Abstract
Myb family proteins are ubiquitously expressed transcription factors. In mammalian cells, they play a critical role in regulating the G1/S cell cycle transition but their role in regulating other cell cycle checkpoints is incompletely defined. Herein, we report experiments which demonstrate that c-Myb upregulates cyclin B1 expression in normal and malignant human hematopoietic cells. As a result, it contributes directly to G2/M cell cycle progression. In cell lines and primary cells, cyclin B1 levels varied directly with c-Myb expression. Chromatin immunoprecipitation assays, mutation analysis, and luciferase reporter assays revealed that c-Myb bound the cyclin B1 promoter preferentially at a site just downstream of the transcriptional start site. The biological significance of c-Myb, versus B-Myb, binding the cyclin B1 promoter was demonstrated by the fact that expression of inducible dominant negative c-Myb in K562 cells accelerated their exit from M phase. In addition, expression of c-Myb in HCT116 cells rescued cyclin B1 expression after B-myb expression was silenced with small interfering RNA. These results suggest that c-Myb protein plays a previously unappreciated role in the G2/M cell cycle transition of normal and malignant human hematopoietic cells and expands the known repertoire of c-myb functions in regulating human hematopoiesis.
The c-myb proto-oncogene is the normal cellular homologue of v-myb, the transforming oncogene of the avian myeloblastosis virus, and avian leukemia virus E26 (33). It is a member of a transcription factor family that consists of at least two other highly homologous proteins encoded by genes designated A-myb and B-myb (31). c-myb is expressed predominantly in hematopoietic tissues. A-myb is expressed preferentially in gonadal tissue and B lymphocytes (50), while B-myb is expressed ubiquitously (44). Located on chromosome 6p in humans, the predominant c-myb transcript encodes an ∼75-kDa nuclear binding protein composed of three distinct functional domains which are a DNA binding domain at the N terminus, a central transactivation domain, and a C-terminal negative regulatory domain (43).
c-Myb's preferential expression in hematopoietic cells suggests that it plays an important role in blood cell development, a supposition which has been proven correct. Silencing the c-myb gene with antisense oligodeoxynucleotides (13) or by homologous recombination (27) has shown that the protein is required for definitive hematopoiesis. In vitro, hematopoietic cells do not proliferate in the absence of c-Myb (13), and in vivo, loss of c-Myb is embryonic lethal due to failure of erythropoiesis (27). Other in vitro experiments have shown that constitutive overexpression of c-Myb blocks differentiation of myeloid and erythroid cells and the associated growth arrest that occurs with maturation (4, 7, 46). T-lymphocyte development is also c-Myb dependent, as shown by the use of antisense oligodeoxynucleotides (12), expression of dominant negative (DN) c-Myb protein (1, 35), and tissue-specific loss of floxed c-Myb in Cre-producing mouse strains (2, 10). Most recently, c-Myb has been demonstrated to play a regulatory role in megakaryocytopoiesis, thrombopoiesis (6, 26), and B-lymphocyte development (49). c-Myb target genes involved in these functions have been identified and include mim-1 (30, 40), c-kit (16, 41, 51), myeloblastin (23), and neuromedin U (47), as well as c-myc (45) and DNA topoisomerase IIa (5). Microarray and subtraction cloning methods have suggested that there are many other candidate c-Myb target genes, the expression of which is likely context dependent (21, 29).
The role of c-Myb in regulating the G1/S transition of human hematopoietic cells is well known (33), but its function, if any, in other phases of the hematopoietic cell cycle has not been well studied. This question has taken on greater interest in light of two recent papers which suggested that the Drosophila Myb and B-Myb proteins are involved in the regulation of cyclin B expression, a major regulator of G2/M cell cycle progression (34, 54). One study used the Drosophila eye imaginal disc model to show that Drosophila Myb upregulated cyclin B expression directly by binding its promoter (34). Another study, with human ganglioblastoma T98G cells, demonstrated that B-Myb, in conjunction with E2F, directly upregulated cdc2 and cyclin B1 expression. Of note with regard to this report, c-Myb was specifically investigated in the ganglioblastoma model and was found not to associate with E2F and therefore not to play a role in cyclin B1 expression or the G2/M cell cycle transition (54). These reports, and our own preliminary data which suggested that c-Myb might regulate cyclin B1 expression (28), prompted the investigations reported in this paper. In distinct contrast to the ganglioblastoma cell studies, we report herein that in normal and malignant human hematopoietic cells, c-Myb directly regulates cyclin B1 expression and, as a result, contributes directly to the regulation of the G2/M transition phase of the cell cycle in human blood cells.
MATERIALS AND METHODS
Cells and culture conditions.
K562 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin. K562 cells expressing either a conditionally responsive c-Myb (K562-c-Myb-ER) (45) or a conditionally active DN c-Myb (K562-MERT) (47) protein upon exposure to 1 μM 4-hydroxytamoxifen (4-OHT) have been previously described. Mo7e cells were maintained in Iscove's modified Dulbecco's medium supplemented with 10% FBS and 20 ng/ml granulocyte-macrophage colony-stimulating factor. HCT116 cells, a human colorectal carcinoma cell line, were maintained in McCoy's 5A medium with 10% FBS. Human CD34+ cells were isolated with CD34 MACS microbeads (Miltenyi Biotec, Auburn, CA) according to the manufacturer's protocol from bone marrow and peripheral blood collected from healthy consenting donors. Freshly isolated cells were cultured in RPMI 1640 medium supplemented with 10% FCS.
De novo protein synthesis was inhibited by culturing the cells in the presence of cycloheximide (Sigma-Aldrich, St. Louis, MO) at a concentration of 10 mg/ml for 6 h. Human peripheral blood T lymphocytes (PBL) were isolated from healthy donors by density gradient centrifugation with Ficoll-Paque Plus (GE Healthcare, Piscataway, NJ).
Chromatin immunoprecipitation (ChIP) assays.
Confluent cells were diluted 1:5 in fresh medium and cultured for an additional 8 to 12 h for K562 and Mo7e cells and 16 h for HCT116 cells prior to cross-linking with 0.4% formaldehyde. Following cross-linking, the cells were lysed, DNA-protein complexes were immunoprecipitated, and the formaldehyde-cross-linked DNA was reverse cross-linked with a ChIP assay kit (Upstate, Charlottesville, VA) according to the manufacturer's protocol. DNA-Myb complexes were immunoprecipitated with anti-c-Myb (clone 1-1; Upstate) and with anti-B-Myb (a mixture of N19 and H115; Santa Cruz Biotechnology). ChIP assays were also completed with anti-acetyl histone H4 as a positive control and with no chromatin, no antibody, or no anti-immunoglobulin G (IgG) as a negative control. The DNA isolated from the ChIP assay was purified with a QIAquick PCR purification kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions. The purified DNA was then amplified by real-time PCR by standard protocols with SYBR green and cyclin B1 promoter-specific primers that flanked both Myb binding sites, −118 to +85 bp relative to the transcription start site (forward, 5′-ATCGCCCTGGAAACGCATTCTCT-3′; reverse, 5′-AGAAGCAGAACACCGGAGGC-3′).
Dual-luciferase reporter assay.
A 992-bp fragment containing the human cyclin B1 promoter was amplified from normal human lymphocyte DNA by reverse transcription-PCR with cyclin B1-specific primers (forward, 5′-GCAAATGACAAAGCAAATGGGG-3′; reverse, 5′-ACAACCAGCAGAAACCAACAGC-3′). The amplified fragment was then ligated into the pGL3-basic vector at the KpnI and BglII sites (Promega). Cyclin B1 promoter constructs with mutated Myb binding sites were generated with the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) as instructed by the manufacturer. The pcDNA3 vector containing full-length c-myb (3 μg) or B-myb (3 μg) was cotransfected with phRL (0.02 μg) and pGL3-cyclin B1 promoter (1 μg) into the K562 cells with cell line nucleofector kit V (Amaxa Biosystems, Gaithersburg, MD) according to the manufacturer's protocol. In some assays, K562 cells were cotransfected with phRL (0.02 μg), pGL3-cyclin B1 promoter (1 μg), and small interfering RNA (siRNA; 4 μg). Posttransfection (48 h), cells were diluted 1:5 in fresh medium and cultured for an additional 6 to 8 h. The cells were then lysed and firefly and Renilla luciferase activities were measured as directed, with a dual-luciferase reporter assay kit (Promega, Madison, WI) and a luminometer (Monolight 2010; Analytical Luminescence Laboratory, San Diego, CA).
siRNA preparation and transfection.
Human c-myb siRNAs were generated as previously described (47), with 5′-UGUUAUUGCCAAGCACUUAAA-3′ and 5′-UAAGUGCUUGGCAAUAACAGAA-3′ for c-myb-1 and 5′-AAGCACUUAAAGGGGAGAAUU-3′ and 5′-UUCUCCCCUUUAAGUGCUUGG-3′ for c-myb-2. Human B-myb siRNA duplex 1 (D-010444-01), control siRNA duplexes, and siRNA pool were purchased from Dharmacon (Louisville, CO). Control, c-myb, and B-myb siRNAs (5 μg) were delivered to Mo7e cells, human T cells, and CD34+ cells by nucleofection (Amaxa Biosystems) (39). Transfection of the expression vector and siRNAs to HCT116 cells was done with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Western blot analysis.
Protein lysates (50 μg) were fractionated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred electrophoretically to polyvinylidene difluoride membranes, and incubated with anti-Myb (clone 1-1; Upstate), anti-B-Myb (N-19; Santa Cruz Biotechnology), anti-cdk1/cdc2 (Upstate), anti-cyclin B1 (Upstate), anti-cyclin A (clone BF683; Upstate), anti-p55 CDC (Santa Cruz Biotechnology), or anti-β-actin (clone AC-15; Sigma-Aldrich) by standard Western blotting procedures.
Quantitative analysis of c-myb, B-myb, cyclin B1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and 18S by real-time PCR.
Total RNA was extracted and isolated with the RNeasy mini kit with an RNase-free DNase set (QIAGEN) and reverse transcribed with an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). The cDNA was subsequently amplified in TaqMan Universal Master Mix (Perkin-Elmer Applied Biosystems, Foster City, CA) supplemented with gene-specific primers (0.5 μM) and the appropriate gene-specific TaqMan probe (0.1 or 0.2 μM). The PCR parameters set on the iCycler (Bio-Rad) consisted of 2 min at 50°C, 10 min at 95°C, and then 50 cycles each of 15 s at 92°C and 1 min at 60°C. All of the probes contained the reporter dye 6-carboxyfluorescein and the appropriate quencher. The sequences for the primers and probes were as follows: c-myb (forward primer, 5′-GAAGGTCGAACAGGAAGGTTATCT-3′; reverse primer, 5′-GTAACGCTACAGGGTATGGAACA-3′; probe, 5′-TCAAAAGCCAGCCAGCCAGCAGTG-3′), B-myb (forward primer, 5′-CAGAGCCCTTGGAGGAATT-3′; reverse primer, 5′-CAGGCTCGTTTCTGGTGG-3′; probe, 5′-CCTGGTCCTCACGCTTCGG-3′), cyclin B1 (forward primer, 5′-CGGGAAGTCACTGGAAACAT-3′; reverse primer, 5′-AAACATGGCAGTGACACCAA-3′; probe, 5′-AGGTTGTTGCAGGAGACCAT-3′), GAPDH (forward primer, 5′-GACAGTCAGCCGCATCTTCTT-3′; reverse primer, 5′-CCAATACGACCAAATCCGTTGAC-3′; probe, 5′-CGTCGCCAGCCGAGCCACATCG-3′), and 18S (forward primer, 5′-GGACATCTAAGGGCATCACAGACC-3′; reverse primer, 5′-TGACTCAACACGGGAAACCTCAC-3′; probe, 5′-TGGCTGAACGCCACTTGTCCCTCTAA-3′). To quantitate the amount of each gene in a given sample, the mean number of copies from a triplicate determination for each gene in the test cDNA was normalized to the mean number of copies of 18S or GAPDH.
Cell cycle analysis.
HCT116 cells were synchronized in the S phase of the cell cycle with aphidicolin (Biomol, Plymouth Meeting, PA) at 2 μg/ml for 24 h. The expression vector (1 μg) and siRNA (3 μg) were then transfected into the cells. K562-MERT cells were first arrested at the G1/S boundary by being subjected to two 24-h culture cycles in 2 mM thymidine (Sigma-Aldrich) with a 12-h thymidine-free interval between these cycles. After the second thymidine block, K562-MERT cells were released into the cell cycle for 3 h and subsequently arrested at the G2/M boundary by treatment with 40 ng/ml nocodazole (Biomol, Plymouth Meeting, PA) for 11 h. The K562-MERT cells were harvested at several different time points after release from the nocodazole block and addition of 4-OHT. Following treatment, the cells were fixed, washed with phosphate-buffered saline, incubated with 5 μg/ml DNase-free RNase (Roche Applied Science, Indianapolis, IN) for 30 min at 37°C, and stained with 40 μg/ml propidium iodide (Sigma-Aldrich, St. Louis, MO). The propidium iodide-stained cells were detected in a FACScan flow cytometer (Becton Dickinson, San Jose, CA) and analyzed with CellQuest software (Becton Dickinson).
Statistical analyses.
Statistical comparisons of the data were completed with Student's t test. The level of significance was set at P values of <0.05 in all cases.
RESULTS
c-Myb directly regulates cyclin B1 expression.
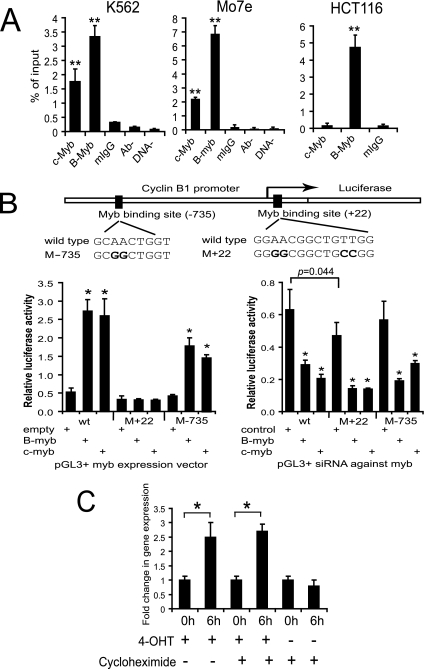
To determine if c-Myb transactivates the cyclin B1 gene directly, we used a ChIP strategy with extracts derived from K562, Mo7e, and HCT116 cells. As shown in Fig. 1A, ChIPs carried out with anti-c-Myb or anti-B-Myb antibody precipitated significant amounts of DNA product compared to the various control reactions used. These controls included ChIP assays performed under identical conditions except for (i) the use of anti-IgG in place of the Myb antibodies, (ii) the use of no input antibody (Ab−), and (iii) the use of no input DNA (DNA−). The specificity of the ChIP reactions was further demonstrated by the fact that that only a B-Myb product was detectable in HCT116 cells. These cells are nonhematopoietic, having been derived from a patient with colon cancer, and are not known to express c-Myb. The results presented in Fig. 1A are also of interest because they suggest that B-Myb is expressed at higher levels than c-Myb in all of the cell types examined and that both forms of the protein can bind the cyclin B1 promoter.
FIG. 1.
Myb directly activates cyclin B1 expression. (A) Purified nucleoprotein complex was obtained from K562, Mo7e, and HCT116 cells via ChIP assays with anti-c-Myb (c-Myb), anti-B-Myb (B-Myb), mouse IgG (mIgG), no antibody (Ab−), and no chromatin (DNA−). The precipitated DNA fractions were analyzed by real-time PCR with primers specific to the cyclin B1 promoter region. **, P < 0.01 compared to the value of mouse IgG. (B) At the top is a diagram of the Myb binding sites within 1,000 bp of the transcriptional start site for cyclin B1, which is represented by the bent arrow. The bold letters indicate the positions of the nucleotide changes in the mutant forms of the cyclin B1 promoter we generated. The two bottom bar graphs show the results of the dual-luciferase reporter assays. The assays were performed with K562 cells and the pGL3-cyclin B1 promoter vector in the presence of c-myb and B-myb expression constructs, a control empty vector, or siRNA against myb 54 to 56 h posttransfection. *, P < 0.05 compared to the value of the empty-vector or siRNA control. wt, wild type. (C) Real-time PCR for cyclin B1 expression in K562-c-MybER cells in the absence or presence of the protein synthesis inhibitor cycloheximide. RNA was analyzed for cyclin B1 expression at 0 and 6 h posttreatment with 4-OHT and/or cycloheximide. *, P < 0.05.
Sequence analysis of the cyclin B1 promoter revealed eight potential Myb binding sites within 1,000 bp of the transcriptional start site. Two of the eight sites were canonical, 5′-pyrimidine AAC (G/T) G-3′ (3). The remaining six sites were noncanonical, 5′-AACNG-3′ (32). Accordingly, we next sought to determine which of these sites c-Myb and B-Myb might bind to and the effect of such binding on cyclin B1 expression. To inform, and make more efficient, our search of likely regulatory elements, we used the GALA database (http://gala.cse.psu.edu). Two of the possible eight Myb binding sites we identified by simple sequence analysis were predicted by the GALA software to be likely candidates for Myb binding. These sites were designated +22 and −735, respectively, on the basis of their locations with reference to the transcriptional start site (Fig. 1B). To investigate the potential biological significance of the +22 and −735 sites, we generated identical mutations in each (AACNG to GGCNG) and then used cyclin B1 promoters containing these mutated binding sites (M+22 and M−735, respectively) in dual-luciferase reporter assays (Fig. 1B). Baseline expression of the wild-type, M+22, and M−735 reporter constructs in K562 cells is shown in the row marked as “empty” in the lower left part of Fig. 1B. While expression of the wild-type promoter construct was low and presumably driven by endogenous Myb expression, one notes no difference in expression from the wild-type or M−735 promoter. A small but consistent decrease with respect to expression in cells transfected with the M+22 construct versus cells transfected with the wild-type promoter suggested that this site might be of biologic significance. Additional support for this interpretation is also shown in the lower left part of Fig. 1B. Cotransfection of the exogenous c-Myb or B-Myb expression vector increased cyclin B1 promoter activity significantly in cells expressing the wild-type cyclin B1 promoter. However, in cells expressing the M+22 mutated promoter, luciferase expression could not be restored by coexpressing either c-Myb or B-Myb (Fig. 1B, lower left part). In contrast, luciferase expression was substantially rescued, and to a similar degree by c-Myb or B-Myb, in cells expressing the mutated M−735 promoter. In aggregate, these results strongly suggested that the Myb binding site most proximal to the transcriptional start site (+22) is biologically significant and used by both the c-Myb and B-Myb proteins.
To provide additional support for the hypothesis that Myb binding to the +22 site was of biologic significance, silencing experiments with c-Myb- and B-myb-specific siRNAs were carried out (Fig. 1B, lower right part). In K562 cells cotransfected with the wild-type or M−735 cyclin B1 promoter, luciferase activity was not affected by control siRNA. A statistically significant decrease was observed in cells transfected with the M+22 construct compared to the control (P = 0.044) but the decrease noted was proportionately similar to that observed in the empty-vector transfection experiments shown in the lower left part of Fig. 1B. This suggested that the decrease in luciferase activity observed was not secondary to the control siRNA but rather to impaired Myb protein binding to the cyclin B1 promoter construct. It is also worth noting that in this set of experiments, luciferase expression was significantly diminished after silencing with either c-Myb- or B-Myb-targeted siRNA.
An additional observation supporting the potential biological significance of the +22 site derives from examination of the cyclin B1 promoter regions from 13 different organisms. In every case, a potential Myb binding site was found in close proximity to the transcriptional start site. Of particular interest, the Myb binding site, GGAACGGCTGTT G/A GT, we detected in the cyclin B1 promoter was found in the same site, completely conserved, in all seven of the mammalian species examined, including humans, rats, and mice (data not shown).
To further support our hypothesis that the Myb proteins bound the +22 site specifically, we carried out a ChIP experiment with primers designed to amplify a 500-bp region of genomic DNA containing four potential Myb binding sites. No DNA was amplified by real-time PCR after immunoprecipitation with either Myb antibody (data not shown). These data, in aggregate, strongly suggest that both c-Myb and B-Myb bind the +22 site specifically and in a biologically relevant manner.
Finally, to provide additional proof for our contention that c-Myb binds to the cyclin B1 promoter and thereby regulates its expression, we used an alternate experimental strategy. We previously reported on K562-c-Myb-ER cells which express a 4-OHT-inducible, conditionally active c-Myb fusion protein (45). Since this construct contains the c-Myb transactivation domain, it is very unlikely to complement B-myb function. Cells were initially treated with cycloheximide to inhibit endogenous protein synthesis. After 6 h, we measured cyclin B1 mRNA expression by real-time PCR (Fig. 1C). Compared to results obtained in the absence of cycloheximide, one can observe that after 6 h, in the presence of tamoxifen, cyclin B1 mRNA expression was restored in K562-c-MybER cells. 4-OHT itself had no effect on cyclin B1 expression (data not shown). Accordingly, this result strongly supports the hypothesis that c-Myb protein directly upregulates cyclin B1 expression in living hematopoietic cells.
Inhibition of Myb expression or activity suppresses cyclin B1 expression.
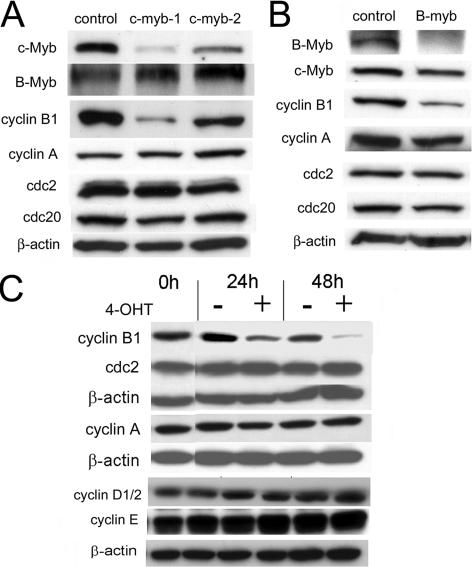
The experiments reported above demonstrated that c-Myb could bind the cyclin B1 promoter and upregulate its expression in vitro and in living cells. We then asked if the opposite might be true, i.e., if inhibition of c-Myb would lead to a decrease in cyclin B1 expression. We addressed this question by two different experimental approaches. We first examined the change in cyclin B1 expression when c-myb expression was suppressed with siRNA. Several siRNAs targeting the human c-myb mRNA were synthesized. One of these, c-myb-1 siRNA, diminished c-myb expression by 85% as measured by densitometry in Mo7e cells 24 h after transfection (Fig. 2A). Another siRNA, c-myb-2, was much less active, yielding only a 45% knockdown in the same time period. When we investigated the effect of these siRNAs on cyclin B1 expression in hematopoietic cells, we found that c-myb-1 decreased cyclin B1 protein expression by 87% while c-myb-2 only partially suppressed cyclin B1 expression (30%) (Fig. 2A). In agreement with a previous study examining the relationship between c-Myb and cell cycle gene expression (16), we found that exposing cells to either of these Myb-targeted siRNAs had no effect on the E2F-dependent, G2/M phase gene cyclin A, cdc2, or cdc20. It is also very important to note that neither one of these siRNA molecules had any effect on B-myb expression so that the effects observed are specific for c-Myb.
FIG. 2.
Silencing c-Myb expression decreases cyclin B1 expression. (A) c-myb-1 and c-myb-2 siRNAs against c-myb were transfected into Mo7e cells as described in Materials and Methods. Protein expression was determined by Western blotting 24 h after transfection. (B) B-myb and control siRNA duplexes (5 μg) were transfected into Mo7e cells as described in Materials and Methods. Protein expression was then determined by Western blotting 24 h posttransfection. (C) Western blot analyses of cyclin B1, cyclin A, cdc2, cyclin D1/2, and cyclin E protein expression in K562-MERT cells following 1 μM 4-OHT treatment for 0, 24, and 48 h.
We next examined the effect of B-myb-targeted siRNA on cyclin B1 expression in Mo7e cells. The B-myb siRNA produced an efficient knockdown of B-Myb expression. The specificity of the knockdown was demonstrated by the absence of an effect on c-Myb or β-actin expression (Fig. 2B). In accord with the ganglioblastoma cell results, silencing of B-Myb also resulted in a decrease in cyclin B1 expression (54). In contrast to the report of Zhu et al., however, silencing of B-myb in Mo7e leukemia cells resulted in no change in cyclin A or cdc2 expression and only a 25% decrease in cdc20 expression (Fig. 2B), suggesting that neither c-Myb nor B-Myb directly regulates the expression of these genes in malignant hematopoietic cells.
To validate the siRNA experiments, we then addressed this question with a tamoxifen-inducible DN Myb expression construct (MERT) (24, 45, 47, 48). We observed a decrease in cyclin B1 protein levels when endogenous Myb activity was inhibited by MERT in K562-MERT cells (Fig. 2C). Addition of 4-OHT to K562-MERT cells was followed by an ∼60% decrease in cyclin B1 expression in about 24 h. By 48 h in the presence of 4-OHT, cyclin B1 was decreased ∼95% compared to that at time zero. In contrast, and consistent with the results reported above, expression of cyclin A and cdc2 at the protein level did not change (Fig. 2C). We also examined the expression of additional G1 cyclins in the same cells after activation of the DN Myb protein (Fig. 2C). As shown, cyclin D1/2 and cyclin E levels did not change, a result which is in agreement with the work of Ku et al. (20) but in contrast to the work of Zhu et al. (54).
c-Myb partially rescues cyclin B1 expression and G2/M transition when B-myb expression is silenced by siRNA.
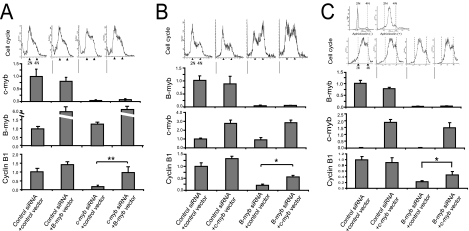
To further explore the potential significance of c-Myb and B-Myb in regulating the S and G2/M phases of the hematopoietic cell cycle, we silenced c-Myb or B-Myb in K562 cells with siRNA and determined the effects of such silencing on cell cycling. We also determined if exogenously expressed B-Myb or c-Myb could rescue cyclin B1 expression and restore cell cycling. In the first set of experiments, a B-Myb expression construct and siRNA against c-myb were transfected into K562 cells. A cell cycle analysis was carried out 48 h later (top of Fig. 3A). In K562 cells cotransfected with control siRNA and a control vector, we determined that 22% of these control cells were in the G2/M phase of the cell cycle. When we cotransfected c-myb siRNA and the control vector into K562 cells, 45% of the control cells were blocked in G2/M. In contrast, only 13% of the cells were blocked in G2/M when c-myb siRNA and a B-Myb expression vector were cotransfected into K562 cells, suggesting that overexpression of B-Myb was able to rescue the blocked cells. We also measured c-myb, B-myb, and cyclin B1 mRNA expression in these cells by real-time PCR (Fig. 3A, bar graphs). When c-myb expression was silenced with c-myb-specific siRNA, cyclin B1 expression fell ∼80% compared to that in controls. Overexpression of B-myb almost completely restored cyclin B1 expression to the control cell level.
FIG. 3.
c-Myb substantially rescues cyclin B1 expression and G2/M transition in K562 and HCT116 cells. (A) K562 cells were transfected with either pcDNA3 (control vector) or pcDNA-B-myb (B-myb vector) expression constructs and either control or c-myb-targeted siRNA as described in Materials and Methods. Forty-eight hours posttransfection, a cell cycle analysis was conducted (four graphs above the bar graphs) and RNA was isolated, reverse transcribed, and amplified by real-time PCR with primers specific to c-myb, B-myb, and cyclin B1 (bar graphs). For cell cycle analyses, the graphs, from left to right, correspond to (i) control siRNA and the control vector, (ii) control siRNA and the B-myb vector, (iii) c-myb siRNA and the control vector, and (iv) c-myb siRNA and the B-myb vector, respectively. The combinations of the expression vector and siRNA that were transfected into K562 cells are indicated below the respective bar graphs. **, P < 0.01. (B) K562 cells were transfected with either the pcDNA3 (control vector) or the pcDNA-c-myb (c-myb vector) expression construct and either control or B-myb-targeted siRNA as described in Materials and Methods. Forty-eight hours posttransfection, a cell cycle analysis was performed (four graphs above the bar graphs) and RNA was isolated, reverse transcribed, and amplified by real-time PCR with primers specific to c-myb, B-myb, and cyclin B1 (bar graphs). For cell cycle analyses, the graphs, from left to right, correspond to (i) control siRNA and the control vector, (ii) control siRNA and the c-myb vector, (iii) B-myb siRNA and the control vector, and (iv) B-myb siRNA and the c-myb vector, respectively. The combinations of the expression vector and siRNA that were transfected into K562 cells are indicated below the respective bar graphs. *, P < 0.05. (C) HCT-116 cells transfected with either the pcDNA3 (control vector) or the pcDNA-c-myb (c-myb vector) expression construct and either control or B-myb-targeted siRNA as described in Materials and Methods. The cells were cultured in medium supplemented with aphidicolin for 24 h to synchronize the cells. The aphidicolin was removed from the medium, and the cells were then cultured for an additional 12 h. Thirty-six hours posttransfection, cell cycle analysis was conducted (four graphs above the bar graphs) and RNA was isolated, reverse transcribed, and amplified by real-time PCR with primers specific to c-myb, B-myb, and cyclin B1 (bar graphs). The combinations of the expression vector and siRNA that were transfected into HCT116 cells are indicated below the respective bar graphs. *, P < 0.05. The six graphs above the bar graphs correspond to cell cycle analyses. The two cell cycle analysis graphs above the four cell cycle analysis graphs correspond to subconfluent HCT116 cells in the absence (top left) and presence (top right) of aphidicolin 24 h prior to transfection. The bottom four cell cycle analysis graphs, from left to right, correspond to (i) control siRNA and the control vector, (ii) control siRNA and the c-myb vector, (iii) B-myb siRNA and the control vector, and (iv) B-myb siRNA and the c-myb vector, respectively. For all of the real-time PCR experiments, the expression of c-myb, B-myb, and cyclin B1 was normalized to that of GAPDH. The gene expression in the control cells for all of the bar graphs was then adjusted to 1.0, except for the c-myb bar graph with HCT-116 cells (C). For this graph (C), the relative expression of c-myb expression in control HCT116 cells was compared to c-myb expression in control K562 cells (B).
In the second set of experiments, a c-Myb expression vector and siRNA targeting B-myb were cotransfected into K562 cells. Cell cycle analyses (Fig. 3B, top) and quantitation of c-myb, B-Myb, and cyclin B1 gene expression by real-time PCR (Fig. 3B, bar graphs) were conducted 48 h posttransfection. When K562 cells were cotransfected with control siRNA and a control expression vector, 24% of the cells were found to be in the G2/M phase of the cell cycle. When we cotransfected B-myb siRNA and the control vector into K562 cells, 67% of the control cells were blocked in G2/M. Cotransfecting B-myb siRNA along with the c-Myb expression vector yielded 20% of the control cells in G2/M, indicating that c-Myb overexpression was able to rescue blocked cells (Fig. 3B, top). In accord with these results, the bar graphs depicted in the lower part of Fig. 3B reveal that c-myb overexpression increased cyclin B1 expression ∼2.7-fold in comparison to that in cells in which B-Myb was silenced with siRNA. It is apparent, however, that the c-myb rescue of cyclin B1, while robust, was not complete as the cyclin B1 expression level was only ∼50% of that observed in cells expressing B-myb alone.
Finally, we sought to determine if c-Myb could rescue B-Myb function in nonhematopoietic cells which do not normally express c-Myb. For these experiments, we used human colon carcinoma cell line HCT116 because of the relative ease with which this cell line could be synchronized and transfected. HCT116 cells were synchronized in the S phase of the cell cycle with aphidicolin (Fig. 3C, top flow cytograms). The cells were then transfected with c-Myb or control expression vectors, as well as control or B-myb siRNA. Posttransfection, the cells were maintained in aphidicolin for a total of 24 h, after which time they were placed in aphidicolin-free medium. Cell cycle analyses 12 h postrelease from aphidicolin revealed that 32% of the control cells moved to the G2/M phase. It also demonstrated that in cells in which endogenous B-Myb expression was not disrupted, expression of exogenous c-Myb did not affect cell cycle progression. In contrast, transfecting HCT-116 cells with siRNA targeted to B-Myb resulted in 37% fewer cells in G2/M compared to control cells 12 h postrelease (Fig. 3C, top). When B-Myb expression was complemented by exogenously expressed c-Myb in HCT-116 cells, 25% more cells were in G2/M compared to when B-Myb was silenced, suggesting that c-myb could substantially compensate for the loss of B-Myb in these cells. In these same cells, we measured c-myb, B-myb, and cyclin B1 gene expression by real-time PCR (Fig. 3C, bottom). Expression of c-Myb had no effect on B-Myb mRNA levels. The specificity of the siRNA used to silence B-Myb is shown by the fact that these molecules had no effect on c-Myb mRNA levels. Interestingly, in cells expressing endogenous B-Myb, the exogenous expression of c-Myb produced no further changes in cyclin B1 expression (Fig. 3B, bottom graph), suggesting that cyclin B1 expression was already being maximally driven by B-Myb or that, at least in this model, c-Myb and B-Myb compete for the same binding sites in the cyclin B1 promoter (as suggested by the results discussed above). By 12 h after the aphidicolin block was released, cyclin B1 expression decreased by ∼75% compared to that of controls in cells treated with the B-Myb-targeted siRNA. In contrast, in cells expressing exogenous c-Myb, cyclin B1 expression was diminished by only ∼50% in B-myb-silenced cells, indicating that a partial rescue had again been accomplished (Fig. 3B, bottom graph). Note that the difference in cyclin B1 expression in cells expressing c-Myb was statistically significant compared to cyclin B1 expression in cells treated with B-Myb siRNA (P < 0.01).
Activation of DN Myb accelerates transit through the M phase of the cell cycle.
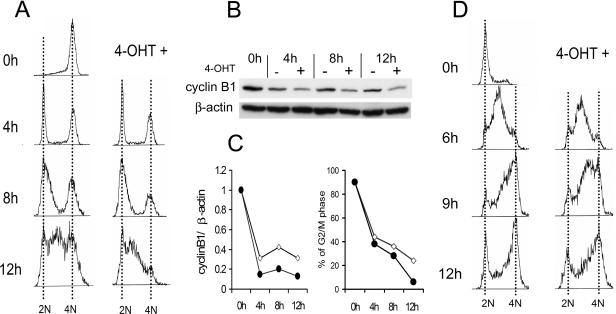
Since cyclin B1 is known to regulate transition through the G2/M phase of the cell cycle, an additional test of the biologic importance of Myb's regulation of cyclin B1 expression was carried out. We examined cell cycle progression in K562-MERT expressing the DN Myb which suppresses both c-Myb and B-Myb function. Cells were synchronized at the G2/M phase by nocodazole treatment following a double-thymidine block (18). Immediately following release from the block, the DN Myb protein was activated by 4-OHT. Cell cycle progression was then analyzed as a function of cellular DNA content by flow cytometry (Fig. 4A). After treatment with nocodazole, ∼90% of the untreated cells accumulated in the G2/M phase (Fig. 4A, time 0 h). At 4 h after release from the block, ∼46% of the untreated cells exited the G2/M phase and moved into the G1 phase. At 8 h after release, 64% of the untreated cells were in G1. In 4-OHT-treated cells, 62% and 72% of the cells exited G2/M and moved into G1 at 4 h and 8 h, respectively. These differences were reproducible in four experiments. By 8 h, ∼70% of the 4-OHT-treated cells moved into G1 or early S phase but no cells transited the late S phase. At 12 h, the percentages of cells in the G2/M phase were 24% and 6% for untreated and 4-OHT-treated cells expressing DN c-Myb, respectively. These results are consistent with the hypothesis that Myb plays a role in regulating transition through the M phase of the cell cycle and that this transition is accelerated in the absence of Myb. Cyclin B1 protein expression was also measured in the same cells (Fig. 4B and C). After addition of 4-OHT to K562-MERT cells, cyclin B1 expression decreased ∼2- to 2.5-fold compared to that in untreated cells at 4, 8, and 12 h. This reduction of cyclin B1 in 4-OHT-treated cells expressing DN Myb was observed consistently in multiple experiments and was also concordant with a decrease in 4-OHT-treated cells in the G2/M phase of the cell cycle (Fig. 4C).
FIG. 4.
Activation of DN Myb accelerates transit through the M phase of the cell cycle. (A) K562-MERT cells were synchronized at G2/M by nocodazole treatment following a double-thymidine block. Subsequently, the cells were released into the cell cycle and then treated with 4-OHT to allow MERT to inhibit endogenous Myb activity. The data provided are representative of four independent experiments. (B) Cyclin B1 protein expression was determined by Western blot analysis with the lysate of the cells used in panel A. A representative Western blot analysis of three independent experiments is shown. (C) On the left is cyclin B1 protein expression in the absence (diamonds) or presence (circles) of 4-OHT from panel B quantitated by densitometry. On the right are the percentages of K562-MERT cells in the G2/M phase of the cell cycle in the absence (open circles) or presence (filled circles) of 4-OHT from panel A. (D) K562-MERT cells were synchronized in the G1 phase by a double-thymidine block and subsequently released into the cell cycle in medium in the absence or presence of 4-OHT.
Finally, we examined the effect of DN Myb on passage through the S phase. K562-MERT cells were synchronized in G1 by a double-thymidine block and were then released in medium without or with 4-OHT (Fig. 4D). Initially, ∼70% of the cells were in the G1 phase and ∼30% were in S phase. Progression through the S phase was delayed in 4-OHT-treated K562-MERT cells compared to untreated K562-MERT cells. By 12 h, ∼70% of the untreated K562-MERT cells moved from G1 into the G2/M phase while ∼30% of the cells were in the S phase. In contrast, ∼50% of the 4-OHT-treated K562-MERT cells remained in the S phase of the cell cycle. Since cyclin B1 is not known to effect passage through the S phase, we think it likely that another, as yet unidentified, Myb-regulated protein is important in this phase of the cycle as well. Our data, in aggregate, lead us to suggest that delay of the S phase and acceleration through G2/M may be the operative mechanism whereby loss of Myb activity inhibits cell proliferation and ultimately leads to accumulation of cells at the G1/S interface.
Cyclin B1 expression and c-myb expression increase concordantly in phytohemagglutinin (PHA)- or interleukin-2 (IL-2)-stimulated normal human PBL.
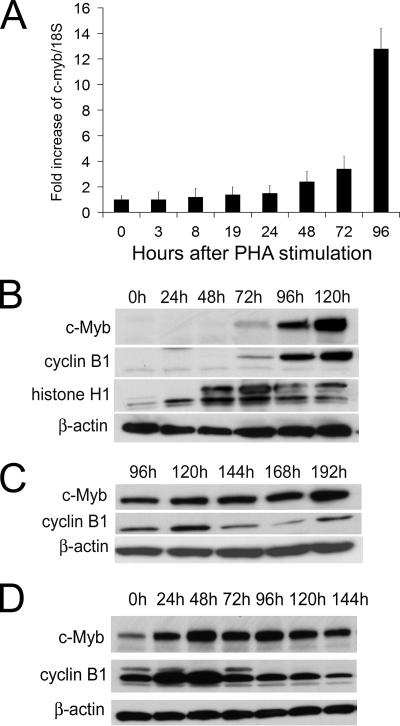
The results reported above were observed in cell lines. To determine the relevance of these results to normal cells, we also examined the potential role of c-myb in regulating the G2/M transition in normal human PBL. We chose these cells because they were readily available in large numbers and are dependent on c-Myb expression for proliferation. Knowing that c-myb mRNA and protein have a very short half-life and that c-Myb would have to be expressed in G2/M if it were playing a role in this phase of the cell cycle, we began by quantitating c-myb expression by real-time PCR in resting and stimulated lymphocytes. Surprisingly, we found that c-myb was constitutively expressed in normal human PBL (Fig. 5A). We also found that after PHA stimulation, in amounts ranging from 1 to 6 μg/ml and with several different lots of PHA, c-Myb expression increased, not after 24 h as some have reported (9, 14, 42) but only slightly at 48 h and then unequivocally and more robustly between 72 and 96 h (Fig. 5A). These results were confirmed by multiple replicates of the same experiments.
FIG. 5.
Cyclin B1 expression and c-myb expression increase concordantly in PHA- or IL-2-stimulated human lymphocytes. (A) Real-time PCR of c-myb expression in human PBL stimulated with 3 μg/ml PHA for 3, 8, 19, 24, 48, 72, and 96 h. c-myb expression was normalized to 18S RNA. (B) Western blot analyses of c-Myb, cyclin B1, and histone H1 in PBL stimulated with PHA for 0, 24, 48, 72, 96, and 120 h. The membranes were first probed with anti-c-myb and then stripped, and cyclin B1 expression and β-actin expression were determined on the same membrane with the respective antibodies. (C) Western blot analyses of Myb and cyclin B1 in PBL stimulated with PHA for 96, 120, 144, 168, and 192 h. For all experiments, PBL were stimulated with 3 μg/ml PHA. (D) Western blot analyses of c-Myb and cyclin B1 in PBL stimulated with IL-2. PBL activated by PHA were starved for 24 h and then stimulated with 30 ng of IL-2/ml for up to 6 days.
We also examined c-Myb protein levels in T lymphocytes at 120 h (Fig. 5B) and at 192 h (Fig. 5C) after PHA stimulation. Since histone H1 expression peaked at 72 h, we assumed that S phase occurred at ∼72 h as well (52). We confirmed that the S phase occurred at ∼72 h by flow cytometric cell cycle analysis of PHA-stimulated lymphocytes and examination of [3H]thymidine uptake after 3 μg/ml PHA stimulation (data not shown). The maximal level of c-Myb and cyclin B1 was detected at 120 h. This is of interest because cyclin B1 expression peaks at the G2/M phase (25, 36, 38). After 120 h, the level of cyclin B1 decreased but began to increase again at 192 h (Fig. 5C). c-Myb expression did not change after 120 h but did increase again at 192 h, concordant with the second observed increase in cyclin B1 expression.
To determine if the results described above might have been due to the type of proliferative stimulus used, we examined c-Myb expression kinetics in normal PBL after IL-2 stimulation. PBL were pretreated with PHA (1 μg/ml) for 24 h in order to prime them for proliferation. After this time, the cells were washed and placed in medium devoid of PHA in order to arrest the cells in G1. After a second 24-h interval in the PHA-free medium, IL-2 (30 ng/ml) was added and c-Myb expression was monitored serially thereafter. The maximal level of c-Myb expression occurred after 48 h of IL-2 stimulation, and it was concordant with the peak levels of cyclin B1 expression (Fig. 5D).
Repression of c-myb decreases cyclin B1 expression in normal human T cells and CD34+ cells.
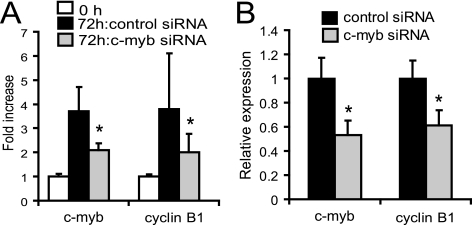
Lastly, we examined cyclin B1 expression in human T lymphocytes and CD34+ cells when c-myb was repressed by siRNA. The c-myb-1 siRNA was transfected into unstimulated human T lymphocytes, and then the cells were cultured for 72 h in the medium with PHA (5 μg/dl). After 72 h, the c-Myb level in the siRNA-treated cells was decreased by ∼43% compared to that in control siRNA-treated cells. In accord with this result and those reported above, cyclin B1 mRNA expression decreased by 47% in these cells (Fig. 6A). We also measured cyclin B1 mRNA expression in human bone marrow CD34+ cells when c-myb was suppressed with siRNA. c-myb-1 siRNA was transfected into the cells, and c-myb expression and cyclin B1 expression were measured at 48 h after the transfection. c-myb expression and cyclin B1 expression also decreased concordantly (Fig. 6B). Therefore, it is highly likely that the relationships among c-Myb, cyclin B1 expression, and cell cycle progression apply to normal hematopoietic cells as well.
FIG. 6.
c-myb repression induces a cyclin B1 decrease in human T cells and CD34+ cells. (A) Real-time PCR analysis for cyclin B1 and c-myb expression in human PBL stimulated with PHA at 5 μg/ml for 72 h following c-myb-1 siRNA transfection. (B) c-myb expression and cyclin B1 expression were measured by real-time PCR 24 h posttransfection of c-myb-1 siRNA into CD34+ cells from human bone marrow. The expression of c-myb and cyclin B1 genes was normalized to that of GAPDH. *, P < 0.05 compared to the value of control siRNA.
DISCUSSION
Recent studies have suggested that B-Myb may be an important link in an E2F-regulated “feed forward loop” (22) whereby genes expressed in G1/S are also important in regulating the expression of genes required for the G2/M transition (54). For example, it has been suggested that in rat embryonic fibroblasts and human ganglioblastoma cells, E2F3 upregulates B-Myb in G1/M and that B-Myb, in turn, upregulates cdc2, cyclin A2, and cyclin B1 in G2/M (54). Germane to this report, however, is the fact that these genes were found to be responsive only to B-Myb, and specifically not c-Myb, when investigated. However, since genes regulated by c-Myb have been reported to be cell type specific (29) and because c-Myb is known to play an important role in hematopoietic cell proliferation, we sought to address the possibility that in hematopoietic cells, c-Myb might also play a role in regulating other phases of the cell cycle.
Perhaps surprisingly, we found that, in contrast to the studies of Zhu et al. (54), c-Myb can regulate cyclin B1 expression not only in hematopoietic cells but in colon cancer-derived cells as well. Further, we developed a great deal of evidence to support the hypothesis that Myb regulation of cyclin B1 expression is direct. Specifically, we demonstrated that c-Myb interacts directly with the cyclin B1 promoter in vivo (Fig. 1A). Reporter assays showed that overexpression of both c-Myb and B-Myb increased cyclin B1 promoter activity through the Myb binding site, GGAACGGCTGTTGG, located 22 bp downstream of the transcriptional start site. In addition, a conditionally active c-Myb protein restored cyclin B1 mRNA expression in the presence of cycloheximide in K562 cells.
Knowing that c-Myb directly regulates cyclin B1 expression in hematopoietic cells, it was reasonable to further propose a role for c-Myb in the G2/M transition phase of these cells. While our data demonstrate that B-Myb, as well as c-Myb, is capable of regulating cyclin B1 expression and the G2/M transition, it is equally clear that in cells in which B-Myb is silenced with siRNA, c-Myb can rescue B-Myb function in both respects (Fig. 3A and B). This relationship was also shown to be functional in HCT116 cells, which do not normally express c-Myb. In cells in which B-Myb had been silenced with siRNA, exogenously expressed c-Myb partially rescued cyclin B1 expression and the associated delay of G2/M transit (Fig. 3C). Finally, in K562-MERT cells expressing a DN Myb protein, we noted an accelerated G2/M transit time (Fig. 4). This is consistent with previous studies which suggest that cyclin B1 destruction in metaphase is essential in order for cells to leave the M phase (8, 17, 37). While this result does not discriminate between the roles of c-Myb and B-Myb (the DN Myb protein suppresses both forms), it does speak to the role of Myb's importance in regulating G2/M transit in hematopoietic cells.
Since the bulk of our experiments were carried out with cell lines, we also attempted to demonstrate that c-myb can regulate cyclin B1 expression in normal human hematopoietic cells. For this purpose, we used PBL and bone marrow-derived CD34+ cells. While it was not possible to perform experiments with synchronized populations of these cells, we were nonetheless able to demonstrate the same relationships between c-Myb and cyclin B1 expression by cytokine stimulation or by using the DN expression system or siRNA-mediated inhibition of c-Myb. Curiously, in the experiments carried out with T lymphocytes, we also showed that c-Myb protein expression peaked in the G2/M phase (Fig. 5), rather than in G1/S, as has been previously reported for PHA-stimulated cells (9, 11, 15, 33, 53). These results are certainly consistent with our finding that c-Myb directly regulates the expression of cyclin B1, but this discrepancy might also be explained by differences in the methods used to detect c-Myb expression and differences in the mitotic stimulants used. Regardless, the results presented clearly demonstrate that the relationship between c-Myb and cyclin B1 expression is operational in normal cells, as well as in cell lines, and support the hypothesis that this relationship is of physiologic significance.
In summary, while recent studies suggest that B-Myb, and specifically not c-Myb, may be important in regulating the G2/M transition in certain cell types (54), we now provide data which suggest that in normal and malignant human hematopoietic cells, and likely in human colonic cells as well, c-Myb and B-Myb are both capable of directly regulating cyclin B1 expression. These data are consistent with the observation that critical Myb protein functions are conserved across the phyla (19, 34, 54). They also provide strong support for the hypothesis that Myb plays an important role in regulating the G2/M, as well as the G1/S, cell cycle transition in hematopoietic cells. Our studies do not allow us to definitively answer the question of whether c-Myb or B-Myb is the more important physiologic regulator of cyclin B1 in hematopoietic cells. The fact that B-Myb is more abundantly expressed than c-Myb might be interpreted to mean that it is of greater physiologic importance. However, since c-Myb can transactivate cyclin B1 to almost the same extent as B-Myb (Fig. 1B and 2) and can at least partially rescue B-Myb-deficient cells, we hypothesize that in the hematopoietic cell context it may well play an important role as a coregulator of cyclin B1 or, perhaps more likely, by providing a measure of redundancy, and the concomitant survival advantage that accompanies such redundancy, for Myb protein function in hematopoietic cells.
Acknowledgments
We thank Martin Carroll (Division of Hematology/Oncology, University of Pennsylvania School of Medicine) for helpful comments in preparing the manuscript.
This work was supported by grants from the NIH to S.E.S. (K01 DK068283) and A.M.G. (RO1 CA101859). A. M. Gewirtz is a Distinguished Clinical Scientist of the Doris Duke Charitable Foundation, which also supported this work.
Footnotes
Published ahead of print on 22 January 2007.
REFERENCES
- 1.Allen, R. D., III, T. P. Bender, and G. Siu. 1999. c-Myb is essential for early T cell development. Genes Dev. 13:1073-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender, T. P., C. S. Kremer, M. Kraus, T. Buch, and K. Rajewsky. 2004. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat. Immunol. 5:721-729. [DOI] [PubMed] [Google Scholar]
- 3.Biedenkapp, H., U. Borgmeyer, A. E. Sippel, and K. H. Klempnauer. 1988. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature 335:835-837. [DOI] [PubMed] [Google Scholar]
- 4.Bies, J., R. Mukhopadhyaya, J. Pierce, and L. Wolff. 1995. Only late, nonmitotic stages of granulocyte differentiation in 32Dcl3 cells are blocked by ectopic expression of murine c-myb and its truncated forms. Cell Growth Differ. 6:59-68. [PubMed] [Google Scholar]
- 5.Brandt, T. L., and D. J. Kroll. 1997. NF-M trans-activates the human DNA topoisomerase II alpha promoter independently of c-Myb in HL-60 cells. Leuk. Res. 21:711-720. [DOI] [PubMed] [Google Scholar]
- 6.Carpinelli, M. R., D. J. Hilton, D. Metcalf, J. L. Antonchuk, C. D. Hyland, S. L. Mifsud, L. Di Rago, A. A. Hilton, T. A. Willson, A. W. Roberts, R. G. Ramsay, N. A. Nicola, and W. S. Alexander. 2004. Suppressor screen in Mpl−/− mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc. Natl. Acad. Sci. USA 101:6553-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke, M. F., J. F. Kukowska-Latallo, E. Westin, M. Smith, and E. V. Prochownik. 1988. Constitutive expression of a c-myb cDNA blocks Friend murine erythroleukemia cell differentiation. Mol. Cell. Biol. 8:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clute, P., and J. Pines. 1999. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1:82-87. [DOI] [PubMed] [Google Scholar]
- 9.Cures, A., C. House, C. Kanei-Ishii, B. Kemp, and R. G. Ramsay. 2001. Constitutive c-Myb amino-terminal phosphorylation and DNA binding activity uncoupled during entry and passage through the cell cycle. Oncogene 20:1784-1792. [DOI] [PubMed] [Google Scholar]
- 10.Emambokus, N., A. Vegiopoulos, B. Harman, E. Jenkinson, G. Anderson, and J. Frampton. 2003. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 22:4478-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganter, B., and J. S. Lipsick. 1999. Myb and oncogenesis. Adv. Cancer Res. 76:21-60. [DOI] [PubMed] [Google Scholar]
- 12.Gewirtz, A. M., G. Anfossi, D. Venturelli, S. Valpreda, R. Sims, and B. Calabretta. 1989. G1/S transition in normal human T-lymphocytes requires the nuclear protein encoded by c-myb. Science 245:180-183. [DOI] [PubMed] [Google Scholar]
- 13.Gewirtz, A. M., and B. Calabretta. 1988. A c-myb antisense oligodeoxynucleotide inhibits normal human hematopoiesis in vitro. Science 242:1303-1306. [DOI] [PubMed] [Google Scholar]
- 14.Golay, J., A. Capucci, M. Arsura, M. Castellano, V. Rizzo, and M. Introna. 1991. Expression of c-myb and B-myb, but not A-myb, correlates with proliferation in human hematopoietic cells. Blood 77:149-158. [PubMed] [Google Scholar]
- 15.Gonda, T. J. 1998. The c-Myb oncoprotein. Int. J. Biochem. Cell. Biol. 30:547-551. [DOI] [PubMed] [Google Scholar]
- 16.Hogg, A., S. Schirm, H. Nakagoshi, P. Bartley, S. Ishii, J. M. Bishop, and T. J. Gonda. 1997. Inactivation of a c-Myb/estrogen receptor fusion protein in transformed primary cells leads to granulocyte/macrophage differentiation and down regulation of c-kit but not c-myc or cdc2. Oncogene 15:2885-2898. [DOI] [PubMed] [Google Scholar]
- 17.Holloway, S. L., M. Glotzer, R. W. King, and A. W. Murray. 1993. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell 73:1393-1402. [DOI] [PubMed] [Google Scholar]
- 18.Isakson, P. C., J. Purkerson, K. Catron, and T. P. Bender. 1991. A novel method for synchronizing a B cell lymphoma. J. Immunol. Methods 145:137-142. [DOI] [PubMed] [Google Scholar]
- 19.Ito, M., S. Araki, S. Matsunaga, T. Itoh, R. Nishihama, Y. Machida, J. H. Doonan, and A. Watanabe. 2001. G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13:1891-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ku, D. H., S. C. Wen, A. Engelhard, N. C. Nicolaides, K. E. Lipson, T. A. Marino, and B. Calabretta. 1993. c-myb transactivates cdc2 expression via Myb binding sites in the 5′-flanking region of the human cdc2 gene. J. Biol. Chem. 268:2255-2259. [PubMed] [Google Scholar]
- 21.Lang, G., J. R. White, M. J. Argent-Katwala, C. G. Allinson, and K. Weston. 2005. Myb proteins regulate the expression of diverse target genes. Oncogene 24:1375-1384. [DOI] [PubMed] [Google Scholar]
- 22.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 23.Lutz, P. G., A. Houzel-Charavel, C. Moog-Lutz, and Y. E. Cayre. 2001. Myeloblastin is an Myb target gene: mechanisms of regulation in myeloid leukemia cells growth-arrested by retinoic acid. Blood 97:2449-2456. [DOI] [PubMed] [Google Scholar]
- 24.Lyon, J. J., and R. J. Watson. 1995. Conditional inhibition of erythroid differentiation by c-Myb/oestrogen receptor fusion proteins. Differentiation 59:171-178. [DOI] [PubMed] [Google Scholar]
- 25.Maity, A., W. G. McKenna, and R. J. Muschel. 1995. Evidence for post-transcriptional regulation of cyclin B1 mRNA in the cell cycle and following irradiation in HeLa cells. EMBO J. 14:603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metcalf, D., M. R. Carpinelli, C. Hyland, S. Mifsud, L. Dirago, N. A. Nicola, D. J. Hilton, and W. S. Alexander. 2005. Anomalous megakaryocytopoiesis in mice with mutations in the c-Myb gene. Blood 105:3480-3487. [DOI] [PubMed] [Google Scholar]
- 27.Mucenski, M. L., K. McLain, A. B. Kier, S. H. Swerdlow, C. M. Schreiner, T. A. Miller, D. W. Pietryga, W. J. Scott, Jr., and S. S. Potter. 1991. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65:677-689. [DOI] [PubMed] [Google Scholar]
- 28.Nakata, Y., S. Shetzline, C. Sakashita, A. Kalota, A. Ptasznik, Y. Zhang, S. G. Emerson, and A. M. Gewirtz. 2005. c-Myb plays a role in G2/M cell cycle transition by direct regulation of cyclin B1 expression in hematopoietic cells. Blood (ASH Annu. Meet. Abstr.) 106:1355. [Google Scholar]
- 29.Ness, S. A. 2003. Myb protein specificity: evidence of a context-specific transcription factor code. Blood Cells Mol. Dis. 31:192-200. [DOI] [PubMed] [Google Scholar]
- 30.Ness, S. A., A. Marknell, and T. Graf. 1989. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 59:1115-1125. [DOI] [PubMed] [Google Scholar]
- 31.Nomura, N., M. Takahashi, M. Matsui, S. Ishii, T. Date, S. Sasamoto, and R. Ishizaki. 1988. Isolation of human cDNA clones of myb-related genes, A-myb and B-myb. Nucleic Acids Res. 16:11075-11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogata, K., S. Morikawa, H. Nakamura, A. Sekikawa, T. Inoue, H. Kanai, A. Sarai, S. Ishii, and Y. Nishimura. 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79:639-648. [DOI] [PubMed] [Google Scholar]
- 33.Oh, I. H., and E. P. Reddy. 1999. The myb gene family in cell growth, differentiation and apoptosis. Oncogene 18:3017-3033. [DOI] [PubMed] [Google Scholar]
- 34.Okada, M., H. Akimaru, D. X. Hou, T. Takahashi, and S. Ishii. 2002. Myb controls G2/M progression by inducing cyclin B expression in the Drosophila eye imaginal disc. EMBO J. 21:675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson, R., and K. Weston. 2000. c-Myb regulates the proliferation of immature thymocytes following beta-selection. EMBO J. 19:6112-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piaggio, G., A. Farina, D. Perrotti, I. Manni, P. Fuschi, A. Sacchi, and C. Gaetano. 1995. Structure and growth-dependent regulation of the human cyclin B1 promoter. Exp. Cell Res. 216:396-402. [DOI] [PubMed] [Google Scholar]
- 37.Pines, J. 1999. Four-dimensional control of the cell cycle. Nat. Cell Biol. 1:E73-E79. [DOI] [PubMed] [Google Scholar]
- 38.Pines, J., and T. Hunter. 1991. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol. 115:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ptasznik, A., Y. Nakata, A. Kalota, S. G. Emerson, and A. M. Gewirtz. 2004. Short interfering RNA (siRNA) targeting the Lyn kinase induces apoptosis in primary, and drug-resistant, BCR-ABL1+ leukemia cells. Nat. Med. 10:1187-1189. [DOI] [PubMed] [Google Scholar]
- 40.Quéva, C., S. A. Ness, F. A. Grasser, T. Graf, B. Vandenbunder, and D. Stehelin. 1992. Expression patterns of c-myb and of v-myb induced myeloid-1 (mim-1) gene during the development of the chick embryo. Development 114:125-133. [DOI] [PubMed] [Google Scholar]
- 41.Ratajczak, M. Z., D. Perrotti, P. Melotti, M. Powzaniuk, B. Calabretta, K. Onodera, D. A. Kregenow, B. Machalinski, and A. M. Gewirtz. 1998. Myb and ets proteins are candidate regulators of c-kit expression in human hematopoietic cells. Blood 91:1934-1946. [PubMed] [Google Scholar]
- 42.Reed, J. C., J. D. Alpers, P. C. Nowell, and R. G. Hoover. 1986. Sequential expression of protooncogenes during lectin-stimulated mitogenesis of normal human lymphocytes. Proc. Natl. Acad. Sci. USA 83:3982-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakura, H., C. Kanei-Ishii, T. Nagase, H. Nakagoshi, T. J. Gonda, and S. Ishii. 1989. Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc. Natl. Acad. Sci. USA 86:5758-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sala, A., and R. Watson. 1999. B-Myb protein in cellular proliferation, transcription control, and cancer: latest developments. J. Cell. Physiol. 179:245-250. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt, M., V. Nazarov, L. Stevens, R. Watson, and L. Wolff. 2000. Regulation of the resident chromosomal copy of c-myc by c-Myb is involved in myeloid leukemogenesis. Mol. Cell. Biol. 20:1970-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selvakumaran, M., D. A. Liebermann, and B. Hoffman-Liebermann. 1992. Deregulated c-myb disrupts interleukin-6- or leukemia inhibitory factor-induced myeloid differentiation prior to c-myc: role in leukemogenesis. Mol. Cell. Biol. 12:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shetzline, S. E., R. Rallapalli, K. J. Dowd, S. Zou, Y. Nakata, C. R. Swider, A. Kalota, J. K. Choi, and A. M. Gewirtz. 2004. Neuromedin U: a Myb-regulated autocrine growth factor for human myeloid leukemias. Blood 104:1833-1840. [DOI] [PubMed] [Google Scholar]
- 48.Taylor, D., P. Badiani, and K. Weston. 1996. A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev. 10:2732-2744. [DOI] [PubMed] [Google Scholar]
- 49.Thomas, M. D., C. S. Kremer, K. S. Ravichandran, K. Rajewsky, and T. P. Bender. 2005. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity 23:275-286. [DOI] [PubMed] [Google Scholar]
- 50.Trauth, K., B. Mutschler, N. A. Jenkins, D. J. Gilbert, N. G. Copeland, and K. H. Klempnauer. 1994. Mouse A-myb encodes a trans-activator and is expressed in mitotically active cells of the developing central nervous system, adult testis and B lymphocytes. EMBO J. 13:5994-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vandenbark, G. R., Y. Chen, E. Friday, K. Pavlik, B. Anthony, C. deCastro, and R. E. Kaufman. 1996. Complex regulation of human c-kit transcription by promoter repressors, activators, and specific myb elements. Cell Growth Differ. 7:1383-1392. [PubMed] [Google Scholar]
- 52.van Wijnen, A. J., F. Aziz, X. Grana, A. De Luca, R. K. Desai, K. Jaarsveld, T. J. Last, K. Soprano, A. Giordano, J. B. Lian, et al. 1994. Transcription of histone H4, H3, and H1 cell cycle genes: promoter factor HiNF-D contains CDC2, cyclin A, and an RB-related protein. Proc. Natl. Acad. Sci. USA 91:12882-12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weston, K. 1998. Myb proteins in life, death and differentiation. Curr. Opin. Genet. Dev. 8:76-81. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, W., P. H. Giangrande, and J. R. Nevins. 2004. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 23:4615-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]