Abstract
Saccharomyces cerevisiae adapts to hypoxia by expressing a large group of “anaerobic” genes. Among these, the eight DAN/TIR genes are regulated by the repressors Rox1 and Mot3 and the activator Upc2/Mox4. In attempting to identify factors recruited by the DNA binding repressor Mot3 to enhance repression of the DAN/TIR genes, we found that the histone deacetylase and global repressor complex, Rpd3-Sin3-Sap30, was not required for repression. Strikingly, the complex was instead required for activation. In addition, the histone H3 and H4 amino termini, which are targets of Rpd3, were also required for DAN1 expression. Epistasis tests demonstrated that the Rpd3 complex is not required in the absence of the repressor Mot3. Furthermore, the Rpd3 complex was required for normal function and stable binding of the activator Upc2 at the DAN1 promoter. Moreover, the Swi/Snf chromatin remodeling complex was strongly required for activation of DAN1, and chromatin immunoprecipitation analysis showed an Rpd3-dependent reduction in DAN1 promoter-associated nucleosomes upon induction. Taken together, these data provide evidence that during anaerobiosis, the Rpd3 complex acts at the DAN1 promoter to antagonize the chromatin-mediated repression caused by Mot3 and Rox1 and that chromatin remodeling by Swi/Snf is necessary for normal expression.
The Saccharomyces cerevisiae DAN/TIR genes are among a large group of genes that are upregulated during adaptation to anaerobic growth (37, 59, 65, 66). These genes code for cell wall mannoproteins, which play a significant role in cell wall permeability. The kinetics of expression of these genes ranges from 30 minutes to 3 hours following the onset of anaerobiosis (1, 59). This suggests that as the cells descend towards anaerobiosis, certain requirements that are necessary for survival in that milieu are incrementally satisfied by specific alterations in gene expression. The importance of the DAN/TIR genes is further underlined by the fact that disruption of some of them, such as TIR1, TIR3, and TIR4, abrogates anaerobic growth (1), indicating that the corresponding proteins play essential functions during anaerobic adaptation. Moreover, it appears that a complex programmed cell wall remodeling occurs during adaptation to anaerobiosis, as shown by the fact that the major aerobic cell wall mannoproteins encoded by CWP1 and CWP2 are replaced by their anaerobic counterparts, encoded by the DAN/TIR genes, under those conditions (1).
The precise mechanisms by which the DAN/TIR genes are regulated are still being elucidated. We showed earlier that these genes are regulated by heme, which is synthesized only in the presence of oxygen, and by three DNA binding transcription factors (2, 13, 59). The activator Upc2 acts through a consensus site termed AR1 to induce the expression of these genes in anaerobiosis. Upc2 was also identified as a regulator of the anaerobic sterol transport system (2, 75). It shares extensive homology with Ecm22 and with a Candida albicans protein (CaUpc2) (71). The repressors Rox1 and Mot3 function synergistically to efficiently repress these genes in aerobic cultures (60).
In attempting to identify factors which might be recruited by Mot3 to enhance repression of the DAN/TIR genes and the hypoxic gene ANB1, we tested the role of the Tup1/Ssn6 and Rpd3 complexes. These global repressors have been shown to be recruited by DNA binding repressors to remodel chromatin at several promoters (10, 15, 24, 26, 39, 44). We previously found that Tup1/Ssn6 is required for Rox1-mediated repression of the hypoxic gene ANB1 but not for repression of ANB1 by Mot3 (60). Tup1/Ssn6 also plays a role in repressing DAN1, but this again appears to occur via a separate mechanism from that used by Mot3 (60). We then tested the role of the histone deacetylase (HDAC) and global repressor Rpd3 in mediating repression of the anaerobic genes. We found that the Rpd3 complex was not required for repression. Surprisingly, it was instead required for the expression of all the DAN/TIR genes and the hypoxic gene ANB1, which are negatively regulated by heme and Mot3.
The S. cerevisiae Rpd3 histone deacetylase is a class I HDAC that has been demonstrated to regulate the expression of a large number of genes (7, 36, 51, 52, 56). Two distinct complexes of Rpd3 have been identified, namely, a small complex of 0.6 MDa (Rpd3S) and a large one of 1.2 MDa (Rpd3L) (12, 31). These two complexes share three subunits, namely, Rpd3, Sin3, and Ume1. Rco1 and Eaf3 were found to be specific for the small Rpd3 complex, which negatively regulates transcriptional elongation by RNA polymerase II (PolII). The large Rpd3 complex, which includes Sap30, Pho23, Rxt1, Rxt2, Dep1, and Sds3, appears to be the promoter-targeting form of the two complexes. Rpd3 does not bind to DNA and is therefore recruited to promoters by DNA-binding transcription factors. For instance, Rpd3 is recruited by Ume6 to directly repress the expression of INO1, a gene involved in inositol biosynthesis (24). Furthermore, repression by Ash1 at the HO promoter is mediated by Rpd3 recruitment at these sites (11). Recently, Rpd3 was shown to activate transcription of the osmoresponsive gene HSP12 when recruited to that promoter in response to activation by the mitogen-activated protein kinase Hog1 transcriptional activator (16). Thus, Rpd3 can function as an activator of transcription in addition to its usual role as a repressor.
Here we show that Rpd3, along with its targets, the N termini of histones H3 and H4, is required for the expression of the anaerobic DAN/TIR genes. We found that the Rpd3 complex is recruited during anaerobic growth. Genetic analysis shows that Rpd3 is dispensable in the absence of the repressor Mot3, implying a functional interaction between these two regulators. Chromatin immunoprecipitation (ChIP) analysis reveals that Rpd3 is required for nucleosome loss at the induced DAN1 promoter and for stable binding of the activator Upc2. The chromatin remodeling complex Swi/Snf also plays a pivotal role in the regulation of these genes. Taken together, these observations define a novel chromatin-mediated regulatory mechanism responsible for DAN/TIR induction.
MATERIALS AND METHODS
Yeast strains.
The strains used in this work are listed in Table 1. Strains Y41 and Y42 were derived from NYS429 and NSY430, respectively, as follows. NYS429 and NSY430 were transformed with a 4.5-kb EcoRI-BglII-PvuII fragment from the Trp blaster plasmid (4). The resulting Trp− Ura+ colonies were screened for resistance to 5-fluoroorotic acid to test for the loss of the URA3 marker.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| NSY429 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1) (hht2-hhf2) pNS329 (CEN TRP1 HHF1-HHT1) | 56 |
| NSY430 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1) (hht2-hhf2) pNS329 (CEN TRP1 HHF1-HHT1) rpd3::LEU2 | 56 |
| NSY438 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1) (hht2-hhf2) pNS338 [CEN TRP1 hhf1-8 (H4Δ2-26) HHT1] | 56 |
| NSY458 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1) (hht2-hhf2) pNS358 [CEN TRP1 HHF1 hht1-2 (H3Δ1-28)] | 56 |
| K699 | leu2-3,112 ura3-1 his3-11 trp1-1 can100 | 16 |
| PAY304 | W303 MATα SIN3-HA-HIS3 | F. Posas |
| NOY847 | MATura3 his3 trp1 leu2-3,112 rrn5Δ::LEU2 Δ(hht1-hhf1) Δ(hht2-hhf2) pNOY434 [HIS3 RRN5-(HA1)3-His6] pNOY436 (TRP1 HHT2 myc-HHF2) | 29 |
| W303-1A | MATaura3-1 leu2-3,112 trp1-1 can1-100 ade2-1 his3-11,15 [PSI+] | 5 |
| MAY6 | sin3::KanMX (W303-1A) | 5 |
| MAY7 | rpd3::KanMX (W303-1A) | 5 |
| MAY8 | sap30::KanMX (W303-1A) | 5 |
| FY1339 | MATaura3-52 his3Δ200 trp1Δ63 | 23 |
| FY2081 | MATaura3-52 his3Δ200 trp1Δ63 MOT3-Myc18::TRP1 | 23 |
| Y4 | MATahis3Δ1 leuΔ0 met15Δ0 ura3Δ0 swi3::KanMX | Resgen |
| Y5 | MATahis3Δ1 leuΔ0 met15Δ0 ura3Δ0 swi10::KanMX | Resgen |
| Y6 | MATahis3Δ1 leuΔ0 met15Δ0 ura3Δ0 snf2::KanMX | Resgen |
| Y7 | MATahis3Δ1 leuΔ0 met15Δ0 ura3Δ0 snf6::KanMX | Resgen |
| Y41 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1) (hht2-hhf2) pMS329 (CEN ARS ura3Δ0 HHT2-HHF1) | This study |
| Y42 | MATα ura3-52 leu2-3,112 trp1-289 his31 (hht1-hhf1) (hht2-hhf2) pMS329 (CEN ARS ura3Δ0 HHT2-HHF1) rpd3::LEU2 | This study |
| Y56 | Y41 mot3::KanMX | This study |
| Y57 | Y42 mot3::KanMX | This study |
| Y72 | Y41 mot3::HIS3 rox1::KanMX | This study |
| Y85 | Y41 mot3::HIS3 | This study |
| Y102 | Y41 MOT3-Myc18 | This study |
| Y103 | Y42 MOT3-Myc18 | This study |
Strains Y56 and Y57 were generated by transformation of Y41 and Y42, respectively, with a PCR fragment obtained from the proprietary mot3::KanMX strain derived from BY4741 (76). The knockout fragment was verified by PCR. Strain Y72 was made by transforming strain Y41 with a PCR fragment that was amplified from a mot3::HIS3 strain (42), using primers homologous at positions −964 and +1880 to generate strain Y85. A rox1::KanMX disruption PCR fragment (76) was then introduced into Y85. Both knockout fragments were verified by PCR. Strains Y102 and Y103 were generated as follows. The MOT3-Myc18 PCR product was amplified from strain FY2081 (23), using primers homologous at +891 and +2330. The PCR fragment was transformed into strains Y41 and Y42 and then verified by PCR.
Plasmids.
The plasmid YCp22UPC2(G888A) was described elsewhere (2). YCp33RPD3 and YEp112RPD3 were generated as follows. An XbaI-PstI PCR fragment containing the RPD3 gene was obtained from NSY429 (56), using primers homologous at −994 and +1386. The PCR product was digested with XbaI and PstI and ligated into YCp33 and YEplac112 which had been digested with the same enzymes, generating YCp33RPD3 and YEp112RPD3, respectively. The plasmids YCp33RPD3H188A and YEp112RPD3H188A were generated by excising a 0.7-kb BglII-MluI fragment from YEplac112LexARPD3H188A (25) and subcloning it into YCp33RPD3 and YEp112RPD3, respectively, which had been digested with the same enzymes.
YCp22 UPC2-HA3 was constructed as follows. YCp22UPC2 (2) was digested with HindIII and BglII and ligated with a linker oligonucleotide, AGCTTTGAGCGAGA or AACTCGCTCTCTAG. The new plasmid was digested with ClaI and ligated with another linker oligonucleotide, CGATGACTACTACTAACTTTAGCGATTTCTCGAG or CGCTCGAGAAATCGCTAAAGTTAGTAGTAGTCAT. The modified plasmid was cut with XhoI and inserted with a three-hemagglutinin (HA3) PCR fragment amplified from template pMPY-HA3 (57), resulting in YCp22UPC2HindIII/BglII-HA3. Finally, a UPC2 fragment was excised from YCp22UPC2, using AgeI and NdeI, and ligated into YCp22UPC2HindIII/BglII-HA3 which was digested with the same enzymes.
Cell growth, yeast transformation, and Northern analysis.
Yeast cells were grown at 30°C under aerobic and anaerobic conditions in yeast extract-peptone-dextrose or synthetic complete medium. Anaerobic cultures were bubbled with high-purity nitrogen and harvested in late log phase after 2 h of anaerobic growth. Yeast transformation was performed according to the LiAc TRAFO (lithium acetate/single-stranded carrier DNA/polyethylene glycol) method as described previously (19). For Northern analysis, RNAs were extracted and analyzed as described previously (78). Briefly, 10 μg of RNA was run in agarose-formaldehyde gels and blotted onto nylon membranes in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate). The membranes were then baked for 2 h and hybridized with probes labeled by random priming. Northern blots were quantitated on a Molecular Dynamics phosphorimager.
Western blotting.
Western analysis was performed with whole-cell extracts that were prepared by glass bead lysis. Protein concentrations were determined by the Bradford assay (Bio-Rad). Twenty-microgram samples of denatured proteins in 5% 2-mercaptoethanol were fractionated in 10 to 12% polyacrylamide gels for 1 h at 120 V and transferred to Millipore polyvinylidene difluoride membranes at 30 V for 1 h. The membrane was blocked for 1 h at room temperature in BLOTTO A solution (100 mM Tris, pH 7.6, 0.1% Tween 20, 5% nonfat dry milk) and incubated overnight at 4°C with new BLOTTO A solution containing primary antibody. The blot was washed three times for 10 min each in TBST (100 mM Tris, pH 7.6, 0.1% Tween 20, 0.9% NaCl) and incubated with the secondary antibody, horseradish peroxidase-linked anti-rabbit immunoglobulin, for 1 h. The blot was again washed three times for 10 min each in TBST. Detection of the appropriate protein was revealed with an ECL detection kit and exposure to Kodak X-Omat radiographic film (Eastman Kodak Co., Rochester, NY).
ChIP.
Chromatin immunoprecipitation was performed as previously described (18, 34). Briefly, formaldehyde cross-linked samples were sonicated to a size range of 0.2 to 0.6 kb. Immunoprecipitated (IP) samples were eluted according to standard protocols using the following antibodies: 20 μl hemagglutinin antibody (Y-11, SC-805; Santa Cruz Biotechnology), 20 μl c-Myc antibody (A-14, SC-789; Santa Cruz Biotechnology), 3 μl histone H3 antibody (06-755; Upstate Cell Signaling Solutions), and 4.0 μl acetylated histone H4 antibody (06-866; Upstate Cell Signaling Solutions). For analysis, 2.0 μl of IP DNA and 1.0 μl of a 1:100 dilution of input DNA were amplified by quantitative radioactive PCR for 22 cycles, and the products were separated in a 7.5% nondenaturing polyacrylamide gel. Each ChIP experiment was repeated two or three times, and multiple repeats of the same PCR were performed to test for pipetting errors. Quantitation was determined by using a Molecular Dynamics phosphorimager. Primers used for amplification are available upon request.
RESULTS
Rpd3 is required for the expression of anaerobic genes.
In our effort to delineate the molecular mechanisms by which the DAN/TIR genes and the hypoxic gene ANB1 are regulated, we reported earlier the isolation of Mot3 as a repressor of these heme-repressed genes (60). To elucidate the mechanism of repression by Mot3, we tested in that study the role of the Tup1/Ssn6 complex. This global repressor has been shown to be recruited by DNA binding repressors to remodel chromatin at several promoters (10, 39, 44, 62, 74, 77). We observed constitutive expression of the DAN1 and ANB1 genes in a tup1Δ/ssn6Δ strain, suggesting that the global repressor Tup1/Ssn6 is indeed required for efficient repression of the genes. However, we found that Tup1/Ssn6 is required for Rox1 repression of the ANB1 gene but not for repression by Mot3 at that promoter (60). Ectopic expression of Rox1 and Mot3 in tup1Δ/ssn6Δ strains in anaerobic cultures showed that Rox1 can no longer repress, while Mot3 is still capable of repressing the expression of the ANB1 gene (60).
We then tested whether the histone deacetylase and global repressor Rpd3 has a role in mediating repression of the anaerobic genes. We found that Rpd3 was not required for repression of the anaerobic genes. We did not observe constitutive expression of any of the anaerobic DAN/TIR genes or the hypoxic gene ANB1 in an rpd3Δ strain in aerobic cultures (Fig. 1A, lanes 1 and 2). Surprisingly, Rpd3 was required for the expression of the anaerobic genes which are negatively regulated by Mot3 (Fig. 1A, lane 4). Interestingly, two anaerobically expressed genes, namely, OLE1 (Fig. 1) and TIP1 (not shown), which are not controlled by either heme or Mot3, were not regulated by Rpd3. These results implicate Rpd3 as part of the DAN/TIR and ANB1 regulatory circuit responding to a heme-deficient milieu by inducing the activation of these anaerobic genes.
FIG. 1.
Rpd3 is required for expression of anaerobic proteins. (A) Wild-type and rpd3Δ strains were grown aerobically or anaerobically for 2 h. RNAs were isolated for a Northern blot, which was hybridized with [32P]dCTP-labeled probes for DAN1, DAN3-4, TIR1, ANB1, OLE1, and ACT1. The TIR1 probe also hybridizes to another band, which corresponds to TIR2 mRNA (2). (B) rpd3Δ yeast cells were transformed with the following plasmids: YCp33 (vector), YCp33Rpd3, and YCp33Rpd3H188A. These cells were grown anaerobically for 2 h in the appropriate medium. Total RNA was isolated and analyzed by Northern blotting, using probes for DAN1 and ACT1. (C) Relative DAN1 mRNA levels were quantified by phosphorimaging (Molecular Dynamics) and normalized to the ACT1 internal loading control. The values represent the averages for two independent experiments, and standard deviations are indicated. Note that yeast cells were grown in Ura-deficient medium for the experiment of panel B, resulting in lower expression levels of DAN1 than those for cells grown in YPD medium, as for panel A.
Enzymatic activity of Rpd3 is necessary for expression of the anaerobic genes.
To determine whether the enzymatic activity of Rpd3 is needed for the expression of the anaerobic genes, we first grew yeast cells in the presence and absence of the histone deacetylase inhibitor trichostatin A (TSA). Northern blot analysis showed a significant reduction in expression of the anaerobic gene DAN1 (data not shown), suggesting that Rpd3 function is indeed required for full activation of the DAN/TIR genes. However, TSA is not a specific inhibitor of Rpd3, as it also affects the activities of all class I and II histone deacetylases (67). Moreover, several studies have demonstrated that TSA affects a wide range of cellular processes, including apoptosis, differentiation, and DNA synthesis (30, 32, 69). Therefore, it is not clear whether the reduction in the expression of DAN1 is due to the loss of deacetylase activity of Rpd3 or through other cellular abnormalities caused by the drug. To resolve the possible indirect effects of trichostatin A on DAN1 expression, we utilized an enzymatically defective form of Rpd3 (Rpd3H188A) (25). rpd3Δ yeast strains transformed with catalytically inactive Rpd3, in contrast with wild-type Rpd3, could not suppress the noninducible phenotype of the rpd3Δ strain (Fig. 1B and C), implying that Rpd3's enzymatic activity is needed for transcriptional activation.
The large Rpd3 complex and its targets are involved in activation of anaerobic genes.
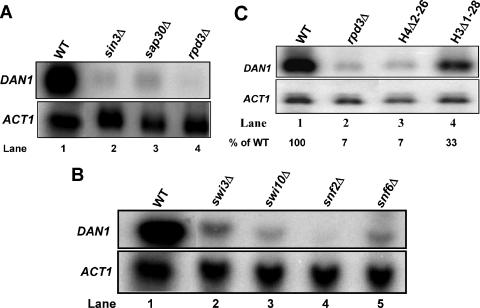
To determine which Rpd3 complex (small or large) contributes to DAN1 activation and to identify other chromatin-related factors that regulate DAN1 expression, we tested more than 50 strains from a yeast knockout library (76). We observed a nearly absolute requirement for some of the known components of the Rpd3 complex (Rpd3, Sin3, and Sap30) (Fig. 2A) and a partial requirement for other factors, such as Pho23, Ume6, Sds3, and Ume1 (not shown). Since Sap30, Pho23, and Sds3 are components of the Rpd3L but not the Rpd3S complex (12, 31), these results strongly suggest that the large Rpd3 complex is the one required for the induction of DAN1. Some of the subunits of the Swi/Snf complex were also needed for DAN1 expression (Fig. 2B). We also observed a partial requirement for a few factors in other categories (Taf14, Ada2, Ada3, Ccr4, and Caf130) (data not shown). These results suggest that Rpd3-mediated activation of DAN1 occurs via the same Rpd3 complex previously identified as a global repressor and underscore the role of chromatin in anaerobic gene expression.
FIG. 2.
The large Rpd3 complex and its targets are required for DAN1 expression. Yeast strains were grown aerobically and shifted to anaerobic growth. (A) Northern blots for wild-type (WT), sap30Δ, sin3Δ, and rpd3Δ strains. (B) Northern blots for wild-type, swi3Δ, swi10Δ, snf2Δ, and snf6Δ strains. (C) Northern blots for wild-type and rpd3Δ strains and strains with N-terminal deletions of histones H4 (hhf1Δ2-26) and H3 (hht1Δ1-28).
Since the amino termini of histones H3 and H4 are known substrates of Rpd3 (26, 53, 54), we hypothesized that these histone tails might be needed for DAN1 expression. Northern blot analysis showed that yeast strains carrying N-terminal deletions of histone H4 (hhf1Δ2-26) and, to a lesser extent, histone H3 (hht1Δ1-28) showed substantial reductions in DAN1 expression (Fig. 2C). We also found substantially reduced anaerobic expression of DAN1 in a yeast strain expressing H4 with K5,8,12,16Q mutations (strain NSY491) (56; data not shown); we did not examine the H3(K4,9,14,18,23,27Q) mutant. These results, which are in contrast to the more typical upregulation caused by these mutations (43, 55, 68), indicate that the H3 and H4 amino termini are needed for activation and that the histone H4 tail is particularly important.
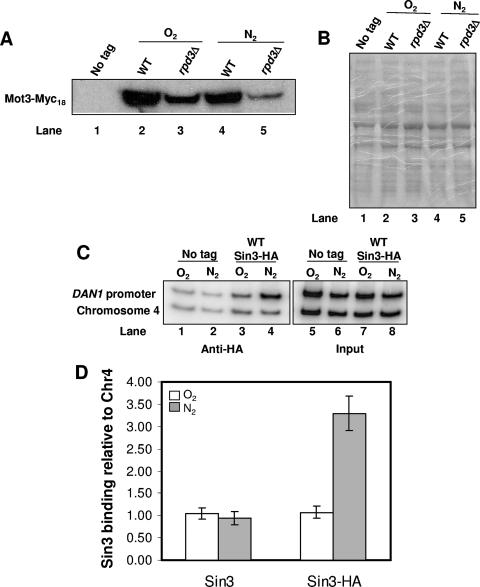
Rpd3 is directly involved in activation of DAN1.
Rpd3 could regulate the expression of the anaerobic genes indirectly by inhibiting the expression of a repressor, particularly Mot3. However, Western blot analysis showed no increase, and in fact a decrease, in Mot3 protein levels during anaerobic growth in an rpd3Δ strain (Fig. 3A and B, compare lanes 4 and 5). Since this result does not rule out regulation by Rpd3 of an unidentified repressor, we next tested whether the Rpd3 complex is recruited to the DAN1 promoter during hypoxia. For this purpose, we performed ChIP experiments, using HA3-tagged Sin3 as a surrogate for Rpd3, which is somewhat refractory to the standard cross-linking protocol used for ChIP (36). Since Sin3 and Rpd3 are intimately associated in the Rpd3 repressor complex, Sin3 binding is expected to provide an accurate reflection of Rpd3 binding (16, 24). We found 3.5-fold enrichment of Sin3 binding and, by inference, the Rpd3 complex at the UAS region of the DAN1 promoter in anaerobic compared to aerobic cultures (Fig. 3C, compare lanes 3 and 4, and Fig. 3D). These results strongly suggest that Rpd3 is directly involved in transcriptional activation of the anaerobic genes.
FIG. 3.
The Rpd3 complex is directly involved in activation of the anaerobic genes. (A) Mot3-Myc18 (Y102 and Y103) and untagged (Y41) cells were grown aerobically or shifted to anaerobic growth for 2 h in the appropriate medium. Extracts prepared from these cells were subjected to immunoblotting with a monoclonal antibody against the Myc epitope. (B) To determine equal loading of the samples, the proteins blotted onto the nylon membrane were stained with Ponceau reagent prior to being blocked with milk. (C) Sin3-HA3 (PAY304) and untagged (K699) cells were grown aerobically or shifted to anaerobic growth for 2 h in the appropriate medium and subjected to ChIP using anti-HA antibodies. Samples were used to amplify the DAN1 regulatory region, using primers that span the activator Upc2 site, together with a control PCR that amplified a region of chromosome IV that is outside any open reading frames. The labeled PCR products were quantitated by using a PhosphorImager. (D) The graph represents the ratios of DAN1 promoter IP samples to input samples relative to the ratio of chromosome IV (Chr4) IP samples to input samples. The values represent the averages for two independent experiments, and standard deviations are indicated.
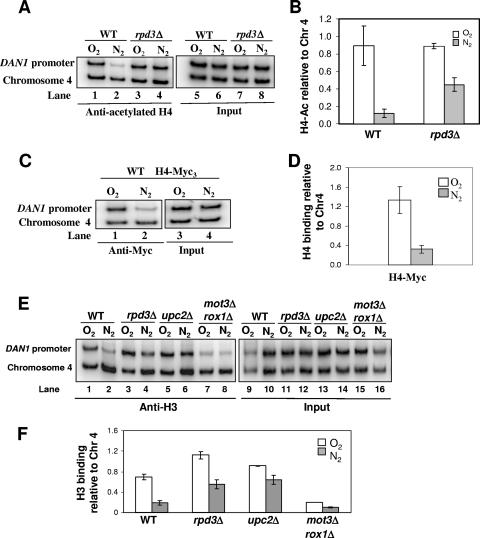
The Rpd3 complex is required for nucleosome loss during DAN1 expression.
Having established that Rpd3 is present at the DAN1 promoter during induction and that both its enzymatic activities and histone targets are needed for transcriptional activation, we hypothesized that one possible mechanism by which Rpd3 participates in the regulation of the anaerobic genes is by deacetylating its histone targets, particularly histone H4. We monitored levels of acetylated histone H4 at the DAN1 promoter in the induced and repressed states by ChIP analysis. We observed a drastic reduction in the level of acetylated histone H4 in anaerobic wild-type cells (Fig. 4A, compare lanes 1 and 2). This reduction was greatly diminished in the rpd3Δ mutant (Fig. 4A, compare lanes 2 and 4). We next investigated whether the reduction in acetylated histone H4 at the DAN1 promoter during anaerobic induction could reflect a loss in overall nucleosome density. For this purpose, we performed ChIP using Myc-tagged histone H4 as well as a pan-histone H3 antibody that recognizes histone H3 regardless of its modification state. These experiments revealed a substantial decrease in histone H4 (Fig. 4C) and histone H3 (Fig. 4E, compare lanes 1 and 2) protein levels at the DAN1 promoter in wild-type cells under anaerobic conditions. This loss in histone density was dependent on Rpd3 (Fig. 4E, compare lanes 2 and 4). This suggests that the Rpd3 complex acts upstream of a requisite chromatin remodeling step during DAN1 activation. We also demonstrated a correlation between DAN1 activation and a loss in histone density, as shown by the constitutive and noninducible mutant mot3Δrox1Δ and upc2Δ strains, respectively (Fig. 4E, lanes 5 to 8).
FIG. 4.
The Rpd3 complex is needed to remodel chromatin at the DAN1 promoter during anaerobic growth. (A) Wild-type and rpd3Δ strains. (C) Myc3-H4 strain. (E) Wild-type, rpd3Δ, upc2Δ, and mot3Δ rox1Δ strains. Yeast strains were grown aerobically or shifted to anaerobic growth for 2 h. Chromatin was cross-linked with formaldehyde and immunoprecipitated with the indicated antibodies. The samples were analyzed as described in the legend to Fig. 3. The graphs (B, D, and F) were generated as described in the legend to Fig. 3. The values represent the averages for two independent experiments, and standard deviations are indicated. Chr4, chromosome IV.
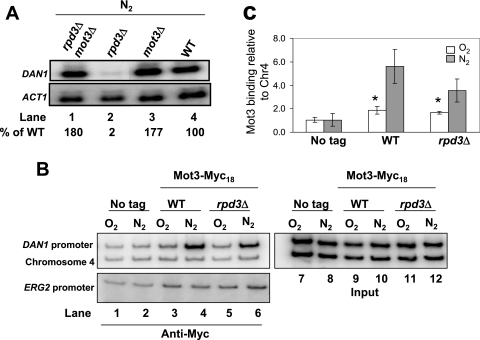
Rpd3 is needed to counteract Mot3-mediated repression of DAN1.
The anaerobic genes that require Rpd3 for activation are negatively regulated by heme and Mot3 (13, 27, 59, 60). Therefore, we decided to probe for genetic interactions between Rpd3, Mot3, and other factors which regulate the expression of the anaerobic genes. We first asked whether the constitutive expression of DAN1 in mot3Δ yeast cells in aerobic cultures required Rpd3. We found that the constitutive phenotype was epistatic, i.e., the DAN1 mRNA level in the double mutant strain (rpd3Δ mot3Δ) was the same as that in the single mutant mot3Δ strain (data not shown), suggesting that Mot3 and Rpd3 are involved in the same transcriptional regulatory function or pathway. In anaerobic cultures, the inhibitory effect of the RPD3 deletion was suppressed in mot3Δ yeast cells, indicating that the Rpd3 complex is required to counteract repression by Mot3 (Fig. 5A). This was surprising because Mot3 had been assumed to repress only under aerobic conditions, and it implies that Rpd3 antagonizes Mot3 repression following the onset of hypoxia.
FIG. 5.
Rpd3 antagonizes Mot3 function. (A) Northern blots from wild-type, rpd3Δ, mot3Δ, and mot3Δ rpd3Δ strains grown aerobically and shifted to anaerobic growth for 2 h. (B and C) Mot3-Myc18 (Y102 and Y103) and untagged (Y41) cells were grown aerobically or shifted to anaerobic growth for 2 h. Chromatin was cross-linked with formaldehyde and immunoprecipitated with the indicated antibodies. The samples were analyzed as described in the legend to Fig. 3. ERG2 was used as a positive control (23). The values represent the averages for four independent experiments, and standard deviations are indicated. Asterisks indicate P values of <0.01 by Student's t test for the difference between the wild-type or rpd3Δ samples and the untagged, uninduced samples. Chr4, chromosome IV.
We next investigated whether the Rpd3 complex antagonizes Mot3 binding at the DAN1 promoter following the onset of hypoxia. We found that in wild-type cells, Mot3 is present at the DAN1 promoter under both repressed and induced conditions (Fig. 5B, compare lanes 3 and 4, and Fig. 5C, where Student's t test indicates that ChIP of Myc-tagged Mot3 in aerobic wild-type or rpd3Δ yeast cells is greater than that in an untagged sample [P < 0.01]). This binding was not affected when RPD3 was deleted (Fig. 5B, compare lanes 4 and 6). We concluded that Rpd3 antagonizes Mot3 function, but not by inhibiting Mot3 binding at the DAN1 promoter.
Rpd3 is required for Upc2 to induce DAN1 and for stable Upc2 binding at the DAN1 promoter.
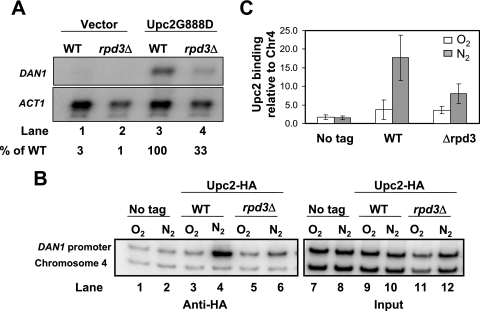
The fact that both Upc2 (2) and the Rpd3 complex are required for the expression of the anaerobic genes suggests some kind of functional interaction between these two factors. It is possible that one factor facilitates the binding of the other. To determine whether they functionally interact, we performed an epistasis test. When combining the dominant constitutive phenotype of the Upc2G888D allele with the poorly inducible phenotype of the rpd3Δ strain, we found that DAN1 expression in the double mutant was strongly reduced compared to that in the single mutant Upc2G888D in aerobic cultures (Fig. 6A, compare lanes 3 and 4). This indicates that Upc2 requires the Rpd3 complex for full activation of DAN1 expression.
FIG. 6.
Rpd3 is needed for stable binding of the activator Upc2. (A) rpd3Δ and wild-type strains were transformed with plasmid YCp33UPC2G888D and grown aerobically. Total RNA was isolated and analyzed by Northern blotting, using probes prepared from DAN1 and ACT1. (B) Wild-type and rpd3Δ strains were transformed with the plasmid YCp22UPC2-HA and grown aerobically or shifted to anaerobic growth for 2 h. Chromatin was cross-linked with formaldehyde and immunoprecipitated with the indicated antibodies. The samples were analyzed as described in the legend to Fig. 3. (C) The graph was generated as described in the legend to Fig. 3. The values represent the averages for two independent experiments, and standard deviations are indicated. The picture in panel A was assembled from images cut from the same blot. Chr4, chromosome IV.
One function of Upc2 is to bind to the DAN1 promoter and recruit the transcriptional machinery to turn on the expression of the anaerobic genes. We hypothesized that the Rpd3 complex is required for stable binding of Upc2 at the DAN1 promoter, based on the observation that Rpd3 is needed for Upc2 function. Using ChIP analysis, we found that the activator Upc2, in wild-type cells, binds preferentially to the DAN1 promoter in anaerobic cultures (Fig. 6B, compare lanes 3 and 4, and Fig. 6C) and that binding is attenuated in cells lacking Rpd3 (Fig. 6B, compare lanes 4 and 6, and Fig. 6C). This experiment further demonstrates a functional relationship between these two regulators and suggests that Rpd3, by counteracting the Mot3-Rox1 repression ensemble, allows stable binding of the activator Upc2 at the DAN1 promoter during anaerobiosis.
DISCUSSION
In this study, we provide evidence that the Rpd3L histone deacetylase complex is not required for repression of the anaerobic DAN/TIR genes but, surprisingly, is required for their activation. Our results indicate that Rpd3 enzymatic activity, as well as its principal targets, the histone H3 and H4 tails, is needed for DAN1 anaerobic expression. Moreover, we found that the Rpd3 complex is present at the DAN1 promoter under inducing conditions and is needed for chromatin remodeling and stable binding of the activator Upc2 at the DAN1 promoter. Finally, we show that a principal function of Rpd3 in DAN1 activation is to overcome repression mediated by Mot3. Together, these results point to a novel mechanism for DAN/TIR gene activation in which Rpd3 acts to antagonize Mot3 repression and facilitate Upc2 binding and activation.
A positive role for Rpd3 in transcriptional regulation was previously reported. Rpd3 has been shown to counteract transcriptional silencing and is required for the activation of genes involved in cell wall biosynthesis (17, 70). However, the previous works did not rule out indirect effects of Rpd3 and provided no mechanistic insights on how Rpd3 functions to facilitate gene expression. More recently, De Nadal et al. (16) provided evidence for a direct role for the Rpd3/Sin3 complex in the activation of the osmoresponsive genes. Rpd3 was shown to physically interact with the activator Hog1, and Sin3 was found to bind directly to the activated HSP12 promoter. Furthermore, reduced levels of promoter-associated acetylated histone H4 and increased RNA PolII binding were observed only in the presence of Rpd3. However, the reduced levels of acetylated histone H4 observed in rpd3Δ yeast may have been due to nucleosome loss, as we observed for DAN1 (Fig. 4). Whether Rpd3 activates osmoregulated genes by counteracting repressor function or facilitating activator binding and nucleosome depletion, as we show for DAN1, is currently unknown. Other histone deacetylases, particularly HDAC1 and HDAC7, have also been shown to play a positive role in transcriptional regulation (28, 49).
The histone deacetylase requirement for the expression of DAN1 appears to be specific for Rpd3, since neither overexpression of other histone deacetylases, such as Hos2 or Sir2, in rpd3Δ yeast nor deleting those factors and others, such as Hda1, Hos1, Hos3, Sir1, and Sir3-4, in a wild-type strain affects the expression level of DAN1 (data not shown). The histone deacetylase Hos2 belongs to the same family as Rpd3 (52, 53) and has been shown to play positive and direct roles in regulation of the GAL genes (72) and in promoting the yeast long terminal repeat-retrotransposon Ty1 integration (45), but it does not play a role in DAN1 regulation.
The histone H3 and H4 tails appear to play essential but distinct roles in the regulation of DAN1. This was inferred from Northern blot results, which show a more pronounced effect on DAN1 expression upon deletion of the histone H4 tail than that with deletion of the histone H3 tail, as well as from epistasis analyses with Upc2 and Mot3 (data not shown). Previous work has shown that, in contrast to the case for DAN1, induction of the anaerobic gene ANB1, which is not in the DAN/TIR family, does not require the histone H3 and H4 amino termini (27), although it does require Rpd3 (Fig. 1). Further work will be needed to clarify the reason for this difference and to determine whether Rpd3 contributes to DAN1 and ANB1 activation by the same or distinct mechanisms.
Although both histone H3 and H4 tails are required for full expression of DAN1 in anaerobic cultures (Fig. 2A), histones are strongly depleted from the DAN1 promoter under inducing conditions (Fig. 4). Similar observations have been made for the induced PHO5 and GAL1-10 promoters (8, 9, 33, 38, 40), and an inverse relationship has been reported between histone density and binding of RNA PolII (58). It is likely that the histone H3 and H4 tails play a role during the early stages of DAN1 activation. For example, they may facilitate binding of Rpd3 and/or the Swi/Snf complex during activation. In addition, it remains possible that deacetylation of nonhistone proteins, such as Mot3, by Rpd3 at these promoters may be involved in turning on the expression of the anaerobic genes.
We observed Mot3 binding at the DAN1 promoter under both induced and repressed conditions, and this binding was independent of Rpd3 (Fig. 5). This finding is consistent with our Western blot data showing that Rpd3 does not indirectly regulate DAN1 by repressing Mot3 expression (Fig. 3A). It appears that more Mot3 protein is bound to the DAN1 promoter in anaerobic than aerobic cultures. This seems counterintuitive but could be due to masking of the epitope by other bound factors, including nucleosomes, under aerobic conditions. These results are in contrast to another report which showed Mot3 bound to the ANB1 promoter only under aerobic conditions (44). At present, we do not understand the reason for these contrasting results. Other reports have shown repressors bound to particular promoters in both induced and repressed states (14, 46, 48, 73). In some instances, these repressors behave as activators that can recruit the transcription machinery. It is unclear at this juncture what role Mot3 might play in anaerobic induction. One possibility is that Mot3 binding under activated conditions could facilitate a rapid and efficient return to the repressed state, as speculated for the Sko1-Cyc8-Tup1 repression complex (48).
A loss of Rpd3 results in a reduced level of Mot3 (Fig. 3A). We currently have no explanation for this finding and do not know whether it is a direct or indirect effect. The drastic decreases in Mot3 protein levels observed in rpd3Δ strains in anaerobic cultures were probably the result of two distinct inhibitory mechanisms affecting Mot3 expression, namely, a lack of oxygen and a lack of Rpd3. We previously demonstrated that oxygen is required for MOT3 expression (60), and in this study, we show a reduction in the Mot3 protein level in the absence of oxygen (Fig. 3, compare lanes 2 and 4). In the absence of both oxygen and Rpd3, these two inhibitory mechanisms function synergistically to efficiently decrease Mot3 production (Fig. 3, compare lanes 2 and 4 with lane 5).
The strong requirement for the Swi/Snf components in DAN1 activation (Fig. 2C), combined with the decrease in histone density, suggests that the Swi/Snf complex may remove nucleosomes in trans from this promoter. Nucleosome disruption upon activation has also been observed at other yeast promoters, but those found previously to undergo nucleosome depletion upon activation are not as strongly Swi/Snf dependent as DAN1 (8, 33, 38, 40, 50, 58). Thus, DAN1 appears to represent the best candidate identified so far for having nucleosome depletion catalyzed by the Swi/Snf complex in vivo. Other promoters (SUC2 and PHO8) undergo Swi/Snf-dependent chromatin remodeling upon activation, but histone loss has not been shown to occur at these promoters (20, 22). We also found that Swi/Snf is needed to antagonize the repressive effects of Mot3 and Rox1 (data not shown). This may reflect a general requirement for Swi/Snf at genes under the control of global repressors (for example, SUC2, INO1, and RNR3) (22, 47, 61).
One question that arises is how Rpd3, which deacetylates histones, and the Swi/Snf remodeling complex can both be required for DAN1 activation in light of evidence that the Swi/Snf complex is recruited to acetylated histones under inducing conditions (3, 21, 41, 64). One possibility is that deacetylation of histones or other factors by Rpd3 may act as a platform for efficient binding by the activator Upc2, which then recruits the Swi/Snf complex to trigger nucleosomal eviction. Such roles for acetylation and deacetylation and other posttranslational modifications in the recruitment of activators have been proposed before (6, 35, 63).
Thus, at the DAN1 promoter, instead of organizing repressive chromatin, Rpd3 is essential for events that lead to dismantling of nucleosomes to facilitate binding of the transcriptional machinery. Based on these results, we propose that Rpd3 is recruited to the DAN1 promoter under strict anaerobic conditions. The presence of Rpd3 at the promoter counteracts the function of the repressor Mot3, which leads to stable binding of the activator Upc2. Upc2 then recruits the chromatin remodeling complex Swi/Snf to reorganize chromatin, thereby facilitating the binding of the transcriptional machinery, which results in the activation of gene expression.
The participation of the Rpd3 complex in the expression of the anaerobic genes reported here is of special interest for two main reasons. First, the fact that it is recruited during anaerobiosis clearly indicates that the response of the anaerobic genes to oxygen depletion is regulated by chromatin. Second, the paradoxical role of an HDAC complex in activation provides new insight into the relationship between histone modifications and gene regulation.
Acknowledgments
We thank Kevin Struhl, Fred Winston, Francesc Posas, Steve Hanes, and Masayasu Nomura for plasmids and yeast strains and Richard Zitomer for a critical reading of the manuscript.
This work was supported by NSF grant MCB0114040 to C.V.L. and by Ruth L. Kirschstein National Research Service Award (NRSA) fellowship F31GM072113 to O.S.
Footnotes
Published ahead of print on 8 January 2007.
We dedicate this paper to our friend and mentor Charles V. Lowry, who is greatly missed.
REFERENCES
- 1.Abramova, N., O. Sertil, S. Mehta, and C. V. Lowry. 2001. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 183:2881-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abramova, N. E., B. D. Cohen, O. Sertil, R. Kapoor, K. J. Davies, and C. V. Lowry. 2001. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics 157:1169-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 4.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arevalo-Rodriguez, M., M. E. Cardenas, X. Wu, S. D. Hanes, and J. Heitman. 2000. Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase. EMBO J. 19:3739-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 9.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. [DOI] [PubMed] [Google Scholar]
- 10.Bone, J. R., and S. Y. Roth. 2001. Recruitment of the yeast Tup1p-Ssn6p repressor is associated with localized decreases in histone acetylation. J. Biol. Chem. 276:1808-1813. [DOI] [PubMed] [Google Scholar]
- 11.Carrozza, M. J., L. Florens, S. K. Swanson, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 1731:77-87. [DOI] [PubMed] [Google Scholar]
- 12.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581-592. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, B. D., O. Sertil, N. E. Abramova, K. J. Davies, and C. V. Lowry. 2001. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 29:799-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conlan, R. S., N. Gounalaki, P. Hatzis, and D. Tzamarias. 1999. The Tup1-Cyc8 protein complex can shift from a transcriptional co-repressor to a transcriptional co-activator. J. Biol. Chem. 274:205-210. [DOI] [PubMed] [Google Scholar]
- 15.Cooper, J. P., S. Y. Roth, and R. T. Simpson. 1994. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 8:1400-1410. [DOI] [PubMed] [Google Scholar]
- 16.De Nadal, E., M. Zapater, P. M. Alepuz, L. Sumoy, G. Mas, and F. Posas. 2004. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427:370-374. [DOI] [PubMed] [Google Scholar]
- 17.De Rubertis, F., D. Kadosh, S. Henchoz, D. Pauli, G. Reuter, K. Struhl, and P. Spierer. 1996. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384:589-591. [DOI] [PubMed] [Google Scholar]
- 18.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 20.Gregory, P. D., A. Schmid, M. Zavari, M. Munsterkotter, and W. Horz. 1999. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 18:6407-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 22.Hirschhorn, J. N., S. A. Brown, C. D. Clark, and F. Winston. 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 6:2288-2298. [DOI] [PubMed] [Google Scholar]
- 23.Hongay, C., N. Jia, M. Bard, and F. Winston. 2002. Mot3 is a transcriptional repressor of ergosterol biosynthetic genes and is required for normal vacuolar function in Saccharomyces cerevisiae. EMBO J. 21:4114-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 25.Kadosh, D., and K. Struhl. 1998. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kastaniotis, A. J., T. A. Mennella, C. Konrad, A. M. Torres, and R. S. Zitomer. 2000. Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast. Mol. Cell. Biol. 20:7088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato, H., S. Tamamizu-Kato, and F. Shibasaki. 2004. Histone deacetylase 7 associates with hypoxia-inducible factor 1alpha and increases transcriptional activity. J. Biol. Chem. 279:41966-41974. [DOI] [PubMed] [Google Scholar]
- 29.Keener, J., J. A. Dodd, D. Lalo, and M. Nomura. 1997. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl. Acad. Sci. USA 94:13458-13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kemp, M. G., M. Ghosh, G. Liu, and M. Leffak. 2005. The histone deacetylase inhibitor trichostatin A alters the pattern of DNA replication origin activity in human cells. Nucleic Acids Res. 33:325-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, N. J. Thompson, C. Boone, A. Emili, J. S. Weissman, T. R. Hughes, B. D. Strahl, M. Grunstein, J. F. Greenblatt, S. Buratowski, and N. J. Krogan. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593-605. [DOI] [PubMed] [Google Scholar]
- 32.Kook, S. H., Y. O. Son, S. K. Han, H. S. Lee, B. T. Kim, Y. S. Jang, K. C. Choi, K. S. Lee, S. S. Kim, J. Y. Lim, Y. M. Jeon, J. G. Kim, and J. C. Lee. 2005. Epstein-Barr virus-infected Akata cells are sensitive to histone deacetylase inhibitor TSA-provoked apoptosis. J. Biochem. Mol. Biol. 38:755-762. [DOI] [PubMed] [Google Scholar]
- 33.Korber, P., T. Luckenbach, D. Blaschke, and W. Horz. 2004. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol. Cell. Biol. 24:10965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 35.Kurdistani, S. K., and M. Grunstein. 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4:276-284. [DOI] [PubMed] [Google Scholar]
- 36.Kurdistani, S. K., D. Robyr, S. Tavazoie, and M. Grunstein. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31:248-254. [DOI] [PubMed] [Google Scholar]
- 37.Lai, L. C., A. L. Kosorukoff, P. V. Burke, and K. E. Kwast. 2005. Dynamical remodeling of the transcriptome during short-term anaerobiosis in Saccharomyces cerevisiae: differential response and role of Msn2 and/or Msn4 and other factors in galactose and glucose media. Mol. Cell. Biol. 25:4075-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 39.Li, B., and J. C. Reese. 2001. Ssn6-Tup1 regulates RNR3 by positioning nucleosomes and affecting the chromatin structure at the upstream repression sequence. J. Biol. Chem. 276:33788-33797. [DOI] [PubMed] [Google Scholar]
- 40.Lohr, D. 1997. Nucleosome transactions on the promoters of the yeast GAL and PHO genes. J. Biol. Chem. 272:26795-26798. [DOI] [PubMed] [Google Scholar]
- 41.Lomvardas, S., and D. Thanos. 2002. Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110:261-271. [DOI] [PubMed] [Google Scholar]
- 42.Madison, J. M., A. M. Dudley, and F. Winston. 1998. Identification and analysis of Mot3, a zinc finger protein that binds to the retrotransposon Ty long terminal repeat (delta) in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1879-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann, R. K., and M. Grunstein. 1992. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 11:3297-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mennella, T. A., L. G. Klinkenberg, and R. S. Zitomer. 2003. Recruitment of Tup1-Ssn6 by yeast hypoxic genes and chromatin-independent exclusion of TATA binding protein. Eukaryot. Cell 2:1288-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mou, Z., A. E. Kenny, and M. J. Curcio. 2006. Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics 172:2157-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papamichos-Chronakis, M., T. Gligoris, and D. Tzamarias. 2004. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 5:368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson, C. L., W. Kruger, and I. Herskowitz. 1991. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell 64:1135-1143. [DOI] [PubMed] [Google Scholar]
- 48.Proft, M., and K. Struhl. 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9:1307-1317. [DOI] [PubMed] [Google Scholar]
- 49.Qiu, Y., Y. Zhao, M. Becker, S. John, B. S. Parekh, S. Huang, A. Hendarwanto, E. D. Martinez, Y. Chen, H. Lu, N. L. Adkins, D. A. Stavreva, M. Wiench, P. T. Georgel, R. L. Schiltz, and G. L. Hager. 2006. HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol. Cell 22:669-679. [DOI] [PubMed] [Google Scholar]
- 50.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 51.Robert, F., D. K. Pokholok, N. M. Hannett, N. J. Rinaldi, M. Chandy, A. Rolfe, J. L. Workman, D. K. Gifford, and R. A. Young. 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16:199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang, N. Suka, and M. Grunstein. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109:437-446. [DOI] [PubMed] [Google Scholar]
- 53.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93:14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 55.Sabet, N., F. Tong, J. P. Madigan, S. Volo, M. M. Smith, and R. H. Morse. 2003. Global and specific transcriptional repression by the histone H3 amino terminus in yeast. Proc. Natl. Acad. Sci. USA 100:4084-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sabet, N., S. Volo, C. Yu, J. P. Madigan, and R. H. Morse. 2004. Genome-wide analysis of the relationship between transcriptional regulation by Rpd3p and the histone H3 and H4 amino termini in budding yeast. Mol. Cell. Biol. 24:8823-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 58.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sertil, O., B. D. Cohen, K. J. Davies, and C. V. Lowry. 1997. The DAN1 gene of S. cerevisiae is regulated in parallel with the hypoxic genes, but by a different mechanism. Gene 192:199-205. [DOI] [PubMed] [Google Scholar]
- 60.Sertil, O., R. Kapoor, B. D. Cohen, N. Abramova, and C. V. Lowry. 2003. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 31:5831-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma, V. M., B. Li, and J. C. Reese. 2003. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev. 17:502-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, R. L., and A. D. Johnson. 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25:325-330. [DOI] [PubMed] [Google Scholar]
- 63.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 64.Syntichaki, P., I. Topalidou, and G. Thireos. 2000. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 404:414-417. [DOI] [PubMed] [Google Scholar]
- 65.Tai, S. L., V. M. Boer, P. Daran-Lapujade, M. C. Walsh, J. H. de Winde, J. M. Daran, and J. T. Pronk. 2005. Two-dimensional transcriptome analysis in chemostat cultures. Combinatorial effects of oxygen availability and macronutrient limitation in Saccharomyces cerevisiae. J. Biol. Chem. 280:437-447. [DOI] [PubMed] [Google Scholar]
- 66.Ter Linde, J. J., H. Liang, R. W. Davis, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 1999. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J. Bacteriol. 181:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thiagalingam, S., K. H. Cheng, H. J. Lee, N. Mineva, A. Thiagalingam, and J. F. Ponte. 2003. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann. N. Y. Acad. Sci. 983:84-100. [DOI] [PubMed] [Google Scholar]
- 68.Thompson, J. S., X. Ling, and M. Grunstein. 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369:245-247. [DOI] [PubMed] [Google Scholar]
- 69.Uchida, H., T. Maruyama, T. Nagashima, H. Asada, and Y. Yoshimura. 2005. Histone deacetylase inhibitors induce differentiation of human endometrial adenocarcinoma cells through up-regulation of glycodelin. Endocrinology 146:5365-5373. [DOI] [PubMed] [Google Scholar]
- 70.Vannier, D., D. Balderes, and D. Shore. 1996. Evidence that the transcriptional regulators SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S. cerevisiae. Genetics 144:1343-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vik, A., and J. Rine. 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6395-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, A., S. K. Kurdistani, and M. Grunstein. 2002. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298:1412-1414. [DOI] [PubMed] [Google Scholar]
- 73.Washburn, B. K., and R. E. Esposito. 2001. Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast. Mol. Cell. Biol. 21:2057-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watson, A. D., D. G. Edmondson, J. R. Bone, Y. Mukai, Y. Yu, W. Du, D. J. Stillman, and S. Y. Roth. 2000. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 14:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilcox, L. J., D. A. Balderes, B. Wharton, A. H. Tinkelenberg, G. Rao, and S. L. Sturley. 2002. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277:32466-32472. [DOI] [PubMed] [Google Scholar]
- 76.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-Macdonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, Z., and J. C. Reese. 2004. Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279:39240-39250. [DOI] [PubMed] [Google Scholar]
- 78.Zitomer, R. S., J. W. Sellers, D. W. McCarter, G. A. Hastings, P. Wick, and C. V. Lowry. 1987. Elements involved in oxygen regulation of the Saccharomyces cerevisiae CYC7 gene. Mol. Cell. Biol. 7:2212-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]