Abstract
Although ribosomal proteins (RPs) are essential cellular constituents in all living organisms, mechanisms underlying regulation of their gene expression in mammals remain unclear. We have established that 22 out of 79 human RP genes contain sequences similar to the human DREF (DNA replication-related element-binding factor; hDREF) binding sequence (hDRE) within 200-bp regions upstream of their transcriptional start sites. Electrophoretic gel mobility shift assays and chromatin immunoprecipitation analysis indicated that hDREF binds to hDRE-like sequences in the RP genes both in vitro and in vivo. In addition, transient luciferase assays revealed that hDRE-like sequences act as positive elements for RP gene transcription and cotransfection of an hDREF-expressing plasmid was found to stimulate RP gene promoter activity. Like that of hDREF, expression of RP genes is increased during the late G1 to S phases, and depletion of hDREF using short hairpin RNA-mediated knockdown decreased RP gene expression and cell proliferation in normal human fibroblasts. Knockdown of the RPS6 gene also resulted in impairment of cell proliferation. These data suggest that hDREF is an important transcription factor for cell proliferation which plays roles in cell cycle-dependent regulation of a number of RP genes.
Promoters of Drosophila melanogaster genes related to DNA replication, such as those for the 180-kDa catalytic subunit of DNA polymerase α and proliferating cell nuclear antigen (PCNA), contain a common 8-bp palindromic sequence (5′-TATCGATA-3′), named the DNA replication-related element (DRE) (12). These DREs are required for promoter activities both in cultured cells and in flies in vivo (41). We have purified the DRE-binding factor (DREF) from cultured Drosophila cells, consisting of an 86-kDa polypeptide homodimer specifically binding to DRE, and isolated a cDNA (12, 13). The importance of Drosophila DREF in development has been demonstrated from studies using transgenic flies (11, 14, 44). For example, ectopic expression of Drosophila DREF in eye imaginal disc cells behind the morphogenetic furrow, which are normally postmitotic, induced ectopic DNA synthesis and apoptosis and abolished photoreceptor specifications (11). More recently, we and Hyun et al. have succeeded in knocking down Drosophila DREF expression in various tissues (16, 45). Decreased levels of DREF in developing wing and eye imaginal discs were associated with reduction in wing size with smaller cells and drastically aberrant small and rough eyes, respectively. These lines of evidence indicate that the Drosophila DRE/DREF system performs important roles in regulation of cell growth as well as cell proliferation during development.
How many and what kind of genes other than those described above are under control of the Drosophila DRE/DREF system? Immunostaining of polytene chromosomes of salivary glands revealed that Drosophila DREF binds to hundreds of loci (8, 10), and recent computational analysis of core promoters in the Drosophila genome showed DRE to be the second most frequent motif apparent in core promoter sequences from −60 to +40, with a frequency higher than those for the TATA box and initiator sequences (4, 24). Already, we and others have demonstrated that the Drosophila DRE/DREF system regulates a number of Drosophila genes involved in DNA replication as well as those involved in cell cycle progression through S (dE2F1) (33) and G2/M (D-raf, D-myb, and cyclin A) phases (25, 29, 34).
Despite much progress in understanding the Drosophila DRE/DREF system, little is known about the corresponding mammalian DRE/DREF system. We have identified a human homologue of DREF (hDREF) and a binding consensus sequence [hDRE; 5′-TGTCG(C/T)GA(C/T)A-3′] (26). Our previous study showed that the expression level of hDREF is elevated during the G1-to-S transition and reaches a maximum at S phase in normal human fibroblasts. Moreover, RNA interference experiments targeting hDREF pointed to an important role in cell cycle progression. We also demonstrated that the histone H1 gene carrying an hDRE is regulated by hDREF (26). However, other functions and target genes of hDREF remain to be clarified.
The ribosome is a vital organelle which is responsible for protein synthesis in all living organisms. Production of mature ribosomes consisting of rRNAs and ribosomal proteins (RPs) requires a highly coordinated multistep process, and many reports have shown that the initiation of rRNA transcription is tightly linked to cell cycle progression, with synthesis of rRNA increasing during G1 phase, becoming maximal in S and G2 phases, and being repressed during mitosis (7, 20, 22). Similarly, coordinated synthesis of all RP genes during the cell cycle, leading to the precise and equimolar production of the 79 RPs necessary for ribosome biogenesis and translation control, is required to support adequate protein synthesis (37). Despite the obvious importance of RP gene expression for cell proliferation, only a limited number of experimental studies of mammalian RP gene promoter structures and transcriptional regulation have been performed so far. Recent determination of a complete map and nucleotide sequences now allows searches for regulatory elements common to a number of human RP genes, and Perry has identified “box A” [5′-TCTCGCGA(G/T)-3′] as one of the most conserved sequences in RP gene promoters (28).
In this report, we document evidence that hDREF is a transcription factor essential for cell proliferation which binds to box A and positively regulates a set of human RP genes. We also show that mammalian RP gene expression is induced during late G1 and S phases and that fluctuation of hDREF expression during the cell cycle is in line with cell cycle-dependent expression of RP genes.
MATERIALS AND METHODS
Plasmids.
The expression plasmid pcDNA3-HA-hDREF, containing a full-length cDNA for hDREF in the pcDNA3-HA vector (where HA is hemagglutinin), was described previously (26). To construct a plasmid expressing a si4 short hairpin RNA (shRNA)-resistant mRNA of hDREF, base substitutions were made by site-specific mutagenesis employing the overlap extension method (32) and confirmed by DNA sequencing. The amplified hDREF cDNA fragment carrying base substitutions was inserted between the NheI and ApaI sites of the pcDNA3-HA vector. To construct pCSII-EF-hDREF-IRES2-Venus, a cDNA encoding full-length hDREF obtained from pcDNA3-HA-hDREF was inserted into the blunt-ended BamHI site of the pCSII-EF-MCS-IRES2-Venus lentivirus vector (where IRES2 is internal ribosome entry site 2) (42).
To create a lentivirus vector expressing HA-hDREF directed by the human metallothionein gene (MTIIA) promoter, the EF-1α gene promoter in pCSII-EF-MCS-IRES2-Venus was swapped with the −765 to +55 region of the MTIIA gene and a cDNA encoding HA-hDREF was inserted. Lentiviruses expressing shRNA against hDREF or RPS6 were prepared as follows. A pair of 48- or 49-base oligonucleotides containing a 20-nucleotide targeting sequence, a spacer sequence which provided a loop structure, and an extra sequence to facilitate cloning were annealed and cloned into an attL-containing pENTER/U6 plasmid (Invitrogen). The cassette containing the U6 promoter and shRNA target sequence was then transferred to a self-inactivating lentivirus vector (pCS-RfA-EG), generating pCS-U6-shRNA-EG.
Luciferase reporter plasmids containing the promoter region of RPS6 (RPS6-310+43/pGL3) were obtained as follows. The promoter region of human RPS6 (−310 to +43) was amplified by PCR with genomic DNA from HFF cells as a template and was ligated between the Asp718 and BglII sites of the pGL3 basic vector (Promega). RPL10A-193+17/pGL3 and RPL12-277+57/pGL3, containing the upstream regions from position −193 to position +17 of RPL10A and from position −277 to position +57 of RPL12, respectively, were created by the same procedures as RPS6. Linker insertion mutants of the luciferase reporter plasmids were created by digesting wild-type reporter plasmids with NruI and then ligating those with a 12-mer EcoRI linker (TaKaRa). Base substitution derivatives (mut1, mut2, mut3, mut4, and mut5) and a deletion derivative (ΔhDRE) of RPS6-310+43/pGL3 were generated as follows. An upstream region of RPS6 was amplified by PCR using primer sets containing a 3-bp substitution mutation or a 10-bp deletion mutation with RPS6-310+43/pGL3 as a template and inserted between the Asp718 and BglII sites of the pGL3 basic vector. All plasmids were isolated and purified using a QIAGEN plasmid Midi kit (QIAGEN).
Oligonucleotides.
Double-stranded oligonucleotides containing hDRE-like sequence in the promoter region of the human RPS6 gene (RPS6) (position at −56 to −22 with respect to the transcriptional start site) and its base substitution derivative sequences (mut1, mut2, mut3, mut4, and mut5) or a deletion derivative sequence (ΔhDRE), shown below, were synthesized. The sequences used are as follows: for RPS6-hDRE, 5′-GTACTTCTGCTCATCTCGCGAGAACTGAAAGCGCC-3′ and 5′-GGCGCTTTCAGTTCTCGCGAGATGAGCAGAAGTAC-3′; for mut1, 5′-GTACTTCTGCctgTCTCGCGAGAACTGAAAGCGCC-3′ and 5′-GGCGCTTTCAGTTCTCGCGAGAcagGCAGAAGTAC-3′; for mut2, 5′-GTACTTCTGCTCA ctcCGCGAGAACTGAAAGCGCC-3′ and 5′-GGCGCTTTCAGTTCTCGCGgagTGAGCAGAAGTAC-3′; for mut3, 5′-GTACTTCTGCTCATCTtatGAGAACTGAAAGCGCC-3′ and 5′-GGCGCTTTCAGTTCTCataAGATGAGCAGAAGTAC-3′; for mut4, 5′-GTACTTCTGCTCATCTCGCagaAACTGAAAGCGCC-3′ and 5′-GGCGCTTTCAGTTtctGCGAGATGAGCAGAAGTAC-3′; for mut5, 5′-GTACTTCTGCTCATCTCGCGAGggtTGAAAGCGCC-3′ and 5′-GGCGCTTTCAaccCTCGCGAGATGAGCAGAAGTAC-3′; and for ΔhDRE, 5′-CCGTACTTCTGCTCAACTGAAAGCGCCTAT-3′ and 5′-ATAGGCGCTTTCAGTTGAGCAGAAGTACGG-3′. The nucleotide sequences corresponding to the 10-bp hDRE are shown in bold letters, with nucleotides substituted for the wild-type sequence in lowercase. These oligonucleotide pairs were used to create base substitution mutant RPS6 promoter-luciferase reporter plasmids (RPS6-310+43/pGL3) and also as competitors in electrophoretic gel mobility shift assays (EMSA). Double-stranded oligonucleotides containing hDRE in the promoter region of the human histone H1 gene were described previously (26).
Oligonucleotide pairs used for PCR amplification of the promoter regions of RPS6 (−310 to +43), RPL10A (−193 to +17), and RPL12 (−277 to +57) were synthesized. The sequences used are as follows: for RPS6, 5′-ATAggtaccGTCAGATGCAAAGTG-3′ and 5′-ATATagatctCTTGAAGCAGCTGAACGCCT-3′; for RPL10A, 5′-ATAggtaccCGACAACTCTGTGGGTTACCG-3′ and 5′-ATATagatctATGGCTTCTCACGCCGCGCTA-3′; and for RPL12, 5′-ATAggtaccGGGTGACACTCACGATAAAGG-3′ and 5′-ATATagatctATTCGGGACGACCGAAGGAAG-3′. Recognition sites for Asp718 and BglII are denoted by lowercase letters.
PCR primer pairs were synthesized and used for semiquantitative reverse transcriptase PCR (RT-PCR) for transcripts of the following genes: hDREF, 5′-TGGTGGAGGAGCTGAGCAACTT-3′ and 5′-AGAAACACCTGCTCG TCCACGT-3′; RPS6, 5′-TGGATGCAAATCTGAGCGTT-3′ and 5′-TTCTTTATTTTTCTTGGTACGCT-3′; RPL10A, 5′-AGCAGCACTGTGACGAGGCTA-3′ and 5′-TTTGGCCACCATGTTTTCGTTG-3′; RPL12, 5′-AGTCGTATACCTGAGGTGCACCGGA-3′ and 5′-GCCATCAACATTACAGCCCACTGAC-3′; and ACTB (β-actin), 5′-CGCTCGTCGTCGACAACGGCTC-3′ and 5′-TCAAACATGATCTGGGTCATCTTCTC-3′. PCR primer pairs for the human histone H1 gene were described previously (26).
PCR primer pairs used in chromatin immunoprecipitation (ChIP) assays are as follows: RPS6 proximal, 5′-ACAACCTCAGACCCACACCCAACCG-3′ and 5′-ATATAGATCTCTTGAAGCAGCTGAACGCCT-3′; RPS6 distal, 5′-ATGTGCCTTTGGAAGGCCTTAGCAC-3′ and 5′-CAAAGTGTGTGGTGCTAATAGTCCCA-3′; RPL10A proximal, 5′-ATAGGTACCCGACAACTCTGTGGGTTACCG-3′ and 5′-ATATAGATCTATGGCTTCTCACGCCGCGCTA-3′; RPL10A distal, 5′-GTGGGATACCAGGCACACAACGTGG-3′ and 5′-TCTAGTCTTCTAGGTCATTCTAGTC-3′; RPL12 proximal, 5′-ATCTCTAGCTTCAGCGCACCGCGGT-3′ and 5′-ATATAGATCTATTCGGGACGACCGAAGGAAG-3′; and RPL12 distal, 5′-AACATTCTTCTTACTTGAAGGGTGC-3′ and 5′-GTGCTGGCTCAGTCCCTCACAATCT-3′.
Oligonucleotide pairs used for constructing lentivirus vectors (pCS-U6-shRNA-EG) expressing shRNAs against hDREF (si3 and si4), shRNAs against RPS6 (RPS6-#1 and RPS6-#2), and scramble shRNAs as a negative control were as follows: for si3, 5′-caccGCAACAACCACCACCTCATGCccacaccGCATGAGGTGGTGGTTGTTG-3′ and 5′-aaaaCAACAACCACCACCTCATGCggtgtggGCATGAGGTGGTGGTTGTTGc-3′; for si4, 5′-caccgCAACTTCAAGTCCCAGAAGGccacaccCCTTCTGGGACTTGAAGTTG-3′ and 5′-aaaaCAACTTCAAGTCCCAGAAGGggtgtggCCTTCTGGGACTTGAAGTTGc-3′; for RPS6-#1, 5′-caccgCGTCTTGTTACTCCACGTccacaccACGTGGAGTAACAAGACGc-3′ and 5′-aaaaGCGTCTTGTTACTCCACGTggtgtggACGTGGAGTAACAAGACGC-3′; for RPS6-#2, 5′-caccgGAACAAATTGCGAAGAGAccacaccTCTCTTCGCAATTTGTTCC-3′ and 5′-aaaaGGAACAAATTGCGAAGAGAggtgtggTCTCTTCGCAATTTGTTCc-3′; and for scramble, 5′-caccgcgcgctttgtaggattcgccacacccgaatcctacaaactacaaa-3′ and 5′-aaaaggcgctttgtaggatacgggtgtggcgaatcctacaaagcgcgc-3′. Sense and antisense target sequences against hDREF or RPS6 mRNAs are shown by capital letters.
To construct a plasmid expressing a si4 knockdown-resistant hDREF, the following oligonucleotides were used: 5′-tggaggagctgagTaaTttTaaAAGccAaaAagtgcttggcctca-3′ and 5′-tgaggccaagcactTttAggCTTttAaaAttActcagctcctcca-3′. The substituted nucleotides are indicated by capital letters.
Cell culture.
HeLa cells were cultured as described previously (42). HFF cells, generated by infection with a retrovirus expressing hTERT (a gift from T. Kiyono) (21), were maintained in Dulbecco's modified Eagle's medium (high glucose) with 10% fetal bovine serum and antibiotic-antimycotic (Sigma) at 37°C under 5% CO2.
Preparation and transduction of recombinant lentiviruses.
The plasmids used in preparation of recombinant lentiviruses were described previously (42). Recombinant lentiviruses were produced as follows. 293FT cells were cultured according to the manufacturer's instructions (Invitrogen). For overexpression of hDREF, 7 μg of pCSII-EF-hDREF-IRES2-Venus, 5 μg of pCAG-HIVgp, and 4 μg of pCMV-VSV-G-RSV-Rev were cotransfected. For knockdown against hDREF or RPS6, 13 μg of pCS-U6-shRNA-EG, 5 μg of pCAG-HIVgp, and 4 μg of pCMV-VSV-G-RSV-Rev were cotransfected. Supernatants containing the recombinant lentiviruses were collected 48 h after DNA transfection, passed through a 0.45-μm filter, and used for transduction experiments. HFF cells were transduced with indicated lentiviruses for 6 h, washed with phosphate-buffered saline (PBS) once, and used for the experiments. Transduction efficiencies were determined by detecting green fluorescent protein (GFP) or Venus, a derivative of yellow fluorescent protein, using an IX-70 fluorescence microscope (Olympus).
Antibodies.
Rabbit polyclonal antibodies against acetyl-histone H3 (catalog no. 06-599) and acetyl-histone H4 (catalog no. 06-866) and a mouse monoclonal antibody against RNA polymerase II (clone CTD4H8; catalog no. 05-0623) were obtained from Upstate. A rat monoclonal anti-HA antibody (clone 3F10) was purchased from Roche, and rabbit polyclonal anti-GFP antibodies (632459) were obtained from Clontech. Polyclonal antibodies specifically reacting with hDREF were raised against glutathione S-transferase-hDREF and affinity purified as described previously (26). Anti-rabbit immunoglobulin G (IgG) species-specific antibodies linked to horseradish peroxidase (NA934) were obtained from GE Healthcare Biosciences.
Western blotting.
Cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to an Immobilon-NC membrane (Millipore). Membranes were blocked with PBS containing 5% skim milk and 0.05% Tween 20 and incubated with the primary antibody. Secondary horseradish peroxidase-conjugated anti-rabbit IgG species-specific antibodies were used for detection with chemiluminescence reagent (ECL; GE Healthcare Biosciences).
BrdU labeling.
Cells were plated at 2.6 × 103 cells/cm2 and transduced with each recombinant lentivirus as described in the figure legends. After each time point, cells were incubated in the presence of 15 μg/ml of bromodeoxyuridine (BrdU) (Roche) for 2 h. Detection of incorporated BrdU was performed as described previously (26). For each sample, at least 150 cells per field were counted and three fields were observed (total counted cell number was more than 500). The percentages of BrdU-positive cells in three fields were averaged.
Quantitative RT-PCR.
Total RNA was prepared from HFF cells by using an RNeasy mini kit (QIAGEN) according to the manufacturer's protocol. Total RNAs from synchronized WI38 cells, kindly provided by M. Fujita, were described previously (5). One microgram of RNA was subjected to synthesis of first-strand cDNAs with oligo(dT)20 primers and a recombinant Moloney murine leukemia virus reverse transcriptase (Invitrogen). PCRs were performed in 25 μl of reaction mixture containing 1× buffer II (Applied Biosystems), 1.25 mM MgCl2, 10 pmol of each of the gene-specific primer pairs, 200 μM each of dATP, dTTP, dCTP, and dGTP, 18.5 kBq [α-32P]dCTP, 0.1 U of AmpliTaq Gold (Applied Biosystems), and cDNA (1 μl of 30× diluted with H2O) as a template. The PCR included heating for 10 min at 95°C, followed by 20 cycles of 96°C for 30 s, 57°C for 30 s, and 72°C for 1 min to amplify quantitatively cDNAs for RPS6, RPL10A, RPL12, and the histone H1 gene. PCR for amplification of hDREF and β-actin cDNA included 25 and 18 cycles, respectively, under the same reaction conditions. PCR products were separated on 6% polyacrylamide gels. After drying, images were taken using FLA-3000 (Fuji Film) and the amount of cDNA was quantified with the Image Gauge program (Fuji Film). For quantitative analysis, each primer set was used to generate a standard curve with serially diluted cDNA as a template.
In vivo protein synthesis.
Cells were seeded to 12-well plates (3 × 104 cells/well) and, the next day, transduced with recombinant lentiviruses. After 48 h, they were washed once with PBS and then replenished with Dulbecco's modified Eagle's medium without methionine and cysteine (Invitrogen) supplemented with 10% dialyzed fetal calf serum, 2 mM glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. One hour later, cells were labeled with 18.5 kBq of EXPRE35S35S (PerkinElmer) per well for 1 h and then washed with PBS containing methionine and cysteine (10 mM each). Cells were lysed in 200 μl of lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% sodium deoxycholate, 0.1% Triton X-100, and 1% NP-40, and aliquots of 10 μl were taken for precipitation in 10% trichloroacetic acid (TCA) containing 10 mM each of methionine and cysteine. The precipitates were spotted onto filters (GF/C) and washed three times with 5% TCA and once with ethanol. After drying, filters were subjected to liquid scintillation counting and protein synthesis was calculated from TCA-insoluble counts per minute per 1 × 104 cells in each sample.
DNA transfection and luciferase assays.
For luciferase assays, 3.5 × 104 HeLa cells were seeded in 24-well plates and cultured overnight. The next day, DNA-calcium phosphate precipitates containing a total of 1.5 μg of plasmids were added (42). The lack of plasmids was compensated for with an empty vector. At 4 h after addition of the precipitates, the precipitates were removed, and cells were cultured for 20 h in fresh medium. Firefly luciferase activities were measured using a dual luciferase reporter assay system (Promega) with a Lumat LB9501 luminometer (Berthold) and normalized to sea pansy luciferase activity by using pRL-TK (Promega). The assays were carried out in triplicate, and the averages are shown, together with standard deviations.
In vitro transcription/translation and EMSA.
In vitro transcription/translation of hDREF was performed using pcDNA3-HA-hDREF as a template and a TNT T7 quick coupled transcription/translation system (Promega) according to the manufacturer's protocol. EMSA were performed as described previously (26), using in vitro-synthesized HA-hDREF proteins.
ChIP assays.
ChIP assays were performed based on a published protocol (36). HFF cells (0.8 × 107 cells) stably expressing HA-tagged hDREF were washed twice with PBS containing 2% fetal calf serum, suspended in 30 ml of PBS, and cross-linked with 1.1% formaldehyde for 5 min at room temperature. After termination of the cross-linking by addition of glycine to a final concentration of 0.125 M, cells were lysed with 3 ml of lysis buffer containing 50 mM Tris-HCl (pH 8.1), 5 mM EDTA, 1% sodium dodecyl sulfate, 1 mM sodium fluoride, 1 mM sodium orthovanadate, and protease inhibitors. Cell lysates were sonicated to yield chromatin fragments of ∼600 bp, as assessed by agarose gel electrophoresis, and then diluted 10-fold with dilution buffer (20 mM Tris-HCl [pH 8.1], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 1 mM sodium fluoride, 1 mM sodium orthovanadate, and protease inhibitors). Diluted lysates were precleared by incubation with 20 μl of protein A-Sepharose and 20 μl of protein G-Sepharose (A/G mix; GE Healthcare Sciences) and sheared salmon sperm DNA. After centrifugation for 5 min at 7,000 × g, the supernatant was transferred to a fresh tube. One-tenth of the precleared lysate was subjected to immunoprecipitation by rocking overnight at 4°C with the appropriate antibody, and immune complexes were then precipitated with A/G mix and sheared salmon sperm DNA. Beads were collected by centrifugation and washed twice sequentially with Paro buffer I, Paro buffer II, Paro buffer III, and Tris-EDTA (27), and immunoprecipitates were eluted by adding 200 μl of elution buffer (1% sodium dodecyl sulfate, 0.1 M NaHCO3), followed by incubation at 65°C overnight. After centrifugation for 5 min at 7,000 × g, DNA was purified using a QIAGEN PCR purification kit. A 0.5-μl DNA sample was then subjected to quantitative amplification by use of gene-specific primer pairs.
RESULTS
hDREF positively regulates human cell proliferation.
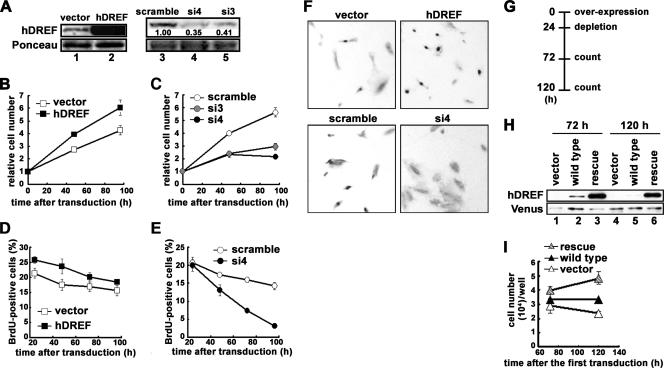
Our previous observation that knockdown of hDREF using small interfering RNA prohibited cells from entering S phase (26) prompted us to ask if hDREF is a rate-limiting factor in human cell proliferation. To test this possibility, we examined the effects of hDREF overexpression and depletion on cell proliferation in HFF cells, a normal human foreskin fibroblast line immortalized with hTERT (21). For overexpression, a lentivirus constitutively expressing hDREF under the control of the human EF-1α gene promoter was used. For depletion experiments, we selected two target sequences (named si3 and si4) in the coding region of hDREF mRNA and constructed lentivirus expressing shRNA against hDREF mRNA under the control of the RNU6A gene promoter. We confirmed that more than 95% of the HFF cells were successfully transduced with lentivirus by a single exposure, as assessed by observing marker fluorescence protein expression (data not shown). Therefore, the transduced cells were directly used for experiments without any selection. Western blotting revealed that transduction with lentivirus overexpressing hDREF resulted in increase of hDREF protein (Fig. 1A, lanes 1 and 2), while expression of si4 and si3 shRNAs reduced the expression level to 35 and 41%, respectively, compared with controls (Fig. 1A, lanes 3, 4, and 5).
FIG. 1.
hDREF regulates human cell proliferation. (A) HFF cells were transduced with lentiviruses carrying an empty vector or expressing hDREF, scramble, si4, or si3 shRNA. Whole-cell extracts at 96 h after transduction were prepared, and protein samples were analyzed by immunoblotting using anti-hDREF antibodies (top). Consistent amounts of proteins loaded were confirmed by staining the transferred membrane with Ponceau dye (bottom). (B) Numbers of HFF cells transduced with lentiviruses carrying an empty vector or expressing hDREF were counted at 0, 48, and 96 h after transduction. Error bars represent the standard deviations with triplicate wells. Representative data from four independent experiments are shown. (C) Numbers of HFF cells transduced with lentiviruses expressing scramble shRNA or si3 and si4 shRNAs against hDREF mRNA. Error bars represent standard deviations with triplicate wells. Representative data from four independent experiments are shown. (D) Increase of BrdU-positive cells with overexpression of hDREF. HFF cells were transduced with lentiviruses carrying an empty vector or expressing hDREF. At 24, 48, 72, and 96 h after transduction, cells were labeled with BrdU for 2 h. Error bars represent the standard deviations with triplicate wells. (E) BrdU-positive cells were decreased by depletion of hDREF. HFF cells were transduced with the lentivirus expressing scramble shRNA or si4 shRNA. The time course and cell labeling with BrdU were the same as those described for panel D. (F) Images of BrdU-labeled HFF cells at 96 h after transduction with the indicated lentiviruses. (G) Rescue experimental design. (H) Whole-cell extracts at 72 and 120 h after the first transduction were prepared, and proteins were analyzed by immunoblotting using anti-hDREF antibodies (top). First-transduction efficiencies were confirmed by immunoblotting using anti-GFP antibodies to detect Venus proteins (bottom). (I) Numbers of HFF cells were counted at 72 and 120 h after the first virus transduction. Error bars represent the standard deviations with triplicate wells. Representative data of three independent experiments are shown.
Strikingly, HFF cells overexpressing hDREF proliferated faster than the control cells (Fig. 1B), with population doubling times of 16.3 h and 27.9 h, respectively. In contrast, HFF cells expressing si3 and si4 shRNAs proliferated significantly more slowly, and the population doubling times calculated with cell numbers at 48 h after virus transduction were 33.8 h and 35.8 h, respectively (Fig. 1C). It should be noted that HFF cells with si4 shRNA appeared to be quiescent after 4 days from transduction. A BrdU labeling experiment also revealed positive effects of hDREF on cell proliferation (Fig. 1D and F), whereas depletion of hDREF with si4 shRNA resulted in failure to incorporate BrdU at 96 h after transduction (Fig. 1E and F). To confirm whether the observed phenotype was specifically induced by depletion of hDREF protein, we performed a rescue experiment (Fig. 1G). We prepared a lentivirus expressing a si4-resistant mRNA of hDREF with an 8-bp substitution mutation in the si4 shRNA targeting region. It should be noted that this mutation did not change the amino acid sequence of hDREF. HFF cells were first transduced with lentiviruses carrying the wild-type or the si4-resistant sequence of hDREF cDNA, and the lentivirus expressing si4 shRNA was subsequently introduced at 24 h after the first transduction. As shown in Fig. 1H, Western blotting of total extract of HFF cells revealed that cells transduced with virus carrying wild-type hDREF cDNA contained a little hDREF at 72 h (Fig. 1H, lane 2) but not at 120 h (Fig. 1H, lane 5) after the first transduction, while cells transduced with lentivirus with empty vector did not express any detectable level of hDREF protein (Fig. 1H, lanes 1 and 4). As expected, cells transduced with lentivirus expressing si4-resistant mRNA for hDREF accumulated hDREF protein (Fig. 1H, lanes 3 and 6). Figure 1I shows cell numbers at 72 and 120 h after the first virus transduction. Neither cells expressing wild-type hDREF mRNA nor cells with lentivirus carrying empty vector proliferated during 72 to 120 h after the first transduction. In contrast, cells expressing si4-resistant hDREF mRNA persisted in a proliferative state. The results indicate that retardation of cell proliferation by si4 shRNA expression is specifically induced as a consequence of hDREF depletion.
hDRE-like sequences are present in the promoter regions of human RP genes.
In order to identify novel target genes of hDREF, we performed a computational search of hDREF-binding sites in a database containing human promoter sequences (DBTSS [http://dbtss.bioinf.med.uni-goettingen.de/]). We found 208 genes carrying a sequence with more than an 8-bp match to 10 bp of the hDREF-binding consensus sequence [hDRE; 5′-TGTCG(C/T)GA(C/T)A-3′] within a 200-bp region upstream from the transcriptional start sites among 8,793 human genes. We noted that 22 out of 79 human RP genes possessed hDRE-like sequences in their promoter regions and that 20 of the relevant hDRE-like sequences were positioned at distances centered around 60 bp upstream of the putative transcriptional start sites (Table 1) . Despite several reports that disruption in one or more of the processes that control protein synthesis is associated with alteration in the cell cycle and cell growth in Saccharomyces cerevisiae (18, 19), the transcriptional regulation of mammalian RP genes in vivo remains unclear (28). Here, we first examined whether RP gene transcription is affected by depletion of hDREF. For this purpose, RPS6, RPL10A, and RPL12 were chosen as representative genes (Table 1).
TABLE 1.
Human RP genes carrying hDRE-like sequence
| Genea | Position | Sequenceb
|
No. of matches | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T | G | T | CG | C/T | G | A | C/T | A | |||
| RPS2 | −38 | T | G | g | CG | T | G | c | T | A | 8 |
| RPS3A | 91 | a | G | T | CG | C | G | A | C | t | 8 |
| 100 | T | G | T | CG | T | G | g | C | g | 8 | |
| RPS4X | −583 | T | G | g | CG | C | G | A | T | c | 8 |
| RPS4Y | −962 | T | G | g | CG | C | G | A | T | c | 8 |
| RPS6* | −538 | T | G | T | CG | T | G | c | C | t | 8 |
| −51 | T | c | T | CG | C | G | A | g | A | 8 | |
| RPS7 | −17 | T | c | T | CG | C | G | A | g | A | 8 |
| RPS11 | −739 | T | G | g | CG | C | G | A | T | c | 8 |
| −275 | T | c | T | CG | C | G | A | T | A | 9 | |
| −192 | T | c | T | CG | C | G | A | g | A | 8 | |
| RPS12 | −279 | T | G | T | CG | C | G | c | T | c | 8 |
| RPS14 | 49 | g | G | g | CG | C | G | A | C | A | 8 |
| RPS15 | −599 | T | G | g | CG | C | G | A | T | c | 8 |
| −38 | T | c | T | CG | C | G | A | T | A | 9 | |
| RPS15A | −53 | T | c | T | CG | C | G | A | T | A | 9 |
| RPS16 | −67 | c | G | T | CG | C | G | A | C | g | 8 |
| RPS18 | −11 | T | G | a | CG | C | G | A | T | A | 9 |
| 63 | T | G | c | CG | T | G | A | T | c | 8 | |
| RPS19 | −865 | T | c | T | CG | C | G | A | g | A | 8 |
| −524 | T | c | T | CG | C | G | A | C | t | 8 | |
| −51 | T | c | T | CG | C | G | A | g | A | 8 | |
| RPS21 | −73 | T | G | g | CG | T | G | A | C | c | 8 |
| RPS26 | −868 | T | G | g | CG | C | G | A | T | c | 8 |
| RPS27A | 150 | T | t | T | CG | T | G | A | a | A | 8 |
| RPS28 | −57 | T | c | T | CG | C | G | A | T | A | 9 |
| RPS30 | −49 | T | G | a | CG | T | G | A | C | A | 9 |
| RPL3 | 21 | T | G | g | CG | T | G | A | T | g | 8 |
| RPL5 | −48 | T | G | g | CG | T | G | A | C | c | 8 |
| RPL6 | −366 | T | G | T | CG | C | G | A | T | c | 9 |
| RPL7A | −51 | T | c | T | CG | C | G | A | T | c | 8 |
| RPL8 | −57 | T | G | T | CG | C | G | g | C | c | 8 |
| RPL9 | −35 | T | t | a | CG | C | G | A | T | A | 8 |
| RPL10 | −233 | T | c | T | CG | C | G | A | C | c | 8 |
| RPL10A* | −41 | T | c | T | CG | C | G | A | T | A | 9 |
| RPL11 | −424 | T | G | g | CG | C | G | A | T | A | 9 |
| −266 | g | c | T | CG | T | G | A | C | A | 8 | |
| RPL12* | −56 | T | c | T | CG | C | G | A | T | A | 9 |
| RPL17 | −51 | T | c | T | CG | C | G | A | g | A | 8 |
| RPL26 | −49 | T | c | T | CG | C | G | A | g | A | 8 |
| RPL28 | −72 | T | G | T | CG | T | G | c | C | c | 8 |
| RPL30 | −408 | T | G | g | CG | C | G | A | T | c | 8 |
| RPL31 | −562 | T | G | T | CG | T | G | t | C | A | 9 |
| −493 | c | G | a | CG | C | G | A | C | A | 8 | |
| RPL35 | −614 | T | G | T | CG | C | G | A | T | c | 9 |
| RPL35A | −985 | T | G | c | CG | T | G | t | C | A | 8 |
| RPLP2 | −182 | T | G | c | CG | C | G | A | a | A | 8 |
*, genes chosen for examination to determine whether RP gene transcription is affected by depletion of hDREF.
The consensus sequence is given at the top. There were 10 matches in the consensus sequence. Sequences corresponding to the consensus sequence are shown in bold, uppercase letters, and nucleotides not matched to the consensus hDREF-binding sequence are shown in lowercase letters.
hDREF depletion reduced transcription of RPS6, RPL10A, and RPL12 genes and in vivo protein synthesis.
Transcripts for RPS6, RPL10A, and RPL12 were assessed by RT-PCR using mRNA extracted from HFF cells expressing si4 shRNA. All three were down-regulated as a consequence of depletion of hDREF (Fig. 2A). Next, we measured the rate of protein synthesis in HFF cells transduced with lentivirus expressing si4 shRNA by monitoring the incorporation of [35S]methionine and [35S]cysteine into total soluble proteins (Fig. 2B). Protein synthesis was diminished by 25%, pointing to insufficiency of ribosome biogenesis caused by down-regulation of plural RP gene expression.
FIG. 2.
Reduction of mRNA levels of RPS6, RPL10A, and RPL12 by hDREF depletion in HFF cells. (A) Total RNA was extracted from HFF cells transduced with lentiviruses expressing scramble shRNA or si4 shRNA against hDREF at 72 h after transduction. A representative gel image for three independent experiments is shown. The relative mRNA levels (RP/ACTB) were determined by densitometry, setting that for scramble as 1.0. Values and error bars in the right histogram represent the average values and standard deviations, respectively, for three independent experiments. (B) Decrease in protein synthesis by depletion of hDREF. HFF cells were transduced with lentiviruses expressing scramble shRNA or si4 shRNA. At 48 h after transduction, protein synthesis rates were measured in triplicate.
hDRE-like sequences in promoter regions of RP genes are positive regulatory elements.
We next investigated whether the hDRE-like sequences indeed act as positive regulatory elements for the RP gene transcriptions. Luciferase reporter plasmids containing promoter regions of RPS6 (−310 to +43), RPL10A (−193 to +17), and RPL12 (−277 to +57) were used for a transient expression assay of luciferase activity in HeLa cells. To test whether hDRE-like sequences are positive elements of the RP gene expression, we destroyed hDRE-like sequences by a 12-bp linker insertion at their centers (Fig. 3A), resulting in reductions of luciferase activity by 38%, 59%, and 46%, respectively, with RPS6, RPL10A, and RPL12. Next, to determine the nucleotide sequence functioning as a positive element, a set of 3-bp substitution mutations was introduced into the outside and inside of the hDRE-like sequence of RPS6. As shown in Fig. 3B, 3-bp substitutions in the hDRE-like sequence (mut2, mut3, mut4, and mut5) reduced luciferase expression, while no significant effect was observed in mut1 with a 3-bp substitution outside of the hDRE-like sequence. In particular, a 3-bp substitution in the center of the hDRE-like sequence (mut3) reduced the luciferase activity by 55% compared to that of the wild type. These results indicate that the 12-bp sequence from −43 to −32 (5′-TCTCGCGAGAAC-3′) is basically important as a positive element of RPS6. To explore whether hDREF can transactivate RP gene promoters, cotransfection experiments were performed. As shown in Fig. 3C, cotransfection of a plasmid expressing hDREF increased luciferase expression directed by each RP gene promoter more than 30% compared with the values obtained without the hDREF-expressing plasmid. To determine whether the increased luciferase expression by hDREF is dependent on the hDRE-like sequence, a cotransfection experiment with the hDREF-expressing plasmid and an RPS6 luciferase reporter plasmid lacking the 10-bp hDRE-like sequence (ΔhDRE) was performed (Fig. 3D). Deletion of the hDRE-like sequence in the RPS6 promoter resulted in reduction of luciferase activity by 49% compared with that of the wild type and complete loss of the stimulation by coexpression of hDREF, indicating that hDREF stimulates luciferase gene expression dependent on the hDRE-like sequence in the RPS6 promoter.
FIG. 3.
hDRE-like sequences act as positive regulatory elements for RP gene promoter activities in vivo. (A) Decrease in promoter activities of RPS6, RPL10A, and RPL12 due to linker insertion mutations in hDRE-like sequences. HeLa cells were cotransfected with 1.4 μg of pGL3 basic vector (basic), wild-type, or linker insertion mutant reporter plasmids containing the indicated RP gene promoters and 0.1 μg of pRL-TK in single wells of 24-well plates. Representative data of two independent experiments are shown. (B) Decrease in promoter activity with base substitution mutations in the hDRE-like sequence of RPS6. HeLa cells were cotransfected with 1.4 μg of wild-type (wt) or base substitution mutant (mut1, -2, -3, -4, and -5) reporter plasmids containing the RPS6 promoter and 0.1 μg of pRL-TK in single wells of 24-well plates. Relative luciferase activities are shown, taking the activity of the wild-type RPS6 promoter as 1.0. Nucleotide sequences from −54 to −40 of the RPS6 promoter and its base substitution mutants are provided, with substituted nucleotides indicated by lowercase letters and sequences corresponding to that of hDRE boxed. Representative data from two independent experiments are shown. (C) Transactivation of the RP gene promoter by hDREF. HeLa cells were cotransfected with 1.0 μg of luciferase reporter plasmids carrying the indicated RP gene promoters (RPS6, RPL10A, and RPL12), 0.1 μg of pcDNA3-HA (−hDREF) or pcDNA3-HA-hDREF (+hDREF), 0.1 μg of pRL-TK, and 0.3 μg of pCMX in single wells of 24-well plates. Representative data of two independent experiments are shown. (D) Transactivation of the RPS6 promoter by hDREF is dependent on the hDRE-like sequence. HeLa cells were cotransfected with 1.0 μg of luciferase reporter plasmids carrying the wild type or the hDRE-like sequence deletion mutant (ΔhDRE) of the RPS6 promoter, 0.2 μg of pcDNA3-HA (−hDREF) or 0.1 μg of pcDNA3-HA-hDREF (+hDREF), 0.1 μg of pRL-TK, and 0.2 or 0.3 μg of pCMX in single wells of 24-well plates. Representative data of two independent experiments are shown.
hDREF binds to hDRE-like sequences of RP genes in vitro and in vivo.
To examine whether hDRE-like sequences in the RP gene promoters can be recognized by hDREF, we carried out a competitive gel mobility shift assay using oligonucleotides harboring the hDRE sequence in the histone H1 gene (H1-hDRE) as a probe with an in vitro-synthesized full-length hDREF protein. Two protein-DNA complexes were formed with hDREF and the radiolabeled probe (Fig. 4, lane 3). These two bands were diminished by adding excess amounts of unlabeled wild-type H1-hDRE (Fig. 4, lane 4) but not by addition of the H1-hDRE mutant oligonucleotide with a 5-bp substitution in the core of hDRE (Fig. 4, lane 5), indicating that these two bands are signals for specific complexes. An oligonucleotide containing the nucleotide sequence from −56 to −22 of the RPS6 (Fig. 4, lane 6) effectively competed for the complex formation between hDREF and H1-hDRE, suggesting that hDREF specifically binds to a 35-bp hDRE-like sequence present in the RPS6 promoter. To determine the nucleotide sequence critical for hDREF binding, a set of oligonucleotides each carrying a 3-bp substitution mutation were used as competitors. DNA-protein complexes were diminished appreciably by adding unlabeled mut1 or mut5 oligonucleotide (Fig. 4, lanes 7 and 11), while mut2, mut3, or mut4 oligonucleotides did not compete for binding (Fig. 4, lanes 8, 9, and 10), indicating that the nucleotide sequence from −43 to −35 is important. The sequence required for hDREF binding almost overlapped with that for transcriptional activation (Fig. 3B), suggesting that hDREF binds to the hDRE-like sequence in the RPS6 promoter and stimulates RPS6 transcription.
FIG. 4.
Complex formation between the hDRE-like sequence in RPS6 and hDREF in vitro. EMSA analysis was performed using [32P]-labeled double-strand oligonucleotides containing histone H1-hDRE as a probe. The probe was incubated with reticulocyte lysate containing no hDREF protein (lane 2) or reticulocyte lysate containing hDREF protein (lanes 3 to 11). The following competitors were added to the binding reaction: no competitor (lanes 2 and 3), oligonucleotide containing H1-hDRE (lane 4) and its base substitution mutant (lane 5), and oligonucleotide containing RPS6-hDRE-like sequence (lane 6) and its base substitution mutants mut1, mut2, mut3, mut4, and mut5 (lanes 7 to 11). The probe was incubated with no reticulocyte lysate and no competitor (lane 1). Two arrowheads denote signals for complexes containing H1-hDRE and hDREF protein. wt, wild type.
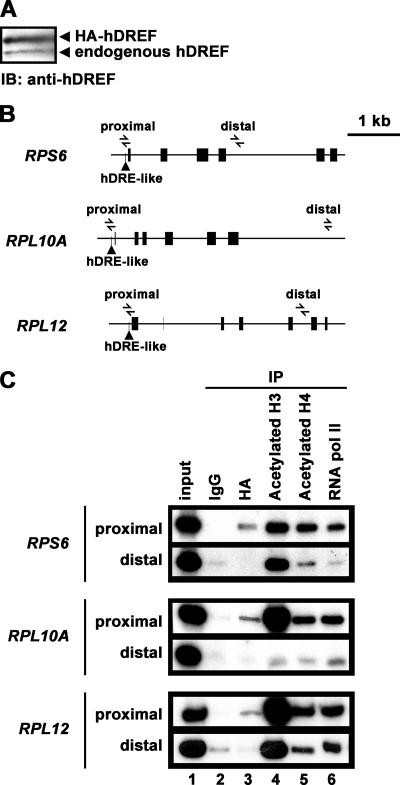
Next, we investigated binding of hDREF to three RP genes in vivo by ChIP (Fig. 5B). Because the polyclonal antibodies against hDREF did not effectively immunoprecipitate chromatin (data not shown), we prepared HFF or HeLa cells stably expressing HA-tagged hDREF by using a lentivirus vector. To avoid artifacts caused by overexpression of hDREF, expression was driven by the human metallothionein gene promoter without metal induction. The expression level of HA-hDREF was estimated to be elevated twofold by Western blotting using anti-hDREF antibody, and immunofluorescence with anti-HA antibody revealed the same subnuclear localization as for endogenous hDREF (Fig. 5A and data not shown). Chromatin from cells expressing HA-hDREF was immunoprecipitated with antibodies recognizing the HA tag, acetylated histone H3, acetylated histone H4, and RNA polymerase II. Results obtained using HFF cells are shown in Fig. 5C. We detected a high level of acetylation of histone H3 inside all three RP genes, even in regions distal to the transcription start sites, whereas no acetylation signals were found outside, as shown distal to RPL10A in Fig. 5C, lane 4. We also observed reproducible gene-specific differences in acetylation level of histone H4 and recruitment of RNA polymerase II (Fig. 5C, lanes 5 and 6), although the significance remains unclear at present. Weak but significant signals for HA-hDREF binding were detected in the promoter regions of all three RP gene promoters but not in the distal region (lane 3), indicating that hDREF is locally present on the promoter regions in vivo. Similar results were obtained using HeLa cells stably expressing HA-hDREF (data not shown).
FIG. 5.
hDREF binds to RP gene promoter regions containing hDRE-like sequences in vivo. (A) Whole-cell extracts of HFF cells transduced with lentiviruses expressing HA-tagged hDREF under the control of the human metallothionein gene promoter were prepared, and protein samples were analyzed by immunoblotting (IB) using anti-hDREF antibodies. Upper and lower bands correspond to exogenous HA-hDREF and endogenous hDREF, respectively. (B) Schematic illustration of the RPS6, RPL10A, and RPL12 genes. Arrowheads represent primers used for PCR. (C) ChIP analysis. PCR was performed using genomic DNA from the input extract and immunoprecipitates (IP) with control IgG and anti-HA, anti-acetylated histone H3, anti-acetylated histone H4, and anti-RNA polymerase II (pol II) antibodies.
Close correlation between expression of hDREF and RP genes in a cell cycle-dependent manner.
We next examined whether fluctuation of hDREF expression during the cell cycle correlates with those of RP genes. RNAs from the synchronized WI38 human diploid fibroblast line were used to determine the levels of mRNAs for hDREF and three RPs (Fig. 6A and B). We checked cell cycle progression after release from serum starvation by detecting histone H1 gene mRNA, which is transiently induced during late G1 to S phases (5, 9, 23). Signals for the histone H1 gene mRNA were first detected in cells at 16 h and reached a maximum at 20 to 24 h after adding serum, indicating that synchronization was successfully performed; this time period corresponds to late G1 to S phases. The hDREF mRNA began to increase after serum addition, reaching a maximum after 6 to 24 h, and then decreased. This cell cycle-dependent fluctuation of mRNA for hDREF is quite similar to that of hDREF protein in HEL cells (26). Signals for mRNAs of the three RP genes were weakly detected in serum-starved cells, and amounts began to increase at 6 h and reached a maximum at 20 to 24 h after serum addition. Considering that RP gene expression was increased subsequent to upregulation of hDREF, an etiological link is conceivable.
FIG. 6.
Cell cycle-dependent fluctuation of hDREF and RP gene expression in WI38 cells. (A) Total RNA was extracted from asynchronous (lane 1) and synchronized (lanes 2 to 9) WI38 cells by use of a serum starvation method. RT-PCR was performed for hDREF, RPS6, RPL10A, RPL12, H1F0 (histone H1), and ACTB (β-actin) genes. (B) Quantitative expression data. The relative mRNA levels for hDREF, RPS6, RPL10A, RPL12, and H1F0 (histone H1) were determined by densitometry, normalized to those for β-actin, and plotted against time after serum addition. Values are given relative to the mRNA levels obtained using WI38 cells at 0 h after serum addition. Average values from three experiments are shown, with standard deviations.
Depletion of RPS6 impairs cell proliferation and mimics the hDREF-depleted phenotype.
Finally, we examined whether depletion of an RP gene containing hDRE in the promoter from human cells caused cell proliferation delay, a phenotype of hDREF-depleted cells. To address this issue, we chose RPS6 as a target for depletion, since a study showed that conditional knockout of mouse rpS6 resulted in defective ribosome biogenesis (38). We made two kinds of lentivirus expressing shRNAs for RPS6 and transduced these lentiviruses into HFF cells. As shown in Fig. 7A, quantitative RT-PCR analysis clearly showed that RPS6-#1 and RPS6-#2 shRNAs reduced RPS6 expression by 49% and 68%, respectively. We therefore investigated effects of RPS6 depletion on cell proliferation and confirmed failure to incorporate BrdU (Fig. 7B and C).
FIG. 7.
Reduction of BrdU incorporation in HFF cells on depletion of the RPS6 gene. (A) Total RNA was extracted from HFF cells transduced with lentiviruses expressing scramble shRNA or RPS6-#1 or -#2 shRNAs against RPS6 at 96 h after transduction. RT-PCR was performed for RPS6 and ACTB (β-actin). (B) HFF cells were transduced with lentiviruses expressing scramble shRNA or RPS6-#1 and -#2 shRNAs. At 48, 72, and 96 h after transduction, cell labeling with BrdU and detection were performed as described in the legend for Fig. 1D. (C) Images of BrdU-labeled HFF cells at 96 h after virus transduction.
DISCUSSION
In the present study, we demonstrated that hDREF positively regulates cell proliferation. We also showed that not only transcription of several human RP genes carrying hDRE-like sequences but also the rate of protein synthesis is reduced by hDREF depletion in human cells. Our evidence from luciferase reporter assays, EMSA, and ChIP analysis provides proof that hDREF is a positive transcription factor binding to hDRE-like sequences in RP genes and strongly indicates that the human DRE/DREF system has an important role in cell proliferation in supporting RP gene expression.
In yeast, it has been demonstrated that deletions of only one of the RP genes can shut down ribosome production and affect critical cell size at the start phase, which occurs late in G1 phase (18). Studies using yeast showed that yeast RP genes are highly expressed in the cycling cells in rich medium but markedly and cooperatively down-regulated in response to a number of environmental stress conditions or during the transition from fermentation to respiration (39). The importance of mammalian RP genes in cell proliferation is clearly evidenced by the finding that adult liver cells of a knockout mouse conditionally deleted for rpS6 fail to proliferate after partial hepatectomy (38). As with yeast, although analyses of the regulation of transcription of mammalian RP genes have been carried out (28), the in vivo functional significance of transcriptional factors, such as GABP, Sp1, and YY1, remains obscure at present. For example, cell-free transcription analysis and EMSA have revealed that an element similar to the GABP-binding consensus sequence of RP genes acts as a positive element and that GABP could bind to the element (1, 43), but GABP binding in vivo has not been confirmed. Similarly, the in vivo contribution of YY1 sites seems obscure, because a functional significance for RP genes could not be demonstrated in a reporter assay (3, 31). hDRE-like sequences found in 22 human RP genes correspond to a sequence named “box A,” one of the most conserved sequences among mammalian RP genes (28), which has been reported to be essential to drive the expression of the mouse rpL7a gene (2). However, this is the first report describing identification and characterization of a transcription factor binding to box A of mammalian RP genes in vivo. It should be emphasized that fluctuation of hDREF expression was here found to be in line with regulation of RP gene expression in a cell cycle-dependent manner.
Many reports have shown that the initiation of rRNA transcription is tightly linked to cell cycle progression and that synthesis of rRNA increases during G1 phase, reaches a maximal level in S and G2 phases, and is repressed in mitosis (7, 20, 22). We found that a 7 out of 10 match with the hDREF-binding sequence is located at the −108 position of the ribosomal DNA genes (26), raising a question of whether the ribosomal DNA genes are coordinately regulated by hDREF. However, the possibility was ruled out by results obtained from ChIP and RT-PCR analyses (data not shown).
We observed that almost all cells ceased proliferation on hDREF depletion associated with reduction in the expression of tested RP genes. We consider there are two possible explanations. The first is that the amount of RP synthesis, in other words, the capacity for protein synthesis in cells, may function as a critical and sensitive sensor to determine whether cells continue to proliferate. This seems likely because we observed that less than 50% depletion of RPS6 expression caused cell proliferation delay. Consistent with our findings, Šulić et al. have recently demonstrated that heterozygotic loss of rpS6 in T cells abrogates the T-cell proliferation normally accompanying receptor stimulation (35). Thus, it is likely that there is a novel checkpoint monitoring whether RP (or ribosome) synthesis is sufficient for continuing cell proliferation in mammals. The second possibility is that down-regulation of another unknown target gene(s) of hDREF may cause cell proliferation to cease. It is worth noting that we have also found hDRE-like sequences in promoter regions of genes required for cell cycle regulation (26), including CDC25C, CDC25A, CDK6, CCNC, CCND3, CCNG1, and CCNT1. Moreover, “M8” [5′-T(C/A)TCGCGAN(A/G)-3′], one of the best-conserved motifs recently found by systematic comparative genomics of the human genome, perfectly matches with hDRE (40). Although binding factors for the M8 motif have yet to be identified, hDREF is likely to recognize several included sequences. Interestingly, tissue-specific gene expression analysis revealed that genes containing M8 motifs exhibit enriched expression in proliferative hematopoietic cells, among 75 human tissues (40). Taking all of the available information into account, we propose that hDREF is involved in coordinating transcription of a subset of genes containing hDRE (or M8), including RP genes, which are related to cell proliferation and/or cell cycle progression. We are currently preparing genome-wide analyses including gene expression analysis by microarrays and ChIP-on-chip to obtain a comprehensive view of the potential target genes under the control of hDREF. Actually, we have performed a microarray-based expression profiling analysis and observed that several ribosomal protein genes were down-regulated by hDREF depletion (H. Osada, unpublished data).
hDREs are positioned at around −60 bp from the transcription start sites in 20 RP genes. Also, M8 motifs (a total of 368 copies in human promoter regions) tend to be present at distances centered at around −62 bp upstream of transcription start sites. Drosophila DRE has been described to exist at a similar position (the average distance to the ATG codon is estimated to be 168 bp) (24). We do not know the biological significance at present, but considering the fact that the Drosophila DREF/TRF2 complex directs core promoter recognition of the PCNA gene (15), we speculate that binding of hDREF at the −60 bp position may have an important role in sequestering uncharacterized factors such as components of the basal transcriptional machinery and initiating transcription. To elucidate this question, it will be necessary to identify factors or the protein complex associated with hDREF. We have already obtained evidence that endogenous hDREF exists in a high-molecular-weight complex in vivo (41a).
It is likely that Drosophila RP genes are under the regulation of the Drosophila DRE/DREF system for the following reasons. (i) Twenty-seven out of 77 Drosophila RP genes carry DREs in their promoters. (ii) Microarray analysis using probes prepared from Drosophila S2 cells depleting DREF revealed expression of some RP genes to be reduced (F. Hirose and A. Matsukage, unpublished data). (iii) Jasper et al. have identified a set of genes that are selectively expressed in the undifferentiated dividing cells but not in the terminally differentiated cells in the Drosophila eye imaginal disc (17). Interestingly, they found that 24 out of those 41 genes contain a perfectly matched or closely resembling DRE sequence within 1 kb of their transcriptional start site and that six RP genes (bonsai, mRpL5, RpP1, sop, RpP2, and RpL12) are included (17). (iv) Some responsibility for the Minute mutant, exhibiting delayed larval development, diminished viability, reduced body size, decreased fertility, and thin bristles, is believed to be carried by RP genes (6, 30). We have observed retarded development of Drosophila expressing an inverted-repeat RNA for the DREF gene in the whole body by use of a GAL4-UAS targeted system with the hsp70-GAL4 driver, resembling a Minute mutant-like phenotype (F. Hirose and M. Yamaguchi, unpublished data). Thus, regulation of RP gene expression, and therefore protein synthesis, might be a common feature of both Drosophila and human DRE/DREF systems, with effects on cell proliferation and cell cycle progression.
In summary, we here demonstrated that hDREF is an important transcriptional factor for cell proliferation involved in up-regulation of plural RP genes during the G1-to-S transition. We also provided evidence that insufficient expression of only one RP gene may impair cell proliferation.
Acknowledgments
We are grateful to S. Taketani for the plasmid carrying the human metallothionein 2A gene promoter, T. Kiyono for HFF cells, M. Fujita for RNAs from WI38 cells, and H. Miyoshi and A. Miyawaki for lentivirus vectors. We thank M. Moore for critical reading of the manuscript.
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) and the 21st Century COE Program. D.Y. was supported by a research fellowship from the JSPS for Young Scientists during the performance of the studies.
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Antoine, M., and P. Kiefer. 1998. Functional characterization of transcriptional regulatory elements in the upstream region and intron 1 of the human S6 ribosomal protein gene. Biochem. J. 336:327-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colombo, P., and M. Fried. 1992. Functional elements of the ribosomal protein L7a (rpL7a) gene promoter region and their conservation between mammals and birds. Nucleic Acids Res. 20:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Rinaldis, E., G. Pisaneschi, O. Camacho-Vanegas, and E. Beccari. 1998. The binding sites for Xenopus laevis FIII/YY1 in the first exon of L1 and L14 ribosomal protein genes are dispensable for promoter expression. Eur. J. Biochem. 255:563-569. [DOI] [PubMed] [Google Scholar]
- 4.Elemento, O., and S. Tavazoie. 2005. Fast and systematic genome-wide discovery of conserved regulatory elements using a non-alignment based approach. Genome Biol. 6:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujita, M., T. Kiyono, Y. Hayashi, and M. Ishibashi. 1996. hCDC47, a human member of the MCM family. J. Biol. Chem. 271:4349-4354. [DOI] [PubMed] [Google Scholar]
- 6.Galloni, M., and B. A. Edgar. 1999. Cell-autonomous and non-autonomous growth-defective mutants of Drosophila melanogaster. Development 126:2365-2375. [DOI] [PubMed] [Google Scholar]
- 7.Grummt, I. 1999. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 62:109-154. [DOI] [PubMed] [Google Scholar]
- 8.Hart, C. M., O. Cuvier, and U. K. Laemmli. 1999. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma 108:375-383. [DOI] [PubMed] [Google Scholar]
- 9.Heintz, N. 1991. The regulation of histone gene expression during the cell cycle. Biochim. Biophys. Acta 1088:327-339. [DOI] [PubMed] [Google Scholar]
- 10.Hirose, F., N. Ohshima, E. J. Kwon, H. Yoshida, and M. Yamaguchi. 2002. Drosophila Mi-2 negatively regulates dDREF by inhibiting its DNA-binding activity. Mol. Cell. Biol. 22:5182-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirose, F., N. Ohshima, M. Shiraki, Y. H. Inoue, O. Taguchi, Y. Nishi, A. Matsukage, and M. Yamaguchi. 2001. Ectopic expression of DREF induces DNA synthesis, apoptosis, and unusual morphogenesis in the Drosophila eye imaginal disc: possible interaction with Polycomb and trithorax group proteins. Mol. Cell. Biol. 21:7231-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirose, F., M. Yamaguchi, H. Handa, Y. Inomata, and A. Matsukage. 1993. Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase α and proliferating cell nuclear antigen. J. Biol. Chem. 268:2092-2099. [PubMed] [Google Scholar]
- 13.Hirose, F., M. Yamaguchi, K. Kuroda, A. Omori, T. Hachiya, M. Ikeda, Y. Nishimoto, and A. Matsukage. 1996. Isolation and characterization of cDNA for DREF, a promoter-activating factor for Drosophila DNA replication-related genes. J. Biol. Chem. 271:3930-3937. [DOI] [PubMed] [Google Scholar]
- 14.Hirose, F., M. Yamaguchi, and A. Matsukage. 1999. Targeted expression of the DNA binding domain of DRE-binding factor, a Drosophila transcription factor, attenuates DNA replication of the salivary gland and eye imaginal disc. Mol. Cell. Biol. 19:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochheimer, A., S. Zhou, S. Zheng, M. C. Holmes, and R. Tjian. 2002. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420:439-445. [DOI] [PubMed] [Google Scholar]
- 16.Hyun, J., H. Jasper, and D. Bohmann. 2005. DREF is required for efficient growth and cell cycle progression in Drosophila imaginal discs. Mol. Cell. Biol. 25:5590-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jasper, H., V. Benes, A. Atzberger, S. Sauer, W. Ansorge, and D. Bohmann. 2002. A genomic switch at the transition from cell proliferation to terminal differentiation in the Drosophila eye. Dev. Cell 3:511-521. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen, P., J. L. Nishikawa, B. J. Breitkreutz, and M. Tyers. 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297:395-400. [DOI] [PubMed] [Google Scholar]
- 19.Jorgensen, P., I. Rupeš, J. R. Sharom, L. Schneper, J. R. Broach, and M. Tyers. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kief, D. R., and J. R. Warner. 1981. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol. Cell. Biol. 1:1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396:84-88. [DOI] [PubMed] [Google Scholar]
- 22.Klein, J., and I. Grummt. 1999. Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. USA 96:6096-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La Bella, F., P. Gallinari, J. Mckinney, and N. Heintz. 1989. Histone H1 subtype-specific consensus elements mediate cell cycle-regulated transcription in vitro. Genes Dev. 3:1982-1990. [DOI] [PubMed] [Google Scholar]
- 24.Ohler, U., G. C. Liao, H. Niemann, and G. M. Rubin. 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3: research0087.1-research0087.12. [DOI] [PMC free article] [PubMed]
- 25.Ohno, K., F. Hirose, K. Sakaguchi, Y. Nishida, and A. Matsukage. 1996. Transcriptional regulation of the Drosophila CycA gene by the DNA replication-related element (DRE) and DRE binding factor (DREF). Nucleic Acids Res. 24:3942-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohshima, N., M. Takahashi, and F. Hirose. 2003. Identification of a human homologue of the DREF transcription factor with a potential role in regulation of the histone H1 gene. J. Biol. Chem. 278:22928-22938. [DOI] [PubMed] [Google Scholar]
- 27.Orlando, V., and R. Paro. 1993. Mapping polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 75:1187-1198. [DOI] [PubMed] [Google Scholar]
- 28.Perry, R. P. 2005. The architecture of mammalian ribosomal protein promoters. BMC Evol. Biol. 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryu, J. R., T. Y. Choi, E. J. Kwon, W. H. Lee, Y. Nishida, Y. Hayashi, A. Matsukage, M. Yamaguchi, and M. A. Yoo. 1997. Transcriptional regulation of the Drosophila-raf proto-oncogene by the DNA replication-related element (DRE)/DRE-binding factor (DREF) system. Nucleic Acids Res. 25:794-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sæbøe-Larssen, S., and A. Lambertsson. 1996. A novel Drosophila Minute locus encodes ribosomal protein S13. Genetics 143:877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Safrany, G., and R. P. Perry. 1995. The relative contributions of various transcription factors to the overall promoter strength of the mouse ribosomal protein L30 gene. Eur. J. Biochem. 230:1066-1072. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., p. 13.36-13.39. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Sawado, T., F. Hirose, Y. Takahashi, T. Sasaki, T. Shinomiya, K. Sakaguchi, A. Matsukage, and M. Yamaguchi. 1998. The DNA replication-related element (DRE)/DRE-binding factor system is a transcriptional regulator of the Drosophila E2F gene. J. Biol. Chem. 273:26042-26051. [DOI] [PubMed] [Google Scholar]
- 34.Sharkov, N. V., G. Ramsay, and A. L. Katzen. 2002. The DNA replication-related element-binding factor (DREF) is a transcriptional regulator of the Drosophila myb gene. Gene 297:209-219. [DOI] [PubMed] [Google Scholar]
- 35.Šulić, S., L. Panić, M. Barkić, M. Merćep, M. Uzelac, and S. Volarević. 2005. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 19:3070-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas, G. 2000. An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2:E71-E72. [DOI] [PubMed] [Google Scholar]
- 38.Volarević, S., M. J. Stewart, B. Ledermann, F. Zilberman, L. Terracciano, E. Montini, M. Grompe, S. C. Kozma, and G. Thomas. 2000. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288:2045-2047. [DOI] [PubMed] [Google Scholar]
- 39.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 40.Xie, X., J. Lu, E. J. Kulbokas, T. R. Golub, V. Mootha, K. Lindblad-Toh, E. S. Lander, and M. Kellis. 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434:338-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi, M., Y. Hayashi, Y. Nishimoto, F. Hirose, and A. Matsukage. 1995. A nucleotide sequence essential for the function of DRE, a common promoter element for Drosophila DNA replication-related genes. J. Biol. Chem. 270:15808-15814. [DOI] [PubMed] [Google Scholar]
- 41a.Yamashita, D., H. Komori, Y. Higuchi, T. Yamaguchi, T. Osumi, and F. Hirose. 5 January 2007. hDREF self-association via LATC domain is necessary for its nuclear accumulation and DNA binding. doi: 10.1074/jbc.M607180200. [DOI] [PubMed]
- 42.Yamashita, D., T. Yamaguchi, M. Shimizu, N. Nakata, F. Hirose, and T. Osumi. 2004. The transactivating function of peroxisome proliferator-activated receptor γ is negatively regulated by SUMO conjugation in the amino-terminal domain. Genes Cells 9:1017-1029. [DOI] [PubMed] [Google Scholar]
- 43.Yoganathan, T., N. K. Bhat, and B. H. Sells. 1992. A positive regulator of the ribosomal protein gene, β factor, belongs to the ETS oncoprotein family. Biochem. J. 287:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshida, H., Y. H. Inoue, F. Hirose, K. Sakaguchi, A. Matsukage, and M. Yamaguchi. 2001. Over-expression of DREF in the Drosophila wing imaginal disc induces apoptosis and a notching wing phenotype. Genes Cells 6:877-886. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida, H., E. J. Kwon, F. Hirose, K. Otsuki, M. Yamada, and M. Yamaguchi. 2004. DREF is required for EGFR signalling during Drosophila wing vein development. Genes Cells 9:935-944. [DOI] [PubMed] [Google Scholar]