Abstract
Human glucocorticoid receptor (hGR) is expressed as two alternately spliced C-terminal isoforms, α and β. In contrast to the canonical hGRα, hGRβ is a nucleus-localized orphan receptor thought not to bind ligand and not to affect gene transcription other than by acting as a dominant negative to hGRα. Here we used confocal microscopy to examine the cellular localization of transiently expressed fluorescent protein-tagged hGRβ in COS-1 and U-2 OS cells. Surprisingly, yellow fluorescent protein (YFP)-hGRβ was predominantly located in the cytoplasm and translocated to the nucleus following application of the glucocorticoid antagonist RU-486. This effect of RU-486 was confirmed with transiently expressed wild-type hGRβ. Confocal microscopy of coexpressed YFP-hGRβ and cyan fluorescent protein-hGRα in COS-1 cells indicated that the receptors move into the nucleus independently. Using a ligand binding assay, we confirmed that hGRβ bound RU-486 but not the hGRα ligand dexamethasone. Examination of the cellular localization of YFP-hGRβ in response to a series of 57 related compounds indicated that RU-486 is thus far the only identified ligand that interacts with hGRβ. The selective interaction of RU-486 with hGRβ was also supported by molecular modeling and computational docking studies. Interestingly, microarray analysis indicates that hGRβ, expressed in the absence of hGRα, can regulate gene expression and furthermore that occupation of hGRβ with the antagonist RU-486 diminishes that capacity despite the lack of helix 12 in the ligand binding domain.
Glucocorticoids are an important class of natural and synthetic steroid hormone signaling molecules whose physiological actions can affect nearly every part of the human body. The effects of glucocorticoids are mediated by the glucocorticoid receptor (GR), a hormone binding transcription factor of the steroid family of nuclear receptors. The gene for human GR (hGR) was originally cloned in 1985 and consists of nine exons (12). This structural organization produces two isoforms of GR, hGRα and hGRβ, by the alternative splicing of exon 9 (22). Exon 9 encodes the carboxy-terminal end of the ligand binding domain (LBD) for GR, as well as the 3′ untranslated region. Thus, the hGRα and -β isoforms are identical up through amino acid 727, at which point they diverge. hGRα has an additional 50 amino acids that encode helices 11 and 12 of the ligand binding domain. In contrast, hGRβ has only an additional 15 distinct amino acids. Consequently, hGRβ is missing helix 12 of the ligand binding domain and possesses a unique sequence in helix 11 compared to hGRα. hGRα is the classical GR and is found in the cytoplasm in the absence of ligand. Upon ligand binding, hGRα translocates to the nucleus, where it affects gene transcription. In contrast, immunohistochemistry studies have shown that hGRβ is constitutively nuclear and does not bind agonists (22, 23). In addition, hGRβ is thought to affect gene transcription only by acting as a dominant negative to hGRα and altering the ability of hGRα to signal (1, 21, 22, 38).
The distribution and relative expression of hGRα versus hGRβ have been examined in a number of human cell lines (23), as well as in both healthy and diseased human cells and tissues. While there is general agreement that the expression of hGRα is greater than the expression of hGRβ in all cells and tissues, the actual extent of hGRβ expression is less clear. Although mRNA for hGRβ has been found in a variety of human tissues (1, 22, 25), hGRβ protein has been shown to have a more restricted cellular distribution. Most frequently, hGRβ protein has been found in healthy T lymphocytes, macrophages, neutrophils, eosinophils, and endogenous peripheral blood mononuclear cells (9, 10, 33). In addition, hGRβ protein has been reported in brain, lung, and heart tissue (23), although there is a contradictory report (25). Interestingly, the expression of hGRβ has also been shown to be increased in glucocorticoid-resistant forms of asthma (3, 9, 16, 30), ulcerative colitis (13), nasal polyposis (10), and chronic lymphocytic leukemia (19, 29). In addition, we have previously shown that the expression of hGRβ can be activated in cells by proinflammatory cytokines (36). Under these conditions, hGRβ is the predominant receptor in the cells and a state of glucocorticoid resistance ensues. These reports suggest a potential physiological consequence to changes in hGRβ expression. Thus, hGRβ may be a key modulator of the progression of certain immune-related glucocorticoid-resistant diseases. In this report, we describe the identification of the antiglucocorticoid/antiprogestin compound RU-486 as a ligand for hGRβ which causes nuclear translocation of both transiently and stably transfected hGRβ in COS-1 and U-2 OS cell lines. In addition, we show that hGRβ introduced into U-2 OS cells in the absence of hGRα can widely regulate gene expression and that this action of the receptor is modulated by the glucocorticoid receptor antagonist RU-486 despite the absence of a helix 12 in hGRβ.
MATERIALS AND METHODS
Plasmids.
The plasmids pEYFP-hGRα and pECFP-hGRα were previously described (27). Plasmid pCMV-hGRβ was previously described (22). The plasmid pEYFP-hGRβ was made by replacing the ClaI/BamHI fragment of pEYFP-hGRα (containing the hGRα-specific 3′ coding sequences) with the ClaI/BamHI fragment from pCMV-hGRβ (containing the hGRβ-specific coding sequences as well as 1,430 bp of hGRβ 3′ untranslated region). The plasmids pTET-OFF and pTRE2hyg were obtained from BD Biosciences Clontech (Mountain View, CA).
Compounds.
The following compounds were purchased from Steraloids, Inc. (Newport, RI): cortexolone (4-pregnen-17,21-diol-3,20-dione), corticosterone (4-pregnen-11β,21-diol-3,20-dione), cortisol (4-pregnen-11β,17,21-triol-3,20-dione), cortisone (4-pregnen-17,21-diol-3,11,20-trione), deltafludrocortisone (1,4-pregnadien-9α-fluoro-11β,17,21-triol-3,20-dione), desoximetasone (1,4-pregnadien-9α-fluoro-16α-methyl-11β,21-diol-3,20-dione), dexamethasone (1,4-pregnadien-9α-fluoro-16α-methyl-11β,17,21-triol-3,20-dione), dexamethasone 21-mesylate (1,4-pregnadien-9α-fluoro-16α-methyl-11β,17,21-triol-3,20-dione-21-methanesulfonate), 17β-estradiol [1,3,5(10)-estratrien-3,17β-diol], prednisolone(1,4-pregnadien-11β,17,21-triol-3,20-dione), progesterone (4-pregnen-3,20-dione), RU-486 (4,9-estradien-17α-propynyl, 11β-[4-dimethylamino]phenyl-17β-ol-3-one), testosterone (4-androsten-17β-ol-3-one), triamcinolone (1,4-pregnadien-9α-fluoro-11β,16α,17,21-tetrol-3,20-dione), and triamcinolone acetonide (1,4-pregnadien-9α-fluoro-11β,16α,17,21-tetrol-3,20-dione-16,17-acetonide).RU-486 was also purchased from Sigma-Aldrich (St. Louis, MO). Compound 3 {2′-(3-pyridyl)-11β,17,21-trihydroxy-16α-methyl-20-oxopregn-4-eno[3,2-c]pyrazole}, compound 6 {2′-(4-iodophenyl)-11β,17,21-trihydroxy-16α-methyl-20-oxopregn-4-eno[3,2-c]pyrazole}, compound 11 {2′-(4-bromophenyl)-11β,17,21-trihydroxy-16α-methyl-20-oxopregn-4-eno[3,2-c]pyrazole}, compound 12 {2′-(4-fluorophenyl)-11β,17,21-trihydroxy-16α-methyl-20-oxopregn-4-eno[3,2-c]pyrazole}, and compound 16b {2′-(2-chloro-3-pyridyl)-11β,17,21-trihydroxy-16α-methyl-20-oxopregn-4-eno[3,2-c]pyrazole} were kind gifts from R. Hochberg (Yale University, New Haven, CT). Deacylcortivazol {2′-(phenyl)-11β,17,21-trihydroxy-6,16α-dimethyl-20-oxopregn-4,6-dieno[3,2-c]pyrazole} was a kind gift from S. Simons (NIDDK, NIH, Bethesda, MD). RU-28362(1,4,6-androstatrien-6-methyl-17α-propynyl, 11β,17-diol-3-one) was a kind gift from P. Housley(University of South Carolina School of Medicine, Columbia, SC). ZK98299 (4,9-gonadien-11β-[4-dimethylamino]phenyl-17α-ol-17β-[3-hydroxypropyl]-13α-methyl-3-one)was a kind gift from T. Archer (NIEHS, NIH, Research Triangle Park, NC). C. E. Cook (Research Triangle Institute, Research Triangle Park, NC) and D. McDonnell (Duke University, Durham, NC) kindly provided RTI compounds as follows: RTI 3021-002, -003, -020, -021, and -023 and RTI 6413-001, -002, -006, -009a, -015, -016, -018, -028, -029E, -029Z, -030, -031, -039, -042, -043, -043ox, -044, -045, -045ox, -046a, -046b, -049b, -050a, -050b, -051a, -051b, -052, -054, -055, -056, -057, and -058. [3H]RU-486 was obtained from American Radiolabeled Chemicals, Inc. (St. Louis, MO). [3H]dexamethasone was obtained from Perkin-Elmer Life Sciences (Woodbridge, Ontario, Canada).
Generation of U-2 OS cell lines stably expressing hGRβ.
U-2 OS cells were transfected with the pTET-OFF regulatory plasmid to establish the U-OFF parental cell line (20). In these cell lines, protein expression can be regulated by the addition of tetracycline to the medium. MluI and EcoRV ends were added onto the coding region (amino acids 1 to 742) of hGRβ by PCR amplification of the pCMVhGRβ plasmid. The pTRE2hyg vector was digested with MluI and EcoRV, and the two DNAs were ligated to form the pTRE2hGRβ plasmid. The pTRE2hGRβ plasmid was then transfected into the U-OFF cells, and clones that stably express hGRβ were selected using 200 μg/ml of Geneticin and 500 μg/ml of hygromycin. Two hundred micrograms of hygromycin per milliliter was used for maintenance. Several clones were obtained, and the hGRβ receptor levels were compared using Western blot analysis with an hGRβ-specific antibody. Clone identity was further confirmed by isolating total RNA and performing reverse transcription-PCR (RT-PCR) with hGRβ-specific primers. The resulting PCR products were sequenced.
Cell culture and transfection.
COS-1 cells were maintained in Dulbecco modified Eagle medium (DMEM) with high glucose (Invitrogen Life Technologies) supplemented with 10% fetal calf serum-calf serum, 50 units/ml penicillin, and 0.05 mg/ml streptomycin (Sigma-Aldrich). U-2 OS cells (wild type) were maintained in DMEM-F-12 medium (Invitrogen Life Technologies) supplemented with 5% heat-inactivated fetal calf serum, 50 units/ml penicillin, 0.05 mg/ml streptomycin, and 2 mM l-glutamine. U-OFF stable cells were maintained in DMEM-F-12 medium supplemented with 10% fetal calf serum-calf serum, 50 units/ml penicillin, 0.05 mg/ml streptomycin, 2 mM l-glutamine, and 0.2 mg/ml Geneticin (Invitrogen Life Technologies). U-2 OSα and U-2 OSβ stable cells were maintained in U-OFF medium with 0.2 mg/ml hygromycin B (Invitrogen Life Technologies). All cells were grown at 37°C and 5% CO2 in a humidified incubator and passaged every 3 to 7 days, as they approached confluence.
One day prior to transfection, COS-1 or U-2 OS cells were transferred to 78.5-cm2 dishes (7.5 × 105 cells/dish). Cells were transfected with TransIt-LT1 reagent (Mirus, Madison, WI) as described by the manufacturer using 20 μl TransIt-LT1 and 1.5 μg DNA per dish. The next day, cells were transferred to 9.6-cm2 glass-bottomed dishes (MatTek Corp., Ashland, MA), 1.5 × 105 cells/dish in medium containing charcoal-stripped serum. Microscope imaging was done the following day.
Confocal microscopy.
On the day of imaging, transfected COS-1 or U-2 OS cells were treated with 1 μM of steroid for 3 to 6 h. Subsequently, cells expressing yellow fluorescent protein (YFP)-hGRα or YFP-hGRβ were observed using a Zeiss LSM 510 confocal laser scanning microscope as previously reported (27). Cells expressing cyan fluorescent protein (CFP)-hGRα were observed on the same microscope, exciting fluorescence with an argon laser at 458 nm and collecting emission with a 470- to 500-nm band-pass filter. Quantitative receptor localization analysis was carried out using the Zeiss LSM 510 software to determine the fluorescence intensity of the receptor in an equivalently sized region in both the nucleus and the cytoplasm of at least 10 cells per treatment condition per experiment. Experiments were repeated at least twice. The average ratio of nuclear to cytoplasmic intensity was then used as a measure of receptor distribution throughout the cell.
Immunocytochemistry and Western blot analysis.
Immunocytochemistry was carried out on COS-1 or U-2 OS cells transfected with CMV-hGRα or CMV-hGRβ and on U-2 OSβ cells, as previously described using affinity-purified anti-GR#57 antibody prepared in our laboratory (37, 38). Western blot assays for hGRα and hGRβ were carried out on protein extracts from U-OFF, U-2 OSα, and U-2 OSβ cells. Cell pellets were sonicated in low-detergent buffer (20 mM Tris-Cl, pH 7.5, 2 mM EDTA, 150 mM NaCl, 0.5% Triton X-100, with one protease inhibitor tablet [Roche; no. 1 836 153] per 10 ml buffer added immediately prior to use) for 30 seconds and then centrifuged for 15 min at 12,800 × g at 4°C. The protein concentration of the supernatants was determined using Bio-Rad protein assay reagent (Bio-Rad Laboratories, Hercules, CA) against bovine serum albumin standards. Protein samples were heated in 1× Laemmli loading buffer plus 2-mercaptoethanol for 5 min at 100°C prior to being electrophoretically resolved on an 8% precast Tris-glycine gel (Invitrogen Life Technologies, Carlsbad, CA). Proteins were transferred to an 0.2-μm nitrocellulose membrane, blocked in 10% nonfat dry milk in TBS-T (50 mM Tris, 150 mM NaCl, 0.5% Tween 20, pH 7.5) overnight at 4°C, washed in TBS-T, and then incubated with anti-β-actin (1:10,000; catalog no. MAB1501; Chemicon, Temecula, CA) plus either anti-GR#57 (1:1,000) or BShGR (1:1,000; catalog no. PA3-514; Affinity BioReagents) in TBS-T for 1 h at room temperature. After being washed in TBS-T, the blot was incubated with goat anti-rabbit peroxidase-conjugated antibody (ECL Western blotting analysis system; Amersham Biosciences, Buckinghamshire, England) diluted 1:10,000 in TBS-T for 1 h at room temperature. After further washing, bands were visualized with enhanced chemiluminescence detection reagents (ECL; Amersham Biosciences) as specified by the manufacturer.
Ligand binding assays. (i) Column binding assay.
One day prior to the assay, cells were plated at 1.5 × 107 cells/145-cm2 dish in medium containing charcoal-stripped serum. On the day of assay, cells were harvested by incubation for 10 min in Versene (2.68 μM KCl, 1.47 μM KH2PO4, 137 mM NaCl, 0.54 μM EDTA, 8.09 mM Na2HPO4 · 7H2O) at 37°C and 5% CO2 followed by scraping. Cells were resuspended in 1 ml ice-cold phosphate-buffered saline, radiolabeled steroid was added with or without 500- to 1,000-fold-excess unlabeled steroid, and cells were incubated on ice for 2 h with gentle agitation. Cells were collected by centrifugation at 4°C, resuspended in an equal volume of ice-cold buffer A (20 mM sodium phosphate, pH 7.0, 50 mM NaCl, 10% glycerol, 2 mM β-mercaptoethanol, 1 mM EDTA, pH 8.0), and homogenized at 4°C using a prechilled homogenizer with three 10-second bursts interspersed with 10-second rests on ice. Extracts were centrifuged at 165,000 × g for 1 h at 2°C. The supernatant was applied at 4°C to a Sephadex G-25 HiTrap desalting column (Amersham Biosciences Corp., Piscataway, NJ) previously equilibrated with buffer A according to the manufacturer's instructions, followed by 7.5 ml buffer A to elute. One-hundred-microliter fractions were collected and counted in a scintillation counter.
(ii) Ethanol extraction assay.
U-2 OSβ cells were plated in medium containing charcoal-stripped serum plus 1 μM dexamethasone 1 day prior to assay at 1 × 106 cells/well in six-well plates. Duplicate wells were incubated with 1, 5, 10, 25, 50, or 100 nM 3H-labeled RU-486 in the absence (total binding) or presence (nonspecific binding) of 10 μM cold RU-486 for 2 h at 37°C. Cells were then washed five times with 1 ml/well ice-cold phosphate-buffered saline and incubated at room temperature for 30 min in 1-ml/well 100% ethanol to extract the RU-486. Ethanol was then removed and counted in 5 ml scintillation fluid in a scintillation counter. For each assay, count data were converted to pmol and normalized to the total protein in one well of cells cultured in parallel. Total and nonspecific pmol/mg data were analyzed using GraphPad Prism 4 (GraphPad Software, Inc., San Diego, CA). Global curve fitting was performed simultaneously on the total and nonspecific binding data. The nonspecific curve was then subtracted from the total curve to obtain a curve of specific ligand binding which was fitted using the nonlinear regression curve-fitting function for a hyperbola (one-site binding) in this program. This generates a best-fit curve from which Bmax and Kd are determined. This assay was repeated four times; Kd values from the four assays were averaged to obtain the reported Kd.
Computational studies: molecular modeling and ligand docking.
The development of a homology molecular model for hGRβ was reported previously (38). This model was employed for computational docking studies. The Schrodinger Glide v.3.5 docking software was used for all ligand docking studies, and GlideScore was used as the docking scoring function for ligand docking evaluation and comparison. The Glide docking method and scoring function for ligands have been found to compare favorably with most docking methods and have been demonstrated capable of predicting ligand-receptor binding interactions (7, 8). Four potential hGRβ ligands which were evaluated experimentally were computationally docked with the hGRβ model receptor: dexamethasone, ZK98299, RTI 6413-001, and RU-486. The starting three-dimensional (3D) structures for the dexamethasone and RU-486 ligands for docking were extracted, respectively, from the solved crystal structures for hGRα, Protein Data Bank files 1M2Z and 1NHZ. The structure for ZK98299 was obtained as a two-dimensional sdf file from PubChem, and the structure for RTI 6413-001 was built and optimized beginning with the 3D structure for RU-486, which was modified using the BioMedCAche software. All starting ligand structures were prepared for docking using the Schrodinger Ligprep software to generate energy-minimized correct 3D ligand structures for docking including tautomeric, stereochemical, and ionization variations. The hGRβ active site was defined for the ligand docking (volume of the receptor searched when attempting to dock a ligand) by superimposing the hGRβ receptor model structure on that of the solved crystal structure of the hGRα receptor with bound ligand (Protein Data Bank files 1M2Z, 1NHZ, and 1P93). The receptor grid binding box for hGRβ was defined as the area superimposed on the ligand binding site within the hGRα receptor crystal structure. Ligand docking of the same four ligands performed with the wild-type hGRβ receptor was repeated with a model of the mutant Q642V hGRβ receptor, and the same computational methodology was used for docking with the mutant receptor model.
Microarray analysis.
U-OFF and U-2 OSβ cells were cultured in charcoal-stripped serum medium for 24 h prior to treatment. Total RNA was extracted from 5 × 10 6 U-OFF or U-2 OSβ cells treated with either ethanol vehicle or 1 μM RU-486 for 6 h using the RNAqueous total RNA isolation kit (Ambion Inc. Austin, TX) according to the manufacturer's instructions. RNA was treated with DNase using the DNA-free DNase treatment and removal reagents (Ambion Inc.) according to to manufacturer's instructions prior to use with the microarray. Four pairs of RNA (vehicle versus RU-486 treated) were harvested for each cell type to yield four biological replicates for gene expression analysis.
Linear amplification label protocol and feature extraction.
Gene expression analysis was conducted using Agilent Human1Av2 arrays (Agilent Technologies, Palo Alto, CA). Total RNA was amplified using the Agilent Low RNA Input Fluorescent Linear Amplification kit protocol. Starting with 500 ng of total RNA, Cy3- or Cy5-labeled cRNA was produced according to the manufacturer's protocol. For each two-color comparison, 750 ng of each Cy3- and Cy5-labeled cRNA was mixed and fragmented using the Agilent In Situ Hybridization kit protocol. Hybridizations were performed for 17 h in a rotating hybridization oven using the Agilent 60-mer oligonucleotide microarray processing protocol. Slides were washed as indicated in this protocol and then scanned with an Agilent scanner. Data were obtained using the Agilent Feature Extraction software (v7.5), using defaults for all parameters.
Rosetta Resolver (v5.0).
Images and GEML files, including error and P values, were exported from the Agilent Feature Extraction software and deposited into Rosetta Resolver (version 5.0) (Rosetta Biosoftware, Kirkland, WA). The resultant ratio profiles were combined into ratio experiments as described in the work of Stoughton and Dai (32). In total, three ratio experiments were built containing eight arrays each (four biological replicates each with dye swaps) for U-OFF vehicle versus U-2 OSβ vehicle, U-2 OSβ vehicle versus RU-486, and U-OFF vehicle versus RU-486. Based on these experiments, lists of genes that were determined to be statistically differentially expressed using Resolver's error model at P < 0.001 were saved. The lists were then combined into a single list and the genes clustered hierarchically using Rosetta Resolver.
In a separate series of experiments, total RNA was isolated from U-OFF and U-2 OSα cells cultured in charcoal-stripped serum medium and processed and analyzed as described above. Three sets of RNA were isolated for three biological replicates.
Quantitative RT-PCR analysis.
U-OFF and stably hGRβ-expressing cell lines were cultured in charcoal-stripped serum medium for 24 hours prior to treatment and then treated with vehicle or 1 μM RU-486 for 6 hours. Total RNA was isolated using the QIAGEN RNeasy minikit. Real-time PCR was performed using the 7900HT Sequence Detection System and predesigned primer/probe sets available from Applied Biosystems (Foster City, CA) following the manufacturer's instructions. The signal obtained from each gene primer/probe set was normalized to that of the unregulated housekeeping gene cyclophilin B primer/probe set (also available from Applied Biosystems). Each primer/probe set was analyzed in triplicate and with at least three different sets of RNA isolated from U-OFF cells, U-2 OSβ vehicle-treated cells, and U-2 OSβ RU-486-treated cells and normalized to cyclophilin B.
Statistical analysis.
Statistical analysis for Fig. 1, 3, and 6 was performed using JMP5.0.1 software (SAS, Cary, NC). One-way analysis of variance (ANOVA) was performed for Fig. 1 and 3, and two-way ANOVA for Fig. 6, to identify the existence of statistically significant treatment differences for each cell and receptor type. Where significance was indicated, post hoc testing using Dunnett's test (comparison versus vehicle control) was carried out. Statistical analysis for Fig. 2 and 4 was performed by Shyamal Peddada of the NIEHS Biostatistics Branch. For each cell and receptor type, we tested the null hypothesis that all three treatment groups have the same probability of observing any given category against the two-sided alternative that each of the treated groups was different from the control (vehicle-treated) group. We tested the above hypothesis using a Dunnett-type test statistic along the lines of the work of Peddada et al. (24). The P values were determined using the bootstrap methodology (6). In all cases statistical significance was accepted at P < 0.05.
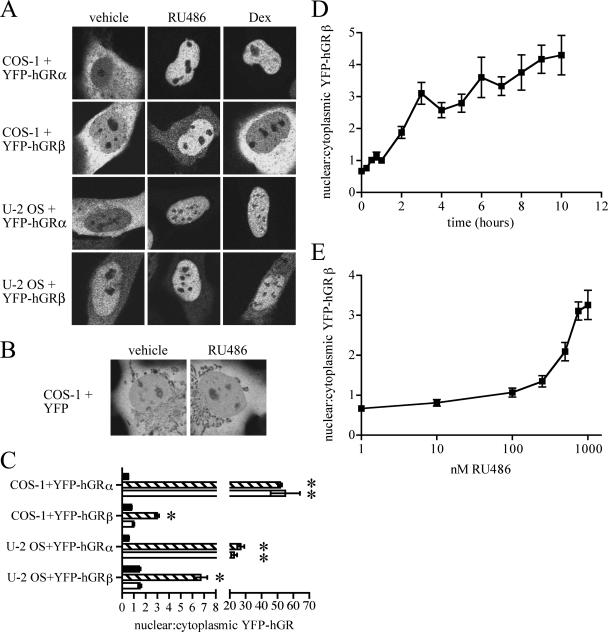
FIG. 1.
YFP-hGRβ translocates into the nucleus of transfected cells in response to RU-486. (A) COS-1 and U-2 OS cells were transiently transfected with plasmids expressing YFP-hGRα or YFP-hGRβ and treated with ethanol vehicle or 1 μM RU-486 or dexamethasone (Dex) for 3 h before being examined with confocal microscopy. Both treatments caused nuclear translocation of YFP-hGRα in both cell lines; only RU-486 caused nuclear translocation of YFP-hGRβ. (B) COS-1 cells were transiently transfected with a plasmid expressing YFP and treated with 1 μM RU-486 for 3 h before imaging. RU-486 had no effect on the cellular localization of YFP. (C) The localization of YFP-hGRα and YFP-hGRβ in response to steroid treatment was quantified by determining the ratio of the fluorescence intensity in an area of the nucleus divided by the fluorescence intensity in a similarly sized area of the cytoplasm. Black bars indicate no treatment; striped bars indicate 1 μM RU-486; white bars indicate 1 μM dexamethasone. This analysis confirmed that RU-486 but not dexamethasone caused nuclear translocation of YFP-hGRβ in both cell lines. Data are means ± SEMs (n ≥ 30 cells per treatment condition); *, significantly different at P < 0.05 versus vehicle for that receptor-cell type combination. (D) COS-1 cells were transfected with a plasmid expressing YFP-hGRβ and treated for 0, 0.25, 0.5, 0.75, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 h with 1 μM RU-486. Cells were then imaged and quantified as in panel C. Nuclear localization was clearly evident by 2 h of treatment. Data are means ± SEMs. (E) COS-1 cells were transfected with a YFP-hGRβ expression plasmid and treated for 3 h with 1, 10, 100, 250, 500, 750, or 1,000 nM of RU-486. Cells were then imaged and quantified as in panel C. Nuclear translocation of YFP-hGRβ first became evident with 250 nM RU-486. Data are means ± SEMs.
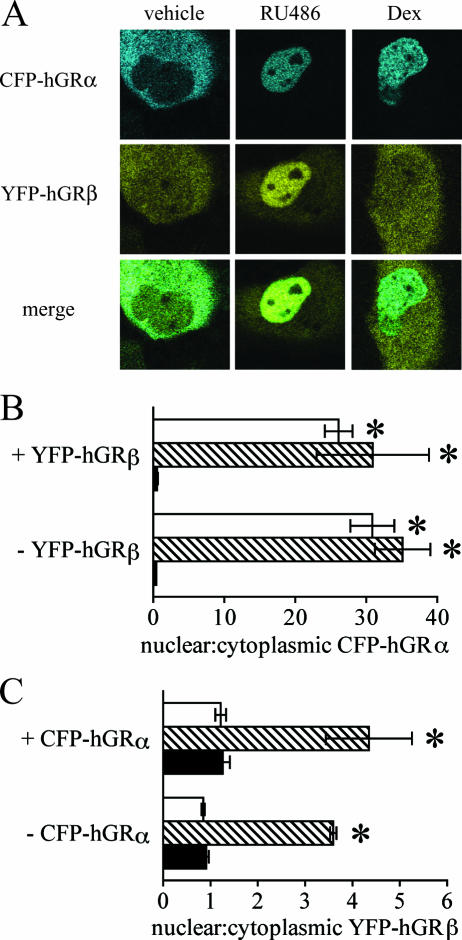
FIG. 3.
RU-486-dependent nuclear translocation of hGRβ is not due to cytoplasmic heterodimerization with hGRα. (A) COS-1 cells were transiently transfected with equal amounts of plasmids expressing CFP-hGRα and YFP-hGRβ and treated with ethanol vehicle or 1 μM RU-486 or dexamethasone (Dex) for 3 h before being examined with confocal microscopy. Blue indicates localization of CFP-hGRα; yellow indicates YFP-hGRβ localization; in the merged image, green indicates areas where both receptors are located. Both RU-486 and dexamethasone caused nuclear translocation of CFP-hGRα; only RU-486 caused translocation of YFP-hGRβ. (B) Localization of CFP-hGRα was quantified by determining the ratio of the fluorescence intensity in an area of the nucleus divided by the fluorescence intensity in a similarly sized area of the cytoplasm in the absence or presence of cotransfected YFP-hGRβ, with or without steroid treatment. Black bars indicate vehicle treatment; striped bars indicate 1 μM RU-486; white bars are 1 μM dexamethasone. Both dexamethasone and RU-486 caused nuclear translocation of CFP-hGRα, regardless of the presence of YFP-hGRβ. Data are means ± SEMs (n ≥ 30 cells per treatment condition); *, significant difference at P < 0.05 versus vehicle treatment for that receptor-treatment combination. ANOVA indicated no statistically significant effect of YFP-hGRβ on the localization of CFP-hGRα. (C) Localization of YFP-hGRβ was quantified as in panel B in the absence or presence of cotransfected CFP-hGRα, with or without steroid treatment. Black bars indicate vehicle treatment; striped bars indicate 1 μM RU-486; white bars are 1 μM dexamethasone. RU-486 caused nuclear translocation of YFP-hGRβ to the same extent, and dexamethasone had no effect on translocation, regardless of the presence of CFP-hGRα. Data are means ± SEMs (n ≥ 30 cells per treatment condition); *, significant difference at P < 0.05 versus vehicle treatment for that receptor-treatment combination. ANOVA indicated no statistically significant effect of CFP-hGRα on the localization of YFP-hGRβ.
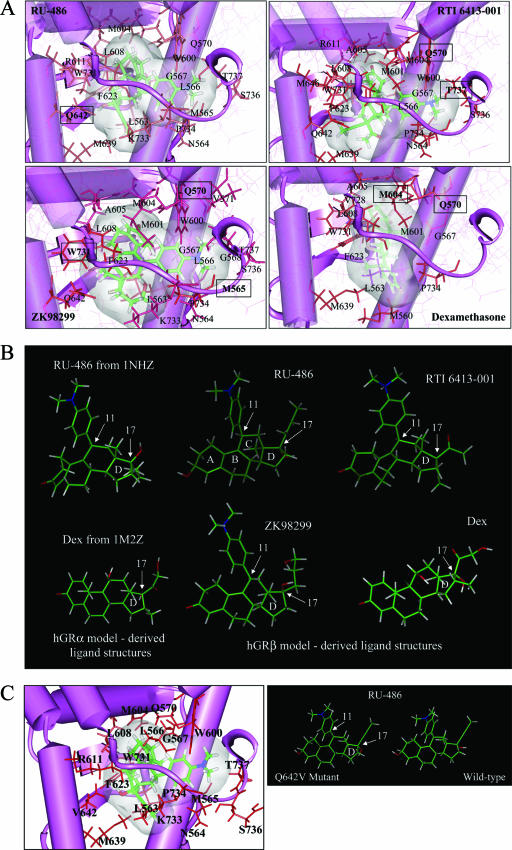
FIG. 6.
Computer modeling of the hGRβ ligand binding domain docked with RU-486, RTI 6413-001, ZK98299, or dexamethasone. (A) Illustrations of the highest GlideScore poses for the ligands RU-486, RTI 6413-001, ZK98299, and dexamethasone computationally docked into the model of the hGRβ ligand binding domain. Amino acids labeled on the diagrams are predicated to be within 3 angstroms of the respective ligands; boxed amino acids are predicted to form hydrogen bonds with the respective ligands. The C-terminal tail of the hGRβ ligand binding domain can be seen to curl snugly around the dimethylaniline substituent of RU-486, RTI 6413-001, and ZK98299, while dexamethasone seems to make little contact with the hGRβ C-terminal tail. (B) Conformation of RU-486, RTI 6413-001, ZK98299, and dexamethasone (Dex) when computationally docked in the hGRβ ligand binding domain (middle and right-hand structures); the conformation of RU-486 and dexamethasone obtained from the solved crystal structures of the hGRα ligand binding domain (Protein Data Bank files 1NHZ and 1M2Z, respectively) is shown for comparison (left-hand structures). The positions of the D ring and carbons 11 and 17 are indicated. (C) (Left) Illustration of the ligand RU-486 computationally docked into the model of a Q642V hGRβ mutant ligand binding domain. Amino acids labeled on the diagrams are predicated to be within 3 angstroms of the ligand. No amino acids are predicted to form hydrogen bonds with RU-486 in the ligand binding domain of this mutant hGRβ. (Right) Conformation of RU-486 when computationally docked in the Q642V versus wild-type hGRβ ligand binding domains. Note that the orientation of the hydroxyl group on carbon 17 (indicated) differs between the two structures.
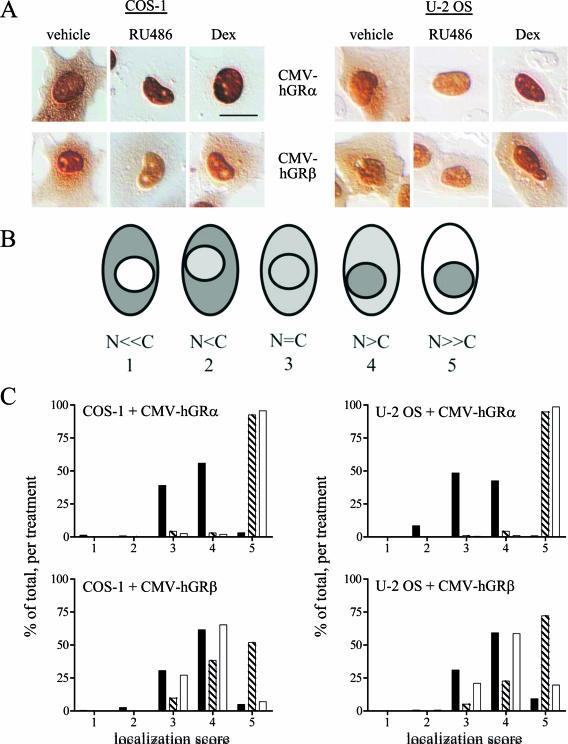
FIG. 2.
Wild-type hGRβ translocates into the nucleus in response to RU-486. (A) COS-1 (left) and U-2 OS (right) cells were transiently transfected with CMV-hGRα or CMV-hGRβ plasmids, which express wild-type hGRα or hGRβ, respectively, and treated with ethanol vehicle or 1 μM RU-486 or dexamethasone (Dex) for 3 h before being processed for immunocytochemistry with the GR#57 antibody, which recognizes both hGRα and hGRβ. (B) Schematic illustration of the scoring system used to quantitate the localization of the receptors with different treatments. A number value was assigned to each cell based on the relative amount of cytoplasmic and nuclear receptor as indicated (N, nuclear receptor; C, cytoplasmic receptor): N ≪ C, 1; N < C, 2; N = C, 3; N > C, 4; N ≫ C, 5. (C) Frequency histograms of the resulting localization scores are plotted (n ≥ 130). Black bars indicate receptor localization with vehicle treatment; striped bars indicate 1 μM RU-486; white bars indicate 1 μM dexamethasone. Both ligands caused statistically significant changes in the frequency histogram of hGRα, reflecting its nuclear translocation: the percentage of cells scored as 5 is higher for the RU-486 and dexamethasone treatments than for the vehicle treatment, while there are more vehicle-treated cells scoring at 4 or below. In contrast, only RU-486 caused nuclear translocation of hGRβ: there was little difference in the number of vehicle-treated cells versus dexamethasone-treated cells at any score, while the number of RU-486-treated cells with a score of 5 was greater than the number of vehicle-treated cells with that score. Bars, 25 μm.
FIG. 4.
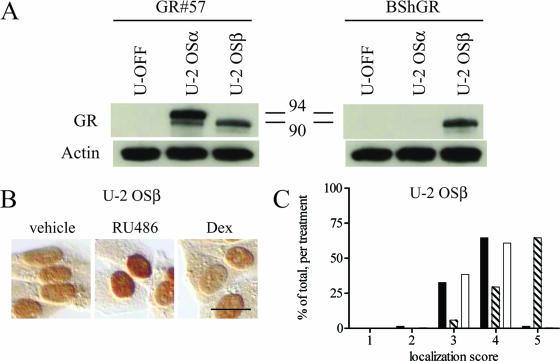
Expression and RU-486-dependent nuclear translocation of wild-type hGRβ in the U-2 OSβ stable cell line. (A) The U-2 OS cell line U-2 OSβ, stably expressing hGRβ under the control of a Tet-OFF promoter system, was created. U-2 OSα, U-2 OSβ, and the U-OFF parental cell line were treated for at least 3 h with 1 μM RU-486 and then harvested for Western blot assays. The GR#57 antibody recognizes both hGRα (94 kDa) and hGRβ (90 kDa); the BShGR antibody recognizes hGRβ only; the actin antibody was used to demonstrate equal loading of the lanes. The U-OFF cells did not express detectable levels of GR, while U-2 OSα and U-2 OSβ exclusively expressed hGRα or hGRβ, respectively. Numbers in the middle are molecular masses in kilodaltons. (B) U-2 OSβ cells were treated with ethanol vehicle or 1 μM RU-486 ordexamethasone (Dex) for 3 h before being processed for immunocytochemistry with the GR#57 antibody. Bar, 25 μm. RU-486 but not dexamethasone caused nuclear translocation of stably expressed hGRβ. (C) Quantification of the immunocytochemical localization of hGRβ receptor was carried out by assigning localization scores and plotting a frequency histogram as in Fig. 2B. This analysis confirmed that RU-486 but not dexamethasone caused the wild-type receptor to undergo nuclear translocation. Black bars indicate vehicle treatment; striped bars indicate 1 μM RU-486; white bars indicate 1 μM dexamethasone; n ≥ 290.
Microarray data accession number.
The microarray data discussed have been deposited in NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (5) and are accessible through GEO Series accession number GSE5310.
RESULTS
The hGRβ glucocorticoid receptor isoform undergoes nuclear translocation in response to RU-486.
Our first indication that RU-486 might interact with hGRβ came from confocal imaging studies. Plasmids expressing hGRα or hGRβ that were tagged with YFP at the amino terminus of GR (YFP-hGRα and YFP-hGRβ) were transiently transfected into COS-1 cells. This cell line was chosen because it does not express detectable levels of endogenous GR. Transfected cells were treated for 3 h with 1 μM dexamethasone or RU-486 and then imaged live using confocal laser scanning microscopy (Fig. 1A). Untreated COS-1 cells expressing YFP-hGRα showed the expected cytoplasmic distribution of unliganded receptor. Also, as predicted, both dexamethasone and RU-486 caused complete nuclear translocation of YFP-hGRα. However, untreated COS-1 cells expressing YFP-hGRβ also showed a largely cytoplasmic receptor distribution. This pattern differs from the primarily nuclear distribution of non-YFP-tagged hGRβ in COS-1 cells that we have previously reported (23). Surprisingly, YFP-hGRβ translocated into the nucleus of cells in response to treatment with RU-486 but not in response to dexamethasone, suggesting for the first time that hGRβ may be able to bind ligand.
Previous reports have suggested a nuclear distribution of the wild-type hGRβ receptor, regardless of the presence of agonist (23). Since it was possible that the RU-486-stimulated nuclear translocation of YFP-hGRβ was a cell-type-specific response, we examined this issue in the human osteosarcoma cell line U-2 OS, which also does not express detectable levels of endogenous GR (Fig. 1A). Again, YFP-hGRα was located in the cytoplasm of U-2 OS cells in the absence of treatment and translocated to the nucleus in response to treatment with RU-486 or dexamethasone. Although YFP-hGRβ was located primarily in the nucleus of these cells in the absence of treatment, the receptor was clearly present in the cytoplasm as well. Nuclear translocation of YFP-hGRβ occurred in response to treatment with RU-486 but not dexamethasone. This RU-486-dependent nuclear translocation of YFP-hGRβ was not due to an effect of RU-486 on the YFP tag since COS-1 cells that transiently expressed YFP alone showed no change in YFP distribution with 3 h of 1 μM RU-486 treatment (Fig. 1B).
To facilitate direct comparison of glucocorticoid receptor distribution in multiple cells across experiments, the subcellular distribution in the two cell lines was quantified. At least 30 cells for each cell, receptor, and treatment type were examined. For each cell, the fluorescence intensity of an area in the nucleus and the fluorescence intensity of an equally sized area in the cytoplasm were used to create a ratio, which was then combined to give an average ± standard error of the mean (SEM) for each cell type and treatment condition (Fig. 1C). With this ratio, numbers less than 1 indicate primarily cytoplasmic receptor localization, numbers equal to 1 indicate equal receptor distribution across the cell, and numbers greater than 1 indicate primarily nuclear localization. This quantification confirmed that both dexamethasone and RU-486 caused complete nuclear translocation of YFP-hGRα in both cell types, while only RU-486 promoted nuclear translocation of YFP-hGRβ in the two cell types examined. Thus, in two different cell lines, treatment with RU-486 promotes nuclear translocation of transiently expressed YFP-hGRβ.
The time course of RU-486-dependent YFP-hGRβ nuclear translocation was next determined by treating COS-1 cells transiently expressing YFP-hGRβ with 1 μM RU-486 for various times and then imaging the cells and quantifying them as in Fig. 1A and C (Fig. 1D). These results indicated that changes in receptor distribution occurred with as little as 30 min of treatment. YFP-hGRβ localization was primarily nuclear by 2 h of treatment and was maximal by 6 h. The RU-486 dose dependence of the YFP-hGRβ nuclear translocation was also determined. YFP-hGRβ-expressing COS-1 cells were treated for 3 h with various concentrations of RU-486 before being imaged and quantified (Fig. 1E). Significant receptor nuclear translocation occurred at a concentration of 100 nM of RU-486 and reached maximum at 750 nM RU-486. Thus, the kinetics of hGRβ translocation are slower than those observed for hGRα with either agonists or antagonists.
We next evaluated if the nuclear translocation that we observed for YFP-hGRβ in response to RU-486 also occurred with wild-type hGRβ. For these experiments, COS-1 and U-2 OS cells were transiently transfected with plasmids expressing wild-type hGRα or hGRβ (cytomegalovirus [CMV]-hGRα or CMV-hGRβ, respectively), treated for 3 h with 1 μM RU-486 or dexamethasone, and then fixed and analyzed by immunocytochemistry for the localization of the receptors using the GR#57 antibody that recognizes both receptor isoforms (Fig. 2A). Results were similar to those obtained with the YFP-tagged receptors. In both cell types, wild-type hGRα was located in the cytoplasm in the absence of treatment and translocated to the nucleus with RU-486 or dexamethasone treatment. Similarly, in both cell types, wild-type hGRβ was located in both the nucleus and the cytoplasm in the absence of treatment. RU-486 facilitated nuclear translocation of hGRβ whereas dexamethasone did not. To facilitate direct comparison of these results in multiple cells across experiments, receptor localization was quantified by assigning a number value to each cell based on the relative amount of cytoplasmic and nuclear receptor: specifically, where N = nuclear and C = cytoplasmic receptor, N ≪ C, 1; N < C, 2; N = C, 3; N > C, 4; N ≫ C, 5 (Fig. 2B). At least 130 cells were analyzed per cell type, receptor, and treatment; localization scores were then plotted as frequency histograms and analyzed for statistical differences (Fig. 2C). This analysis confirmed that, in both cell types, wild-type hGRα responded with nuclear translocation to both RU-486 and dexamethasone: the percentage of cells scored as 5 is higher for the RU-486 (striped bars) or dexamethasone (white bars) treatment than for the vehicle treatment (black bars). In contrast, the localization of wild-type hGRβ changed only in response to RU-486: there was little difference in the number of vehicle- versus dexamethasone-treated cells (black versus white bars, respectively) at any score, while the number of RU-486 (striped bars)-treated cells increased as the scores indicated progressively nuclear localization.
hGRβ moves into the nucleus independently of hGRα.
Since the hGRβ isoform of hGR has not been previously demonstrated to bind ligand, the mechanism underlying the observed RU-486-dependent nuclear translocation of hGRβ was unclear. One possibility was an RU-486-dependent heterodimerization of hGRβ with a small amount of hGRα in the cytoplasm, followed by nuclear translocation of the receptor dimer. Alternatively, hGRβ might bind RU-486, followed by conformational changes in hGRβ that stimulate nuclear translocation, similar to the canonical response of hGRα to ligand binding.
If cytoplasmic heterodimerization of hGRβ with hGRα is the mechanism by which nuclear translocation of hGRβ occurs, in COS-1 and U-2 OS cells it does so in the presence of limiting quantities of hGRα, since the hGRα protein is not detected in these cells by Western blotting (see Fig. 4A and data not shown). This could explain the incomplete nuclear translocation of hGRβ observed in Fig. 1A and 2A: more hGRα protein might be needed to obtain complete hGRβ translocation. To determine if increased hGRα expression could result in complete nuclear translocation of hGRβ, COS-1 cells were transiently cotransfected with equal amounts of plasmids expressing CFP-hGRα and YFP-hGRβ, treated with 1 μM RU-486 or dexamethasone for 3 h, and then imaged live using confocal microscopy (Fig. 3). Tagging the two receptors with different spectral variants of GFP made it possible to simultaneously determine the localization of the two receptors. As with YFP-hGRα, CFP-hGRα was present in the cytoplasm in the absence of treatment and translocated to the nucleus upon treatment with RU-486 or dexamethasone (Fig. 3A, top). Similarly, YFP-hGRβ was found in the cytoplasm in the absence of treatment and underwent nuclear translocation in response to RU-486 but not dexamethasone (Fig. 3A, middle). The merged images show where in the cell the two receptor isoforms colocalized, as indicated by the green color (Fig. 3A, bottom).
To facilitate direct comparison of these receptor distributions in multiple cells across experiments, at least 30 cells for each receptor and treatment type were quantified by determining the ratio of nuclear to cytoplasmic fluorescence intensity, as before (Fig. 1C). To determine the effect of YFP-hGRβ on the cellular localization of CFP-hGRα, the localization ratios for CFP-hGRα were compared in the presence and absence of YFP-hGRβ (Fig. 3B). Both RU-486 and dexamethasone caused complete nuclear translocation of CFP-hGRα, in the presence and absence of YFP-hGRβ. ANOVA indicated no statistically significant effect of YFP-hGRβ on the localization of CFP-hGRα. Similarly, the effect of CFP-hGRα on the cellular localization of YFP-hGRβ was determined (Fig. 3C). RU-486, but not dexamethasone, caused nuclear translocation of YFP-hGRβ in both the presence and absence of CFP-hGRα. Again, nuclear translocation of YFP-hGRβ was not complete, even in the presence of similar amounts of CFP-hGRα. ANOVA indicated no statistically significant difference in the extent of this receptor localization due to the presence of CFP-hGRα. Thus, nuclear translocation of hGRβ in response to RU-486 is not likely due to heterodimerization of the receptor with hGRα in the cytoplasm, followed by hGRα-facilitated nuclear translocation. Rather, hGRβ is able to undergo nuclear translocation on its own in response to RU-486.
hGRβ is selective for RU-486.
The observation that hGRβ is able to undergo nuclear translocation in response to the ligand RU-486 suggests that other ligands may cause nuclear translocation as well. Since nuclear translocation of the receptor is an easily observed biological process, we used this method to screen for other potential ligands of hGRβ. Accordingly, COS-1 cells were transfected with the YFP-hGRβ expression plasmid, treated with various ligands, and then observed live using fluorescence microscopy for nuclear localization of YFP-hGRβ (Table 1). Several classes of ligands were examined, including 10 glucocorticoids; four antiglucocorticoids; six analogs of cortivasol (14, 15); and ligands for the estrogen, progesterone, and androgen receptors. In addition, 37 antiprogestins with structural similarities to RU-486 were also tested (26, 35). Of the 57 compounds tested, only RU-486 promoted nuclear translocation of YFP-hGRβ, suggesting that hGRβ is highly selective for RU-486.
TABLE 1.
Observation of ligand-dependent nuclear translocation of receptor
| Ligand (reference) | YFP-hGRβ | YFP-hGRα |
|---|---|---|
| Glucocorticoids | ||
| Corticosterone | No | Yes |
| Cortisol | No | Yes |
| Cortisone | No | Yes |
| Deltafludrocortisone | No | Yes |
| Desoximetasone | No | Yes |
| Dexamethasone | No | Yes |
| Prednisolone | No | Yes |
| RU-28362 | No | NDa |
| Triamcinolone | No | Yes |
| Triamcinolone acetonide | No | Yes |
| Antiglucocorticoids | ||
| Cortexolone | No | Yes |
| Dexamethasone-21-mesylate | No | Yes |
| RU-486 | Yes | Yes |
| ZK98299 | No | Yes |
| Other nuclear receptor ligands | ||
| 17β-Estradiol | No | ND |
| Progesterone | No | ND |
| Testosterone | No | ND |
| Cortivasol analogs | ||
| Deacylcortivazol | No | ND |
| 3 (15) | No | ND |
| 6 (14) | No | ND |
| 11 (14) | No | ND |
| 12 (14) | No | ND |
| 16b (15) | No | ND |
| Antiprogestins | ||
| RTI 3021-002 (35) | No | |
| RTI 3021-003 (35) | No | |
| RTI 3021-020 (35) | No | |
| RTI 3021-021 (35) | No | |
| RTI 3021-023 (35) | No | |
| RTI 6413-001 (26) | No | |
| RTI 6413-002 (26) | No | |
| RTI 6413-006 (26) | No | |
| RTI 6413-009a (26) | No | |
| RTI 6413-015 (26) | No | |
| RTI 6413-016 (26) | No | |
| RTI 6413-018 (26) | No | |
| RTI 6413-028 (26) | No | |
| RTI 6413-029E (26) | No | |
| RTI 6413-029Z (26) | No | |
| RTI 6413-030 (26) | No | |
| RTI 6413-031 (26) | No | |
| RTI 6413-039 (26) | No | |
| RTI 6413-042 (26) | No | |
| RTI 6413-043 (26) | No | |
| RTI 6413-043ox (26) | No | |
| RTI 6413-044 (26) | No | |
| RTI 6413-045 (26) | No | |
| RTI 6413-045ox (26) | No | |
| RTI 6413-046a (26) | No | |
| RTI 6413-046b (26) | No | |
| RTI 6413-049b (26) | No | |
| RTI 6413-050a (26) | No | |
| RTI 6413-050b (26) | No | |
| RTI 6413-051a (26) | No | |
| RTI 6413-051b (26) | No | |
| RTI 6413-052 (26) | No | |
| RTI 6413-054 (26) | No | |
| RTI 6413-055 (26) | No | |
| RTI 6413-056 (26) | No | |
| RTI 6413-057 (26) | No | |
| RTI 6413-058 (26) | No |
ND, not determined.
RU-486 is a ligand for hGRβ.
To determine if hGRβ is able to bind RU-486, a stable cell line, U-2 OSβ, was created that expresses wild-type hGRβ in U-2 OS cells under the control of a tetracycline-responsive promoter system. Specific expression of hGRβ in this cell line was confirmed by Western blot comparison of hGRβ expression with expression of hGRα in a U-2 OSα stable cell line and endogenous GR expression in the U-OFF parental cell line used to make the U-2 OSβ cells (20) (Fig. 4A). As expected, endogenous hGR was not detected in the U-OFF parental cell line by this method. Only hGRα was detected in the U-2 OSα cell line, while only hGRβ was detected in the U-2 OSβ cell line. Treatment of U-2 OSβ cells for 3 h with 1 μM RU-486 or dexamethasone, followed by immunocytochemical localization of hGRβ using the GR#57 antibody indicated that hGRβ undergoes RU-486-dependent but not dexamethasone-dependent nuclear translocation in this cell line (Fig. 4B). Quantification of these results was carried out as in Fig. 2 on at least 290 cells per treatment and confirmed that only RU-486 caused statistically significant nuclear translocation of hGRβ in the U-2 OSβ cells (Fig. 4C). These results were also confirmed in a second, independent clone of this stable cell line (data not shown).
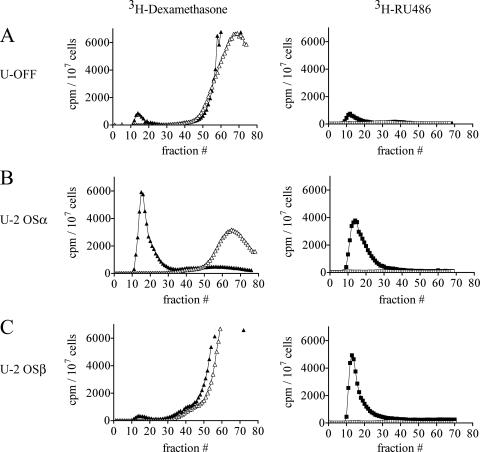
The U-OFF, U-2 OSα, and U-2 OSβ stable cell lines were then used to determine the ability of hGRα and hGRβ to bind RU-486 versus dexamethasone. Whole-cell ligand binding assays were performed by incubating each cell type with 3H-labeled RU-486 or [3H]dexamethasone for 2 h at 0°C in the presence or absence of excess unlabeled steroid. Cell lysates were then prepared and applied to a Sephadex G-50 column, and fractions were collected to separate the early-eluting bound steroid from the late-eluting free steroid (Fig. 5). The U-OFF cells exhibited a small amount of binding to both [3H]dexamethasone and [3H]RU-486, which was competed by the presence of excess cold ligand, indicating that the binding was saturable (Fig. 5A). This small amount of binding may be due to trace amounts of endogenous hGR or perhaps progesterone receptor expression in these cells that is not detectable by other methods such as Western blotting (for example, Fig. 4A). The U-2 OSα cells also exhibited binding to both [3H]dexamethasone and [3H]RU-486 (Fig. 5B). However, the extent of binding in U-2 OSα cells was 5 to 7.5 times that seen in the U-OFF cell line, indicating that the binding was due to the greatly increased expression of hGRα in these cells compared to the U-OFF cells. Finally, the U-2 OSβ cells exhibited a small amount of [3H]dexamethasone binding comparable to that seen with the U-OFF parental cell line (Fig. 5C). In contrast, in two clones of the U-2 OSβ cell line (Fig. 5C and data not shown), the U-2 OSβ cells exhibited six times the amount of [3H]RU-486 binding that was observed with the U-OFF cells, indicating that hGRβ is able to bind the ligand RU-486. This binding was confirmed with a second type of whole-cell ligand binding assay in which the bound ligand was extracted from the cells with ethanol (see Materials and Methods; data not shown). Using the ethanol extraction assay, we determined the Kd of RU-486 binding to hGRβ to be approximately 138 nM (L. Lewis-Tuffin and J. Cidlowski, unpublished results), whereas the Kd of RU-486 binding to hGRα has been reported to be between 5 and 10 nM (27). Thus, the affinity of RU-486 for hGRβ is clearly lower than that for hGRα.
FIG. 5.
Wild-type hGRβ binds RU-486. (A) Whole-cell ligand binding assays were performed with U-OFF cells treated with 100 nM [3H]dexamethasone (closed triangles) or 100 nM [3H]RU-486 (closed squares) for 2 h in the presence or absence of excess cold steroid (50 μM dexamethasone [open triangles] or 100 μM RU-486 [open squares]). Cells were disrupted and applied to a Sephadex G-50 column, and fractions were collected to separate early-eluting bound steroid from late-eluting free steroid. The small elution between fractions 10 and 20 indicates ligand binding to the small amount of endogenous hGR or progesterone receptor (PR) present in these cells; this binding was effectively competed away with excess cold ligand, indicating that it was not nonspecific binding. (B) The experiment in panel A was repeated using U-2 OSα cells treated with 20 nM [3H]dexamethasone (closed triangles) or 100 nM [3H]RU-486 (closed squares) with and without 20 μM and 10 μM unlabeled steroid (open symbols), respectively. Both [3H]dexamethasone and [3H]RU-486 bound to hGRα in these cells; this binding was effectively competed away with excess cold ligand. (C) The experiment in panel A was repeated using U-2 OSβ cells treated with 100 nM [3H]dexamethasone (closed triangles) or 100 nM [3H]RU-486 (closed squares), with or without 50 μM or 100 μM unlabeled steroid (open symbols), respectively. As with the U-OFF cells, the small elution between fractions 10 and 20 with [3H]dexamethasone treatment indicates ligand binding to the small amount of endogenous hGR or PR present in these cells. In contrast, [3H]RU-486 showed sixfold-higher amounts of binding, which were effectively competed away with excess cold ligand in these cells, indicating binding of [3H]RU-486 to hGRβ.
Computational docking studies of hGRβ ligand binding.
To understand the molecular interactions that might underlie the selective nature of the hGRβ-RU-486 interaction, computational docking studies were performed. Four of the potential hGRβ ligands that were experimentally evaluated (dexamethasone, RU-486, RTI 6413-001, and ZK98299) were successfully computationally docked into the hGRβ binding site. Figure 6A illustrates the highest score pose and conformation for each of the four ligands docked into the hGRβ model structure. All docked poses and conformations of the dexamethasone ligand had significantly lower docked GlideScores than did the other three ligands and significantly higher total energy values for the ligand and receptor combined (data not shown). In addition, there were significantly fewer good van der Waals contacts between the docked dexamethasone ligand and hGRβ receptor than obtained between the docked RU-486 ligand and hGRβ receptor (data not shown). A comparison of the conformations of the docked ligands shows that, in the case of RU-486, ZK98299, and RTI 6413-001, the dimethylaniline substituent on position 11 of the steroid ring C structure fills the binding pocket space and causes the hGRβ C terminus to fold around it (Fig. 6A). This position 11 substituent is absent in dexamethasone, and the volume occupied by this ligand is of a substantially different shape and size than those for the other three ligands (Fig. 6A). Dexamethasone also demonstrates substantially greater conformational flexibility within the binding pocket due to its smaller shape and size and less-complete filling of the binding cavity (data not shown).
In addition to the dimethylaniline on position 11, the substituents present on the 17 position of the steroid ring D system and their stereochemical orientation also appear to be of significant importance for the hGRβ ligand. Computational docking indicates that the RU-486 substituent groups in the 17 alpha and beta positions on the steroid ring D participate in favorable H-bonding interactions with hGRβ residue Q642 (Fig. 6A). In contrast, ZK98299 and RTI 6413-001 have different chemical substituents present on position 17 compared with RU-486 (Fig. 6B). It appears that the larger 17β-C-O-CH3 substituent present in RTI 6413-001 compared with the smaller 17β-OH in RU-486 causes these substituents to dock into the receptor (in the highest ranked docked poses, Fig. 6A) with opposite stereochemical orientations (Fig. 6B), precluding formation of an H bond between hGRβ residue Q642 and RTI 6413-001. Thus, the computational docking studies suggest that a combination of the dimethylaniline group on position 11 of the C ring and the 17β-OH in a stereochemical orientation that facilitates its interaction with Q642 may underlie the productive interaction of RU-486 with the hGRβ ligand binding domain.
It has previously been shown that the glutamine 642 valine (Q642V) point mutation in hGRα dramatically decreases the interaction of the receptor with ligands containing a 17α-OH group (such as dexamethasone) (18) but does not affect interaction with ligands that do not have this group. To assess the effect of this mutation on the interaction of hGRβ with the 17β-OH group of RU-486, computational docking studies were repeated with a molecular model of a mutant Q642V hGRβ receptor (Fig. 6C). The size and chemical composition differences between the glutamine and valine side chains indicate that the terminal oxygen atom of the glutamine side chain, which is 2.0 angstroms from the hydrogen of the 17β-OH group of RU-486 and forms a hydrogen bond with it, has been replaced with one of the terminal hydrogens on the valine side chain. Consequently, the orientation of the OH substituent on the RU-486 D ring changes so that now it is the O of this group that is closest to the terminal hydrogen of the V642 side chain. The distances between the three terminal V642 hydrogens and the RU-486 OH group are now 2.6, 3.0, and 3.6 angstroms, respectively. These distances are longer and less favorable for hydrogen bond formation between RU-486 and the Q642V mutant residue than is the distance between the wild-type hGRβ glutamine oxygen and the RU-486 ligand hydrogen. Additionally, this mutation alters the conformation of the RU-486 substituent 17β-OH group within the hGRβ ligand binding pocket. Our data suggest that the orientation of this group is critical for ligand docking with hGRβ. Thus, the modeling results suggest that a Q642V hGRβ receptor would not bind RU-486 while the Q642V hGRα receptor should.
Regulation of gene expression by hGRβ.
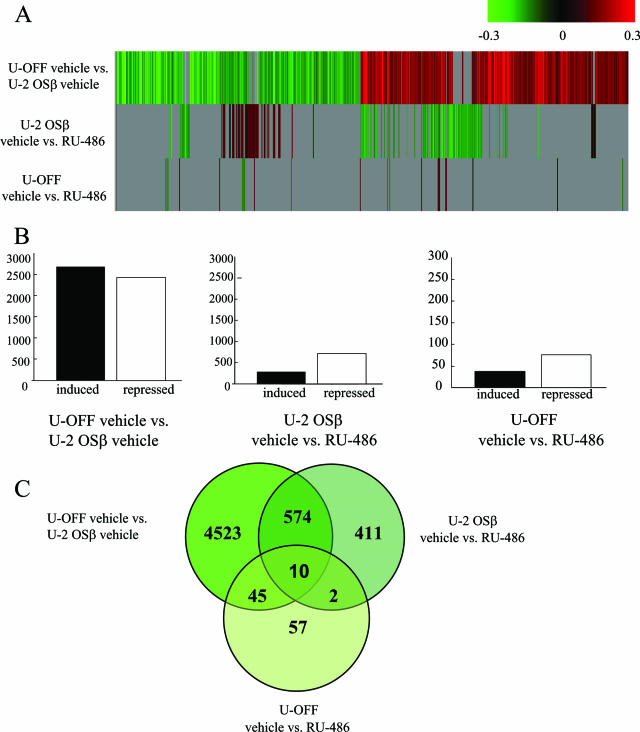
The ability of hGRβ to regulate gene expression is unknown, and reporter assays have consistently suggested that hGRβ can regulate expression only by antagonizing the action of hGRα, regardless of the presence of RU-486 (1, 22). However, the regulation of transiently transfected reporter constructs may not reflect the regulation of endogenous genes in the context of chromatin. Based on our results indicating that RU-486 can bind to hGRβ, we hypothesized that RU-486 might modulate the functional activity of hGRβ and perhaps even be an agonist for this receptor isoform. To determine if hGRβ was capable of regulating gene expression on its own in a chromatin context, we performed a total genome microarray analysis. U-OFF and U-2 OSβ cells were cultured in charcoal-stripped serum medium for 24 h prior to treatment and subsequent RNA isolation. Total RNA was prepared from U-OFF and U-2 OSβ cells treated with vehicle or 1 μM RU-486 for 6 h, and microarray analysis was performed on four biological replicates for each cell and treatment type. For each biological replicate, three comparisons were made: U-OFF vehicle versus U-2 OSβ vehicle, U-2 OSβ vehicle versus U-2 OSβ RU-486, and U-OFF vehicle versus U-OFF RU-486. The results from each of the biological replicates were combined, and the data were then examined for genes that were differentially regulated at P < 0.001 in any one of the three comparisons (5,622 genes total combined). Figure 7A shows a cluster analysis of these three comparison groups. Genes shown in green are repressed down to −0.3 (twofold repressed), and genes shown in red are induced up to 0.3 (twofold induced). Those genes represented in gray had a P value of >0.001 and therefore did not pass our stringency requirement for significance. The top cluster analysis shown in Fig. 7A, the comparison of U-OFF vehicle with U-2 OSβ vehicle, reveals the ability of hGRβ to regulate gene expression on its own, that is, in the absence of hGRα and in the absence of ligand. In this comparison, 5,152 genes met our criteria for significant regulation. Of these genes, 2,685 were induced and 2,467 were repressed by hGRβ expression (Fig. 7B). These results indicate that hGRβ is able to regulate gene expression in the absence of hGRα, a property of hGRβ that was not previously known. The middle cluster analysis, comparing U-2 OSβ vehicle with U-2 OSβ RU-486, shows that in hGRβ-expressing cells treated with RU-486 only 997 genes were significantly regulated (Fig. 7A). This is far less than the number of genes regulated by hGRβ alone. Of the 997 genes, only 260 were induced while a larger number of genes (737) were repressed (Fig. 7B). The third comparison, U-OFF vehicle versus U-OFF RU-486, shows that in the absence of exogenous glucocorticoid receptor RU-486 treatment significantly regulated 114 genes: 44 genes were induced and 70 genes were repressed. Thus, RU-486 modulates the ability of hGRβ to regulate gene expression in a manner consistent with its binding to hGRβ. Interestingly, RU-486 appears to behave as an antagonist to hGRβ-mediated gene regulation.
FIG. 7.
hGRβ can regulate gene expression. (A) Microarray analysis was performed on RNA from four biological replicates of U-OFF and U-2 OSβ cells treated for 6 h with either ethanol vehicle or 1 μM RU-486. Genes that were significantly different at P < 0.001 were identified for each comparison: U-OFF vehicle versus U-2 OSβ vehicle, U-2 OSβ vehicle versus RU-486, and U-OFF vehicle versus RU-486. These genes were combined into one list that was subjected to cluster analysis. Red indicates genes that were induced for each comparison; green indicates genes that were repressed. Fold changes are presented on a log-scale continuum with 0 (black) indicating no change for a given comparison. Any genes with P > 0.001 did not meet our stringency requirements for significance and are shown in gray. (B) Genes that were identified as being significantly regulated in panel A for each comparison were separated into induced versus repressed. Of the 5,152 genes that were statistically significantly regulated by hGRβ expression, 2,685 were induced and 2,467 were repressed (left graph). Of the 997 genes that met our criteria for being statistically significantly regulated by hGRβ plus RU-486 treatment, 260 were induced and 737 were repressed (middle graph). U-OFF cells treated with RU-486 yielded 114 significantly regulated genes with 44 genes induced and 77 repressed (right graph). (C) Genes from the combined gene lists are depicted using Venn diagrams. Genes that are common between each of the three comparison groups are represented in the overlapping circles.
To further compare the three gene lists, we constructed a Venn diagram. Figure 7C shows the number of genes unique to each analysis: those regulated by hGRβ on the left, hGRβ genes further regulated by treatment with RU-486 on the right, and genes regulated by RU-486 in the absence of receptor (U-OFF) on the bottom. The numbers in the overlapping circles represent the subset of genes that were common between the three gene lists. Thus, the microarray data indicate that hGRβ can regulate a unique set of genes and that RU-486 is an antagonist of this endogenous, unliganded hGRβ activity.
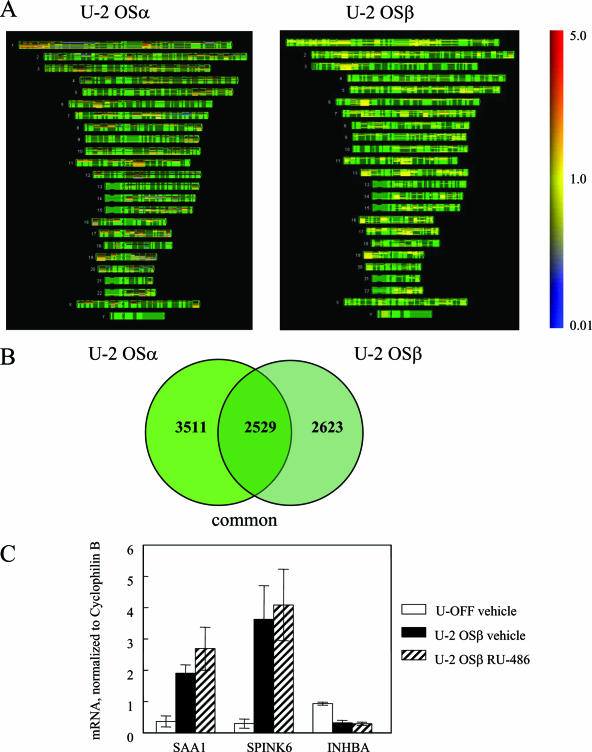
We next compared genes that were regulated by hGRβ to those regulated by hGRα. For this comparison, we isolated RNA from both the U-OFF parental cells and those stably expressing hGRα (U-OFF versus U-2 OSα), which were cultured for 24 h in charcoal-stripped serum medium. However, prior to experimental analysis, these cells were continuously cultured in medium containing non-charcoal-stripped fetal calf serum; therefore, we cannot rule out a residual effect of glucocorticoids from the fetal calf serum in the case of hGRα. Microarray analysis was performed on three biological replicates, and the results from the combined gene list showed that a total of 6,040 genes were regulated by hGRα at P < 0.001. These 6,040 genes were then compared to the 5,152 genes that were regulated by the hGRβ-expressing cells (U-OFF versus U-2 OSβ) using human chromosome mapping (Fig. 8A). These maps show the physical position of the genes with known loci. The structure of each chromosome is depicted in green, induced genes are red, and repressed genes are blue. The color bar on the right shows the expression level of these genes ranging from 5.0 (highly induced) to 0.01 (highly repressed). This analysis illustrates that there are significant differences in the genes that are affected by the expression of these two glucocorticoid receptors across the human genome. Analysis of the lists by Venn diagram (Fig. 8B) illustrates that a significant number of genes were both commonly and uniquely regulated by these two glucocorticoid receptor isoforms. Additionally, we analyzed gene regulation by the two active forms of the receptor, hGRα treated with 100 nM dexamethasone and hGRβ vehicle treated. Although these experiments were performed at different times with slightly different parameters, we found ∼1,000 genes commonly regulated (data not shown).
FIG. 8.
Comparison of gene regulation between hGRα and hGRβ. (A) Microarray analysis was performed on RNA from three biological replicates of U-OFF and U-2 OSα cells. Genes that were significantly different at P < 0.001 were identified and combined into one list. This gene list was compared to the combined gene list of the hGRβ-expressing cells (U-OFF versus U-2 OSβ) using human chromosome mapping. These maps show the physical positions of the genes with known loci. The structure of each chromosome is depicted in green with induced genes in red and repressed genes in blue. The color bar on the right shows the expression level of these genes ranging from 5.0 (highly induced) to 0.01 (highly repressed). (B) The Venn diagram illustrates the genes that are commonly regulated by both hGRα and hGRβ. (C) Quantitative RT-PCR was performed using the 7900HT sequence detection system using predesigned primer/probe sets for SAA1, SPINK6, and INHBA and a custom primer/probe for cyclophilin B available from Applied Biosystems. Each primer/probe set was analyzed in triplicate and with at least three different sets of RNA isolated from U-OFF cells, U-2 OSβ vehicle-treated cells, and U-2 OSβ RU-486-treated cells and normalized to cyclophilin B.
Finally, we used quantitative RT-PCR to confirm some of the genes uniquely regulated by hGRβ (Fig. 8C). Two genes that were highly up-regulated by the expression of hGRβ were serum amyloid A-1 (SAA1, increased 5.3-fold), which is increased in plasma concentration during acute inflammatory reactions, and serine peptidase inhibitor, Kazal type 6 (SPINK6, increased 12.4-fold). In contrast, the tumor suppressor gene inhibin beta A (INHBA) was down-regulated 2.9-fold by hGRβ. In general, genes that were up-regulated by hGRβ in the absence of hormone were down-regulated by treatment with RU-486, demonstrating that RU-486 has an antagonistic effect on gene regulation (data deposited in GEO, http://www.ncbi.nlm.nih.gov/geo/ [5], accessible through GEO Series accession number GSE5310).
DISCUSSION
The canonical view of hGRβ has been that it is barely expressed, does not bind ligand, and controls transcription only via a dominant-negative effect on hGRα-induced transcription. Therefore, early studies suggested that it was generally of little physiological importance. However, increasing evidence, both previously published and presented here, suggests that this view is limited and incorrect. Our work presented here indicates that hGRβ is able to control transcription without hGRα involvement. We also demonstrate that hGRβ can interact with at least one ligand (RU-486) and shed some light on factors that may govern ligand interaction with the hGRβ LBD. Furthermore, the increased expression of hGRβ in the development of glucocorticoid-resistant forms of immune-related diseases is increasingly well documented, particularly in asthma and ulcerative colitis (17). Importantly, these studies together suggest that it is the relative ratio of hGRα to hGRβ that is the critical factor. Thus, although hGRβ expression may be low in noninflammatory cells, increases in its expression during inflammation may produce significant effects on glucocorticoid sensitivity. The development of glucocorticoid resistance is a serious complication for diseases such as asthma in which the most effective treatments exploit the anti-inflammatory and immunomodulatory actions of glucocorticoids. A ligand for hGRβ could potentially reverse its contribution to glucocorticoid insensitivity and restore the effectiveness of glucocorticoid treatments.
The LBD of hGRβ is identical to that of hGRα up through amino acid 727, which corresponds to the end of helix 10 in the hGRα LBD. From there, the two receptors differ significantly. Although the crystal structure of the hGRβ LBD has not yet been determined, a comparative model of the hGRβ LBD was developed based on the X-ray crystal structure of the hGRα LBD and the structures of previously solved homologous nuclear receptor LBDs (38). This model indicates that in addition to being truncated prior to the hGRα LBD helix 12, the last 15 amino acids of the hGRβ LBD form a somewhat flexible structure that does not resemble the highly ordered helix 11 of hGRα. Together, these features are thought to underlie the inability of hGRβ to bind ligand. Although ligands may be able to enter the LBD of both hGRα and hGRβ, the resulting conformational changes that serve to retain ligands in the hGRα LBD cannot occur with the hGRβ LBD. Thus, ligands that enter the hGRβ LBD may occupy it for such a brief time that they would be considered to have low affinity and not affect hGRβ activity. We report here for the first time that hGRβ can bind a ligand, RU-486, in such a way as to change the cellular localization of the receptor and alter its ability to regulate gene expression.
Given the predicted structural differences between the hGRβ and hGRα LBDs, the observation that hGRβ binds RU-486 and changes its cellular localization was completely unexpected. Indeed, previous work in our laboratory failed to observe binding of RU-486 to hGRβ (22). However, this early work was done using cells transiently transfected with hGRβ and with a low concentration of RU-486 (50 nM), the combination of which may explain the lack of binding observed in that study. Our dose-response results for the nuclear translocation of YFP-hGRβ indicate that at least 100 nM RU-486 is necessary for translocation, which is consistent with the estimated Kd of 138 nM for RU-486 binding to hGRβ. It is therefore likely that in our original experiment we did not observe binding of RU-486 to hGRβ because the concentration of 3H-ligand was too low (18). Because the RU-486 affinity for hGRβ is low compared to the interaction of compounds such as dexamethasone or RU-486 with hGRα, we were concerned that the effects of RU-486 on hGRβ might instead be due to an unidentified contaminant in the RU-486 stock. To address this issue, we confirmed that freshly prepared RU-486 from two different sources (Steraloids and Sigma) had identical effects on hGRβ cellular localization (data not shown). In addition, mass spectrometry analysis confirmed that the only appreciable impurity in either stock solution was 0.5% of an 11β-[4-monomethylamino] phenyl version of RU-486 (data not shown). Therefore, it is unlikely that a contaminating compound caused the effects on hGRβ localization seen with RU-486 treatment.
The interaction between RU-486 and hGRβ was initially suggested by studies of the cellular localization of transiently transfected YFP-hGRβ in COS-1 cells. YFP-hGRβ is found primarily in the cytoplasm in COS-1 cells, in contrast to the primarily nuclear localization of YFP-hGRβ in another cell line, U-2 OS (Fig. 1), as well as of wild-type hGRβ in COS-1 cells (Fig. 2) and other cell types (23). This unusual localization of YFP-tagged hGRβ in COS-1 facilitated the observation of nuclear translocation in response to RU-486 but not dexamethasone. This translocation may not necessarily indicate a ligand-hGRβ interaction. As was noted, one possibility is that RU-486 facilitates heterodimer formation between hGRα and hGRβ as a result of binding to hGRα, which results in cotranslocation of the receptors. While this may be possible, two observations make this mechanism unlikely to account for the RU-486-dependent hGRβ nuclear translocation. First, transiently transfected hGRβ is expressed far in excess of any endogenous hGRα in COS-1 and U-2 OS cells. It is hard to explain how the very small amounts of endogenous hGRα in these cells could heterodimerize with and cause translocation of such an excess of hGRβ over a time course of minutes to hours, especially in COS-1 cells, where the majority of YFP-hGRβ is initially found in the cytoplasm. If such a mechanism did exist, it would be expected that the extent of nuclear translocation of YFP-hGRβ would increase in the presence of increased hGRα expression. This was not the case, however; the same, incomplete nuclear translocation of YFP-hGRβ occurred in response to RU-486 in the presence of equivalent levels of CFP-hGRα (Fig. 3). Furthermore, CFP-hGRα did undergo complete nuclear translocation in response to RU-486 in these cells: the differential distribution of the two receptors in the same cell is highlighted in the merged images in Fig. 3. Together, these results strongly indicate that heterodimerization of hGRβ with hGRα does not occur in the cytoplasm and is not responsible for the RU-486-dependent nuclear translocation of hGRβ.
Although ligands may exist for hGRβ that do not cause cytoplasmic-to-nuclear translocation of YFP-tagged hGRβ in COS-1 cells, this is nevertheless a convenient approach to screen for other candidate hGRβ ligands. Accordingly, we examined a total of 57 compounds for their ability to cause nuclear translocation of hGRβ that might indicate receptor-ligand binding. RU-486 has several features that make it a unique glucocorticoid receptor ligand (as seen in Fig. 6B). In particular, both the nuclear localization studies and the computational docking studies support the idea that the 11β-dimethylaniline group, the 17α-propynyl group, and the placement of the hydroxyl group in the 17β (rather than 17α) position all may contribute to the unique properties of RU-486 as a ligand for hGRβ. Several of the ligands that we examined contained these features, though only RU-486 contained them all. For example, ZK98299 has the 11β-dimethylaniline group, RU-28362 has the 17α-propynyl and 17β-hydroxl groups, RTI 6413-001 has the 11β-dimethylaniline and 17α-propynyl groups, and RTI 3021-002 has the 11β-dimethylaniline and 17β-hydroxl groups. When all of these groups are present in the ligand, they may form unique interactions with amino acids lining the pocket of the hGRβ LBD that could facilitate prolonged occupation of the LBD even in the absence of helix 12.
Indeed, our computational docking studies support the importance of these substituents in potential hGRβ ligands, as evidenced by the good van der Waals interactions exhibited between the RU-486 ligand and hGRβ receptor, which were absent in the case of the docked dexamethasone ligand. To begin with, the hGRβ C terminus seems to wrap around and interact with the 11β-dimethylaniline group when this substituent is present in a potential ligand. Furthermore, it appears that the highest-ranked docked pose for RU-486 in the hGRβ structure has the stereochemistry on position 17 reversed from that of docked RU-486 in the solved hGRα crystal structure (Protein Data Bank 1NHZ) (Fig. 6B). This finding can be attributed to differences in the ligand interactions with the vastly different hGRα and hGRβ C-terminal structures. The 17α-propynyl substituent may facilitate this specific stereochemical orientation of the RU-486 17-OH group within the hGRβ LBD, permitting hydrogen bond interactions with Q642 in the computational docking studies with the hGRβ model structure. The importance of hydrogen bond interactions between ligand and Q642 has already been established for hGRα, both with modeling data (18) and experimentally (2, 18, 28). Computational docking studies of a Q642V mutant of hGRβ support the importance of this residue in hydrogen bond formation and the stereochemistry of substituent 17 on the RU-486 ligand. Experimentally, only RU-486 was able to bind to hGRβ and cause nuclear translocation of the receptor. Such ligand specificity on the part of a receptor is a highly sought feature in drug development. It will be important to learn more about the basis for this specificity, as well as how it can be exploited to produce higher-affinity hGRβ ligands.
Previous work with transiently overexpressed hGRβ in COS-1 cells and endogenous hGRβ in HeLa S3 cells indicates that hGRβ is primarily found in the nucleus (23). It is not known if this localization is the result of an endogenous ligand or is a property inherent to hGRβ. It is interesting, however, that substitution of amino acids Lys733 and Pro734 found in helix 11 of hGRβ into hGRα also leads to a nuclear localization of hGRα in the absence of ligand (38). Of greater importance is the ability of the receptor to mediate changes in gene expression, which might be modulated by binding RU-486. Accordingly, we performed microarray analysis on cells that stably express hGRβ in the absence of hGRα to determine the ability of hGRβ to affect gene expression directly. Our results indicate that hGRβ can regulate gene expression by itself, a novel finding that may be especially important in disease states in which hGRβ expression is up-regulated. Furthermore, this regulatory activity of hGRβ can be altered by RU-486. Additionally, the resemblance that is emerging between hGRβ and two other members of the nuclear receptor superfamily of proteins—CAR and ERRα—is interesting (34). All three of these receptors appear to be ligand-independent activators of gene expression. As of yet, they do not appear to have endogenous ligands but can bind synthetic drugs such as TCPOBOP for CAR (34), tamoxifen for ERRα (4), and now RU-486 for hGRβ.
When used as an abortifacient, RU-486 is administered as a single 600-mg dose (31). Other uses for this potent antiprogestin, antiglucocorticoid agent are also being explored, which would likely entail different doses and administration plans (31). Due to its low rate of metabolic clearance in humans, even a single 100-mg dose of RU-486 persists at micromolar concentrations in the blood up to 72 h after administration (11). Thus, although the affinity of hGRβ for RU-486 is relatively low, it is possible that standard use of this drug results in modification of the activity of endogenous hGRβ. Furthermore, our demonstration of the ability of hGRβ to bind a ligand suggests that it may be possible to design other ligands with higher affinity for this receptor. Taken together, this work suggests that hGRβ may have a more important physiological role than has previously been thought, both as an endogenous manipulator of gene expression and as a pharmaceutical target.
Acknowledgments
We thank Paul Housley, Nick Lu, and Brian Necela for invaluable discussions of this work. We thank Richard Hochberg, Stoney Simons, Paul Housley, Donald McDonnell, and Ed Cook for kindly providing many of the ligands screened in Table 1. We thank Danica Ducharme of the NIEHS Microarray Facility for performing the microarray hybridizations. We thank Fred Lih of the NIEHS Mass Spectrometry Work Group for analysis of the purity of our RU-486 stocks.
Footnotes
Published ahead of print on 22 January 2007.
REFERENCES
- 1.Bamberger, C. M., A.-M. Bamberger, M. de Castro, and G. P. Chrousos. 1995. Glucocorticoid receptor β, a potential endogenous inhibitor of glucocorticoid action in humans. J. Clin. Investig. 95:2435-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bledsoe, R. K., V. G. Montana, T. B. Stanley, C. J. Delves, C. J. Apolito, D. D. McKee, T. G. Consler, D. J. Parks, E. L. Stewart, T. M. Willson, M. H. Lambert, J. T. Moore, K. H. Pearce, and H. E. Xu. 2002. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110:93-105. [DOI] [PubMed] [Google Scholar]
- 3.Christodoulopoulos, P., D. Y. M. Leung, M. W. Elliott, J. C. Hogg, S. Muro, M. Toda, S. Laberge, and Q. Hamid. 2000. Increased number of glucocorticoid receptor-β-expressing cells in the airways in fatal asthma. J. Allergy Clin. Immunol. 106:479-484. [DOI] [PubMed] [Google Scholar]
- 4.Coward, P., D. Lee, M. V. Hull, and J. M. Lehmann. 2001. 4-Hydroxytamoxifen binds to and deactivates the estrogen-related receptor gamma. Proc. Natl. Acad. Sci. USA 98:8880-8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Efron, B., and R. Tibshirani. 1993. An introduction to the bootstrap. Chapman and Hall, Inc., New York, NY.
- 7.Friesner, R. A., J. L. Banks, R. B. Murphy, T. A. Halgren, J. J. Klicic, D. T. Mainz, M. P. Repasky, E. H. Knoll, M. Shelley, J. K. Perry, D. E. Shaw, P. Francis, and P. S. Shenkin. 2004. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47:1739-1749. [DOI] [PubMed] [Google Scholar]
- 8.Halgren, T. A., R. B. Murphy, R. A. Friesner, H. S. Beard, L. L. Frye, W. T. Pollard, and J. L. Banks. 2004. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47:1750-1759. [DOI] [PubMed] [Google Scholar]
- 9.Hamid, Q., S. E. Wenzel, P. J. Hauk, A. Tsicopoulos, B. Wallaert, J.-J. Lafitte, G. P. Chrousos, S. J. Szefler, and D. Y. M. Leung. 1999. Increased glucocorticoid receptor β in airway cells of glucocorticoid-insensitive asthma. Am. J. Respir. Crit. Care Med. 159:1600-1604. [DOI] [PubMed] [Google Scholar]
- 10.Hamilos, D. L., D. Y. M. Leung, S. Muro, A. M. Kahn, S. S. Hamilos, S. E. Thawley, and Q. A. Hamid. 2001. GRbeta expression in nasal polyp inflammatory cells and its relationship to the anti-inflammatory effects of intranasal fluticasone. J. Allergy Clin. Immunol. 108:59-68. [DOI] [PubMed] [Google Scholar]
- 11.Heikinheimo, O., K. Kontula, H. Croxatto, I. M. Spitz, T. Luukkainen, and P. Lahteenmaki. 1987. Plasma concentrations and receptor binding of RU 486 and its metabolites in humans. J. Steroid Biochem. 26:279-284. [DOI] [PubMed] [Google Scholar]
- 12.Hollenberg, S. M., C. Weinberger, E. S. Ong, G. Cerelli, A. Oro, R. Lebo, E. B. Thompson, M. G. Rosenfeld, and R. M. Evans. 1985. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature 318:635-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honda, M., F. Orii, T. Ayabe, S. Imai, T. Ashida, T. Obara, and Y. Kohgo. 2000. Expression of glucocorticoid receptor β in lymphocytes of patients with glucocorticoid-resistant ulcerative colitis. Gastroenterology 118:859-866. [DOI] [PubMed] [Google Scholar]
- 14.Hoyte, R. M., D. C. Labaree, J. M. Fede, C. Harris, and R. B. Hochberg. 1998. Iodinated and fluorinated steroid 2′-aryl-[3,2-c] pyrazoles as potential glucocorticoid receptor imaging agents. Steroids 63:595-602. [DOI] [PubMed] [Google Scholar]
- 15.Hoyte, R. M., J. X. Zhang, R. Lerum, A. Oluyemi, P. Persaud, C. O'Connor, D. C. Labaree, and R. B. Hochberg. 2002. Synthesis of halogen-substituted pyridyl and pyrimidyl derivatives of [3,2-c]pyrazolo corticosteroids: strategies for the development of glucocorticoid receptor mediated imaging agents. J. Med. Chem. 45:5397-5405. [DOI] [PubMed] [Google Scholar]
- 16.Leung, D. Y. M., Q. Hamid, A. Vottero, S. J. Szefler, W. Surs, E. Minshall, G. P. Chrousos, and D. J. Klemm. 1997. Association of glucocorticoid insensitivity with increased expression of glucocorticoid receptor β. J. Exp. Med. 186:1567-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis-Tuffin, L. J., and J. A. Cidlowski. 2006. The physiology of human glucocorticoid receptor beta (hGRbeta) and glucocorticoid resistance. Ann. N. Y. Acad. Sci. 1069:1-9. [DOI] [PubMed] [Google Scholar]
- 18.Lind, U., P. Greenidge, M. Gillner, K. F. Koehler, A. Wright, and J. Carlstedt-Duke. 2000. Functional probing of the human glucocorticoid receptor steroid-interacting surface by site-directed mutagenesis. Gln-642 plays an important role in steroid recognition and binding. J. Biol. Chem. 275:19041-19049. [DOI] [PubMed] [Google Scholar]
- 19.Longui, C. A., A. Vottero, P. C. Adamson, D. E. Cole, T. Kino, O. Monte, and G. P. Chrousos. 2000. Low glucocorticoid receptor alpha/beta ratio in T-cell lymphoblastic leukemia. Horm. Metab. Res. 32:401-406. [DOI] [PubMed] [Google Scholar]
- 20.Lu, N. Z., and J. A. Cidlowski. 2005. Translational regulatory mechanisms generate N-terminal glucocorticoid receptor isoforms with unique transcriptional target genes. Mol. Cell 18:331-342. [DOI] [PubMed] [Google Scholar]
- 21.Oakley, R. H., C. M. Jewell, M. R. Yudt, D. M. Bofetiado, and J. A. Cidlowski. 1999. The dominant negative activity of the human glucocorticoid receptor beta isoform. Specificity and mechanisms of action. J. Biol. Chem. 274:27857-27866. [DOI] [PubMed] [Google Scholar]
- 22.Oakley, R. H., M. Sar, and J. A. Cidlowski. 1996. The human glucocorticoid receptor beta isoform. Expression, biochemical properties, and putative function. J. Biol. Chem. 271:9550-9559. [DOI] [PubMed] [Google Scholar]
- 23.Oakley, R. H., J. C. Webster, M. Sar, C. R. Parker, Jr., and J. A. Cidlowski. 1997. Expression and subcellular distribution of the beta-isoform of the human glucocorticoid receptor. Endocrinology 138:5028-5038. [DOI] [PubMed] [Google Scholar]
- 24.Peddada, S. D., K. E. Prescott, and M. Conaway. 2001. Tests for order restrictions in binary data. Biometrics 57:1219-1227. [DOI] [PubMed] [Google Scholar]
- 25.Pujols, L., J. Mullol, J. Roca-Ferrer, A. Torrego, A. Xaubet, J. A. Cidlowski, and C. Picado. 2002. Expression of glucocorticoid receptor alpha- and beta-isoforms in human cells and tissues. Am. J. Physiol. Cell Physiol. 283:C1324-C1331. [DOI] [PubMed] [Google Scholar]
- 26.Sathya, G., M. S. Jansen, S. C. Nagel, C. E. Cook, and D. P. McDonnell. 2002. Identification and characterization of novel estrogen receptor-beta-sparing antiprogestins. Endocrinology 143:3071-3082. [DOI] [PubMed] [Google Scholar]
- 27.Schaaf, M. J., and J. A. Cidlowski. 2003. Molecular determinants of glucocorticoid receptor mobility in living cells: the importance of ligand affinity. Mol. Cell. Biol. 23:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaaf, M. J., L. J. Lewis-Tuffin, and J. A. Cidlowski. 2005. Ligand-selective targeting of the glucocorticoid receptor to nuclear subdomains is associated with decreased receptor mobility. Mol. Endocrinol. 19:1501-1515. [DOI] [PubMed] [Google Scholar]
- 29.Shahidi, H., A. Vottero, C. A. Stratakis, S. E. Taymans, M. Karl, C. A. Longui, G. P. Chrousos, W. H. Daughaday, S. A. Gregory, and J. M. D. Plate. 1999. Imbalanced expression of the glucocorticoid receptor isoforms in cultured lymphocytes from a patient with systemic glucocorticoid resistance and chronic lymphocytic leukemia. Biochem. Biophys. Res. Commun. 254:559-565. [DOI] [PubMed] [Google Scholar]
- 30.Sousa, A. R., S. J. Lane, J. A. Cidlowski, D. Z. Staynov, and T. H. Lee. 2000. Glucocorticoid resistance in asthma is associated with elevated in vivo expression of the glucocorticoid receptor β-isoform. J. Allergy Clin. Immunol. 105:943-950. [DOI] [PubMed] [Google Scholar]
- 31.Spitz, I. M., and C. W. Bardin. 1993. Mifepristone (RU 486)—a modulator of progestin and glucocorticoid action. N. Engl. J. Med. 329:404-412. [DOI] [PubMed] [Google Scholar]
- 32.Stoughton, R. S., and H. Dai. 26February2002. Statistical combining of cell expression profiles. U.S. patent 6,351,712.
- 33.Strickland, I., K. Kisich, P. J. Hauk, A. Vottero, G. P. Chrousos, D. J. Klemm, and D. Y. M. Leung. 2001. High constitutive glucocorticoid receptor β in human neutrophils enables them to reduce their spontaneous rate of cell death in response to corticosteroids. J. Exp. Med. 193:585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suino, K., L. Peng, R. Reynolds, Y. Li, J. Y. Cha, J. J. Repa, S. A. Kliewer, and H. E. Xu. 2004. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol. Cell 16:893-905. [DOI] [PubMed] [Google Scholar]
- 35.Wagner, B. L., G. Pollio, P. Giangrande, J. C. Webster, M. Breslin, D. E. Mais, C. E. Cook, W. V. Vedeckis, J. A. Cidlowski, and D. P. McDonnell. 1999. The novel progesterone receptor antagonists RTI 3021-012 and RTI 3021-022 exhibit complex glucocorticoid receptor antagonist activities: implications for the development of dissociated antiprogestins. Endocrinology 140:1449-1458. [DOI] [PubMed] [Google Scholar]
- 36.Webster, J. C., R. H. Oakley, C. M. Jewell, and J. A. Cidlowski. 2001. Proinflammatory cytokines regulate human glucocorticoid receptor gene expression and lead to the accumulation of the dominant negative beta isoform: a mechanism for the generation of glucocorticoid resistance. Proc. Natl. Acad. Sci. USA 98:6865-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yudt, M. R., and J. A. Cidlowski. 2001. Molecular identification and characterization of a and b forms of the glucocorticoid receptor. Mol. Endocrinol. 15:1093-1103. [DOI] [PubMed] [Google Scholar]
- 38.Yudt, M. R., C. M. Jewell, R. J. Bienstock, and J. A. Cidlowski. 2003. Molecular origins for the dominant negative function of human glucocorticoid receptor beta. Mol. Cell. Biol. 23:4319-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]