown by phase-contrast microscopy (B). Western blot analysis of PPARγ (C), TRα1PV/+ (D, panel a), and TRα1 (D, panel b) protein abundance was carried out as described in Materials and Methods. −, absence of; +, presence of.
DISCUSSION
In spite of the demonstration of the expression of TRs in WAT (20, 34), the role of TRs in WAT homeostasis is not well understood. The striking phenotype of reduced WAT mass exhibited by TRα1PV/+ mice provided us with a tool to understand how mutations of TRα1 affect WAT homeostasis in vivo. One of the mechanisms uncovered in the present study is that TRα1PV acted to interfere with the expression and activity of the master regulator of adipogenesis, PPARγ. We showed that PPARγ is a T3 positively regulated gene, and its expression at the mRNA and protein levels is significantly repressed in the WAT of TRα1PV/+ mice. Importantly, we also found that the transcription activity of PPARγ was repressed by TRα1PV. The dual repression effects of TRα1PV reduce the expression of several PPARγ downstream target genes involved in adipogenesis, resulting in reduced fat mass. In addition to these in vivo findings, we showed directly that the overexpression of TRα1PV blocked the T3-dependent adipogenesis of 3T3-L1 cells. At present, whether PPARγ is directly or indirectly regulated by T3 and how a mutated TRα1 represses the expression of PPARγ mRNA are unclear. However, we have demonstrated that the repression of the transcription activity of PPARγ was due to the competition of TRα1PV with PPARγ for binding to PPRE and for heterodimerization with RXR. The former results in the recruitment of a corepressor, NCoR, to TRα1PV-bound PPRE as shown in vivo by the ChIP assays, and the latter results in the reduced PPRE-bound PPARγ/RXR heterodimers, both of which contribute to the repression of PPARγ transcription activity.
However, intriguingly, no significant reduction in BAT mass was observed in TRα1PV/+ mice. This was not due to the lack of expression of TRs in BAT because, consistent with reports by others (20, 34), we found that both TR isoforms were expressed in BAT (our unpublished results). However, in spite of the reduced expression of PPARγ mRNA (∼50% compared with that of wild-type mice) detected in the BAT of TRα1PV/+ mice, the expression levels of the PPARγ downstream target genes (for LpL, adipsin, and aP2) were not affected (our unpublished results). These observations suggest that there are other compensatory pathways, independent of TRs and specific to BAT, to ameliorate the deleterious effects of TRα1PV/+ mice in the expression and activity of PPARγ and thereby maintain the lipid homeostasis in BAT. Alternatively, it is also possible that PPARγ has both unique and partially redundant functions in WAT and BAT and that some genes, once activated, do not depend on continued PPARγ expression. Such differential effects of PPARγ on gene expression in WAT and BAT are not without precedent. The differential gene expression profiles in WAT and BAT of TRα1PV/+ mice are reminiscent of those observed in BAT of mice with targeted deletions of PPARγ in adipocytes in that there are differences in gene expression profiles of BAT and WAT deficient in PPARγ (18). While the PPARγ downstream genes, such as aP2 and LpL, are repressed in the WAT of PPARγ knockout mice, there are no changes in the expression of these two genes in BAT (18).
In addition to TRα1PV mice, there are two other reported TRα1 knock-in mice harboring different mutations. Liu et al. reported a knock-in mouse with TRα1P398H mutation and (27), and Tinnikov et al. created a knock-in mouse with a TRα1R384C mutation (40). The availability of TRα1 knock-in mice harboring different mutations provides a valuable tool to address an important biological question of whether phenotypic expression of TRα1 knock-in mice is mutation site dependent. Indeed, while there are similar phenotypic expressions, such as embryonic lethality in homozygous mutation and very mild thyroid function disruption, increased mortality, and decreased fertility, shown in heterozygous mice (18, 22, 27), there are distinctive differences among these three TRα1 knock-in mice. TRα1PV mice are dwarfs and TRα1R384C mice exhibit delayed development in young mice, but the growth abnormalities are overcome in adult mice (40). However, TRα1P398H mice have normal sizes for the first 2 to 3 months and then the males display increased body weights (27). Although it is not known whether TRα1R384C mice have metabolic abnormalities, the present study shows that in contrast to TRα1P398H male mice that show age-dependent visceral adiposity, TRα1PV mice exhibit persistent reduced WAT mass in both males and females up to more than 1 year of age. The differences in the phenotypes exhibited by these three knock-in mice could reflect the different effects of the TRα1 mutants on different PPARγ isoforms expressed in tissue-dependent manners. Thus, TRα1 knock-in mice with different mutations share several common phenotypes but also exhibit distinct target tissue- and sex-dependent phenotypes.
The tissue-dependent distinct phenotypic expression among these three mutant mice harboring different mutations could reflect their differences in the degree of the loss of T3 binding and the potency of dominant negative activity of TRα1 mutants. TRα1PV mice completely lose T3 binding and exhibit potent dominant negative activity (22). In contrast, TRα1P398H and TRα1R384C mice only partially lose T3 binding activity (27, 40). The weaker dominant negative activity of TRα1R384C is consistent with the restoration of the target gene expression and the rescue of growth retardation in TRα1R384C mice when serum T4 concentration was increased 10-fold (40). These different degrees of dominant negative activity in these three TRα1 mutants could reflect the different responses to thyroid hormone in a tissue-dependent manner. Indeed, we compared the extent of the effect of troglitazone-dependent PPARγ-mediated transcription activity by TRα1P398H, TRα1R384C, and TRα1PV and found that the mutant-mediated repression correlated with the extent of the loss of T3 binding activity (H. Ying and S.-Y. Cheng, unpublished). In addition, even though all three of these mutant mice have very similar thyroid function profiles, it is known that regional differences of thyroid hormone can differ in a target tissue due to differential expression and activity of deiodinases. Alternatively, the tissue-dependent distinct phenotype could also be due to the different three-dimensional structures of these three TRα1 mutants. TRα1PV has a frameshift mutation in the C-terminal 16 amino acids and is also 1 amino acid shorter than wild-type TRα1 (22). This PV mutation is located in helix 12. TRα1P398H and TRα1R384C have only one mutated amino acid and are located in helices 12 and 11, respectively (41). It is conceivable that these three mutants have different structures such that their interactions with corepressors, coactivators, and heterodimeric partners could differ in tissue-dependent manners, resulting in different abnormal regulations of target genes and different phenotypic expressions. The validation of this possibility will await crystallographic analysis of these three TRα1 mutants.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
We thank J. Wong of Baylor College of Medicine for anti-NCoR antisera, F. Gonzalez of NCI for pPPRE-TK-Luc and pSG5/stop-mPPARγ plasmids, and O. Gavrilova of NIDDK for valuable discussion and expert assistance in some preliminary studies. We are also grateful to D. Berrigan and S. Hursting of NCI for assistance in determining the fat mass in the early phase of the study.
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Almouzni, G., S. Khochbin, S. S. Dimitrov, and A. P. Wolffe. 1994. Histone acetylation influences both gene expression and development of Xenopus laevis. Dev. Biol. 165:654-669. [DOI] [PubMed] [Google Scholar]
- 2.Araki, O., H. Ying, F. Furuya, X. Zhu, and S.-Y. Cheng. 2005. Thyroid hormone β receptor mutants: dominant negative regulators of peroxisome proliferator-activated receptor γ action. Proc. Natl. Acad. Sci. USA 102:16251-16626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassett, J. H., C. B. Harvey, and G. R. Williams. 2003. Mechanisms of thyroid hormone receptor-specific nuclear and extra nuclear actions. Mol. Cell. Endocrinol. 213:1-11. [DOI] [PubMed] [Google Scholar]
- 4.Bhat, M. K., P. McPhie, Y. T. Ting, X.-G. Zhu, and S.-Y. Cheng. 1995. Structure of the carboxy-terminal region of thyroid hormone nuclear receptors and its possible role in hormone-dependent intermolecular interactions. Biochemistry 34:10591-10599. [DOI] [PubMed] [Google Scholar]
- 5.Brucker-Davis, F., M. C. Skarulis, M. B. Grace, J. Benichou, P. Hauser, E. Wiggs, and B. D. Weintraub. 1995. Genetic and clinical features of 42 kindreds with resistance to thyroid hormone. The National Institutes of Health Prospective Study. Ann. Intern. Med. 123:572-583. [DOI] [PubMed] [Google Scholar]
- 6.Carvalho S. D., N. Negrao, and A. C. Bianco. 1993. Hormonal regulation of malic enzyme and glucose-6-phosphate dehydrogenase in brown adipose tissue. Am. J. Physiol. 264:E874-E881. [DOI] [PubMed] [Google Scholar]
- 7.Charlier, T. D., and J. Balthazart. 2005. Modulation of hormonal signaling in the brain by steroid receptor coactivators. Rev. Neurosci. 16:339-357. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, S. Y. 2004. Abnormalities of nuclear receptors in thyroid cancer. Cancer Treat. Res. 122:165-178. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, S.-Y. 2004. New development in thyroid hormone resistance. Hot Thyroidol. www.hotthyroidology.com/editorial_126.html.
- 10.Cheng, S.-Y. 2000. Multiple mechanisms for regulation of the transcriptional activity of thyroid hormone receptors. Rev. Endocr. Metab. Disord. 1:9-18. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, S.-Y. 2003. Thyroid hormone receptor mutations in cancer. Mol. Cell. Endocrinol. 213:23-30. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, S.-Y. 2005. Thyroid hormone receptor mutations and disease: beyond thyroid hormone resistance. Trends Endocrinol. Metab. 16:176-182. [DOI] [PubMed] [Google Scholar]
- 13.Engfeldt, P., P. Arner, J. Bolinder, and J. Ostman. 1982. Phosphodiesterase activity in human subcutaneous adipose tissue in insulin- and noninsulin-dependent diabetes mellitus. J. Clin. Endocrinol. Metab. 55:983-988. [DOI] [PubMed] [Google Scholar]
- 14.Forrest, D., and B. Vennstrom. 2000. Functions of thyroid hormone receptors in mice. Thyroid 10:41-52. [DOI] [PubMed] [Google Scholar]
- 15.Fu, M., T. Sun, A. L. Bookout, M. Downes, R. T. Yu, R. M. Evans, and D. J. Mangelsdorf. 2005. A nuclear receptor atlas: 3T3-L1 adipogenesis. Mol. Endocrinol. 19:2437-2450. [DOI] [PubMed] [Google Scholar]
- 16.Furumoto, H., H. Ying, G. V. Chandramouli, L. Zhao, R. L. Walker, P. S. Meltzer, M. C. Willingham, and S.-Y. Cheng. 2005. An unliganded thyroid hormone receptor β activates the cyclin D1/cyclin-dependent kinase/retinoblastoma/E2F pathway and induces pituitary tumorigenesis. Mol. Cell. Biol. 25:124-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauner, H. 2005. Secretory factors from human adipose tissue and their functional role. Proc. Nutr. Soc. 64:163-169. [DOI] [PubMed] [Google Scholar]
- 18.He, W., Y. Barak, A. Hevener, P. Olson, D. Liao, J. Le, M. Nelson, E. Ong, J. M. Olefsky, and R. M. Evans. 2003. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. USA 100:15712-15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helledie, T., L. Grontved, S. S. Jensen, P. Kiilerich, L. Rietveld, T. Albrektsen, M. S. Boysen, J. Nohr, L. K. Larsen, J. Fleckner, H. G. Stunnenberg, K. Kristiansen, and S. Mandrup. 2002. The gene encoding the Acyl-CoA-binding protein is activated by peroxisome proliferator-activated receptor gamma through an intronic response element functionally conserved between humans and rodents. J. Biol. Chem. 277:26821-26830. [DOI] [PubMed] [Google Scholar]
- 20.Hernández, A., and M. J. Obregon. 1996. Presence and mRNA expression of T3 receptors in differentiating rat brown adipocytes. Mol. Cell. Endocrinol. 121:37-46. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, W., T. Miyamoto, T. Kakizawa, T. Sakuma, S. Nishio, T. Takeda, S. Suzuki, and K. Hashizume. 2004. Expression of thyroid hormone receptor alpha in 3T3-L1 adipocytes; triiodothyronine increases the expression of lipogenic enzyme and triglyceride accumulation. J. Endocrinol. 182:295-302. [DOI] [PubMed] [Google Scholar]
- 22.Kaneshige, M., H. Suzuki, K. Kaneshige, J. Cheng, H. Wimbrow, C. Barlow, M. C. Willingham, and S.-Y. Cheng. 2001. A targeted dominant negative mutation of the thyroid hormone α1 receptor causes increased mortality, infertility and dwarfism in mice. Proc. Natl. Acad. Sci. USA 98:15095-15100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneshige, M., K. Kaneshige, X.-G. Zhu, A. Dace, L. Garrett, T. A. Carter, R. Kazlauskaite, D. G. Pankratz, A. Wynshaw-Boris, B. Weintraub, M. C. Willingham, C. Barlow, and S.-Y. Cheng. 2000. Mice with a targeted mutation in the thyroid hormone β receptor gene exhibit impaired growth and resistance to thyroid hormone. Proc. Natl. Acad. Sci. USA 97:13209-13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleiber, M., and T. A. Rogers. 1961. Energy metabolism. Annu. Rev. Physiol. 23:5-36. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, K. 2005. Adipokines: therapeutic targets for metabolic syndrome. Curr. Drug Targets 6:525-529. [DOI] [PubMed] [Google Scholar]
- 26.Lin, K.-H., K. Ashizawa, and S.-Y. Cheng. 1992. Phosphorylation stimulates the transcriptional activity of the human b1 thyroid hormone nuclear receptor. Proc. Natl. Acad. Sci. USA 89:7737-7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, Y. Y., J. J. Schultz, and G. A. Brent. 2003. A thyroid hormone receptor alpha gene mutation (P398H) is associated with visceral adiposity and impaired catecholamine-stimulated lipolysis in mice. J. Biol. Chem. 278:38913-38920. [DOI] [PubMed] [Google Scholar]
- 28.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meier, C. A., B. M. Dickstein, K. Ashizawa, J. H. McClaskey, P. Muchmore, S. C. Ransom, J. B. Menke, E. H. Hao, S. J. Usala, B. B. Bercu, S.-Y. Cheng, and B. D. Weintraub. 1992. Variable transcriptional activity and ligand binding of mutant β1 3,5,3′-triiodothyronine receptors from four families with generalized resistance to thyroid hormone. Mol. Endocrinol. 6:248-258. [DOI] [PubMed] [Google Scholar]
- 30.O'Malley, B. W. 2003. Sequentiality and processivity of nuclear receptor coregulators in regulation of target gene expression. Nucl. Recept. Signal. 1:e010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono, S., I. D. Schwartz, O. T. Mueller, A. W. Root, S. J. Usala, and B. B. Bercu. 1991. Homozygosity for a dominant negative thyroid hormone receptor gene responsible for generalized resistance to thyroid hormone. J. Clin. Endocrinol. Metab. 73:990-994. [DOI] [PubMed] [Google Scholar]
- 32.O'Shea, P. J., J. H. Bassett, S. Sriskantharajah, H. Ying, S. Y. Cheng, and G. R. Williams. 2005. Contrasting skeletal phenotypes in mice with an identical mutation targeted to thyroid hormone receptor α1 or β. Mol. Endocrinol. 19:3045-3059. [DOI] [PubMed] [Google Scholar]
- 33.Parrilla, R., A. J. Mixson, J. A. McPherson, J. H. McClaskey, and B. D., Weintraub. 1991. Characterization of seven novel mutations of the c-erbAβ gene in unrelated kindreds with generalized thyroid hormone resistance: evidence for two “hot spot” regions of the ligand binding domain. J. Clin. Investig. 88:2123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reyne, Y., J. Nougues, B. Cambon, N. Viguerie-Bascands, and L. Casteilla. 1996. Expression of c-erbAα, c-erbAβ and Rev-erbAα mRNA during the conversion of brown adipose tissue into white adipose tissue. Mol. Cell. Endocrinol. 116:59-65. [DOI] [PubMed] [Google Scholar]
- 35.Rodell, M. 1964. Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. J. Biol. Chem. 239:375-380. [PubMed] [Google Scholar]
- 36.Schoonjans, K., J. Peinado-Onsurbe, A. M. Lefebvre, R. A. Heyman, M. Briggs, S. Deeb, B. Staels, and J. Auwerx. 1996. PPARα and PPARγ activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 15:5336-5348. [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz, H. L., K. A. Strait, N. C. Ling, and J. H. Oppenheimer. 1992. Quantitation of rat tissue thyroid hormone binding receptor isoforms by immunoprecipitation of nuclear triiodothyronine binding capacity. J. Biol. Chem. 267:11794-11799. [PubMed] [Google Scholar]
- 38.Smas, C. M., and H. S. Sul. 1993. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 73:725-734. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki, H., M. C. Willingham, and S.-Y. Cheng. 2002. Mice with a mutation in the thyroid hormone receptor β gene spontaneously develop thyroid carcinoma: a mouse model of thyroid carcinogenesis. Thyroid 12:963-969. [DOI] [PubMed] [Google Scholar]
- 40.Tinnikov, A., K. Nordstrom, P. Thoren, J. M. Kindblom, S. Malin, B. Rozell, M. Adams, O. Rajanayagam, S. Pettersson, C. Ohlsson, K. Chatterjee, and B. Vennstrom. 2002. Retardation of post-natal development caused by a negatively acting thyroid hormone receptor α1. EMBO J. 21:5079-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner, R. L., J. W. Apriletti, M. E. McGrath, B. L. West, J. D. Baxter, and R. J. Fletterick. 1995. A structural role for hormone in the thyroid hormone receptor. Nature 378:690-697. [DOI] [PubMed] [Google Scholar]
- 42.Wahrenberg, H., F. Lonnqvist, and P. Arner. 1989. Mechanisms underlying regional differences in lipolysis in human adipose tissue. J. Clin. Investig. 84:458-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss, R. E., and S. Refetoff. 2000. Resistance to thyroid hormone. Rev. Endocr. Metab. Disord. 1:97-108. [DOI] [PubMed] [Google Scholar]
- 44.Wolfrum, C., D. Q. Shih, S. Kuwajima, A. W. Norris, C. R. Kahn, and M. Stoffel. 2003. Role of Foxa-2 in adipocyte metabolism and differentiation. J. Clin. Investig. 112:345-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yen, P. M. 2003. Molecular basis of resistance to thyroid hormone. Trends Endocrinol. Metab. 14:327-333. [DOI] [PubMed] [Google Scholar]
- 46.Ying, H., F. Furuya, L. Zhao, O. Araki, B. L. West, J. A. Hanover, M. C. Willingham, and S. Y. Cheng. 2006. Aberrant accumulation of PTTG1 induced by a mutated thyroid hormone β receptor inhibits mitotic progression. J. Clin. Investig. 116:2972-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ying, H., H. Suzuki, H. Furumoto, R. Walker, P. Meltzer, M. C. Willingham, and S.-Y. Cheng. 2003. Alterations in genomic profiles during tumor progression in a mouse model of follicular thyroid carcinoma. Carcinogenesis 24:1467-1479. [DOI] [PubMed] [Google Scholar]
- 48.Zavacki, A. M., H. Ying, M. A. Christoffolete, G. Aerts, E. So, J. W. Harney, S. Y. Cheng, P. R. Larsen, and A. C. Bianco. 2005. Type 1 iodothyronine deiodinase is a sensitive marker of peripheral thyroid status in the mouse. Endocrinology 146:1568-1575. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, X.-Y., M. Kaneshige, Y. Kamiya, K. Kaneshige, and S.-Y. Cheng. 2002. Differential expression of thyroid hormone receptor isoforms dictates the dominant negative activity of mutant receptor. Mol. Endocrinol. 16:2077-2092. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, H., S. Yakar, O. Gavrilova, H. Sun, Y. Zhang, H. Kim, J. Setser, W. Jou, and D. LeRoith. 2004. Phloridzin improves hyperglycemia but not hepatic insulin resistance in a transgenic mouse model of type 2 diabetes. Diabetes 53:2901-2909. [DOI] [PubMed] [Google Scholar]