Abstract
Hypoxia-inducible factor (HIF) is a heterodimeric transcription factor, consisting of an alpha subunit and a beta subunit, that controls cellular responses to hypoxia. HIFα contains two transcriptional activation domains called the N-terminal transactivation domain (NTAD) and the C-terminal transactivation domain (CTAD). HIFα is destabilized by prolyl hydroxylation catalyzed by EglN family members. In addition, CTAD function is inhibited by asparagine hydroxylation catalyzed by FIH1. Both hydroxylation reactions are linked to oxygen availability. The von Hippel-Lindau tumor suppressor protein (pVHL) is frequently mutated in kidney cancer and is part of the ubiquitin ligase complex that targets prolyl hydroxylated HIFα for destruction. Recent studies suggest that HIF2α plays an especially important role in promoting tumor formation by pVHL-defective renal carcinoma cells among the three HIFα paralogs. Here we dissected the relative contribution of the two HIF2α transactivation domains to hypoxic gene activation and renal carcinogenesis and investigated the regulation of the HIF2α CTAD by FIH1. We found that the HIF2α NTAD is capable of activating both artificial and naturally occurring HIF-responsive promoters in the absence of the CTAD. Moreover, we found that the HIF2α CTAD, in contrast to the HIF1α CTAD, is relatively resistant to the inhibitory effects of FIH1 under normoxic conditions and that, perhaps as a result, both the NTAD and CTAD cooperate to promote renal carcinogenesis in vivo.
HIF (hypoxia-inducible factor) is a master transcriptional regulator of hypoxia-inducible genes and is composed of the α subunit HIF1α, or one of its paralogs HIF2α or HIF3α, and a HIFβ subunit, such as HIF1β (also called the aryl hydrocarbon receptor nuclear translocator [ARNT]) (12, 14, 17, 20, 50, 53). Whereas HIF1β is constitutively present, the HIFα members are highly unstable except under low-oxygen conditions (hypoxia). Under hypoxic conditions, the HIFα subunits accumulate, bind to a HIFβ subunit, and transcriptionally activate hypoxia-inducible genes bearing canonical HIF DNA-binding sites called hypoxia-responsive elements (HRE). These genes include genes that control angiogenesis (such as vascular endothelial growth factor [VEGF]), energy metabolism (for example, the glucose transporter 1 [GLUT1] and carbonic anhydrase IX [CAIX]), erythropoiesis (such as erythropoietin [EPO]), and mitogenesis (for example, transforming growth factor α and platelet-derived growth factor B) (18, 49). In the presence of oxygen, HIFα proteins are hydroxylated on conserved prolyl residues by members of the egg-laying-defective nine (EglN) prolyl hydroxylases (also called PHD or HPH hydroxylases) (2, 13, 26-28, 55). Prolyl hydroxylation of HIFα creates a binding site for the von Hippel-Lindau (VHL) ubiquitin ligase complex, which then targets HIFα for polyubiquitination and proteasomal degradation (30). In cells lacking functional pVHL, HIFα is not degraded properly, leading to activation of HIF target genes. More than half of clear cell renal cell carcinomas (RCC) are pVHL-defective as a result of VHL mutations or hypermethylation of the VHL locus (30). Accordingly, overproduction of HIF, and its downstream targets, are hallmarks of this disease.
HIF2α, also known as EPAS1, HLF, HRF, or MOP2, shares 48% overall identity with HIF1α (12, 14, 20, 53). Both HIF1α and HIF2α are capable of regulating a variety of broadly expressed hypoxia-inducible genes (21). Recent studies suggest that HIF2α is more important for renal carcinogenesis than HIF1α (4, 33, 40, 41). For example, tumor suppression by pVHL in RCC can be overridden by an HIF2α variant that escapes control by pVHL but not by a similar HIF1α variant (33, 41). Conversely, elimination of HIF2α in VHL−/− RCC cells is sufficient to suppress their ability to form tumors in vivo (32, 56). Therefore, inhibition of HIF2α is both necessary and sufficient for pVHL to suppress tumor formation by RCC. The importance of HIF2α is also suggested by careful analysis of preneoplastic lesions arising from the kidneys of patients who carry germ line VHL mutations (VHL disease) (40). In this setting, there is an apparent switch from HIF1α to HIF2α in association with increased cellular atypia.
HIFα contains two transcriptional activation domains: the N-terminal transactivation domain (NTAD) and the C-terminal transactivation domain (CTAD) (11, 29, 42, 43, 46, 48). In addition to inducing HIFα stability, hypoxia promotes the interaction of the HIFα CTAD with coactivators such as p300/CBP and allows HIF to recruit a larger transcriptional apparatus to hypoxia responsive genes (8, 11). During normoxic conditions, this interaction between p300/CBP and HIFα CTAD is blocked when a conserved asparaginyl residue within the CTAD is hydroxylated by factor inhibiting HIF1 (FIH1) (19, 37, 39). This modification inhibits HIFα CTAD activity by sterically blocking the interaction between HIFα and p300/CBP (5, 6, 9, 15, 19, 36, 38). Like the EglN prolyl hydroxylases, FIH1 is an Fe(II) and 2-oxoglutarate-dependent dioxygenase, and its activity can be blocked with competitive inhibitors of 2-oxoglutarate and with iron chelators (19, 36). Downregulation of FIH1 expression with small interfering RNA leads to activation of HIF target genes, while FIH1 overexpression leads to suppression of these genes (7, 51). In summary, HIF is regulated at the level of protein turnover by prolyl hydroxylation and at the level of coactivator recruitment by asparaginyl hydroxylation.
The role of HIF in tumor development is not restricted to VHL−/− cells. Most solid tumors contain hypoxic regions in which HIFα is stabilized and active, as determined by the induction of HIF target genes that promote survival in a hypoxia environment (18). In most but not all models, genetic disruption of HIF1α or its partner, ARNT1, leads to decreased tumor formation in vivo (44, 47, 52), as does blocking the interaction of HIFα with the coactivator proteins p300/CBP (34, 35).
The fact that HIF target genes are activated upon pVHL loss under normoxic conditions (24) creates a paradox if, however, the HIFα CTAD is inactivated by FIH1 in the presence of oxygen. Clearly, one explanation would be that activation of HIF target genes in this setting is solely due to the HIFα NTAD. In this regard, a naturally occurring HIF1α splice variant that encodes the NTAD, but not the CTAD, is capable of activating at least some HIF target genes (16). To address the relative contribution of HIF2α NTAD and CTAD to hypoxic gene activation and tumor growth, we created a panel of HIF2α mutants in which either the NTAD, the CTAD, or both were altered. Comparison of these mutants showed that the NTAD is indeed sufficient to transcriptionally activate a variety of HIF-responsive promoters. We also discovered that the HIF2α CTAD, in contrast to the HIF1α CTAD, is relatively resistant to FIH1 and cooperates with the HIF2α NTAD to promote tumor growth in vivo.
MATERIALS AND METHODS
Plasmids.
pcDNA3.0-HA-HIF2α P405A/P531A/N847A was generated from pcDNA3.0-HA-HIF2α P405A/P531A plasmid (32) by site-directed mutagenesis using sense primer A (GACTGTGAGGTGGCCGTGCCCGTGCTGG) and the QuikChange site-directed mutagenesis kit (Stratagene). The HIF2α P405A/P531A/N847A cDNA was excised by digestion with BamHI and MfeI and ligated into the pBABE-puro-HA vector (33) cut with BamHI and EcoRI to make pBABE-puro-HA-HIF2α P405A/P531A/N847A.
pBABE-puro-HA-HIF2α P405A/Δ450-572 and pBABE-puro-HA-HIF2α P405A/Δ450-572/N847A were generated by two-step PCR. pcDNA3.0-HA-HIF2α P405A/P531A and pcDNA3.0-HA-HIF2α P405A/P531A/N847A were first amplified with primer B (ATTGACGCAAATGGGCGGTAG) and primer C (CTACAGGGGCCAGTGGGCTCTGGGTGCTGTGG) or primer D (CCACAGCACCCAGAGCCCACTGGCCCCTGTAG) and primer E (GGCAAACAACAGATGGCTGG). Aliquots of these two PCR mixtures were then mixed and amplified with primer B and primer E. The resulting PCR product was digested with BamHI and MfeI and ligated into pBABE-puro-HA vector cut with BamHI and EcoRI. Similarly, pBABE-puro-HA-HIF2α P405A/Δ450-572/Δ820-870 was generated by two-step PCR. pcDNA3.0-HA-HIF2α P405A/P531A was first amplified with primer B and primer C or primer D and primer F (CCGGGCAATTGTCACACCTTGTGGGCTGACGAC). Aliquots of these two PCR mixtures were then mixed and amplified with primer B and primer F. The resulting PCR product was digested with BamHI and MfeI and ligated into pBABE-puro-HA vector cut with BamHI and EcoRI.
pcDNA3.0-HA-HIF2α P405A/P531A was PCR amplified with primers B and F, digested with BamHI and MfeI, and ligated into pBABE-puro-HA vector cut with BamHI and EcoRI to make pBABE-puro-HA-HIF2α P405A/P531A/Δ820-870. pBABE-puro-HA-HIF2α bHLH*/P405A/P531A/Δ820-870 was generated by two-step PCR. pcDNA3.0-HA-HIF2α P405A/P531A was first amplified with primer B and primer G (CATAGAACACCTCCGTCTCAGCGCTAGCAGCGCAAGCCGCAGCATCCCGGGAC) or primer H (GTCCCGGGATGCTGCGGCTTGCGCTGCTAGCGCTGAGACGGAGGTGTTCTATG) and primer F. Aliquots of these two PCRs were then mixed and amplified with primer B and primer F. The resulting PCR product was digested with BamHI and MfeI and ligated into pBABE-puro-HA vector cut with BamHI and EcoRI.
pcDNA3.0-HA-HIF2α P405A/Δ450-572, pcDNA3.0-HA-HIF2α P405A/Δ450-572/N847A, pcDNA3.0-HA-HIF2α P405A/Δ450-572/Δ820-870, pcDNA3.0-HA-HIF2α P405A/P531A/Δ820-870, and pcDNA3.0-HA-HIF2α bHLH*/P405A/P531A/Δ820-870 were made by excising the cDNAs from the corresponding pBABE vectors with BamHI and BstXI and ligating them into pcDNA3-HA (33) cut with these two enzymes.
pcDNA3.0-HA-HIF2α P405A/P531A/N847Q was made by site-directed mutagenesis of pcDNA3.0-HA-HIF2α P405A/P531A (32) with sense primer I (GACTGTGAGGTGCAAGTGCCCGTGCTGG) using the QuikChange site-directed mutagenesis kit (Stratagene). pcDNA3.0-HA-HIF1α P402A/P564A/N803A and pcDNA3.0-HA-HIF1α P402A/P564A/N803Q were made from pcDNA3.0-HA-HIF1α P402A/P564A using the QuikChange site-directed mutagenesis kit (Stratagene) with the primers described previously (19). All plasmids were verified by DNA sequencing.
The pGL2-VEGF promoter (VEGF-Luc) plasmid was a kind gift from Deb Mukhopadhyay (Harvard Medical School). 3×HRE luciferase (3×HRE-Luc) plasmid (a kind gift from Andrew Kung, Dana-Farber Cancer Institute) was created by inserting 3 copies of the canonical hypoxia-response element from the EPO gene and a minimal thymidine kinase promoter into pGL3 (Promega).
Cell culture.
786-O and RCC4 renal carcinoma cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal clone I (HyClone) in a 37°C, 10% CO2 incubator. 786-O renal carcinoma cell subclones stably transfected with pRc-CMV-HA-VHL (WT7) (27) were grown in DMEM containing 10% fetal clone I (HyClone) containing 1 mg/ml G418 in a 37°C, 10% CO2 incubator. Retrovirally infected cells were selected and maintained in the presence of 1.5 μg/ml puromycin.
Phoenix cells (a generous gift of Gary Nolan, Department of Molecular Pharmacology, Stanford University) and U2OS cells were grown in DMEM containing 10% fetal bovine serum (HyClone) in a 37°C, 10% CO2 incubator.
Retroviruses.
Retroviral plasmids were transfected into the Phoenix packaging cell line using FuGene 6 reagent (Roche Molecular Biochemicals) according to the manufacturer's instructions. Tissue culture supernatant was harvested 48 h later, passed though a 0.45-μm filter, and added to cells in the presence of 4 μg/ml Polybrene.
Immunoblot analysis.
Cells were lysed in EBC lysis buffer (50 mM Tris [pH 8.0], 120 mM NaCl, 0.5% NP-40) supplemented with Complete protease inhibitor cocktail (Roche Molecular Biochemicals). Approximately 30 μg of cell extract per lane, as determined by the Bradford method, was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Bio-Rad). After blocking in Tris-buffered saline with 4% nonfat milk, the membranes were probed with antihemagglutinin (anti-HA) mouse monoclonal antibody (HA.11; Covance), anti-HIF2α mouse monoclonal antibody (NB100-132; Novus Biologicals), anti-GLUT1 rabbit polyclonal antibody (GT11-A; Alpha Diagnostic), anti-green fluorescent protein (anti-GFP) monoclonal antibody (BD Biosciences), antivinculin mouse monoclonal antibody (clone hVIN-1; Sigma-Aldrich), anti-α-tubulin mouse monoclonal antibody (clone B-5-1-2; Sigma-Aldrich), or anti-HIFα rabbit polyclonal antibody (a kind gift from Jacques Pouysségur) diluted in Tris-buffered saline with 4% nonfat milk. Bound antibody was detected with the appropriate horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G or goat anti-mouse immunoglobulin G (Pierce) and SuperSignal West Pico or Dura chemiluminescent substrate (Pierce) according to the manufacturer's instructions.
Luciferase assays.
For transcriptional transactivation experiments, U2OS or WT7 cells were transfected in six-well plates using FuGene 6 reagent (Roche Molecular Biochemicals). At 24 h after transfection, cells were lysed and luciferase activity was measured using the dual-luciferase reporter assay system (Promega). Relative luciferase units were determined by dividing the luciferase activity of the VEGF or 3×HRE reporters by that of the pRL-CMV Renilla luciferase control reporter.
Microarray experiments.
Total RNA was isolated using the RNeasy mini kit with on-column DNase digestion (QIAGEN) and processed as described previously (22) for hybridization to microarrays containing approximately 25,000 oligonucleotides. Ratio hybridizations were performed with fluorescent label reversal to eliminate dye bias. Microarrays were purchased from Agilent Technologies. The error models for data processing have been previously described (22). Data were analyzed using Rosetta Resolver version 6.0 software.
In vitro proliferation assays.
Cell proliferation was measured colorimetrically using cell proliferation kit II (Roche Diagnostics) according to the manufacturer's protocol. Briefly, 2,000 cells per well were cultured in 96-well cell culture plates. At the indicated time points, sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy-6-nitro) benzene sulfonic acid hydrate (XTT) was added to the cells. Four hours later, the spectrophotometric absorbance at 450-nm wavelength was measured using a microtiter plate reader.
Nude mouse xenograft assays.
Nude mouse xenograft assays were performed as described previously (33). In brief, cells were released by trypsinization and resuspended in phosphate-buffered saline. Viable cells (107), as determined by trypan blue staining, were injected subcutaneously into the flanks of Swiss nude mice. Both flanks were used for each mouse. The animals were sacrificed 5 to 6 weeks after injection. Tumors were weighed and fixed in formalin for immunohistochemical analysis.
Real-time reverse transcription-PCR.
Total RNA was isolated using the RNeasy mini kit with on-column DNase digestion (QIAGEN). First-strand cDNA was generated using the StrataScript First-Stand synthesis system (Stratagene). Real-time PCR was performed in triplicate using QuantiTect SYBR green PCR master mix (QIAGEN) and the Mx3000P QPCR system (Stratagene). All values were normalized to the level of β-actin mRNA abundance. Primers specific for actin were actinF (CTCTTCCAGCCTTCCTTCCT) and actinR (AGCACTGTGTTGGCGTACAG). Primers specific for EglN3 were EglN3F (TCTCCCGAGAGTTGCGAGAAA) and EglN3R (AGAGGGAACGATCTACACGAG) (54). Primers specific for VEGF were VEGFF (AAGGAGGAGGGCAGAATCAT) and VEGFR (CACACAGGATGGCTTGAAGA). Primers specific for Glut1 were as described previously (3). Primers specific for CITED were CITEDF (GGGCGAGCACATACACTACG) and CITEDR (ACCCATGAACTGGGAGTTGTTA) (54). Primers specific for BNIP3 were BNIP3F (CAGGGCTCCTGGGTAGAACT) and BNIP3R (CTCCGTCCAGACTCATGCTG) (54). Primers specific for ENO1 were ENO1F (CTGGTGCCGTTGAGAAGGG) and ENO1R (GGTTGTGGTAAACCTCTGCTC) (54). Primers specific for PGK1 were PGK1F (TTAAAGGGAAGCGGGTCGTTA) and PGK1R (TCCATTGTCCAAGCAGAATTTGA) (54).
RESULTS
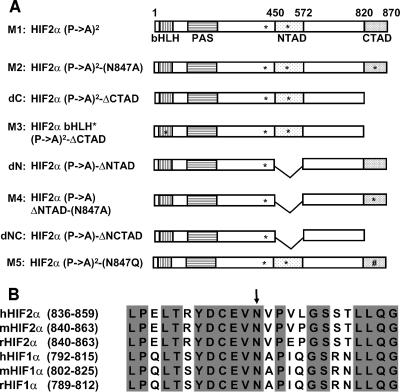
We showed before that an HIF2α variant in which both prolyl hydroxylation sites (proline 405 and 531) were replaced by alanine (Fig. 1A, M1) is no longer recognized by pVHL and is therefore stable, yet it can still bind to DNA and activate transcription (32). Moreover, this variant can override pVHL's ability to inhibit tumor growth by VHL−/− RCC in vivo (32). We mutagenized the NTAD and CTAD within this HIF2α variant to explore the relevant contributions of these two domains to transcriptional activation and tumor promotion by HIF2α (Fig. 1A).
FIG. 1.
HIF2α mutants used in the study. (A) Schematic of the HA-HIF2α mutants used in this study. The bHLH, PAS, NTAD, and CTAD domains are indicated by boxes. The bHLH mutation, Pro405-to-Ala, Pro531-to-Ala, and Asp847-to-Ala substitutions are indicated by asterisks in the corresponding regions. The Asp847-to-Gln substitution is indicated (#) in the corresponding region. (B) Alignment of the C-terminal regions of HIF1α and HIF2α from human (h), mouse (m), and rat (r). The fully conserved amino acids are indicated in gray, and the Asn hydroxylation site is indicated by the arrow.
The HIF1α CTAD can be hydroxylated on asparagine 803 by FIH1. The region surrounding asparagine 803 is highly conserved across species and between HIF1α and HIF2α (Fig. 1B). Substitution of the corresponding asparagine of HIF2α in the context of a Gal4-HIF2α CTAD fusion protein leads to enhanced transcriptional activity that is no longer sensitive to changes in oxygen availability (36, 37). We created two HIF2α mutants in which the HIF2α CTAD was altered. One mutant lacked the CTAD because of a premature stop codon (Fig. 1A, dC). In the other mutant, Asn847 was converted to Ala (Fig. 1A, M2), which we predicted would lead to enhanced transcriptional activation capability. As an additional control, we introduced a 4-amino-acid loss-of-function mutation in the basic helix-loop-helix (bHLH) DNA binding domain of the HIF2α dC mutant (Fig. 1A, M3).
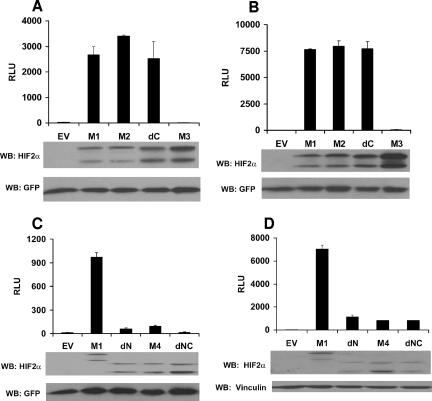
The HIF2α CTAD mutants were next introduced into VHL+/+ U2OS osteosarcoma cells by transient transfection in the presence of a luciferase reporter plasmid containing either the VEGF promoter (Fig. 2A) or a minimal artificial promoter containing 3 copies of the HIF-response element from the EPO gene (Fig. 2B). Surprisingly, neither deletion of the CTAD nor replacement of Asn847 with alanine had a marked effect on HIF2α-dependent transcriptional activation function. The similarities between the M1, M2, and dC mutants were seen over a wide range of plasmid input concentrations (data not shown; see Fig. 7B). Transcriptional activation by the dC mutant was dependent upon DNA-binding capability because it was abolished by the HIF2α bHLH mutation (Fig. 2A and B, M3). All of the mutants were produced at comparable levels, as determined by Western blot analysis. This suggested that the HIF2α NTAD is sufficient, and the HIF2α CTAD unnecessary, for transcriptional activation by HIF2α.
FIG. 2.
Transcriptional activation by the HIF2α mutants. Normalized firefly luciferase values of U2OS cells transiently transfected to produce the indicated HIF2α variants along with a reporter plasmid containing firefly luciferase under the control of the VEGF promoter (VEGF-Luc) (A and C) or 3 tandem HIF-response elements and a minimal promoter (3×HRE-Luc) (B and D). Transfection mixes also contained plasmids encoding Renilla luciferase and, in some experiments, GFP for normalization purposes. Data are the averages of results from duplicate experiments ± standard errors. Production of the HIF2α mutants and GFP was assessed by Western blot (WB) analysis (lower panel). EV, empty vector control; RLU, relative luciferase unit.
FIG. 7.
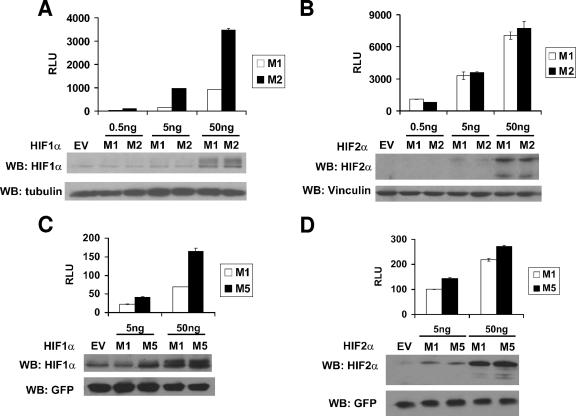
Transcriptional activation by the HIF1α and HIF2α mutants. Normalized firefly luciferase values of U2OS cells transiently transfected to produce the indicated HIF1α (A and C) or HIF2α (B and D) variants along with a reporter plasmid containing firefly luciferase under the control of 3 tandem HIF-response elements upstream of a minimal promoter. Transfection mixes also contained plasmids encoding Renilla luciferase and, in some experiments, GFP for normalization purposes. Data are the averages of results from duplicate experiments ± standard errors. Production of the HIFα mutants, GFP, vinculin, and tubulin was assessed by Western blot (WB) analysis (lower panel). RLU, relative luciferase unit; EV, empty vector control.
To pursue this further, we generated an HIF2α variant in which the residues that are essential for the NTAD function were eliminated by an in-frame deletion (Fig. 1A, dN) (11, 42). The HIF2α dN mutant was impaired with respect to transcription activation of the VEGF (Fig. 2C) or 3×HRE reporters (Fig. 2D) relative to the M1 variant, suggesting that NTAD is necessary for transcriptional activation of HRE-regulated genes. A caveat in these experiments is that the NTAD deletion might affect the folding of the CTAD or its proper orientation once bound to DNA.
These results left open the possibility that the CTAD might substitute for the NTAD were it not for the repressive effects of asparagine hydroxylation. To address this, we introduced the Asn847Ala substitution into the dN mutant (Fig. 1A, M4). However, transcriptional activation by this mutant was not significantly enhanced relative to dN itself (Fig. 2C and D). Nonetheless, the HIF2α dN variant did activate reporters weakly. To determine whether CTAD contributes to that residual activity, we combined the NTAD deletion mutation with the CTAD truncation mutation (Fig. 1A, dNC). This mutant retained weak transcriptional activation capability on the 3×HRE reporter but not the VEGF promoter (Fig. 2C and D). Although the significance of this finding is not clear, it suggests the presence of a cryptic transcriptional activation domain within HIF2α, or perhaps ARNT, that can contribute to the activation of certain genes.
Reintroduction of wild-type pVHL into 786-O VHL−/− cells blocks their ability to form tumors in nude mice (23). In such cells (wild-type [WT] pVHL restored), introduction of an HIF2α M1 variant promotes tumor formation (32). Transcriptional activation by the HIF2α variants described above after introduction into WT7 cells by transient transfection was qualitatively similar to the results obtained with U2OS cells (Fig. 3A). In WT7 cells, the amount of HIF2α produced was below the limit of detection due to a lower transfection efficiency (data not shown).
FIG. 3.
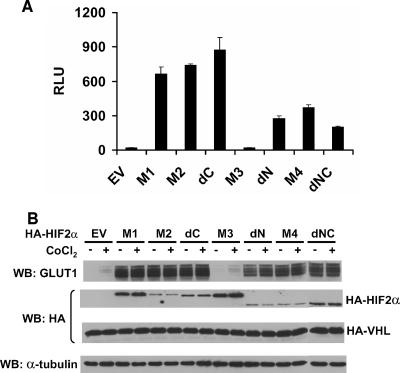
Behavior of HIF2α variants when introduced into VHL−/− renal carcinoma cells stably transfected to produce wild-type pVHL. (A) Normalized firefly luciferase values of WT7 cells transiently transfected to produce the indicated HIF2α variants along with a reporter plasmid containing firefly luciferase under the control of 3 tandem HIF-response elements upstream of a minimal promoter. Transfection mixes also contained plasmids encoding Renilla luciferase for normalization purposes. Data are the averages of results from duplicate experiments ± standard errors. RLU, relative luciferase unit. (B) Western blot (WB) analysis of WT7 cells after stable infection with retroviruses encoding the indicated HIF2α mutants. Where indicated, media contained 100 μM CoCl2 for 16 h prior to harvest. EV, empty vector control.
Next, WT7 cells were infected with retroviruses encoding the HIF2α variants or with the empty vector. Successfully infected cells were selected with puromycin and maintained as pools. In contrast to the U2OS transient-transfection assays, the levels of M2, dC, dN, and M4 were somewhat lower than that of M1 after stable infection of WT7 cells (Fig. 3B). This might, among several possibilities, reflect selection against cells producing higher levels of these variants. As expected, because of the proline-to-alanine substitutions, none of the variants was further induced by the hypoxia mimetic cobalt chloride, which inhibits EglN function (Fig. 3B). All of the variants, with the exception of the bHLH mutant M3, induced the expression of the endogenous HIF-responsive gene GLUT1.
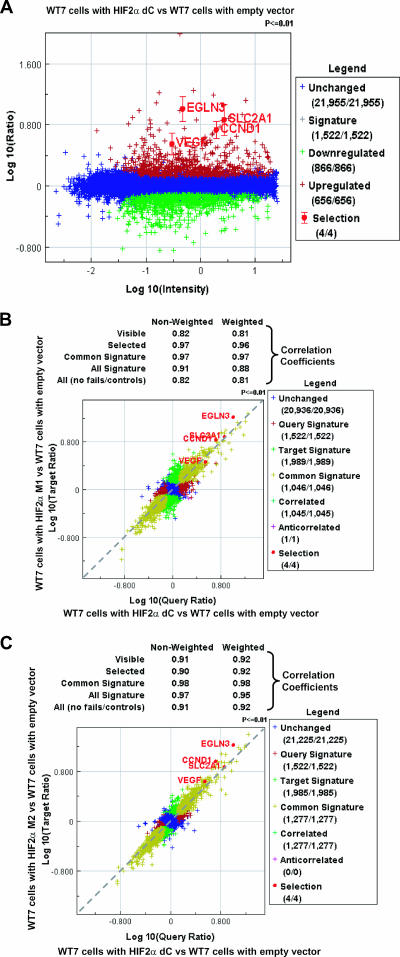
To look more globally at the behavior of endogenous HIF-responsive genes, we performed microarray experiments. In keeping with the experiments described above, the HIF2α dC variant, which lacks the CTAD, retained the ability to activate a wide variety of canonical HIF targets, including EglN3, VEGF, cyclin D1, and GLUT1 (SLC2A1), relative to cells infected with an empty retrovirus (Fig. 4A; also see Table S1 in the supplemental material). Remarkably, the induction of HIF targets by dC was highly similar to the induction observed with the M1 variant, which contains both the NTAD and CTAD (Fig. 4B; also see Table S1 in the supplemental material). This similarity was not merely due to neutralization of the M1 CTAD by FIH1 because the dC profile was also highly similar to the profile observed with M2, which lacks the FIH1 hydroxylation site (Fig. 4C; also see Table S1 in the supplemental material).
FIG. 4.
The NTAD is sufficient for HIF2α regulation of target gene expression. (A) The HIF2α dC variant up-regulates expression of canonical HIF target genes. RNA isolated from WT7 cells expressing HIF2α dC was compared to WT7 cells with the empty vector. Significantly up-regulated (red) or down-regulated (green) genes were determined as described previously (22). Canonical HIF target genes such as cyclin D1 (CCND1), GLUT1 (SLC2A1), VEGF, and EglN3 were highlighted. (B) The HIF2α dC and M1 variants up-regulate expression of canonical HIF target genes to similar levels. RNA isolated from WT7 cells expressing HIF2α dC or M1 variants were compared to WT7 cells with the empty vector, and these two data sets were compared to each other. Genes regulated in both the HIF2α dC and M1 experiments are yellow, those in the dC but not the M1 experiment are red, those in the M1 but not the dC experiment are green, and those in neither experiment are blue. Canonical HIF target genes such as cyclin D1 (CCND1), GLUT1 (SLC2A1), VEGF, and EglN3 were similarly up-regulated in HIF2α dC-expressing cells and M1-expressing cells. (C) HIF2α dC and M2 variants up-regulate expression of canonical HIF target genes to similar levels. RNA isolated from WT7 cells expressing HIF2α dC or M2 variants were compared to WT7 cells with the empty vector, and these two data sets were compared to each other. Genes regulated in both the dC experiment and the M2 experiment are yellow, those in the dC but not the M2 experiment are red, those in the M2 but not the dC experiment are green, and those in neither experiment are blue. Canonical HIF target genes such as cyclin D1 (CCND1), GLUT1 (SLC2A1), VEGF, and EglN3 were similarly up-regulated in HIF2α dC-expressing cells and M2-expressing cells.
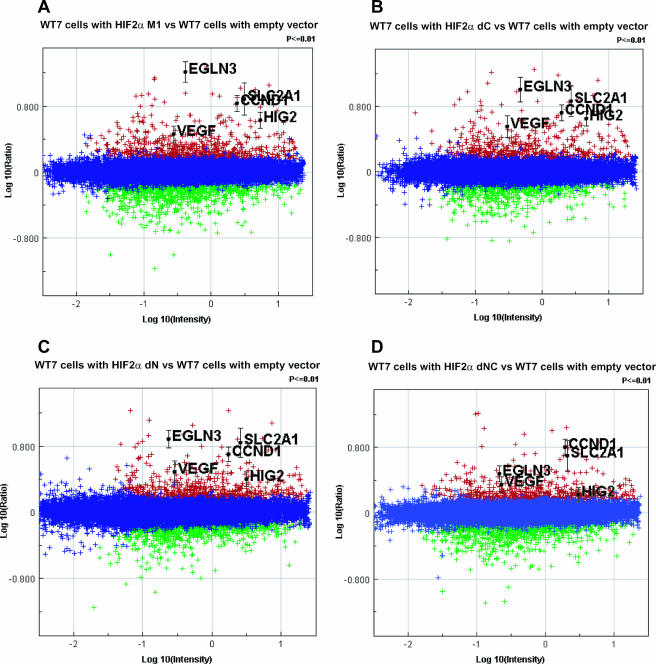
In keeping with the data shown in Fig. 3, the HIF2α dN and dNC variants, which lack the NTAD, also retained the ability to activate many HIF target genes (Fig. 5C and D; also see Fig. S1 and Table S2 in the supplemental material), although the dNC variant was clearly impaired relative to variants containing the NTAD (Fig. 5A and B, note that Fig. 4A is reproduced as Fig. 5B for comparative purposes) or the CTAD (Fig. 5C). Upon careful inspection, loss of either the NTAD or CTAD resulted in subtle quantitative differences in gene expression relative to M1, as exemplified by genes such as EglN3, cyclin D1, and HIG2 (also see Fig. S1 and Table S2 in the supplemental material).
FIG. 5.
Comparison of gene expression profiles of WT7 cells expressing various HIF2α mutants. RNA isolated from WT7 cells expressing HIF2α M1 (A), dC (B), dN (C), and dNC (D) variants were compared to WT7 cells with the empty vector. Significantly up-regulated (red) or down-regulated (green) genes were determined as described previously (22). Canonical HIF target genes, such as cyclin D1 (CCND1), GLUT1 (SLC2A1), VEGF, EglN3, and HIG2, are highlighted.
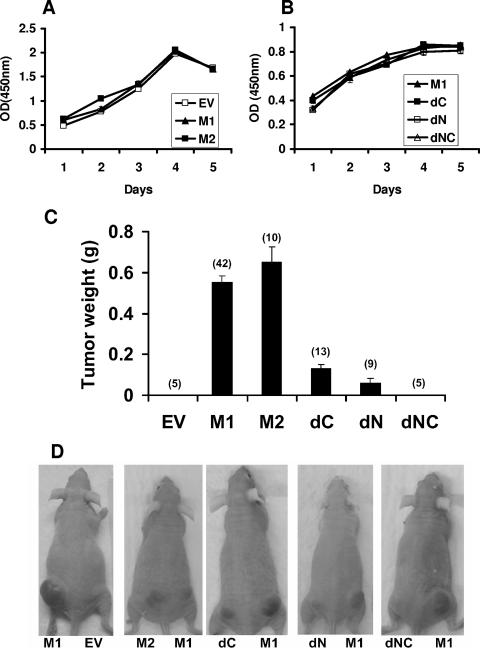
None of the HIF2α variants substantially altered the proliferation of WT7 cells in tissue culture in the presence of serum (Fig. 6A and B), in keeping with earlier results after introduction of M1 into WT8 cells (32). Also as expected, WT7 cells producing HIF2α M1 formed large tumors 5 to 6 weeks after subcutaneous injections in the flanks of nude mice, whereas WT7 cells infected with an empty retrovirus did not (Fig. 6C and D). The behavior of M2, lacking the Asn847 site, in these assays was similar to that of M1, in keeping with the results of our transcriptional assays. Surprisingly, deletion of either the NTAD or the CTAD dramatically reduced tumor forming capability and simultaneous deletion of both essentially eliminated tumor formation (Fig. 6C and D). The tumors formed by the cells expressing the different HIF2α variants were all very similar histologically and expressed similar numbers of vessels, as determined by CD34 staining (data not shown).
FIG. 6.
Proliferation of WT7 cells expressing various HIF2α mutants. (A and B) XTT assay of WT7 cells stably expressing the indicated HIF2α mutants after seeding in 96 well plates. Optical density (OD) at 450 nm values reflect the number of viable cells. Data are the averages of results from triplicate experiments ± standard errors (SE). (C) Tumor weights approximately 5 to 6 weeks after subcutaneous injection of WT7 cells stably expressing the indicated HIF2α mutants indicated in flanks of the nude mice. The number of tumors analyzed is indicated in parentheses. Error bars indicate ±SE. (D) Representative photographs of nude mice analyzed in panel C. EV, empty vector control.
The decreased oncogenicity of the dN and dC HIF2α variants presumably reflects the many small, quantitative, differences with respect to the induction of HIF target genes we documented by gene expression profiling (Fig. 5; also see Fig. S1 and Table S2 in the supplemental material). It is also possible that these differences in gene expression would be magnified under in vivo conditions, although this remains to be tested.
Several groups have shown that mutation of Asn803 in the HIF1α CTAD and the corresponding Asn in the HIF2α CTAD to Ala or Gln renders these transactivation domains, as GAL4 DNA-binding domain fusions, constitutively active and insensitive to oxygen (19, 36, 37). Furthermore, FIH1 inhibits some HIF transcriptional target genes, presumably through inactivation of HIFα CTAD by asparaginyl hydroxylation (7, 51). In our studies, however, replacement of Asn847 with Ala in the context of full-length HIF2α had no effect on HIF2α's ability to activate transcription (Fig. 2A and B) and promote tumor growth (Fig. 6C and D). To address this paradox, we created HIF1α variants analogous to HIF2α M1 and M2 and conducted HIF-responsive reporter gene assays in U2OS cells. Replacement of Asn803 with Ala in HIF1α, in contrast to the results observed for HIF2α, markedly enhanced transcriptional activation by HIF1α (Fig. 7A and B). Similar results were obtained when HIF1α Asn803 and HIF2α Asn847 were replaced with Gln, which more closely resembles Asn but cannot be hydroxylated by FIH1 (Fig. 1A, M5, and Fig. 7C and 7D). Therefore, HIF2α is relatively inured to the effect of FIH1 relative to HIF1α.
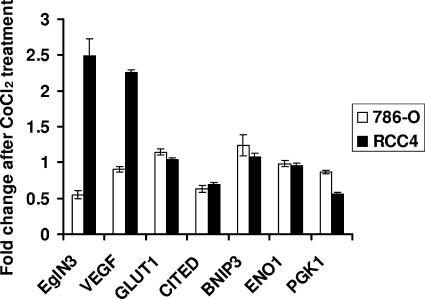
Dayan and coworkers recently reported that some HIF target genes, such as EglN3, VEGF, GLUT1, and CITED, are sensitive to changes in FIH1 activity, whereas others, such as BNIP3, ENO1, and PGK1, are not (7). Our findings predict that hypoxia and hypoxia mimetics should have little effect on FIH1-dependent gene expression in VHL−/− cells that exclusively produce HIF2α, in contrast to VHL−/− cells that produce both HIF1α and HIF2α. Indeed, the hypoxia mimetic CoCl2 did not lead to further induction of HIF target genes in 786-O cells, which exclusively produce HIF2α, but did induce the FIH1 targets EglN3 and VEGF in RCC4 cells, which produce both HIF1α and HIF2α (Fig. 8). The two other FIH1 targets examined, GLUT1 and CITED, were also not induced (Fig. 8), which might, among several possibilities, reflect differences between renal carcinoma cells and the colorectal cells employed by Dayan and colleagues. Although 786-O and RCC4 are not isogenic, these results are consistent with the idea that HIF2α is relatively inured to the effects of FIH1.
FIG. 8.
Induction of HIF target genes in 786-O and RCC4 cells by CoCl2 treatment. 786-O and RCC4 cells were grown in the presence or absence of 200 μM CoCl2 for 16 h. Expression levels of the indicated HIF target genes were determined by quantitative, real-time reverse transcription-PCR and normalized to β-actin mRNA levels. Shown are relative changes after CoCl2 treatment. Data are the averages of results from triplicate experiments ± standard errors.
DISCUSSION
In this study, we began to dissect the relative contribution of the two HIF2α transactivation domains to hypoxic gene activation and renal carcinogenesis. We created a panel of HIF2α mutants in which either the NTAD, CTAD, or both were altered. Comparison of these mutants showed that that the HIF2α NTAD is capable of activating HIF-responsive promoters in the absence of the CTAD. We also investigated the regulation of the HIFα CTAD by FIH1. Interestingly, we found that the HIF2α CTAD, in contrast to the HIF1α CTAD, is relatively resistant to the inhibitory effects of FIH1 under normoxic conditions and that, perhaps as a result, both the HIF2α NTAD and CTAD cooperate to promote renal carcinogenesis in vivo.
Our finding that the HIF2α NTAD is sufficient to activate at least some HIF-responsive genes is consistent with earlier work that showed that a naturally occurring HIF1α splice variant that lacks the HIF1α CTAD, but retains the HIF1α NTAD, is likewise able to activate transcription (16). Transcriptional activation by this HIF1α variant, however, appears to be impaired relative to full-length HIF1α. The same might be true of the corresponding HIF2α variant at lower protein concentrations than used here.
Previous studies showed that the HIF1α and HIF2α CTADs, as GAL4 DNA-binding domain fusions, can be hydroxylated by FIH1, which impairs their ability to activate transcription. Mutation of the relevant hydroxylation sites renders these GAL4 fusions constitutively active and insensitive to oxygen (19, 36, 37). We found, however, that the HIF2α CTAD is less sensitive to FIH1 modification than HIF1α CTAD in the context of their respective full-length proteins. This observation is consistent with a recent study that showed that HIF2α is a poor FIH1 substrate in vitro relative to HIF1α, in part because of differences in residues immediately following their respective Asn hydroxylation sites (1).
At least some HIF targets genes are activated upon pVHL inactivation, even under well-oxygenated conditions. The activation of these targets presumably, based on the considerations outlined above, reflects the fact that the HIFα NTAD is active under normoxic conditions coupled with the relative insensitivity of the HIF2α CTAD to inhibition by FIH1. The latter finding, coupled with the fact that HIF1α, but not HIF2α, is still subject to proteasomal degradation in pVHL-defective cells (25, 31), might explain why HIF2α plays a more prominent oncogenic role in VHL−/− renal carcinoma cells than does HIF1α.
The EglN family members and FIH1 are iron- and 2-oxoglutarate-dependent dioxygenases and can be inhibited with drug-like small molecules. Inhibition of these enzymes might be useful for the treatment of ischemic diseases and anemia. Theoretically, drugs that inhibited EglN alone would not induce genes that require the HIF1α CTAD for activation but would activate genes for which either the HIF1α or HIF2α NTAD is sufficient for activation. In addition, such drugs would induce genes activated by the HIF2α CTAD in cells that produce HIF2α, which is more restricted in its expression than HIF1α (12, 14, 20, 53). In summary, such small molecules might differentially regulate HIF targets depending on the degree to which they are regulated by HIF1α versus HIF2α and the degree to which they depend upon the NTAD, CTAD, or both for full activation. Support for this general idea comes from gain-of-function and loss-of-function studies involving EglN1 (PHD2), which is the primary HIF prolyl hydroxylase under resting conditions, and FIH1 (7, 51). For example, some HIF-responsive genes, such as EglN3 and VEGF, appear to be inhibited by FIH1, but others do not (7, 51). It is also increasingly clear that some genes are preferentially regulated by different HIFα proteins (3, 10, 21, 45).
Deletion of the HIF2α NTAD grossly impaired HIF2α-dependent transcription activation in our reporter gene assays, whereas deletion of the HIF2α CTAD did not. This raises the possibility that HIF2α-dependent transcription is largely driven by the NTAD. A caveat, however, is that the in-frame deletion of the NTAD we introduced might alter HIF2α folding in a way that alters, for example, the folding or presentation of the CTAD. This deletion mutant was not completely unfolded, however, because it retained at least partial transcriptional activation capability, as measured in reporter gene assays and by gene expression profiling. Perhaps more importantly, we found that deletion of either the NTAD or the CTAD impaired tumor promotion by HIF2α. This strongly argues that both the NTAD and the CTAD contribute to HIF2α-dependent transcription in the context of the full-length protein in vivo. Two, non-mutually exclusive, possibilities can explain this cooperativity. The NTAD and CTAD may act synergistically to activate a common set of genes that promote tumorigenesis in vivo. Here the difference between full-length HIF2α and the two deletion mutants would be largely quantitative. In addition, there may be some tumor-relevant genes that are uniquely activated by the NTAD and some by the CTAD. In this scenario, the difference between full-length HIF2α and the two deletion mutants would be qualitative. Our gene expression profiling studies are thus far most consistent with the former. A caveat, however, is that our gene expression studies were performed on cells grown under standard culture conditions. It remains possible that some tumor-relevant differences in HIF target gene expression will only manifest themselves under the conditions arising in tumors in vivo.
Supplementary Material
Acknowledgments
We thank members of the Kaelin laboratory for kind help and valuable discussions, Sabina Signoretti, Mirna Lechpammer, and the DF/HCC Specialized Histopathology Core for excellent help with the immunohistochemical studies.
W.G.K. is an Investigator of the Howard Hughes Medical Institute and is supported by the National Institutes of Health and the Murray Foundation.
Rosetta Inpharmatics, LLC., is a wholly owned subsidiary of Merck & Co.
Footnotes
Published ahead of print on 12 January 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bracken, C. P., A. O. Fedele, S. Linke, W. Balrak, K. Lisy, M. L. Whitelaw, and D. J. Peet. 2006. Cell-specific regulation of hypoxia-inducible factor (HIF)-1α and HIF-2α stabilization and transactivation in a graded oxygen environment. J. Biol. Chem. 281:22575-22585. [DOI] [PubMed] [Google Scholar]
- 2.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294:1337-1340. [DOI] [PubMed] [Google Scholar]
- 3.Carroll, V. A., and M. Ashcroft. 2006. Role of hypoxia-inducible factor (HIF)-1α versus HIF-2α in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-I, or loss of von Hippel-Lindau function: implications for targeting the HIF pathway. Cancer Res. 66:6264-6270. [DOI] [PubMed] [Google Scholar]
- 4.Covello, K. L., M. C. Simon, and B. Keith. 2005. Targeted replacement of hypoxia-inducible factor-1α by a hypoxia-inducible factor-2α knock-in allele promotes tumor growth. Cancer Res. 65:2277-2286. [DOI] [PubMed] [Google Scholar]
- 5.Dames, S. A., M. Martinez-Yamout, R. N. De Guzman, H. J. Dyson, and P. E. Wright. 2002. Structural basis for HIF-1α/CBP recognition in the cellular hypoxic response. Proc. Natl. Acad. Sci. USA 99:5271-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dann, C. E. III, R. K. Bruick, and J. Deisenhofer. 2002. Structure of factor-inhibiting hypoxia-inducible factor 1: an asparaginyl hydroxylase involved in the hypoxic response pathway. Proc. Natl. Acad. Sci. USA 99:15351-15356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dayan, F., D. Roux, M. C. Brahimi-Horn, J. Pouyssegur, and N. M. Mazure. 2006. The oxygen sensor factor-inhibiting hypoxia-inducible factor-1 controls expression of distinct genes through the bifunctional transcriptional character of hypoxia-inducible factor-1α. Cancer Res. 66:3688-3698. [DOI] [PubMed] [Google Scholar]
- 8.Ebert, B. L., and H. F. Bunn. 1998. Regulation of transcription by hypoxia requires a multiprotein complex that includes hypoxia-inducible factor 1, an adjacent transcription factor, and p300/CREB binding protein. Mol. Cell. Biol. 18:4089-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elkins, J. M., K. S. Hewitson, L. A. McNeill, J. F. Seibel, I. Schlemminger, C. W. Pugh, P. J. Ratcliffe, and C. J. Schofield. 2003. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1α. J. Biol. Chem. 278:1802-1806. [DOI] [PubMed] [Google Scholar]
- 10.Elvidge, G. P., L. Glenny, R. J. Appelhoff, P. J. Ratcliffe, J. Ragoussis, and J. M. Gleadle. 2006. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1α, HIF-2α, and other pathways. J. Biol. Chem. 281:15215-15226. [DOI] [PubMed] [Google Scholar]
- 11.Ema, M., K. Hirota, J. Mimura, H. Abe, J. Yodoi, K. Sogawa, L. Poellinger, and Y. Fujii-Kuriyama. 1999. Molecular mechanisms of transcription activation by HLF and HIF1α in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 18:1905-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ema, M., S. Taya, N. Yokotani, K. Sogawa, Y. Matsuda, and Y. Fujii-Kuriyama. 1997. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. USA 94:4273-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43-54. [DOI] [PubMed] [Google Scholar]
- 14.Flamme, I., T. Frohlich, M. von Reutern, A. Kappel, A. Damert, and W. Risau. 1997. HRF, a putative basic helix-loop-helix-PAS-domain transcription factor is closely related to hypoxia-inducible factor-1α and developmentally expressed in blood vessels. Mech. Dev. 63:51-60. [DOI] [PubMed] [Google Scholar]
- 15.Freedman, S. J., Z. Y. Sun, F. Poy, A. L. Kung, D. M. Livingston, G. Wagner, and M. J. Eck. 2002. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1α. Proc. Natl. Acad. Sci. USA 99:5367-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gothie, E., D. E. Richard, E. Berra, G. Pages, and J. Pouyssegur. 2000. Identification of alternative spliced variants of human hypoxia-inducible factor-1α. J. Biol. Chem. 275:6922-6927. [DOI] [PubMed] [Google Scholar]
- 17.Gu, Y. Z., S. M. Moran, J. B. Hogenesch, L. Wartman, and C. A. Bradfield. 1998. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3α. Gene Expr. 7:205-213. [PMC free article] [PubMed] [Google Scholar]
- 18.Harris, A. L. 2002. Hypoxia-a key regulatory factor in tumour growth. Nat. Rev. Cancer 2:38-47. [DOI] [PubMed] [Google Scholar]
- 19.Hewitson, K. S., L. A. McNeill, M. V. Riordan, Y. M. Tian, A. N. Bullock, R. W. Welford, J. M. Elkins, N. J. Oldham, S. Bhattacharya, J. M. Gleadle, P. J. Ratcliffe, C. W. Pugh, and C. J. Schofield. 2002. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 277:26351-26355. [DOI] [PubMed] [Google Scholar]
- 20.Hogenesch, J. B., W. K. Chan, V. H. Jackiw, R. C. Brown, Y. Z. Gu, M. Pray-Grant, G. H. Perdew, and C. A. Bradfield. 1997. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J. Biol. Chem. 272:8581-8593. [DOI] [PubMed] [Google Scholar]
- 21.Hu, C. J., L. Y. Wang, L. A. Chodosh, B. Keith, and M. C. Simon. 2003. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol. Cell. Biol. 23:9361-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 23.Iliopoulos, O., A. Kibel, S. Gray, and W. G. Kaelin, Jr. 1995. Tumour suppression by the human von Hippel-Lindau gene product. Nat. Med. 1:822-826. [DOI] [PubMed] [Google Scholar]
- 24.Iliopoulos, O., A. P. Levy, C. Jiang, W. G. Kaelin, Jr., and M. A. Goldberg. 1996. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc. Natl. Acad. Sci. USA 93:10595-10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isaacs, J. S., Y. J. Jung, and L. Neckers. 2004. Aryl hydrocarbon nuclear translocator (ARNT) promotes oxygen-independent stabilization of hypoxia-inducible factor-1α by modulating an Hsp90-dependent regulatory pathway. J. Biol. Chem. 279:16128-16135. [DOI] [PubMed] [Google Scholar]
- 26.Ivan, M., T. Haberberger, D. C. Gervasi, K. S. Michelson, V. Gunzler, K. Kondo, H. Yang, I. Sorokina, R. C. Conaway, J. W. Conaway, and W. G. Kaelin, Jr. 2002. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl. Acad. Sci. USA 99:13459-13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 28.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, B. H., J. Z. Zheng, S. W. Leung, R. Roe, and G. L. Semenza. 1997. Transactivation and inhibitory domains of hypoxia-inducible factor 1α. Modulation of transcriptional activity by oxygen tension. J. Biol. Chem. 272:19253-19260. [DOI] [PubMed] [Google Scholar]
- 30.Kaelin, W. G., Jr. 2002. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2:673-682. [DOI] [PubMed] [Google Scholar]
- 31.Kim, W. Y., M. Safran, M. R. Buckley, B. L. Ebert, J. Glickman, M. Bosenberg, M. Regan, and W. G. Kaelin, Jr. 2006. Failure to prolyl hydroxylate hypoxia-inducible factor α phenocopies VHL inactivation in vivo. EMBO J. 25:4650-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kondo, K., W. Y. Kim, M. Lechpammer, and W. G. Kaelin, Jr. 2003. Inhibition of HIF2α is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1:E83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondo, K., J. Klco, E. Nakamura, M. Lechpammer, and W. G. Kaelin, Jr. 2002. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell 1:237-246. [DOI] [PubMed] [Google Scholar]
- 34.Kung, A. L., S. Wang, J. M. Klco, W. G. Kaelin, and D. M. Livingston. 2000. Suppression of tumor growth through disruption of hypoxia-inducible transcription. Nat. Med. 6:1335-1340. [DOI] [PubMed] [Google Scholar]
- 35.Kung, A. L., S. D. Zabludoff, D. S. France, S. J. Freedman, E. A. Tanner, A. Vieira, S. Cornell-Kennon, J. Lee, B. Wang, J. Wang, K. Memmert, H. U. Naegeli, F. Petersen, M. J. Eck, K. W. Bair, A. W. Wood, and D. M. Livingston. 2004. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell 6:33-43. [DOI] [PubMed] [Google Scholar]
- 36.Lando, D., D. J. Peet, J. J. Gorman, D. A. Whelan, M. L. Whitelaw, and R. K. Bruick. 2002. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lando, D., D. J. Peet, D. A. Whelan, J. J. Gorman, and M. L. Whitelaw. 2002. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295:858-861. [DOI] [PubMed] [Google Scholar]
- 38.Lee, C., S. J. Kim, D. G. Jeong, S. M. Lee, and S. E. Ryu. 2003. Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau. J. Biol. Chem. 278:7558-7563. [DOI] [PubMed] [Google Scholar]
- 39.Mahon, P. C., K. Hirota, and G. L. Semenza. 2001. FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15:2675-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandriota, S. J., K. J. Turner, D. R. Davies, P. G. Murray, N. V. Morgan, H. M. Sowter, C. C. Wykoff, E. R. Maher, A. L. Harris, P. J. Ratcliffe, and P. H. Maxwell. 2002. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell 1:459-468. [DOI] [PubMed] [Google Scholar]
- 41.Maranchie, J. K., J. R. Vasselli, J. Riss, J. S. Bonifacino, W. M. Linehan, and R. D. Klausner. 2002. The contribution of VHL substrate binding and HIF1-α to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell 1:247-255. [DOI] [PubMed] [Google Scholar]
- 42.O'Rourke, J. F., Y. M. Tian, P. J. Ratcliffe, and C. W. Pugh. 1999. Oxygen-regulated and transactivating domains in endothelial PAS protein 1: comparison with hypoxia-inducible factor-1α. J. Biol. Chem. 274:2060-2071. [DOI] [PubMed] [Google Scholar]
- 43.Pugh, C. W., J. F. O'Rourke, M. Nagao, J. M. Gleadle, and P. J. Ratcliffe. 1997. Activation of hypoxia-inducible factor-1; definition of regulatory domains within the α subunit. J. Biol. Chem. 272:11205-11214. [DOI] [PubMed] [Google Scholar]
- 44.Rankin, E. B., D. F. Higgins, J. A. Walisser, R. S. Johnson, C. A. Bradfield, and V. H. Haase. 2005. Inactivation of the arylhydrocarbon receptor nuclear translocator (Arnt) suppresses von Hippel-Lindau disease-associated vascular tumors in mice. Mol. Cell. Biol. 25:3163-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raval, R. R., K. W. Lau, M. G. Tran, H. M. Sowter, S. J. Mandriota, J. L. Li, C. W. Pugh, P. H. Maxwell, A. L. Harris, and P. J. Ratcliffe. 2005. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell. Biol. 25:5675-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruas, J. L., L. Poellinger, and T. Pereira. 2002. Functional analysis of hypoxia-inducible factor-1 α-mediated transactivation. Identification of amino acid residues critical for transcriptional activation and/or interaction with CREB-binding protein. J. Biol. Chem. 277:38723-38730. [DOI] [PubMed] [Google Scholar]
- 47.Ryan, H. E., J. Lo, and R. S. Johnson. 1998. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 17:3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sang, N., J. Fang, V. Srinivas, I. Leshchinsky, and J. Caro. 2002. Carboxyl-terminal transactivation activity of hypoxia-inducible factor 1α is governed by a von Hippel-Lindau protein-independent, hydroxylation-regulated association with p300/CBP. Mol. Cell. Biol. 22:2984-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schofield, C. J., and P. J. Ratcliffe. 2004. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell. Biol. 5:343-354. [DOI] [PubMed] [Google Scholar]
- 50.Semenza, G. L. 1999. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15:551-578. [DOI] [PubMed] [Google Scholar]
- 51.Stolze, I. P., Y. M. Tian, R. J. Appelhoff, H. Turley, C. C. Wykoff, J. M. Gleadle, and P. J. Ratcliffe. 2004. Genetic analysis of the role of the asparaginyl hydroxylase factor inhibiting hypoxia-inducible factor (HIF) in regulating HIF transcriptional target genes. J. Biol. Chem. 279:42719-42725. [DOI] [PubMed] [Google Scholar]
- 52.Tang, N., L. Wang, J. Esko, F. J. Giordano, Y. Huang, H. P. Gerber, N. Ferrara, and R. S. Johnson. 2004. Loss of HIF-1α in endothelial cells disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis. Cancer Cell 6:485-495. [DOI] [PubMed] [Google Scholar]
- 53.Tian, H., S. L. McKnight, and D. W. Russell. 1997. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11:72-82. [DOI] [PubMed] [Google Scholar]
- 54.Wang, X., and B. Seed. 2003. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 31:e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, F., S. B. White, Q. Zhao, and F. S. Lee. 2001. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. USA 98:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zimmer, M., D. Doucette, N. Siddiqui, and O. Iliopoulos. 2004. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL−/− tumors. Mol. Cancer Res. 2:89-95. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.