Abstract
Mutations of the neurofibromatosis 2 (NF2) tumor suppressor gene have frequently been detected not only in schwannomas and other central nervous system tumors of NF2 patients but also in their sporadic counterparts and malignant tumors unrelated to the NF2 syndrome such as malignant mesothelioma, indicating a broader role for the NF2 gene in human tumorigenesis. However, the mechanisms by which the NF2 product, merlin or schwannomin, is regulated and controls cell proliferation remain elusive. Here, we identify a novel GTP-binding protein, dubbed NGB (referring to NF2-associated GTP binding protein), which binds to merlin. NGB is highly conserved between Saccharomyces cerevisiae, Caenorhabditis elegans, and human cells, and its GTP-binding region is very similar to those found in R-ras and Rap2. However, ectopic expression of NGB inhibits cell growth, cell aggregation, and tumorigenicity in tumorigenic schwanomma cells. Down-regulation and infrequent mutation of NGB were detected in human glioma cell lines and primary tumors. The interaction of NGB with merlin impairs the turnover of merlin, yet merlin does not affect the GTPase nor GTP-binding activity of NGB. Finally, the tumor suppressor functions of NGB require merlin and are linked to its ability to suppress cyclin D1 expression. Collectively, these findings indicate that NGB is a tumor suppressor that regulates and requires merlin to suppress cell proliferation.
Neurofibromatosis type 2 (NF2) is a dominantly inherited disorder that is characterized by predisposition to the development of multiple benign tumors of the central nervous system. The most common tumors found in NF2 are vestibular schwannomas, meningiomas, and ependymomas (3, 56). These tumors grow slowly, but their location predominantly within the central nervous system has catastrophic effects on sensitive intracranial and intraspinal structures, thus resulting in a high rate of morbidity and mortality. The NF2 gene encodes a 595-amino-acid protein called merlin (58) or schwannomin (44), which is a member of the band 4.1 superfamily of cytoskeleton-associated proteins. This family consists of a large group of membrane-associated cytoplasmic proteins including protein 4.1, talin, ezrin, radixin, and moesin (ERM) proteins, several protein-tyrosine phosphatases, and at least two nonmuscle myosins (31). The distinct feature of this superfamily is a conserved region of 200 to 300 amino acids located in the N terminus of each protein. This region is highly conserved among the ERM proteins and merlin (62% identity in the first 350 amino acids). The ERM proteins function as molecular linkers by binding to transmembrane proteins through the amino-terminal domain and linking them to the cortical actin cytoskeleton through a carboxyl-terminal actin-binding domain (1, 42, 43). Therefore, merlin might form a link between the cell membrane and cytoskeletal proteins and thereby participate in cell-cell and cell-matrix contact signaling (2, 17, 29, 35). Studies of cultured cells indicate that expressed merlin protein accumulates in certain actin-rich membrane regions, such as membrane ruffles at the leading edge of migrating cells, consistent with the localization of ERM proteins. However, several lines of evidence demonstrate that merlin has functions that are clearly distinct from those of ERM proteins. First, merlin lacks the well-defined C-terminal actin-binding domain found in ERM proteins. In addition, the ERM proteins are functionally redundant, whereas there is no evidence for redundancy between merlin and the ERM proteins. Homozygous knockout of the Nf2 gene is lethal in mice (34). Heterozygous knockout of Nf2, but not of genes encoding ERM proteins, develops a variety of highly metastatic malignant tumors (12, 33). Finally, expression of merlin, but not ERM proteins, in NIH 3T3 cells significantly inhibits cell growth and Ras transformation (30, 57).
The NF2 gene is composed of 17 exons. Alternative splicing events lead to expression of several isoforms of merlin (19, 28). The two most abundant isoforms lack residues corresponding to either exon 16 (isoform I, 595 amino acids) or exon 17 (isoform II, 590 amino acids), leading to variant C-terminal ends of the proteins. It has been shown that isoform I, but not isoform II, is able to form intramolecular interactions between a central region spanning residues 288 to 400 and the C terminus encoded by exon 17 and is essential for inhibition of cell growth and tumorigenicity in rat schwannoma cell lines (52). A previous study demonstrated that merlin interacts with an actin-binding protein βII-spectrin. As merlin itself lacks the actin binding sites found in ERM protein, signaling from merlin to the actin cytoskleton could be mediated via actin-binding sites on βII-spectrin (48). In addition, several merlin-associated proteins have been identified, including NHE-RF, HRS, and SCHIP-1 (13, 14, 49, 54). NHE-RF is a regulatory cofactor of Na+-H+ exchanger and possesses two PDZ domains that are thought to mediate protein-protein interaction. NHE-RF interacts with both merlin and ERM proteins via downstream PDZ domains (13). HRS refers to the hepatocyte growth factor-regulated tyrosine kinase substrate, whose tyrosine phosphorylation is induced by growth factors and cytokines. Moreover, HRS also interacts with STAM, which is associated with JAK2 and the JAK3/Stat pathway, suggesting a possible role of NF2 in sorting of membrane receptors and inhibition of the Jak/Stat pathway (49, 50). SCHIP-1 is a serine-rich protein that contains coil-coil domains that allow SCHIP-SCHIP dimerization. Interestingly, SCHIP-1 only interacts with some naturally occurring mutants of NF2 or with an NF2-spliced isoform lacking exons 2 and 3 but not with the NF2 isoforms exhibiting growth-suppression activity (14). A recent study showed that merlin also interacts with Ral guanine nucleotide dissociation stimulator (RalGDS), a downstream molecule of Ras and inhibits RalGDS-induced RalA activation, colony formation, and cell migration in mammalian cells (45).
In addition, the phosphorylation status of merlin is related to its tumor suppressor function, i.e., hyperphosphorylation inactivates merlin, whereas the dephosphorylated form of merlin is associated with its growth inhibitory function (53). It has been shown that, in logarithmically growing low-density cell cultures, CD44 interacts with phosphorylated merlin. Upon treating cells with CD44 antibody or CD44 ligand hyaluronate, merlin becomes hypophosphorylated, which results in disassociation with CD44 and inhibition of cell growth. Thus, CD44 and merlin form a molecular switch to specify cell growth arrest or proliferation (37, 46). Constitutively active Rac has been shown to induce merlin phosphorylation and decreases the association of merlin with the cytoskeleton through PAK1 and PAK2 (24, 60, 61), and overexpression of merlin inhibits Rac-induced signaling, such as activation of JNK and PAK1, leading to downregulation of cyclin D1 (21, 24, 47, 51, 62). Although these findings greatly enhance our knowledge of merlin function, the mechanisms by which merlin is regulated and contributes to tumor suppression remain elusive.
With the goal to understand the normal cellular function of merlin and how it acts as a tumor suppressor, we utilized the yeast two-hybrid system to isolate potential interacting proteins. In this report, we describe the identification of a novel merlin-interacting protein, NGB, which contains five sequence motifs, G1 to G5 that are well conserved in the small G-protein family. NGB interacts with merlin both in vitro and in vivo in mammalian cells. The G-protein homology domain of NGB and the N and C termini of merlin are required for their interaction. Expression of NGB prevents degradation of merlin and inhibits cell growth in rat schwannoma and human glioma cells. Our findings suggest that NGB is itself a tumor suppressor that plays an important role in merlin regulation.
MATERIALS AND METHODS
Reagents and antibodies.
Cell culture media, protein A/G beads, and Lipofectamine Plus were from Invitrogen (Carlsbad, CA). [γ-32P]ATP, Tran-35S label, and [methyl-3H]thymidine were from ICN (Irvine, CA). NGB polyclonal antibodies were generated by immunizing rabbits with glutathione S-transferase (GST)-NGB (amino acids [aa] 338 to 501) fusion protein. The serum was collected and affinity purified by passing it over a protein A-Sepharose column (Pharmacia) according to the manufacturer's procedure. Anti-HA (hemagglutinin) and -Flag antibodies were purchased from Roche (Indianapolis, IN) and Sigma (St. Louis, MO), respectively. Anti-cyclin D1 and -β-actin antibodies were from Santa Cruz (CA). Anti-NF2 antibody was described previously (9). The EnzChek caspase-3 assay kit was purchased from Molecular Probes.
Cell culture, transfection, and tumor DNAs.
JS-1 rat schwannoma cells (a gift of D. Gutmann), which express low levels of NF2 (52), and 82HTB rhabdomyosarcoma cells (American Type Culture Collection), which express abundant NF2, and Nf2-deficient (−/−) mouse embryonic fibroblasts (MEFs) (62) as well as glioma cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. Cells were transfected with NGB, NF2, and RNA interference (RNAi) expression plasmids using Lipofectamine Plus. Stably transfected clonal cell lines were established following G418 selection. Primary glioma DNAs were obtained from patients who underwent surgery at H. Lee Moffitt Cancer Center and at Community Hospital, Braunschweig, Germany.
Cell proliferation, viability, DNA synthesis, and colony formation assays.
NF2-, NGB-, or NF2/NGB-transfected cells were plated in 35-mm dishes at a density of 1.0 × 105 cells/dish. Cell numbers were measured with a Coulter Counter (Coulter Electronics, FL) daily for up to 3 days following seeding. MTS assays were performed according to the manufacturer's recommendations (Promega, Madison, WI). The cells were plated in 96-well microtiter plates at a density of 1.0 × 103 cells/well in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. The number of cells at 1, 2, and 3 days was determined using cell counter and the colorimetric CellTiter96 AQueous (MTS) assay (Promega). Results were depicted as absorbance at 490 nm as a function of time. Cell viability was examined with Trypan blue staining and caspase-3 activity following treatment of cells with VP16 (5 μM), taxol (100 nM), or doxorubicin (2 μM) for 12 h.
Thymidine incorporation was used to investigate the effect of NGB and/or NF2 on DNA synthesis. The cells were grown to 80% confluence in 96-well plates, and during the last 16 h of growth, they were subjected to 5 μCi/ml of [3H]thymidine. After rinsing twice with ice-cold serum-free medium, the cells were incubated twice with 5 ml of 10% trichloroacetic acid for 10 min on ice and lysed in 500 μl 1% sodium dodecyl sulfate (SDS) in 0.3 N NaOH for 30 min at 37°C. Incorporated radioactivity was quantitated with a spectrometer. Colony formation in soft agar was examined as previously described (8).
Plasmid constructions.
For yeast two-hybrid screening, the N-terminal portion of merlin encoding amino acids 1 to 374 (4, 6, 44, 58) was cloned into the EcoRI and BamHI sites of pJK202 to create the bait pNLexA-NF2N. For mapping of interaction domains, a series of deletion fragments of NF2 and NGB were prepared by PCR and inserted into pJK202 or pJG4-5. The oligonucleotide primers used were as follows: for NF2, 5′-ATGGCCGGGGCCATCGCTTC-3′, 5′-CCCAAGACGTTCACCGTGAG-3′, 5′-CGGACTCTGGGGCTCCG-3′, 5′-CTAGAGCTCTTCAAAGAAGGC-3′, 5′-AGCGAGGCGCTGCCGCTC-3′, 5′-TGAGGAGTTAAACTTGAAGAC-3′, and 5′-GCCTCGGTGCTCTGCGTACC-3′; for NGB, 5′-ATGGCACATTACAACTTCAAG-3′, 5′-ACCCTGCTTTTGTGTGGGTAC-3′, 5′-AATAAAGTGAATGAGGTGCTG-3′, 5′-TTGGAAGAATTAGAAAAAG-3′, 5′-CTATCTCCTGTCCTTTTTACC-3′, 5′-TTTCTTCATGATGGCTGG-3′, 5′-TCCCTTCATTTTGGTTTC-3′, and 5′-CCTGGTATTCGGATCAATG-3′. The coding sequence of NGB was ligated to pGEX to generate a GST-NGB fusion protein (Pharmacia). A fused HA-NGB was constructed by first creating an EcoRI site just 5′ of the start codon using PCR and inserting this coding sequence into the EcoRI and XbaI sites of a cytomegalovirus (CMV)-based expression vector, pcDNA3 (Invitrogen). A double-stranded oligonucleotide adapter encoding the HA epitope tag was then inserted between the engineered EcoRI site of the cDNA and HindIII sites of the vector to create CMV-Flag-NGB and CMV-HA-NGB. A similar strategy was used to create CMV-Flag-NF2 and CMV-HA-NF2. The constructs prepared using PCR products were confirmed by nucleotide sequencing. Adenovirus-cyclin D1 was kindly provided by S. Chellappan and I. Cozar-Castellano. The lentivirus of pLKO.1-shRNA/Nf2 (5′-GCTCTTAGAAATCGCCACCAA-3′) was obtained from Sigma.
Yeast two-hybrid screen.
A genetic screen using the yeast two-hybrid system was performed as previously described (19). Briefly, yeast strain EGY191, which harbors the LexAop-Leu2 reporter gene, was transformed with bait plasmid pNLexA-NF2N and reporter plasmid pSH18-34 and subsequently transformed with a human brain interaction library. Approximately 2 × 106 primary library transformants were obtained and plated on Ura− His− Trp− Leu− galactose-raffinose plates. Candidate clones were identified by their ability to grow on Ura− His− Trp− Leu− galactose-raffinose plates, but not on Ura− His− Trp− Leu− glucose plates, and their ability to yield blue colonies on Ura− His− Trp− X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-galactose-raffinose plates, but not on Ura− His− Trp− glucose plates. KC8 cells were transformed with plasmids isolated from positive colonies. The specificity of interaction of candidate plasmids with pNLexA-NF2N was tested by retransformation of positive clones into yeast harboring either the NF2 bait or several unrelated bait plasmids. Nucleotide sequence analysis of cDNA inserts was performed using an Applied Biosystems automated sequencer.
Northern blot analysis and cDNA library screening.
A northern filter containing poly(A)+ RNAs prepared from various human tissues (Clontech) was hybridized with a radiolabeled NF2 or NGB probe. NGB clones were isolated by plaque hybridization from a human skeletal muscle cDNA library (Clontech). DNA probes were labeled using [32P]dCTP and a random primer kit (Amersham). Filters were preincubated for 3 h at 60°C in hybridization solution (0.5 M sodium phosphate [pH 7.2], 7% SDS, 2.0 mM EDTA, 0.1% sodium pyrophosphate, and 100 μg/ml of denatured salmon sperm DNA). The filters were then incubated overnight at 60°C with a 32P-labeled NGB probe in hybridization solution, washed two times with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at 65°C and analyzed by autoradiography.
In vitro protein binding assays.
A solution binding assay is designed to examine the interaction of GST fusion proteins with endogenous cellular protein, in vitro-translated protein, or protein overexpressed in the cells. Expression and purification of GST fusion proteins were performed as previously described (8). Cell extracts were precleared with glutathione-agarose beads and then incubated with GST fusion protein-conjugated glutathione-agarose beads for 2 h at 4°C. The beads were then washed five times with TNN buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.5% Nonidet P-40) containing 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 2 μg/ml leupeptin, and 1 mM sodium orthovanadate. The samples were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to the membrane, and detected with the antibody.
Immunoblotting and immunoprecipitation.
A2780 human ovarian cancer cells were synchronized at different stages of the cell cycle using a double thymidine block (28). DNA content measured by flow cytometry was used to monitor cell synchronization. Immunoblotting was carried out as described previously (8). Briefly, equivalent amounts of protein were separated on a SDS-PAGE gel and transferred onto a Hybond-C Super membrane (Amersham). Following overnight incubation in 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) at 4°C, the filters were exposed for 1 h at room temperature to antibody in blocking solution (1% BSA in PBS) and then detected with the ECL Western blotting analysis system (Amersham). For immunoprecipitation, cells were washed in ice-cold PBS and lysed in 1 ml of lysis buffer. Cell debris was removed by centrifugation at 14,000 rpm for 10 min at 4°C. Cell lysate was precleared with protein A-G (2:1) agarose (Gibco BRL) beads for 20 min, centrifuged to remove the beads, and incubated with antiserum for 1 h. Protein A-G beads were then added, and the suspension was incubated at 4°C for 2 h with constant rotation. The beads were washed four times with incubation buffer, and proteins were eluted with Laemmli sample buffer. Eluted proteins were analyzed by SDS-PAGE and immunoblotting with appropriate antibodies.
Immunofluorescence.
82HTB cells were grown on glass chamber slides for 2 to 3 days. Cells were fixed for 1 h at 37°C using 2% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.4) and permeabilized with cold (−20°C) ethanol for 15 min. After washing three times with PBS-0.1% Triton X-100 (PBST), the cells were treated for 1 h with blocking buffer (0.9% NaCl, 1% BSA, 10 mM Tris-HCl, pH 8.0, 0.05% Tween 20, and 0.02% NaN3), washed three times with PBST, and then incubated for 1 h with 150 μl of PBST and 1% BSA containing either anti-NGB polyclonal antibody or anti-NF2 monoclonal antibody at a dilutions of 1:20 to 1:40. After three washes with PBST, the cells were incubated for 1 h with 150 μl of PBST and 1% BSA containing fluorescein isothiocyanate-conjugated goat anti-rabbit or anti-mouse immunoglobulin G secondary antibody (Sigma) (1:200 dilution) and rhodamine-conjugated phalloidin (Sigma) (1:200 dilution). The cells were then washed three times with PBST and mounted using Vectashield mounting fluid (Vector). Imaging was analyzed using a confocal microscope.
Cell migration, attachment, and aggregation assays.
A membrane invasion culture system (MICS) was used to assess the migratory ability of the JS1 cells as previously described (18). Briefly, a 10-μm-pore-size polycarbonate membrane (Osmonics, CA) soaked in 0.1% gelatin was placed between the upper and lower plates of the MICS chamber with 1 × 105 cells and incubated for 4 h at 37°C. Those cells that had migrated through the pores to the lower wells were harvested with 2 mM EDTA in PBS and stained with a LeukoStat staining kit (Fisher Scientific), and cells in 6 to 8 random high-power microscopic fields were counted visually to calculate the migration rate. The migration rates for the stable NGB and NF2 cell lines were determined by normalizing values for the pcDNA3 vector JS1 clones to 100% and comparing its relative migration values with those of the experimental NGB- and NF-expressing cell lines. Cell attachment was measured by seeding 10,000 pcDNA3-, NGB-, or NF-transfected JS1 cells into 96-well plates precoated with 10 μg/ml fibronectin, collagen IV, or collagen I (Gibco Life Sciences). Twelve wells were used for each condition. After 1 or 3 h, the plates were gently washed in 1× PBS, and the number of adherent cells was determined by incubation with 0.5% crystal violet for 30 min, followed by extraction in 1% SDS overnight and spectrophotometric analysis at 540 nm. For cell aggregation assay, subconfluent cultures of pcDNA3-, NGB-, or NF-transfected JS1 cells were detached from tissue culture dishes and washed with medium. Each well of a 24-well, low-binding-affinity tissue culture plate (Costar, Cambridge, MA) contained 500 μl of single-cell suspension at a concentration of 5 × 104 cells/ml in DMEM. Plates were incubated at 4°C or 37°C on a rotating platform for 45 min as described previously (31). Cell aggregation was assessed by counting single cells, small aggregates of 5 to 10 cells, and large aggregates of >10 cells using an inverted phase-contrast microscope. For each measurement, 100 observations were collected and classified into the three groups (single cells, small aggregates of 5 to 10 cells, and large aggregates of >10 cells). Aggregated cell mixtures were fixed with 2% glutaraldehyde. The aggregates were defined as cell clumps containing more than five cells. Aggregates in four randomly selected high-power fields were counted using light microscopy.
GTP binding assay.
GTP binding to NGB was determined with the rapid filtration method (10, 41). Flag-NGB and HA-NF2 were immunoprecipitated from the transfected-COS7 cells with anti-Flag or anti-HA antibody in the presence of protein A-G beads. After being washed three times with lysis buffer, NGB and NF2 proteins were eluted from the beads with Flag or HA peptide in a buffer containing 50 mM Tris (pH 7.4) and 150 mM NaCl. NGB (1 μg) or bovine serum albumin (1 μg) was incubated in binding buffer (20 mM Tris [pH 7.4], 50 mM NaCl, 0.1% Triton X-100, 1 mM dithiothreitol, 40 μg/ml bovine serum albumin, 1 mM EDTA, 10 mM MgCl2, and 1 μM GTPγS [0.45 mCi/sample]) at 22°C. At the indicated times, aliquots of 100 μl were withdrawn in duplicate, and the reaction was stopped by the addition of 150 μl of ice-cold wash buffer (20 mM Tris [pH 7.4], 50 mM NaCl, and 5 mM MgCl2) and immediately filtration through BA 85 nitrocellulose filters equipped in a slot blotting device, followed by four washed with ice-cold wash buffer. The filters that were taken from slot blotting were washed twice with wash buffer for 10 min at room temperature. After air drying, the membranes were exposed to X-ray film and then quantitated by scintillation counting. Nonspecific background was determined by the assay in the absence of added protein. To examine the specificity of GTP binding, nonlabeled ribonucleotides at a final concentration of 20 μM were added to the reaction and incubated for 1 h prior to filtration.
GTPase assay.
Immunopurification of NGB and NF2 was described as above. GTP hydrolysis assays were performed in buffer containing 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM dithiothreitol, 1 mM MgCl2, NGB and/or NF2 protein (1 μg/reaction), 10 μM [32P]GTP, and either 30 mM UTP or 20 mM UTP and 10 mM GTP (11). Reaction mixtures were incubated at 30°C, and 1-μl aliquots were removed at the indicated times and spotted directly on poly(ethyleneimine)-cellulose plates (EM Separations). Chromatograms were developed in 1 M LiCl-1 M formic acid and exposed to X-OMAT AR film (Kodak) for 15 h. The migration of authentic GTP and GDP standards was detected with UV (254 nm). The plate was quantified with a Molecular Dynamics PhosphorImager. The percentage of GTP hydrolysis was calculated by dividing the amount of radioactivity in the GDP region by that in the sum of the GDP and GTP regions.
Pulse-chase experiments.
Prior to the radioactive labeling, normal culture medium was removed, and cells were washed twice with PBS and refed with modified Eagle's medium lacking methionine but supplemented with 10% dialyzed fetal bovine serum. After 30 min, this starvation medium was removed and replaced with 3 ml of the same medium supplemented with 300 μCi of Tran35S label (ICN) per plate. For pulse-chase experiments, cells were incubated in labeling medium for 30 min, and then one set of plates was harvested (pulse) and the other plates were washed twice with warm PBS, refed with normal culture medium supplemented with 1 mM cold methionine, and incubated for various lengths of time (chase). To harvest the cells, plates were washed twice in cold PBS, and the cells were then scraped in PBS and transferred to 15-ml conical tubes. Cells were pelleted at 4°C, resuspended in 1 ml of cold PBS, and counted. Equal numbers of cells (approximately 5 × 105) were then pelleted and resuspended in 100 μl of cold PBS. Three hundred microliters of RIPA buffer (50 mM Tris [pH 7.4], 120 mM NaCl, 0.1% deoxycholic acid, 2% Nonidet P-40, 0.2% SDS, aprotinin and leupeptin [2 μg/ml each], and dithiothreitol and phenylmethylsulfonyl fluoride [1 mM each]) was added. After vortexing for 10 min, the lysate was cleared with a 16,000 × g spin and transferred to a fresh tube containing an additional 600 μl of RIPA buffer. After gentle mixing, an aliquot was removed for analysis of total merlin concentration prior to incubation with antibody. The remaining lysate was mixed with our anti-NF2 antibody for 2 h at 4°C in the presence of 40 μl of a 50% protein A-Sepharose. The beads were then washed three times in RIPA buffer. Proteins were eluted from the beads and, after boiling, were electrophoresed through an SDS-10% polyacrylamide gel. Gels were stained and then subjected to fluorography, dried, and autoradiographed. Quantitation of bands was performed with a PhosphorImager (Molecular Dynamics).
Nude mouse tumor xenograft model.
NGB and NF2 stably transfected JS1 clonal cells as well as pcDNA3-transfected cells were harvested, resuspended in PBS, and injected subcutaneously into 8-week-old female nude mice (106 cells/mouse) as reported previously (8). Tumors were measured every 2 days.
RESULTS
Isolation of NGB using the yeast two-hybrid system.
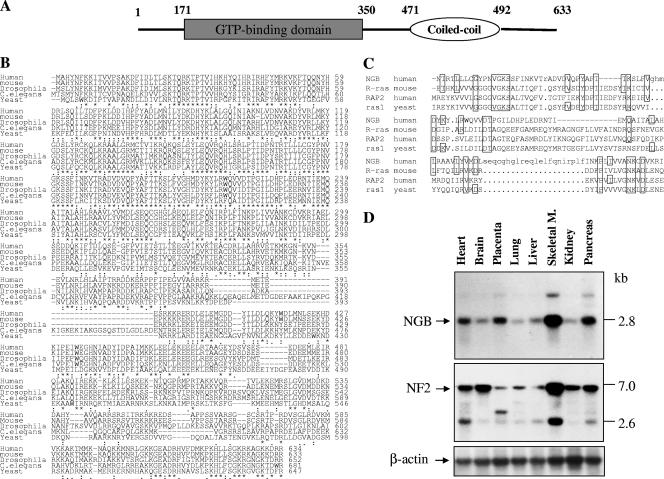
A segment of the merlin N-terminal region that shares high homology with ERM proteins was used as bait in the yeast two-hybrid system to identify merlin-interacting proteins. A human brain cDNA library was used in this screen because merlin is highly expressed in the brain (44, 58). An N-terminal portion (amino acids 1 to 374) of merlin was cloned into the EcoRI and BamHI sites of pJK202 to create the bait pNLexA-NF2N. Three clones that specifically interacted with the bait were identified. Sequence analysis revealed that two of the clones contained overlapping sequences of a cDNA (26). The largest clone contained a 243-amino-acid open reading frame with a conserved GTP-binding domain named NGB (NF2-associated GTP binding protein) (Fig. 1A). Additional cDNA clones were isolated from a human skeletal muscle cDNA library by plaque hybridization using the largest clone as the radiolabeled probe. Sequence analysis revealed that the full-length open reading frame of NGB encoded a 633-amino-acid protein (accession number AF120334) (Fig. 1B). NGB contains coiled-coil domains and five sequence motifs, G1 to G5, which are conserved in the GTPase superfamily (5, 20). Protein analyses by the “All-in-One-SeqAnalyser-SMART3” program showed that the structure and sequence homology of G1 to G5 of NGB are very similar to those found in the Ras and RAP small G-protein families (Fig. 1C). Apart from these characteristic motifs, the amino acid sequence differs substantially from those of the well characterized G-proteins but is similar to uncharacterized Caenorhabditis elegans (similarity, 70%; identity, 58%), Drosophila (similarity, 67%; identity, 54%), and yeast (similarity, 67%; identity, 48%) (accession number U43281) (Fig. 1B) proteins, suggesting the existence of a new subfamily of GTP-binding proteins. The expression pattern of NGB is similar to that of NF2, being abundant in the skeletal muscle, pancreas, and heart (Fig. 1D). These data suggest the potential importance of NGB in NF2 signaling. In addition, we noted the different expression levels between NGB and NF2 in the brain, placenta, and kidney, implying that they may have distinct functions in different cell types.
FIG. 1.
Putative domain structure, protein sequence alignment, and expression pattern of NGB. (A) Schematic representation of domain structure of NGB protein, which is composed of a GTP binding domain at the N terminus and a coiled-coil region at the C terminus. (B) Alignment of amino acid sequence of NGB from human, mouse, Drosophila, C. elegans, and yeast cells. Conserved residues are indicated by asterisks. (C) Sequence comparison of GTP-binding domain between NGB, R-Ras, RAP2, and rasl. Conserved amino acids are boxed. (D) Northern blot analysis. A human multiple-tissue mRNA blot was hybridized with [32P]dCTP-labeled NGB, NF2, and β-actin cDNA probes. M., muscle.
Merlin interacts with NGB in vitro and in vivo.
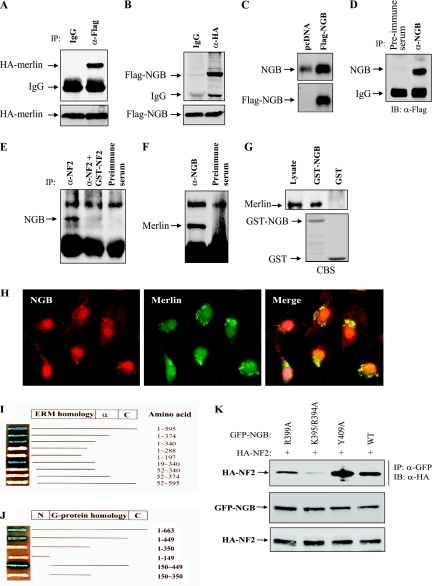
To confirm the association of merlin with NGB that was identified by the yeast two-hybrid system, HA-tagged NF2 and Flag-tagged NGB were cotransfected into HEK293 cells. After 48 h of the transfection, the cells were lysed and immunoprecipitated with anti-HA (NF2) or anti-Flag (NGB) antibody. The immunoprecipitates were subjected to Western blot analysis. As shown in Fig. 2A and B, Flag-NGB and HA-NF2 were readily detected in anti-HA and anti-Flag immunoprecipitates, respectively. To demonstrate that endogenous NGB and NF2 interact, a coimmunoprecipitation experiment was performed using both 82HTB cells and fresh skeletal muscle tissues with antibodies against NF2 and NGB. The specificity of anti-NGB antibody was demonstrated by Western and immunoprecipitation (IP)-Western blot analysis (Fig. 2C and D). Figure 2E shows that NGB was immunoprecipitated with anti-NF2 antibody but not with preimmune serum. Moreover, the immunoprecipitation of NGB was competed by GST-NF2 fusion protein, which was used as an antigen to generate the anti-NF2 antibody. Further, merlin was detected in NGB immunoprecipitates (Fig. 2F). In addition, a GST pull-down assay shows that GST-NGB, but not GST, binds to merlin (Fig. 2G). These data indicate that the association between NGB and merlin is specific in vivo and in vitro.
FIG. 2.
Interaction between NF2 and NGB. (A and B) NGB binds to NF2/merlin in HEK293 cells cotransfected with HA-NF2 and Flag-NGB. After 36 h of transfection, cells were lysed, immunoprecipitated with anti-Flag (α-Flag), and detected with anti-HA (α-HA) antibody (A) or vice versa (B). Bottom panels show expression of transfected plasmids. (C and D) Specificity of anti-NGB antibody. HeLa cells were transiently transfected with Flag-NGB, lysed, and then subjected to immunoblotting (C) or IP-Western blotting analysis (D) with indicated antibodies. (E and F) NGB interacts with NF2 at physiological protein levels. 82HTB rhabdomyosarcoma cells were lysed and immunoprecipitated with anti-NF2 (α-NF2) antibody or the antibody was preincubated with GST-NF2 antigen or preimmune serum. The immunoprecipitates were detected with anti-NGB (α-NGB) antibody (E). Conversely, the NGB immunoprecipitates were blotted with anti-NF2 antibody (F). (G) GST-NGB pull down of merlin. A GST pull-down assay was performed by incubation of GST and GST-NGB with HeLa cell lysate. After being washed four times, pull-down products were immunoblotted with anti-NF2 antibody (top). The bottom panel is an SDS-PAGE gel stained with Coomassie blue (CBS). (H) NGB colocalizes with NF2 in the perinuclear region. 82HTB rhabdomyosarcoma cells were cultured on glass coverslips, probed with anti-NGB and -NF2 antibodies, incubated with fluorescein isothiocyanate- and rhodamine-conjugated secondary antibodies, and analyzed by confocal microscopy. (I and J) Yeast two-hybrid mapping of interaction domains between NF2 and NGB. EGY191 yeast was cotransformed with plasmids encoding a fusion protein between the GAL4 activation domain and full-length or NF2 deletion constructs and plasmids encoding a fusion protein between LexA-DNA binding domain and full-length or deletion NGB constructs. All yeast clones grew on SD-Ura/Trp/His medium containing Gal. The positive interaction was indicated by blue. (K) Identification of the binding site(s) of NGB. HEK293 cells were transfected with the indicated plasmids and lysed, and then the protein was immunoprecipitated with anti-GFP (α-GFP) antibody and detected with anti-HA (α-HA) antibody (top). The middle and bottom panels show expression of transfected plasmids.
The subcellular localization of NGB and merlin was examined by immunofluorescence and confocal microscopy using anti-NGB polyclonal and anti-merlin monoclonal antibodies. NGB localized to the nucleus and cytoplasm (Fig. 2H). NGB and merlin predominantly colocalized in the perinuclear cytoplasm (Fig. 2H). These data support the interaction between merlin and NGB observed in yeast two-hybrid and co-IP experiments.
G-protein homology domain of NGB and amino and carboxyl termini of merlin are required for their interaction.
To identify the domains within merlin and NGB that are required for their binding, we prepared a series of deletion constructs. Yeast two-hybrid tests of interaction revealed that the amino (aa 1 to 52) and carboxyl (aa 288 to 344) termini of merlin are required for binding to NGB (Fig. 2I). Neither the amino terminus nor the carboxyl region alone binds to NGB, suggesting that interdomain association between the amino and carboxyl termini of merlin, which is essential for its tumor suppressor activity (17, 52, 61), is necessary for interaction with NGB (Fig. 2I). The binding region of NGB was mapped to the G-protein homology domain (Fig. 2J). To define the binding site(s) of NGB that interacts with merlin, we created a series of mutant constructs within the G-protein homology domain of NGB. Immunoprecipitation experiments showed that mutation of lysine-395/arginine-394 to alanine (NGB-K395/R394A) significantly reduced the ability of NGB to bind to merlin (Fig. 2K), indicating that the lysine-395 and arginine-394 are essential for their interaction, which could be important for NGB function.
Expression of NGB inhibits cell proliferation and tumorigenicity.
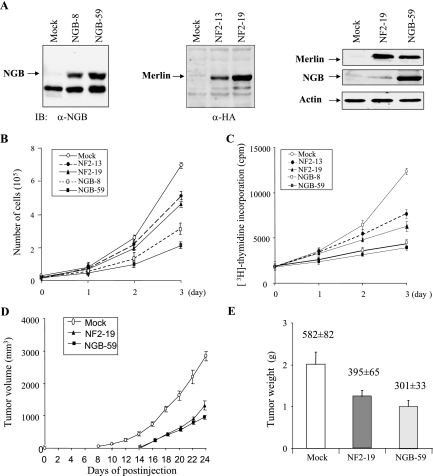
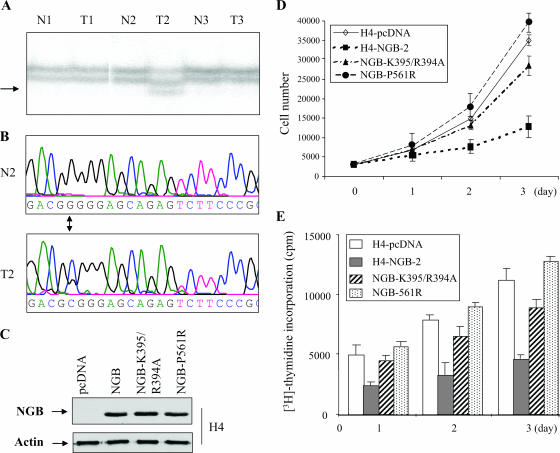
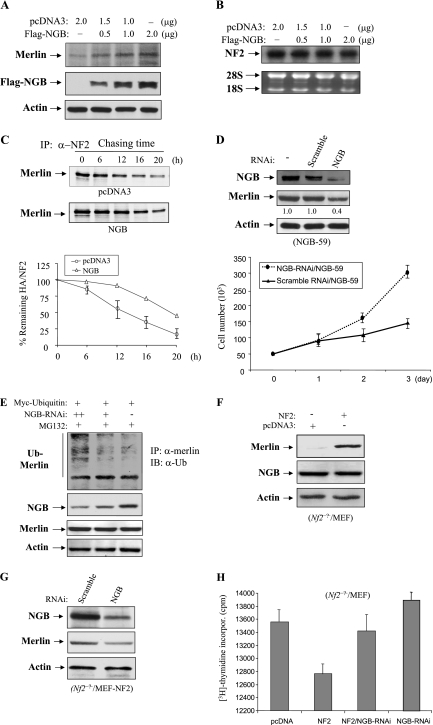
We next examined the phenotypic changes of cells expressing ectopic NGB. Since JS1 rat schwannoma cells have been commonly used for NF2 function studies (18, 52) and express low levels of NGB (Fig. 3A), we stably transfected the NGB into JS1 cells. As controls, NF2 and pcDNA vector were also introduced into JS1 cells. Eight clonal cell lines from each transfectant were established after G418 selection. Expression of NGB and NF2 was confirmed by Western blot analyses. Interestingly, we observed that expression of merlin was elevated in NGB-transfected JS1 cells (Fig. 3A), implying the possible regulation of merlin by NGB. Cell growth, DNA synthesis, and tumorigenicity were evaluated with these clonal cell lines. As shown in Fig. 3B, cell growth was significantly inhibited by ectopic expression of NGB. Furthermore, [3H]thymidine incorporation experiments revealed that NGB represses DNA synthesis (Fig. 3C). Notably, NGB exhibited more inhibitory effects than NF2 on cell proliferation and DNA synthesis (Fig. 3B and C).
FIG. 3.
NGB inhibits JS1 cell proliferation and tumor growth in xenograft mouse model. (A) Immunoblotting (IB) analysis of NGB and NF2 stably transfected JS1 schwannoma clonal cell lines with anti-NGB (α-NGB, left panel), anti-HA (α-HA, middle panel) and indicated antibodies (right panel). (B) NGB inhibits cell growth. Stable NGB- and NF2-transfected cells as well as pcDNA3 vector-transfected cells were seeded in a 48-well plate at 0.2 × 105/well. The cell number was counted daily for 3 days. The experiment was performed three separate times. (C) NGB inhibits DNA synthesis. The stably transfected cells were labeled with [3H]thymidine (3 plates/clone cell/time point) and assayed as described in Materials and Methods. Tumor growth (D) and tumor weight (E) were reduced by NGB. The stably transfected clonal cells were subcutaneously injected into nude mice (3 × 106 cells/mouse). Tumor volume was measured every 2 days, and tumor weight was recorded at time of euthanasia. Data are representative of two independent experiments carried out with 24 mice each (8 mice/clonal cell line). The number above each column represents the proliferative index revealed by Ki-67 immunostaining. Mean values ± standard errors of the proliferative index are given as the counts of Ki-67-positive tumor cell nuclei in 10 consecutive high-power (magnification, ×400) fields.
To determine whether NGB influences schwannoma cell growth in vivo, NGB-, pcDNA-, and NF2-transfected JS1 cells were subcutaneously injected into nude mice (8 mice for each clonal cell line), and their tumor volumes were measured every 2 days. The tumors appeared between 8 to 10 days in all injected mice. Tumors from cells expressing ectopic NGB grew much slower than tumors expressing vector alone (Fig. 3D). Moreover, the tumor weight of NGB transfectants was 50% less than that of vector transfectants (Fig. 3E). In addition, tumor growth and weight as well as the proliferative index of NGB-expressing cells were slightly lower than those of the cells expressing NF2 (Fig. 3D and E). Taken collectively, NGB has tumor suppressor activity, and its ability to inhibit cell and tumor growth is even greater than that of NF2.
NGB is down-regulated in human glioma cell lines, and reconstitution of NGB induces cell growth arrest but not cell death.
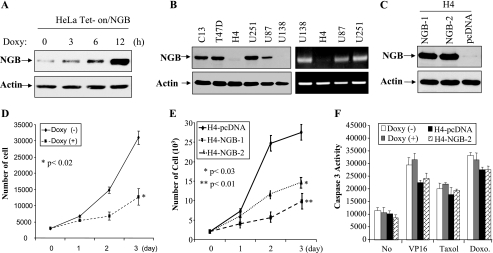
To further demonstrate the tumor suppressor activity of NGB, we established doxycycline-inducible NGB in HeLa cells (Fig. 4A) and examined the NGB protein levels in a dozen human cancer cell lines. NGB was greatly down-regulated in 2 glioma cell lines, one (H4) of which was low at both mRNA and protein levels and the other (U138) only exhibited a low level of NGB protein (Fig. 4B), suggesting that different mechanisms could be involved in the downregulation of NGB in these 2 cell lines. In fact, treatment with 5-azacytidine, a demethylation agent, increased NGB protein expression in H4 but not U138 cells (data not shown).
FIG. 4.
NGB is down-regulated in glioma cells, and reconstitution of NGB induces cell growth arrest but not cell death. (A-C) Immunoblotting analysis of doxycycline (Doxy)-inducible NGB HeLa cells (A), tumor cell lines (B), and stably transfected NGB H4 clonal cell lines (C) with indicated antibodies. Right panels of panel B show mRNA levels of NGB in glioma cell lines examined by reverse transcription-PCR. (D and E) NGB inhibits cell growth. HeLa Tet-on/NGB and H4-NGB cells were seeded in a 48-well plate in triplicate and treated with (+) or without (−) doxycycline (D) as indicated on the figure. The cell number was counted daily for 3 days. The experiment was performed three separate times. (F) NGB does not sensitize cells to chemotherapeutic drug-induced apoptosis. The cells were seeded and treated as described for panels D and E and then administered VP16 (5 μM), taxol (100 nM), or doxorubicin (Doxo., 2 μM). After 12 h of incubation, cells were assayed for caspase-3 activity using the EnzChek Caspase-3 assay kit.
Stably transfected clonal cell lines were established by introducing NGB into H4 cells (Fig. 4C). The cells transfected with pcDNA vector alone were used as a control. Cell proliferation and survival were examined with cell counting, caspase-3 activity analysis, and trypan blue staining. Figures 4D and E show that ectopic expression of NGB represses cell proliferation. However, NGB exhibited no effect on cell survival in response to treatment with VP16, taxol, or doxorubicin (Fig. 4F and data not shown). These findings provided further support that NGB is a tumor suppressor gene and exerts its tumor suppressor function largely through inhibition of cell proliferation.
Expression of NGB suppresses cell migration, attachment, and aggregation.
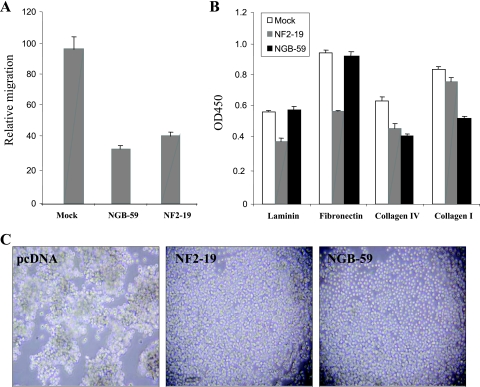
Cell adhesion is crucial for maintaining the structural integrity of tissues. Cell matrix adhesion (cell attachment) is mediated by heterophilic interactions between cell surface receptors and their matrix ligands, whereas cell-cell adhesion (cell aggregation) primarily involves direct homophilic interactions between cell surface molecules (16, 32, 55). As merlin decreases cell migration, adhesion, and attachment (18, 27), we next assessed the effect of NGB on cell migration using a MICS. As shown in Fig. 5A, JS1 cells stably expressing NGB migrate much slower than pcDNA-transfected cells. To determine the effects of NGB on cell attachment and aggregation, pcDNA- and NGB-transfected JS1 cells were trypsinized and equal numbers of cells were seeded onto laminin-, fibronectin-, collagen IV-, and collagen I-coated 96-well plates (cell attachment) or 24-well low-binding-affinity tissue culture plates (cell aggregation) by following the procedures described in Materials and Methods. We observed that cells overexpressing NGB had decreased ability to attach to collagen but not fibronectin or laminin (Fig. 5B). However, both NGB and NF2 significantly inhibited cell aggregation (Fig. 5C). Collectively, these data suggest that NGB may play a role in inhibiting metastasis by regulation of cell migration, attachment and aggregation.
FIG. 5.
Ectopic expression of NGB reduces cell mobility, attachment, and aggregation. (A) Reduced cell mobility was observed in cells overexpressing NGB. An MICS assay was used to directly measure cell migration. The migration is normalized to values obtained with pcDNA3 vector controls and is expressed in μM/h. Standard deviations for each cell line are shown. (B) NGB inhibits cell adhesion. NGB-59 and pcDNA3-rat schwannoma cells were incubated in the presence or absence of 100 μM zinc chloride overnight and then harvested by trypsinization. Equal numbers of cells were added to laminin-, fibronectin-, collagen IV-, and collagen I-coated 96-well plates. After 1 h, the number of adherent cells was determined using crystal violet staining and a spectrophotometric assay. NF2-transfected cells (NF2-19) were used as controls. The experiment was repeated three times. OD450, optical density at 450 nm. (C) JS1 cell aggregation was inhibited by NGB. Single-cell suspensions (500 μl; 5 × 104 cells/ml in DMEM containing 0.5% BSA) were plated into each well of a 24-well low-binding plate. The plate was incubated at 4°C or 37°C on a rotating platform for 30 min. Photographs were taken using a Nikon N6006 camera (magnification, ×100).
Infrequent NGB mutations are detected in human glioblastoma.
As NGB exhibited tumor suppressor activity and is located at human chromosome band 10p15, a region frequently deleted in human glioma (22, 59), we screened 17 paired glioma tumor/normal DNAs for mutations of NGB. Single-strand conformation polymorphism and sequence analyses of all 17 exons of the NGB gene revealed point mutations in exon 1 (C65T) or 16 (C1755G) in two different tumor specimens but not in matched normal DNAs (Fig. 6A and B). The former mutation is located in a noncoding region, and the latter results in an amino acid change from proline to arginine (P561R). No NGB mutation was detected in 10 glioma cell lines examined.
FIG. 6.
Mutation of NGB in a human glioma and loss of tumor suppressor activity of NGB-P561R and NGB-K395/R394A. (A) Single-strand conformation polymorphism analysis of exon 16 of the NGB gene. An abnormal band shift (arrow) was detected in tumor DNA of patient 2 (T2) but not matched DNA (N2). (B) Sequence analyses revealed that a mutation from guanine to cytosine occurs in tumor DNA (arrow between middle and bottom panels), resulting in a change of amino acid from proline to serine. (C) Immunoblotting analysis of H4 cells, which were transfected with wild-type NGB, a naturally occurring mutant NGB-P561R, or the binding site mutant NGB-K395/R394A, with indicated antibodies. (D and E) NGB-P561R and NGB-K395/R394A failed to inhibit cell growth (D) and DNA synthesis (E). H4-NGB-2 is a wild-type NGB stably transfected clonal cell line (see Fig. 4C).
NGB-P561R and NGB-K395/R394A fail to inhibit cell proliferation.
Having found a mutation in NGB in a glioma (NGB-P561R) and that the NGB-K395A/R394A mutant is compromised in its interaction with merlin (Fig. 2K), we sought to determine if these mutations alter NGB tumor suppressor activity. H4 glioma cells, in which NGB is down-regulated (Fig. 4B), were stably transfected with pcDNA-NGB-P561R and NGB-K395/R394A as well as pcDNA vector alone (Fig. 6C). Compared to wild-type NGB stably transfected H4 (H4-NGB-2) (Fig. 4C), NGB-P561R or NGB-K395/R394A had significantly decreased ability to inhibit cell growth and DNA synthesis (Fig. 6D and E), suggesting that neither the mutated NGB in glioma nor NGB-K395/R394A retains tumor suppressor activity. These results also indicate that NGB binding to merlin is essential for its tumor suppressor function.
NGB possesses GTP-binding and GTPase activities.
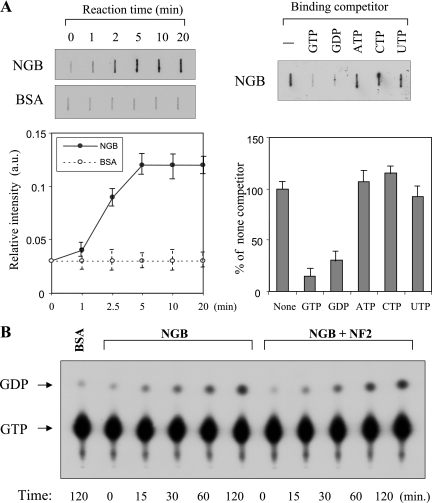
To determine whether the NGB cDNA coded for a protein possessing GTP-binding and GTPase activities, Flag-tagged NGB was expressed, affinity purified, with anti-Flag antibody, and eluted from protein A-G beads with excess Flag peptides. The released Flag-NGB was then subjected to GTP binding assays. Following incubation with [35S]GTPγS, the bound GTP was separated by rapid filtration on nitrocellulose. As seen in Fig. 7A, NGB binds to GTPγS within 2 min. The ability of various ribonucleotides to compete for [35S]GTPγS binding to NGB was also examined. The NGB-[35S]GTPγS binding activity was completely blocked by an excess (20-fold) of unlabeled GTP or GDP but not ATP, UTP, or CTP, indicating that NGB is a specific guanine nucleotide-binding protein (Fig. 7A). GTPase activity of NGB was determined by incubating Flag-NGB with [α-32P]GTP, and the products were analyzed by thin-layer chromatography. Repeated experiments revealed that NGB was capable of hydrolyzing [α-32P]GTP in the presence of an excess of unlabeled UTP but not GTP (Fig. 7B and data not shown), suggesting that NGB has intrinsic GTPase activity.
FIG. 7.
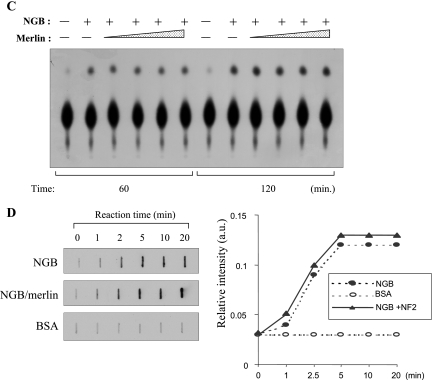
NGB possesses GTP-binding and GTPase activities that are not regulated by merlin. (A) NGB specifically binds to GTP. NGB (1 μg) or BSA (1 μg) was incubated with [35S]GTPγS (1 μM) at 22°C for the indicated times (left panels) or for 1 h in the absence or presence of the indicated nucleotides at concentration of 20 μM (right panels). The amount of [35S]GTPγS binding was determined in a filter binding assay as described in Materials and Methods. Each value is the average of the results from duplicate incubations and is representative of three separate experiments. (B and C) [α-32P]GTP hydrolysis by NGB and merlin does not affect NGB GTPase activity. BSA (control) and NGB were incubated with [α-32P]GTP (10 μM; 2 Ci/mmol). Aliquots were removed from the reaction mixture at the indicated times, analyzed by poly(ethyleneimine) thin-layer chromatography plates, and exposed to film for 5 min. (D) Merlin does not regulate the GTP-binding activity of NGB. A GTP-binding assay was performed as described above except that incubation of NGB was in the presence or absence of merlin.
Merlin does not influence NGB GTPase and GTP-binding activity.
GTP-binding proteins have two interconvertible forms, GTP-bound active and GDP-bound inactive forms. The GTP-bound form is converted to the GDP-bound form by the GTPase reaction, which is regulated by GTPase-activating proteins (GAPs), and the GDP-bound form is converted to the GTP-bound form by the GDP/GTP exchange reaction, which is regulated by GDP/GTP exchange proteins, i.e., GDS/GEF (GDP dissociation stimulator/guanine nucleotide exchange factor) and GDI (GDP dissociation inhibitor). GDS/GEF stimulates the dissociation of GDP and the subsequent binding of GTP to its substrate G proteins, whereas GDI inhibits both reactions. The protein neurofibromin, encoded by the neurofibromatosis type I (NF1) gene, contains the domains conserved in GAP proteins and functions as a RasGAP. While merlin does not have GAP domain, it could act as a GDS/GEF or GDI to regulate NGB. To test this hypothesis, HA-merlin was immunopurified from HA-NF2-transfected COS7 cells. NGB hydrolysis reaction mixtures were incubated with the purified merlin. However, recombinant merlin did not inhibit the GTPase activity of NGB (Fig. 7B and C). In addition, the GTP-binding activity of NGB is not affected by merlin (Fig. 7D). These results suggest that NGB acts upstream rather than downstream of merlin.
NGB impairs the ubiquitination and turnover of merlin.
To address whether NGB might affect the regulation of merlin, HEK293 cells were transfected with increasing amounts of Flag-NGB. Western blot analysis shows that endogenous merlin was upregulated by NGB in a dose-dependent manner (Fig. 8A). However, mRNA levels of the NF2 did not differ between NGB-transfected and untransfected HEK293 cells (Fig. 8B), suggesting that NGB stabilizes merlin at the protein level. To demonstrate NGB inhibition of merlin degradation, pulse-chase assays were carried out in NIH 3T3 cells. After 36 h of transfection with NGB and 60 min of [35S]methionine labeling, NF2 was immunoprecipitated, separated by SDS-PAGE, and then exposed and quantitated with a Phosphoimager. Triplicate experiments revealed that the half-life of merlin was extended from about 12 h in pcDNA-transfected cells to nearly 20 h in NGB-transfected cells (Fig. 8C). Furthermore, RNA interference (RNAi) knockdown of NGB in JS1/NGB-59 cells was found to decrease the expression of merlin and resulted in cell regrowth (Fig. 8D). To explore how NGB regulates turnover of merlin, HeLa cells, which express both NGB and merlin, were transfected with RNAi against NGB and Myc-ubiquitin and then treated with proteasome inhibitor MG132. As shown in Fig. 8E, the ubiquitination of merlin was increased by knockdown of NGB. However, no band shift (phosphorylation) (23) of merlin was observed in the cells expressing ectopic NGB (Fig. 8A) or in cells where we knocked down endogenous NGB (Fig. 8E). Therefore, we conclude that NGB regulates merlin ubiquitination rather than phosphorylation, leading to inhibition of merlin turnover.
FIG. 8.
NGB up-regulates merlin through inhibition of merlin ubiquitination. (A) The protein level of merlin is increased by ectopic expression of NGB. HEK 293 cells were transfected with increasing amounts of Flag-NGB. After 48 h of incubation, cells were lysed and immunoblotted with indicated antibodies. (B) NGB does not regulate merlin at the transcriptional level. Total RNA was isolated from NGB-transfected HEK293 cells and subjected to Northern blot analysis with radiolabeled NF2 cDNA as a probe (top). The bottom panel depicts the ethidium bromide-stained gel showing equal loading of total RNA. (C) NGB inhibits degradation of merlin. Pulse-chase assays were carried out in NIH 3T3 cells transfected with NGB or pcDNA3 vector alone. After 60 min of [35S]methionine labeling, merlin was immunoprecipitated, separated by SDS-PAGE, exposed (top and medium) and quantified with a PhosphorImager (bottom). (D) Knockdown of NGB decreases merlin expression and results in cell growth. NGB-59 JS1 cells were transfected with NGB-RNAi or control RNAi. After 48 h of incubation, cells were lysed and immunoblotted with indicated antibodies (upper). The bottom panel shows the cell growth curve. (E) Knockdown of NGB increases merlin ubiquitination and MG132 inhibits merlin degradation resulting from NGB knockdown. HeLa cells were transfected with Myc-ubiquitin and NGB-RNAi and then treated with MG132 (10 μM) for 6 h. Cells were lysed, immunoprecipitated with anti-merlin antibody, and detected with anti-ubiquitin antibody (top). Panels 2 to 4 show expression of NGB, merlin, and actin. (F and G) Reconstitution of NF2 in Nf2-deficient MEFs does not affect NGB expression, and knockdown of NGB decreases the expression of reintroduced merlin. Results of Western blot analysis of Nf2−/− MEFs, which were transfected with NF2 (F) or NF2/NGB-RNAi (G), with indicated antibodies are shown. (H) Knockdown of NGB largely abrogated merlin-inhibited DNA synthesis. Nf2-null MEFs were transfected with indicated plasmids and/or NGB-RNAi. After 36 h culture, [3H]thymidine incorporation was assayed as described in Materials and Methods.
To further define the link between NGB and merlin and examine if merlin regulates NGB expression, we stably expressed NF2 in Nf2-null MEFs. As shown in Fig. 8F, we found that NF2 did not alter the expression of NGB. In contrast, knockdown of NGB decreased the expression of ectopically transfected merlin in Nf2−/− MEFs (Fig. 8G), providing further support that NGB up-regulates merlin at a posttranslational level. In addition, DNA synthesis in Nf2-deficient MEFs was inhibited by reexpression of merlin, which was largely abrogated by knockdown of NGB (Fig. 8H). However, cell survival was not affected in the MEFs treated with NGB-RNAi (data not shown). Collectively, these findings suggest that merlin acts downstream of NGB to suppress cell proliferation.
NGB down-regulates cyclin D1 and exerts tumor suppressor function through merlin.
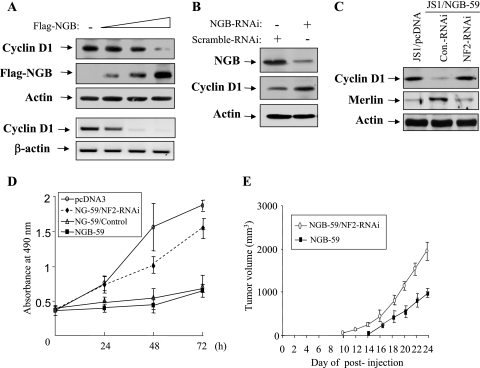
Our previous study has shown that merlin inhibits cell growth largely through reduction of cyclin D1 expression (62). Since NGB stabilizes merlin, we reasoned that NGB could inhibit cyclin D1 and tumor cell growth through merlin. To test this hypothesis, JS1 cells were transiently transfected with increasing amounts of NGB and HeLa cells were treated with NGB-RNAi. Figures 9A and B show that cyclin D1 protein and/or mRNA levels were reduced by ectopic expression of NGB and increased by knockdown of NGB. Furthermore, stable blockage of merlin in JS1/NGB-59 cells by infection with lentivirus expressing short hairpin RNA (shRNA) against NF2 abrogated NGB-reduced cyclin D1 expression (Fig. 9C). Accordingly, NGB-inhibited cell proliferation and tumor growth in nude mice were also considerably reduced by stable knockdown of NF2 in JS1/NGB-59 cells (Fig. 9D and E). Thus, we concluded that NGB exerts its cellular function, at least to some extent, via the stabilization of merlin, which leads to down-regulation of cyclin D1 (62).
FIG. 9.
NGB reduces cyclin D1 expression and inhibits cell growth via merlin. (A) Protein and mRNA levels of cyclin D1 are inhibited by NGB. JS1 cells were transfected with increasing amounts of Flag-NGB and immunoblotted with indicated antibodies (panels 1 to 3). Total RNA was isolated from the cells and subjected to semiquantitative reverse transcription-PCR using primers specific for cyclin D1 and β-actin (panels 4 and 5). (B) Knockdown of NGB increases cyclin D1 level. HeLa cells were transfected with NGB-RNAi using Lipofectamine. Following 72 h of incubation, cells were lysed and immunoblotted with anti-NGB (top), -cyclin D1 (middle), and -actin (bottom) antibodies. (C) NGB-reduced cyclin D1 is abrogated by knockdown of merlin. JS1/NGB-59 cells were infected with lentivirus pLKO.1-shRNA/Nf2 and pLKO.1-puro vector and immunoblotted with indicated antibodies. (D and E) Merlin mediates NGB-inhibited cell growth and tumorigenicity. Indicated cells were seeded in 48-well plates at 0.2 × 105/well. The cell number was counted daily for 3 days. The experiment was performed in triplicate (D). The cells were subcutaneously injected into nude mice (3 × 106 cells/mouse). Tumor volume was measured every 2 days. Data shown are representative of results from two independent experiments carried out with 16 mice each (8 mice/cell line) (E).
Ectopic expression of cyclin D1 largely overrides the tumor suppressor functions of NGB and merlin.
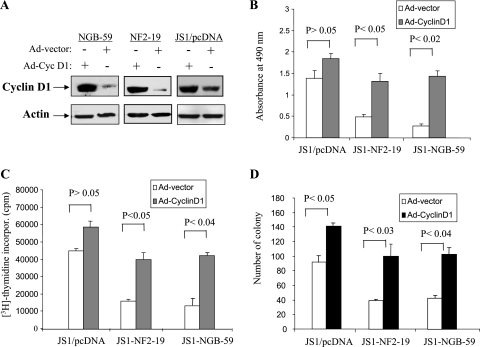
To further explore the role for cyclin D1 as a downstream target of NGB/merlin-mediated tumor suppression, JS1/NGB-59, JS1/NF2-19, and JS1/pcDNA cells were infected with adenovirus expressing cyclin D1 or adenoviral vector (Fig. 10A). Ectopic expression of cyclin D1 in JS1/NGB-59 and JS1/NF2-19 cells considerably abrogated NGB- and merlin-inhibited cell growth and DNA synthesis compared to adeno-cyclin D1-infected JS1/pcDNA cells (Fig. 10B and C). As expected, expression of NGB and NF2 in JS1 cells inhibited colony formation in soft agar. However, adeno-cyclin D1 infection largely overrode the inhibitory effects of NGB and merlin, although overexpression of cyclin D1 in JS1/pcDNA cells also increased colony formation (Fig. 10D). Collectively, these data indicate that cyclin D1 is a major downstream target of NGB/merlin and mediates NGB/merlin tumor suppressor function.
FIG. 10.
Overexpression of cyclin D1 largely overrides the tumor suppressor functions of NGB and merlin. (A) Immunoblotting analysis of JS1/NGB-59, JS1/NF2-19, and JS1/pcDNA cells, which were infected with adeno-cyclin D1 or adeno-vector viruses, with anti-cyclin D1 (upper) and -β-actin (bottom) antibodies. (B and C) Ectopic expression of cyclin D1 significantly reduced the inhibitory effects of NGB and merlin on cell proliferation and DNA synthesis. Cells were seeded in 24-well plates (0.8 × 105/well) in triplicate. Following 24 h of incubation, cell proliferation (B) and DNA synthesis (C) were examined by MTS and [3H]thymidine incorporation assays. (D) Cyclin D1 overrode NGB- and merlin-inhibited cell anchorage-independent growth. Colony formation assays were performed in 100-mm soft agar plates. The number of colonies was counted after 21 days.
DISCUSSION
Using the amino-terminal region of merlin as bait in a yeast two-hybrid screen, we have identified a novel GTP binding protein, NGB. NGB colocalizes and immunoprecipitates with merlin. The interaction occurs between the G-protein homology domain of NGB and the amino and carboxyl termini of merlin. The binding sites of NGB to merlin were mapped to lysine-395/ariginine-394. NGB is well conserved between yeast, C. elegans, and human cells. Down-regulation and infrequent mutation of NGB were observed in human glioma cell lines and primary tumors. Expression of NGB inhibited tumor cell growth in vitro and in vivo without significant effect on cell survival. The mutated NGB in glioma and the mutation of the binding sites of NGB lost the tumor suppressor activity. NGB possesses GTPase and GTP-binding activity and inhibits cyclin D1 expression and tumorigenicity through the stabilization of merlin.
The high homology between merlin and the ERM proteins suggests that these proteins function analogously. The ERM proteins localize to cortical actin structures, particularly in specialized or dynamic regions, such as microvilli, membrane ruffles, or the cleavage furrow, and bind directly to actin through a highly conserved domain at their carboxyl termini (17). Although merlin lacks the carboxyl-terminal actin-binding domain, it does localize to cortical actin structures and is particularly enriched in membrane ruffles. The fact that isoform 2 of merlin interacts with an actin-binding protein βII-spectrin suggests that signaling from merlin to the actin cytoskeleton is mediated via actin-binding sites on βII-spectrin (48). Unlike βII-spectrin, however, NGB does not contain an actin-binding domain and primarily colocalizes with merlin in the perinuclear cytoplasm, suggesting that NGB is not a cytoskeleton protein and could mediate merlin function through a different mechanism.
A number of studies have demonstrated that ERM proteins occupy a crucial position as protein linkers that both respond to and participate in reorganization of membrane-cytoskeleton interactions. The ERM proteins have also been implicated in linked regulation of the activities of particular membrane proteins (29). As expected from its structural motifs, merlin behaves in a manner similar to the ERM proteins but with some notable differences (6, 27, 32). Like the adenomatous polyposis coli tumor suppressor protein, merlin may be involved with multiple partners and signaling pathways, some of which are shared with the ERM proteins. Defining merlin's tumor suppressor function will likely require identifying those differences. However, the fact that inactivation of merlin in the mouse by targeted mutagenesis produces a variety of malignant tumors with a high rate of metastasis (12, 33) suggests that merlin's suppression of tumor formation may involve different partners and pathways in different cell types and genetic backgrounds. To determine whether NGB interacts with ERM proteins, co-IP experiments were performed with anti-ezrin, -radixin, and -moesin antibodies. The results showed that NGB does not bind to ERM proteins (data not shown), suggesting that NGB specifically interacts with merlin and may be an important component in the NF2 tumor suppressor pathway. Previous studies revealed that, like the ERMs, merlin localizes mainly to the interface between the plasma membrane and the actin cytoskeleton, with enrichment in ruffled edges (18, 32, 61). However, merlin, but not the ERMs, is also found in the perinuclear region, sometimes in uncharacterized granules (18, 25, 32). In this report, we show that NGB and merlin colocalize predominantly in the perinuclear cytoplasm (Fig. 2), providing further support that NGB associates with merlin but not ERM proteins.
The NF2 gene consists of 17 exons. Two major alternatively spliced NF2 variants are expressed in normal tissues. Isoform I lacks exon 16 and encodes a 595-amino-acid protein, whereas the isoform II product is five amino acids shorter than that of isoform I due to the insertion of 45-bp exon 16 introducing a premature stop codon and preventing translation of exon 17. It has been demonstrated that intramolecular binding between the amino terminus and carboxyl terminus of isoform I, but not isoform II, of merlin is essential for inhibition of cell growth (17, 52). Isoform II of merlin interacts with the actin-binding protein βII-spectrin. Unlike βII-spectrin, which interacts weakly with isoform I, NGB strongly binds to both isoform I and isoform II of merlin. Both the amino (aa 1 to 52) and carboxyl (aa 288 to 344) termini of merlin are required for its binding to NGB (Fig. 2). Neither the amino terminus nor the carboxyl region alone binds to NGB, suggesting that intermolecular domain association between the amino and carboxyl termini of merlin is necessary for association with NGB.
GTP-binding proteins constitute a large superfamily of regulatory molecules. This superfamily comprises at least three subfamilies (5, 15), including the small GTP-binding proteins (e.g., Ras), the α subunits of large heterotrimeric G-proteins (e.g., transducin), and the GTPases involved in protein synthesis (e.g., elongation and initiation factors). Based on the size of NGB, it resembles the α subunits of the heterotrimeric G-proteins that are known to act as molecular switches (15). Although the conserved GTP-binding sequence of NGB shows the most homology to small GTP-binding proteins, especially R-Ras, overall, NGB does not display significant similarity with the well characterized G-proteins. However, it is highly homologous to an uncharacterized protein found in C. elegans and yeast cells, suggesting the existence of a new subfamily within the GTP-binding protein superfamily. Interestingly, the GTP-binding domain of NGB is required for its interaction with merlin, implying a functional interaction between NGB and merlin. While merlin does not affect GTP-binding and GTPase activity of NGB, ectopic expression of NGB inhibits the degradation of merlin protein, and knockdown of NGB decreases merlin expression. Moreover, blockage of merlin expression largely abrogates NGB tumor suppressor function. In addition, our data showed that the level of merlin induced by expression of NGB (NGB-59) is slightly lower than that of NF2-transfected JS1 (NF-19) cells (Fig. 3A), but NGB exhibits somewhat greater tumor suppressor activity than does merlin (Fig. 3B to E). Therefore, although merlin is a major target of NGB and is required for NGB function, NGB must also regulate other molecules that modulate tumor cell growth.
Finally, loss of chromosome band 10p15, encompassing the NGB locus, has frequently been observed in human glioma and prostate cancer cells. Infrequent mutations of the tumor suppressor gene KLF6 in 10p15 have been detected in these tumor types (7, 36, 38-40). In this report, we show that NGB, which resides about 2 Mb telomeric to KLF6, is mutated in 2 of 17 primary gliomas, 1 of which is a missense mutation. Down-regulation of NGB was also detected in glioma cell lines. Moreover, ectopic expression of NGB inhibits tumor cell growth in vitro and in a xenograft model. These findings suggest that NGB is a candidate tumor suppressor gene, although larger studies of these and other tumor types are required. Further studies are needed to unravel the association of NGB function with its GTPase activity as well as cross talk between NGB and signaling molecules that regulate merlin.
Acknowledgments
We thank Martina Schiebe for the paired glioma/normal DNAs. We also thank Lin Yang for his technical assistance.
This work was supported by grants from the National Institutes of Health (CA77935, CA89242, and CA107078 to J.Q.C.; CA45745, CA114047, and CA06927 to J.R.T.; and CA63366 to E.A.G.), Department of Defense (W81XWH-05-1-0021 and DAMD17-02-1-0671 [J.Q.C.]), and American Cancer Society (institutional grant 8102-13 to J.Q.C.).
Footnotes
Published ahead of print on 8 January 2007.
REFERENCES
- 1.Adams, J. C. 1997. Cell adhesion-spreading frontiers, intricate insights. Trends Cell Biol. 7:107-110. [DOI] [PubMed] [Google Scholar]
- 2.Arpin, M., M. Algrain, and D. Louvard. 1994. Membrane-actin microfilament connections: an increasing diversity of players related to band 4.1. Curr. Opin. Cell Biol. 6:136-141. [DOI] [PubMed] [Google Scholar]
- 3.Baser, M. E. R., D. G. Evans, and D. H. Gutmann. 2003. Neurofibromatosis 2. Curr. Opin. Neurol. 16:27-33. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi, A. B., S. I. Mitsunaga, J. Q. Cheng, W. M. Klein, S. C. Jhanwar, B. Seizinger, N. Kley, A. J. Klein-Szanto, and J. R. Testa. 1995. High frequency of inactivating mutations in the neurofibromatosis type 2 gene (NF2) in primary malignant mesotheliomas. Proc. Natl. Acad. Sci. USA 92:10854-10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117-127. [DOI] [PubMed] [Google Scholar]
- 6.Brault, E., A. Gautreau, M. Lamarine, I. Callebaut, G. Thomas, and L. Goutebroze. 2001. Normal membrane localization and actin association of the NF2 tumor suppressor protein are dependent on folding of its N-terminal domain. J. Cell Sci. 114:1901-1912. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C., E. R. Hyytinen, X. Sun, H. J. Helin, P. A. Koivisto, H. F. Frierson, R. L. Vessella, and J. T. Dong. 2003. Deletion, mutation, and loss of expression of KLF6 in human prostate cancer. Am. J. Pathol. 162:1349-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, J. Q., D. A. Altomare, M. A. Klein, W. C. Lee, G. D. Kruh, N. A. Lissy, and J. R. Testa. 1997. Transforming activity and mitosis-related expression of the AKT2 oncogene: evidence suggesting a link between cell cycle regulation and oncogenesis. Oncogene 14:2793-2801. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, J. Q., W. C. Lee, M. A. Klein, G. Z. Cheng, S. C. Jhanwar, and J. R. Testa. 1999. Frequent mutations of NF2 and allelic loss from chromosome band 22q12 in malignant mesothelioma: evidence for a two-hit mechanism of NF2 inactivation. Genes Chromosomes Cancer 24:238-242. [PubMed] [Google Scholar]
- 10.Finlin, B. S., and D. A. Andres. 1997. Rem is a new member of the Rad- and Gem/Kir Ras-related GTP-binding protein family repressed by lipopolysaccharide stimulation. J. Biol. Chem. 272:21982-21988. [DOI] [PubMed] [Google Scholar]
- 11.Finlin, B. S., H. Shao, K. Kadono-Okuda, N. Guo, and D. A. Andres. 2000. Rem2, a new member of the Rem/Rad/Gem/Kir family of Ras-related GTPases. Biochem. J. 347:223-231. [PMC free article] [PubMed] [Google Scholar]
- 12.Giovannini, M., E. Robanus-Maandag, M. Niwa-Kawakita, M. van der Valk, J. M. Woodruff, L. Goutebroze, P. Merel, A. Berns, and G. Thomas. 1999. Schwann cell hyperplasia and tumors in transgenic mice expressing a naturally occurring mutant NF2 protein. Genes Dev. 13:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Agosti, C., T. Wiederhold, M. E. Herndon, J. Gusella, and V. Ramesh. 1999. Interdomain interaction of merlin isoforms and its influence on intermolecular binding to NHE-RF. J. Biol. Chem. 274:34438-34442. [DOI] [PubMed] [Google Scholar]
- 14.Goutebroze, L., E. Brault, C. Muchardt, J. Camonis, and G. Thomas. 2000. Cloning and characterization of SCHIP-1, a novel protein interacting specifically with spliced isoforms and naturally occurring mutant NF2 proteins. Mol. Cell. Biol. 20:1699-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govek, E. E., S. E. Newey, and L. Van Aelst. 2005. The role of the Rho GTPases in neuronal development. Genes Dev. 19:1-49. [DOI] [PubMed] [Google Scholar]
- 16.Gumbiner, B. M. 1996. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84:345-357. [DOI] [PubMed] [Google Scholar]
- 17.Gusella, J. F., V. Ramesh, M. MacCollin, and L. B. Jacoby. 1999. Merlin: the neurofibromatosis 2 tumor suppressor. Biochem. Biophys. Acta 1423:M29-M36. [DOI] [PubMed] [Google Scholar]
- 18.Gutmann, D. H., L. Sherman, L. Seftor, C. Haipek, K. Hoang Lu, and M. Hendrix. 1999. Increased expression of the NF2 tumor suppressor gene product, merlin, impairs cell motility, adhesion and spreading. Hum. Mol. Genet. 8:267-275. [DOI] [PubMed] [Google Scholar]
- 19.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 20.Hall, A. 1990. The cellular functions of small GTP-binding proteins. Science 249:635-640. [DOI] [PubMed] [Google Scholar]
- 21.Hirokawa, Y., A. Tikoo, J. Huynh, T. Utermark, C. O. Hanemann, M. Giovannini, G. H. Xiao, J. R. Testa, J. Wood, and H. Maruta. 2004. A clue to the therapy of neurofibromatosis type 2: NF2/merlin is a PAK1 inhibitor. Cancer J. 10:20-26. [DOI] [PubMed] [Google Scholar]
- 22.Kimmelman, A. C., D. A. Ross, and B. C. Liang. 1996. Loss of heterozygosity of chromosome 10p in human gliomas. Genomics 34:250-254. [DOI] [PubMed] [Google Scholar]
- 23.Kissil, J. L., K. C. Johnson, M. S. Eckman, and T. Jacks. 2002. Merlin phosphorylation by p21-activated kinase 2 and effects of phosphorylation on merlin localization. J. Biol. Chem. 277:10394-10399. [DOI] [PubMed] [Google Scholar]
- 24.Kissil, J. L., E. W. Wilker, K. C. Johnson, M. S. Eckman, M. B. Yaffe, and T. Jacks. 2003. Merlin, the product of the Nf2 tumor suppressor gene, is an inhibitor of the p21-activated kinase, Pak1. Mol. Cell 12:841-849. [DOI] [PubMed] [Google Scholar]
- 25.Koga, H., N. Araki, H. Takeshima, T. Nishi, T. Hirota, Y. Kimura, M. Nakao, and H. Saya. 1998. Impairment of cell adhesion by expression of the mutant neurofibromatosis type 2 (NF2) genes which lack exons in the ERM-homology domain. Oncogene 17:801-810. [DOI] [PubMed] [Google Scholar]
- 26.Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaJeunesse, D. R., B. M. McCartney, and R. G. Fehon. 1998. Structural analysis of Drosophila merlin reveals functional domains important for growth control and subcellular localization. J. Cell Biol. 141:1589-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao, H., R. J. Winkfein, G. Mack, J. B. Rattner, and T. J. Yen. 1995. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell Biol. 130:507-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luna, E., and A. L. Hitt. 1992. Cytoskeleton-plasma membrane interactions. Science 258:955-964. [DOI] [PubMed] [Google Scholar]
- 30.Lutchman, M., and G. A. Rouleau. 1995. The neurofibromatosis type 2 gene product, schwannomin, suppresses growth of NIH 3T3 cells. Cancer Res. 55:2270-2274. [PubMed] [Google Scholar]
- 31.McCartney, B. M., and R. G. Fehon. 1997. The ERM family proteins and their roles in cell-cell interactions, p. 200-210. In P. Cowijn and M. W. Klymkowsky (ed.), Cytoskeletal-membrane interactions and signal transduction. R. G. Landes Bioscience, Austin, TX.
- 32.McClatchey, A. I. 2003. Merlin and ERM proteins: unappreciated roles in cancer development? Nat. Rev. Cancer 3:877-883. [DOI] [PubMed] [Google Scholar]
- 33.McClatchey, A. I., I. Saotome, K. Mercer, D. Crowley, J. F. Gusella, R. T. Bronson, and T. Jacks. 1998. Mice heterozygous for a mutation at the Nf2 tumor suppressor locus develop a range of highly metastatic tumors. Genes Dev. 12:1121-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClatchey, A. I., I. Saotome, V. Ramesh, J. F. Gusella, and T. Jacks. 1997. The Nf2 tumor suppressor gene product is essential for extraembryonic development immediately prior to gastrulation. Genes Dev. 11:1253-1265. [DOI] [PubMed] [Google Scholar]
- 35.Meng, J. J., D. J. Lowrie, H. Sun, E. Dorsey, P. D. Pelton, A. M. Bashour, J. Groden, N. Ratner, and W. Ip. 2000. Interaction between two isoforms of the NF2 tumor suppressor protein, merlin, and between merlin and ezrin, suggests modulation of ERM proteins by merlin. J. Neurosci. Res. 62:491-502. [DOI] [PubMed] [Google Scholar]
- 36.Montanini, L., L. Bissola, and G. Finocchiaro. 2004. KLF6 is not the major target of chromosome 10p losses in glioblastomas. Int. J. Cancer 111:640-641. [DOI] [PubMed] [Google Scholar]
- 37.Morrison, H., L. S. Sherman, J. Legg, F. Banine, C. Isacke, C. A. Haipek, D. H. Gutmann, H. Ponta, and P. Herrlich. 2001. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 15:968-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muhlbauer, K. R., H. J. Grone, T. Ernst, E. Grone, R. Tschada, M. Hergenhahn, and M. Hollstein. 2003. Analysis of human prostate cancers and cell lines for mutations in the TP53 and KLF6 tumour suppressor genes. Br. J. Cancer 89:687-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narla, G., K. E. Heath, H. L. Reeves, D. Li, L. E. Giono, A. C. Kimmelman, M. J. Glucksman, J. Narla, F. J. Eng, A. M. Chan, A. C. Ferrari, J. A. Martignetti, and S. L. Friedman. 2001. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294:2563-2566. [DOI] [PubMed] [Google Scholar]
- 40.Narla, G., A. Difeo, H. L. Reeves, D. J. Schaid, J. Hirshfeld, E. Hod, W. B. Katz, A. S. Hebbring, A. Komiya, S. K. McDonnell, K. E. Wiley, S. J. Jacobsen, S. D. Isaacs, P. C. Walsh, S. L. Zheng, B. L. Chang, D. M. Friedrichsen, J. L. Stanford, E. A. Ostrander, A. M. Chinnaiyan, M. A. Rubin, J. Xu, S. N. Thibodeau, S. L. Friedman, and J. A. Martignetti. 2005. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 65:1213-1222. [DOI] [PubMed] [Google Scholar]
- 41.Polakis, P. G., R. F. Weber, B. Nevins, J. R. Didsbury, T. Evans, and R. Snyderman. 1989. Identification of the ral and rac1 gene products, low molecular mass GTP-binding proteins from human platelets. J. Biol. Chem. 264:16383-16389. [PubMed] [Google Scholar]
- 42.Rader, C., E. T. Stoeckli, U. Ziegler, T. Osterwalder, B. Kunz, and P. Sonderegger. 1993. Cell-cell adhesion by homophilic interaction of the neuronal recognition molecule axonin-1. Eur. J. Biochem. 215:133-141. [DOI] [PubMed] [Google Scholar]
- 43.Ramesh, V. 2004. Merlin and the ERM proteins in Schwann cells, neurons and growth cones. Nat. Rev. Neurosci. 5:462-470. [DOI] [PubMed] [Google Scholar]
- 44.Rouleau, G. A., P. Merel, M. Lutchman, M. Sanson, J. Zucman, C. Marineau, K. Hoang-Xuan, S. Demczuk, C. Desmaze, B. Plougastel, S. M. Pulst, G. Lenoir, E. Bijlsma, R. Fashold, J. Dumanski, P. de Jong, D. Parry, R. Eldridge, A. Aurias, O. Delattre, and G. Thomas. 1993. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature 363:515-521. [DOI] [PubMed] [Google Scholar]
- 45.Ryu, C. H., S. W. Kim, K. H. Lee, J. Y. Lee, H. Kim, W. K. Lee, B. H. Choi, Y. Lim, Y. H. Kim, K. H. Lee, T. K. Hwang, T. Y. Jun, and H. K. Rha. 2005. The merlin tumor suppressor interacts with Ral guanine nucleotide dissociation stimulator and inhibits its activity. Oncogene 24:5355-5364. [DOI] [PubMed] [Google Scholar]
- 46.Sainio, M., F. Zhao, L. Heiska, O. Turunen, M. den Bakker, E. Zwarthoff, M. Lutchman, G. A. Rouleau, J. Jaaskelainen, A. Vaheri, and O. Carpen. 1997. Neurofibromatosis 2 tumor suppressor protein colocalizes with ezrin and CD44 and associates with actin-containing cytoskeleton. J. Cell Sci. 110:2249-2260. [DOI] [PubMed] [Google Scholar]
- 47.Schulze, K. M., C. O. Hanemann, H. W. Muller, and H. Hanenberg. 2002. Transduction of wild-type merlin into human schwannoma cells decreases schwannoma cell growth and induces apoptosis. Hum. Mol. Genet. 11:69-76. [DOI] [PubMed] [Google Scholar]
- 48.Scoles, D. R., D. P. Huynh, P. A. Morcos, E. R. Coulsell, N. G. Robinson, F. Tamanoi, and S. M. Pulst. 1998. Neurofibromatosis 2 tumour suppressor schwannomin interacts with betaII-spectrin. Nat. Genet. 18:354-359. [DOI] [PubMed] [Google Scholar]
- 49.Scoles, D. R., D. P. Huynh, M. S. Chen, S. P. Burke, D. H. Gutmann, and S. M. Pulst. 2000. The neurofibromatosis 2 tumor suppressor protein interacts with hepatocyte growth factor-regulated tyrosine kinase substrate. Hum. Mol. Genet. 9:1567-1574. [DOI] [PubMed] [Google Scholar]
- 50.Scoles, D. R., V. D. Nguyen, Y. Qin, C. X. Sun, H. Morrison, D. H. Gutmann, and S. M. Pulst. 2002. Neurofibromatosis 2 (NF2) tumor suppressor schwannomin and its interacting protein HRS regulate STAT signaling. Hum. Mol. Genet. 11:3179-3189. [DOI] [PubMed] [Google Scholar]
- 51.Shaw, R. J., J. G. Paez, M. Curto, A. Yaktine, W. M. Pruitt, L. Saotome, J. P. O'Bryan, V. Gupta, N. Ratner, C. J. Der, T. Jacks, and A. I. McClatchey. 2001. The Nf2 tumor suppressor, merlin, functions in Rac-dependent signaling. Dev. Cell 1:63-72. [DOI] [PubMed] [Google Scholar]
- 52.Sherman, L., H. M. Xu, R. T. Geist, S. Saporito-Irwin, N. Howells, H. Ponta, P. Herrlich, and D. H. Gutmann. 1997. Interdomain binding mediates tumor growth suppression by the NF2 gene product. Oncogene 15:2505-2509. [DOI] [PubMed] [Google Scholar]
- 53.Surace, E. I., C. A. Haipek, and D. H. Gutmann. 2004. Effect of merlin phosphorylation on neurofibromatosis 2 (NF2) gene function. Oncogene 23:580-587. [DOI] [PubMed] [Google Scholar]
- 54.Takeshima, H., I. Izawa, P. S. Lee, N. Safdar, V. A. Levin, and H. Saya. 1994. Detection of cellular proteins that interact with the NF2 tumor suppressor gene product. Oncogene 9:2135-2144. [PubMed] [Google Scholar]
- 55.Tan, M., R. Grijalva, and D. Yu. 1999. Heregulin beta1-activated phosphatidylinositol 3-kinase enhances aggregation of MCF-7 breast cancer cells independent of extracellular signal-regulated kinase. Cancer Res. 59:1620-1625. [PubMed] [Google Scholar]
- 56.Thomas, G., P. Merel, M. Sanson, K. Hoang-Xuan, J. Zucman, C. Desmaze, T. Melot, A. Aurias, and O. Delattre. 1994. Neurofibromatosis type 2. Eur. J. Cancer 30A:1981-1987. [DOI] [PubMed] [Google Scholar]
- 57.Tikoo, A., M. Varga, V. Ramesh, J. Gusella, and H. Maruta. 1994. An anti-Ras function of neurofibromatosis type 2 gene product (NF2/Merlin). J. Biol. Chem. 269:23387-23390. [PubMed] [Google Scholar]
- 58.Trofatter, J. A., M. M. MacCollin, J. L. Rutter, J. R. Murrell, M. P. Duyao, D. M. Parry, R. Eldridge, N. Kley, A. G. Menon, K. Pulaski, V. Hasse, C. M.Ambrose, D. Munroe, C. Bove, J. L. Haines, R. L. Martuza, M. E. MacDonald, B. R. Seizinger, M. P. Short, A. J. Buckler, and J. F. Gusella. 1993. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell 72:791-800. [DOI] [PubMed] [Google Scholar]
- 59.Voesten, A. M., E. H. Bijleveld, A. Westerveld, and T. J. Hulsebos. 1997. Fine mapping of a region of common deletion on chromosome arm 10p in human glioma. Genes Chromosomes Cancer 20:167-172. [PubMed] [Google Scholar]
- 60.Xiao, G. H., A. Beeser, J. Chernoff, and J. R. Testa. 2002. p21-activated kinase links Rac/Cdc42 signaling to merlin. J. Biol. Chem. 277:883-886. [DOI] [PubMed] [Google Scholar]
- 61.Xiao, G. H., J. Chernoff, and J. R. Testa. 2003. NF2: the wizardry of merlin. Genes Chromosomes Cancer 38:389-399. [DOI] [PubMed] [Google Scholar]
- 62.Xiao, G. H., R. Gallagher, J. Shetler, K. Skele, D. A. Altomare, R. G. Pestell, S. Jhanwar, and J. R. Testa. 2005. The NF2 tumor suppressor gene product, merlin, inhibits cell proliferation and cell cycle progression by repressing cyclin D1 expression. Mol. Cell. Biol. 25:2384-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]