Abstract
Cell cycle checkpoints are evolutionarily conserved signaling pathways that uphold genomic integrity. Complete inactivation of the mouse checkpoint gene Hus1 results in chromosomal instability, genotoxin hypersensitivity, and embryonic lethality. To determine the functional consequences of partial Hus1 impairment, we generated an allelic series in which Hus1 expression was incrementally reduced by combining a hypomorphic Hus1 allele, Hus1neo, with either wild-type or null (Hus1Δ1) alleles. Primary Hus1neo/Δ1 embryonic fibroblasts exhibited spontaneous chromosomal abnormalities and underwent premature senescence, while higher Hus1 expression in Hus1neo/neo cells allowed for normal proliferation. Antioxidant treatment almost fully suppressed premature senescence in Hus1neo/Δ1 cultures, suggesting a critical role for Hus1 in oxidative stress responses. Treatment of Hus1neo/neo and Hus1neo/Δ1 cells with the DNA adducting agent benzo(a)pyrene dihydrodriol epoxide resulted in a loss of cell viability that was associated with S-phase DNA damage checkpoint failure. Likewise, the DNA polymerase inhibitor aphidicolin triggered increased cell death, chromosomal aberrations, and H2AX phosphorylation, a marker for double-stranded DNA breaks, in Hus1neo/neo and Hus1neo/Δ1 cultures compared to controls. Despite these pronounced genome maintenance defects in cultured Hus1neo/Δ1 and Hus1neo/neo cells, mice of the same genotypes were born at expected frequencies and appeared grossly normal. A significant increase in micronucleus formation was observed in peripheral blood cells from Hus1neo/Δ1 mice, but reduced Hus1 expression did not cause an elevated predisposition to spontaneous tumor development or accelerate tumorigenesis in p53-deficient mice. These results identify differential effects of altered Hus1 gene dosage on genome maintenance during in vitro culture, genotoxic stress responses, embryonic development, and adult homeostasis.
The ability to accurately duplicate the genome and correctly segregate damage-free chromosomes to daughter cells is crucial to the health and longevity of all organisms. To ensure the fidelity of these processes, cells respond to genome damage by arresting the cell cycle, stabilizing replication forks, and inducing DNA repair. Cell cycle checkpoint pathways mediate these DNA damage responses and additionally can induce apoptosis if the damage is beyond repair. By preventing the accumulation of mutations that drives carcinogenesis, checkpoints can be important tumor suppressor mechanisms (3, 21, 27).
Replication inhibitors and bulky DNA lesions activate a checkpoint pathway headed by the phosphatidylinositol kinase-like protein kinase Atr. Stalling of replication forks or processing of DNA lesions causes accumulation of single-stranded DNA that becomes coated with replication protein A (RPA) and attracts Atr in association with its binding partner Atrip (12). Atr then transmits the checkpoint signal by phosphorylating Chk1, Brca1, and other targets, which in turn regulate the cell cycle, replication, and repair machineries. Efficient DNA damage signaling through Atr also requires several accessory factors, including TopBP1/Cut5, Claspin, and the Rad9-Rad1-Hus1 (9-1-1) complex (44).
With predicted structural similarity to proliferating cell nuclear antigen (PCNA), the 9-1-1 complex is believed to function as a checkpoint sliding clamp and molecular scaffold (57). Loading of the 9-1-1 trimer onto chromatin is stimulated by genotoxic stress and is mediated by a clamp loader composed of Rad17 and associated replication factor C subunits (8, 41, 67). Once on chromatin, the 9-1-1 complex participates in DNA damage signaling, promoting phosphorylation of Atr substrates such as Chk1, Rad17, and Rad9 itself (2, 40, 65, 67). Signaling defects in cells lacking 9-1-1 components are associated with failure of an S-phase checkpoint that represses late origin firing in response to DNA damage (2, 40, 64). The mammalian 9-1-1 complex also directly associates with a number of DNA repair proteins, including factors involved in base excision repair (10, 20, 46, 48, 55, 58, 59) and translesion DNA synthesis (26, 42), and additionally is required for homologous recombinational repair (37, 61) as well as telomere maintenance (19, 37). Given these key roles in checkpoint signaling and DNA repair, it is not surprising that cells defective for 9-1-1 function are hypersensitive to a wide variety of genotoxins, including replication inhibitors and DNA damaging agents (23, 28, 40, 60, 61, 63, 64).
The mammalian 9-1-1 complex not only responds to extrinsic stresses but also is required for embryonic development. Targeted inactivation of mouse Hus1 (63) or Rad9 (23) results in widespread cell death and midgestational embryonic lethality. Rad17 is also essential for murine embryogenesis (7). Loss of Atr (6, 14) or Chk1 (31, 51) has even more severe consequences and causes peri-implantation lethality. In all of these mouse models, embryonic lethality is associated with spontaneous chromosomal instability, indicating that these gene products are essential for genome maintenance during normal cell proliferation. Even in the absence of extrinsic stresses, mammalian checkpoints regulate Cdc25A turnover (50), the timing of origin firing (45), and replication fork progression through fragile sites in the genome (11).
The analysis of hypomorphic alleles that are partially impaired for gene function is a traditional genetic strategy that can yield important insights into the activity of a given gene. This approach is particularly appropriate for study of the Atr-dependent checkpoint pathway, as its complete inactivation causes severe phenotypes. Moreover, understanding the impact of partial impairment of this essential checkpoint mechanism has significant biomedical implications. For instance, a hypomorphic ATR mutation is known to cause the human developmental disorder Seckel syndrome (36). Furthermore, components of this pathway have been suggested to represent a new class of tumor suppressors that may be only partially inactivated in cancers (31). Here, we describe a new mouse model based on a hypomorphic Hus1 allele that expresses reduced levels of wild-type Hus1. Partial reduction of Hus1 expression in cultured cells was found to cause premature senescence, spontaneous chromosomal abnormalities, and DNA damage hypersensitivity. Unexpectedly, mice with the same genetic alteration were born at the predicted frequency and, despite indications of genomic instability, developed normally without heightened tumor predisposition.
MATERIALS AND METHODS
Mouse strains.
Previously described Hus1neo and Hus1Δ1 mouse strains were maintained on a 129S6 inbred genetic background (30, 63). Trp53tm1Tyj mice were maintained on a C57BL/6 inbred genetic background (25). All mice were housed in accordance with institutional animal care and use guidelines. For analysis of tumor cohorts, animals were aged until they showed clinical signs of disease. Tumors identified grossly at necropsy were fixed in 10% neutral-buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Histopathological evaluations and tumor classifications were performed blind with respect to genotype. Kaplan Meier survival curves were generated and compared by log rank test using SPSS statistical software.
Cell culture, proliferation measurements, and genotoxin survival assays.
Mouse embryonic fibroblasts (MEFs) were generated from 13.5-dpc embryos from timed matings between Hus1+/neo and Hus1+/Δ1 mice or Hus1+/neo and Hus1neo/Δ1 mice. Briefly, following removal of the heart, liver, and head, embryos were mechanically disaggregated and the resulting single-cell suspension was cultured in culture medium (Dulbecco minimal essential medium [DMEM] supplemented with 10% fetal bovine serum, 1.0 mM l-glutamine, 0.1 mM MEM nonessential amino acids, 100 μg/ml of streptomycin sulfate, and 100 U/ml of penicillin). Serial MEF cultures were maintained according to the 3T3 protocol in which 106 cells were passaged onto 10-cm dishes every 3 days (54). The initial plating was considered passage zero. When primary cultures entered senescence, the cells were provided fresh culture medium but passaged only upon reaching confluence, until immortalized clones emerged. Population doublings were calculated with the formula ΔPDL = log(nf/no)/log2, where no is the initial number of cells and nf is the final number of cells (4). The effect of antioxidant treatment was assessed by culturing cells beginning at passage one in culture medium containing 5 mM N-acetyl-alanine (NAA) (Sigma) or N-acetyl-cysteine (NAC) (Sigma), which was changed daily. For short-term proliferation assays, 105 cells were plated per six-well dish well and, at 24-h intervals, triplicate samples were harvested by trypsinization. The number of viable cells in individual samples was determined after incubation with trypan blue dye. For short-term genotoxin survival assays, 105 cells were plated per six-well dish well, treated with genotoxin, cultured for 3 days, and then harvested by trypsinization, incubated with trypan blue dye, and individually counted. For benzo(a)pyrene dihydrodriol epoxide (BPDE) (NCI carcinogen repository) treatment, the genotoxin was added directly to the medium and cells were then incubated for 1 h, after which time the genotoxin-containing medium was removed, cells were washed once with phosphate-buffered saline (PBS), and fresh medium was added. Treatments with aphidicolin (Sigma) were done similarly, except that genotoxin exposure to cells was for 24 h.
Radioresistant DNA synthesis assay.
Cells were plated in triplicate at a density of 2 × 105 per well of a gelatinized six-well dish. The next day, the cells were treated with BPDE for 1 h, washed with PBS, incubated for 30 min in culture medium without genotoxin, and labeled for 1 h in medium containing 2.5 μCi/ml methyl-[3H]thymidine (2.0 Ci/mmol; NEN Life Science Products, Inc). The radioactive medium was then removed, and unincorporated nucleotides were removed by washing the cells three times in ice-cold 5.0% trichloroacetic acid. The cells were then solubilized in 0.3 N NaOH, followed by neutralization with glacial acetic acid. Radioactivity was quantitated with a liquid scintillation counter.
Northern blot hybridization and reverse transcriptase (RT) PCR.
Total RNA was prepared from passage-one MEF cultures with RNA STAT-60 reagent (Tel-Test). Northern blot hybridization was performed as described previously (63). The signal intensity for wild-type Hus1 transcripts was determined by phosphorimager and normalized to the signal intensity for Gapdh for the same RNA sample. cDNA was prepared from 3 μg of DNase-treated RNA by random priming with the SuperScript preamplification system (Gibco BRL). PCR amplification was performed with the following primers (see also Fig. 2): 1 (5′ BamH1), 5′-CTCGGATCCATGAAGTTTCGCGCCAAG-3′; 2 (3′ EcoRI), 5′-CTCGAATTCCTAGGACAAGGCTGGGAT-3′; 3 (3.224), 5′-TCTTCAGAGACTCCTTCCATT-3′; 4 (Neo2), 5′-TTCGTCCAGATCATCCTGATC-3′; 5 (Neo1), 5′-AGAGGCTATTCGGCTATGACTG-3′; 6 (Neo 3′), 5′-GGTATCGCCGCTCCCGATTCGCAG-3′. PCR products generated with primers 1 and 2 were digested with BamHI and EcoRI and cloned into pBluescript II (Stratagene). All other PCR products were cloned into vector pCR2.1 by TOPO TA cloning (Invitrogen). cDNA inserts were fully sequenced.
FIG. 2.
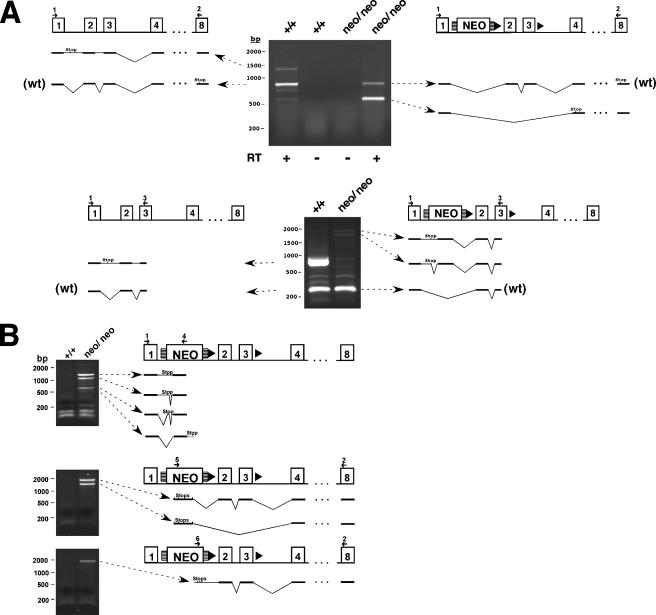
Hus1neo expresses both wild-type and aberrant noncoding transcripts. DNase-treated total RNA from MEFs of the indicated genotypes was reverse transcribed and subjected to PCR analysis using either two Hus1-specific primers (A) or one Hus1-specific and one neo-specific primer (B). Control reactions were performed in the absence of RT as indicated. Primer locations are indicated and correspond to the sequences listed in Materials and Methods. The resulting cDNAs were cloned and sequenced, and the structure of each transcript is diagrammed. Note that Hus1 transcripts from the wild-type allele often show retention of introns one and/or two. In-frame stop codons are indicated for transcripts originating in Hus1 coding sequences. In panel B, the smallest of the sequenced PCR products generated by primers 1 plus 4 does not contain an in-frame stop codon. For this cDNA, predicted sequence adjacent to the PCR product is depicted in gray and includes an in-frame stop codon. Stop codons for all three reading frames are indicated for transcripts originating in the neo cassette.
Metaphase spread preparation.
MEFs were incubated in culture medium containing 0.15 μg/ml Colcemid for 1 h. Cells were then harvested by trypsinization, swollen for 12 min at 37°C in hypotonic buffer (0.034 M KCl, 0.017 M Na3C6H5O7), and fixed for at least 20 min at 4°C in 75% methanol-25% acetic acid. Cells in fixative were then spotted onto microscope slides and stained with 2.0% Giemsa in Gurr buffer (pH 6.8). Chromosomal abnormalities were scored based on standard guidelines (34, 43). For analysis of aphidicolin-induced chromosomal abnormalities, MEFs were treated with 0.05 μM or 0.10 μM aphidicolin for 24 h and then immediately subjected to metaphase spread preparation as described above.
Indirect immunofluorescence.
Cells grown on glass slides were fixed with 2% paraformaldehyde in Tris-buffered saline (TBS) for 35 min at 4°C. The cells were then permeabilized and blocked with 3% bovine serum albumin, 0.2% TritonX-100, and 0.01% nonfat dried milk in TBS for 20 min at room temperature. Cells were incubated sequentially with primary anti-γ-H2AX antibody (JBW301; Upstate) at 1:500 for 45 min, secondary goat anti-mouse immunoglobulin G (H+L)-fluorescein isothiocyanate (FITC) (Southern Biotechnology) at 1:60 for 35 min, and DAPI (4′,6′-diamidino-2-phenylindole) (33 ng/ml) for 1 min. Slides were viewed with a Leica DMRE fluorescence microscope.
Micronucleus assay.
Analysis of micronucleus formation in peripheral blood cells was performed as described by others (39). Briefly, 50 μl of peripheral blood was collected from the mandibular vein into a microcentrifuge tube containing 200 μl of heparin solution (500 USP heparin/ml PBS) and fixed in methanol at −80°C overnight. After washing with ice-cold bicarbonate buffer (0.9% NaCl, 5.3 mM NaHCO3 [pH 7.5]), the cells were incubated in bicarbonate buffer containing RNase A and anti-CD71:FITC antibody (Biodesign International) at 4°C for 45 min. The samples were then washed with bicarbonate buffer, resuspended in propidium iodide (1.25 μg/ml in bicarbonate buffer), and analyzed on a FACSCalibur flow cytometer (Becton-Dickinson, San Jose, CA) as described by others (15).
RESULTS
Hus1neo is a hypomorphic allele that expresses a reduced level of wild-type Hus1.
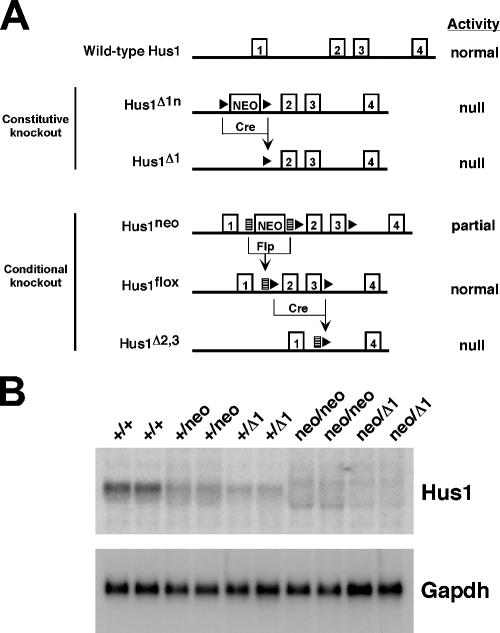
In the process of producing a loxP site-flanked conditional Hus1 allele, Hus1flox (30), we generated the Hus1neo allele, which contains a frt-flanked neo cassette in intron one as well as loxP sites surrounding exons two and three (Fig. 1A). Often the presence of a neo cassette in a targeted gene interferes with the expression of that gene (29). Given that complete Hus1 inactivation results in embryonic lethality and severe proliferation defects in cultured cells, we reasoned that a reduced level of Hus1 expression from the Hus1neo allele might be useful for testing the impact of a partial impairment of this essential checkpoint gene. Therefore, we used Hus1neo, in combination with wild-type Hus1 and the previously described null Hus1Δ1 allele (63), to produce a Hus1 allelic series (Fig. 1A). MEFs of all possible genotypes except Hus1Δ1/Δ1 were generated. Hus1Δ1/Δ1 MEFs cannot be produced because of the early embryonic lethality associated with this genotype (63). Northern blot analysis revealed a striking incremental reduction in Hus1 expression through the series (Fig. 1B). The graded pattern of Hus1 expression observed (Hus1+/+ > Hus1+/neo > Hus1+/Δ1 > Hus1neo/neo > Hus1neo/Δ1) suggested that Hus1neo produced a significantly reduced level of Hus1. Quantification of the Northern blot signal by phosphorimager analysis indicated each Hus1neo allele expressed roughly 40% of the wild-type level of Hus1 (Hus1+/+, 100%; Hus1+/neo, 71.4%; Hus1+/Δ1, 43.5%; Hus1neo/neo, 47.4%; Hus1neo/Δ1, 20.8%).
FIG. 1.
Hus1 allelic series. (A) Schematic representation of the Hus1 alleles generated in systems for constitutive and conditional Hus1 inactivation. The first four Hus1 exons are shown as boxed numbers. A neomycin resistance cassette (NEO), frt sites (lined rectangles), and loxP sites (solid triangles) are shown. Exposure of alleles containing frt sites to the Flp recombinase or alleles containing loxP sites to the Cre recombinase excises the intervening sequence, generating a new allele as indicated. (B) Northern blot analysis of primary MEF cultures. Total RNA was prepared from individual MEF cultures of the indicated genotypes and hybridized with a 32P-labeled Hus1 or Gapdh cDNA probe. RNA prepared from each MEF culture was loaded in duplicate.
Examination of the Northern blot signal for Hus1neo/neo samples suggested the presence of wild-type Hus1 transcripts as well as novel RNA species, including both larger and smaller transcripts. Therefore, a thorough analysis of Hus1 expression from Hus1neo was performed by RT-PCR (Fig. 2). Control reactions from which RT was omitted did not yield detectable PCR products, confirming the absence of contaminating genomic DNA (Fig. 2A and data not shown). Hus1neo contains all wild-type Hus1 sequences and, as expected, produced wild-type Hus1 transcripts (Fig. 2A). Although these RT-PCR assays are not quantitative, a reduced amount of wild-type Hus1 product was amplified from Hus1neo/neo cells compared to Hus1+/+ cells, consistent with the results of the Northern blot analysis. The presence of the neo cassette also induced skipping of exons two and three with increased frequency. Due to a frameshift, the resulting transcript in which exon one is spliced to exon four has the capacity to encode only the first 19 Hus1 amino acids and is functionally inactive (30). The neo cassette used in this study is reported to contain cryptic splice acceptor and donor sites (33), and accordingly some Hus1neo transcripts also spliced from Hus1 sequences into the neo cassette. Other Hus1neo transcripts appeared to originate from within the neo cassette and then spliced into Hus1 sequences (Fig. 2B). Unfortunately, endogenous mouse Hus1 protein could not be detected from primary MEFs with the available antibody reagents, and it was not possible to characterize Hus1 expression from Hus1neo in this manner. However, all aberrant transcripts produced from Hus1neo contained premature stop codons and had the capacity to produce only highly truncated protein fragments containing little Hus1 sequence. As detailed below, Hus1+/neo cells and mice were completely normal in all assays in which they were tested, indicating that Hus1neo has no detectable dominant activities, although we cannot rule out subtle effects due to the production of aberrant transcripts from this allele. Together, these results indicate that Hus1neo is a hypomorphic allele that expresses a reduced level of wild-type Hus1.
Partial reduction of Hus1 expression in cultured cells results in proliferation defects and spontaneous chromosomal abnormalities.
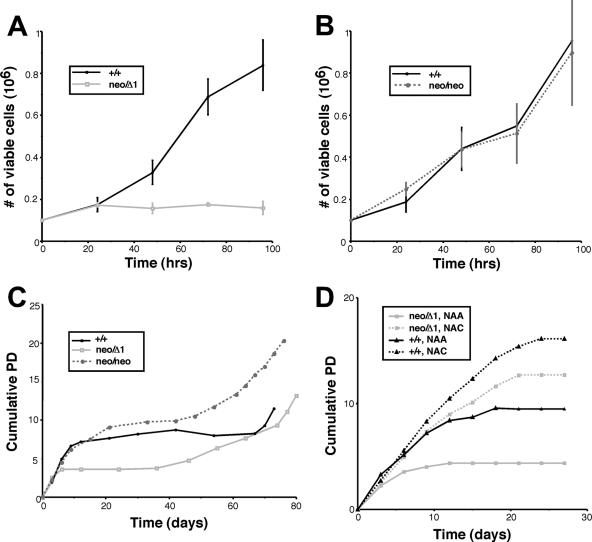
Although the gross morphology of embryos of all genotypes in the allelic series was indistinguishable, Hus1neo/Δ1 fibroblasts appeared to grow poorly once in culture. To quantify these observations, we initially performed short-term proliferation assays with MEFs at passage three. Hus1neo/Δ1 MEFs showed a severely limited growth capacity compared to control Hus1+/+ MEFs (Fig. 3A). Hus1neo/neo MEFs, on the other hand, grew as well as Hus1+/+ cells (Fig. 3B). Hus1+/neo and Hus1+/Δ1 MEFs also showed normal growth (data not shown). The impaired growth of Hus1neo/Δ1 MEFs observed in these short-term assays prompted us to examine the growth capacity of these cells over a longer time frame. Hus1+/+, Hus1neo/neo, and Hus1neo/Δ1 MEFs were cultured according to the 3T3 protocol (54), and cumulative population doublings were calculated. When cultured in this manner, primary mouse fibroblasts typically grow rapidly for 5 to 10 passages before undergoing growth arrest, a process termed senescence. Later, highly proliferative immortalized clones emerge spontaneously. As shown in Fig. 3C, MEFs of all Hus1 genotypes proliferated to similar extents for the first 3 to 6 days in culture. However, Hus1neo/Δ1 MEFs then underwent a premature growth arrest, ceasing proliferation at an earlier passage than Hus1+/+ or Hus1neo/neo MEFs. Eventually, cells of all three genotypes, including Hus1neo/Δ1, became immortalized and resumed rapid growth.
FIG. 3.
Impaired proliferation of cells expressing reduced levels of Hus1. (A and B) Short-term proliferation of Hus1neo/Δ1 and Hus1neo/neo MEFs along with matched Hus1+/+ control cells. MEF cultures at passage three were seeded into six-well dishes in triplicate, and the number of viable cells was determined at the indicated times postplating. Plots show the mean, with error bars representing the standard deviation. (C) Long-term proliferation of Hus1+/+, Hus1neo/neo, and Hus1neo/Δ1 cells. Cumulative population doublings were calculated for MEF cultures of the indicated genotypes maintained according to a 3T3 passaging protocol, as described in Materials and Methods. (D) Effects of antioxidant treatment on the proliferation of Hus1+/+ and Hus1neo/Δ1 cells. Hus1+/+ and Hus1neo/Δ1 MEFs were cultured in culture medium containing 5 mM NAA or NAC, which was changed daily. Cumulative population doublings were calculated as described for panel C.
The senescence of MEFs is believed to be a response to extrinsic cellular stresses. In particular, cells cultured under standard conditions experience high, nonphysiological oxygen levels, and the resulting oxidative DNA damage is a primary driving force for senescence in MEFs (38). We hypothesized that the premature senescence of Hus1neo/Δ1 MEFs might be due to a heightened sensitivity to oxidative stress, as Hus1 associates with several base excision repair proteins that promote the repair of oxidative lesions (10, 20, 46, 48, 55, 58, 59). Therefore, we tested the impact of the antioxidant NAC on the growth of Hus1+/+ and Hus1neo/Δ1 MEFs (Fig. 3D). Cells treated with the control compound NAA behaved in the same manner as the untreated cells described above, with Hus1neo/Δ1 cultures undergoing a proliferative arrest much earlier than their Hus1+/+ counterparts. Remarkably, treatment with the antioxidant NAC nearly fully suppressed the premature senescence of Hus1neo/Δ1 MEFs. Although NAC also permitted sustained proliferation of Hus1+/+ MEFs at later time points, it significantly enhanced the proliferation of Hus1neo/Δ1 MEFs at early time points, several passages before it had any detectable effect on Hus1+/+ cells, suggesting that premature senescence in Hus1neo/Δ1 MEFs is due at least in part to an impaired response to oxidative stress.
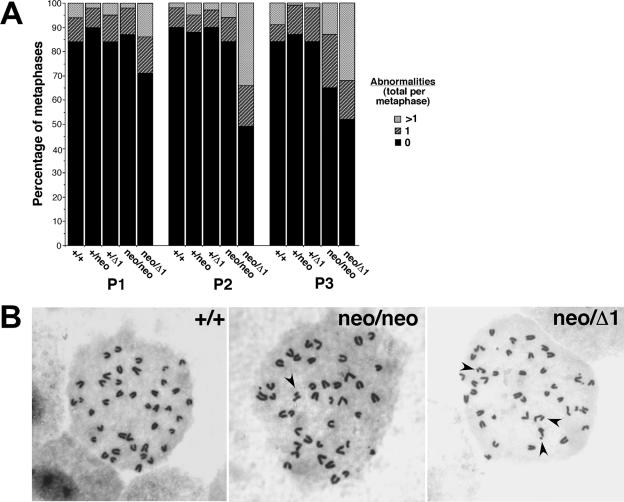
We next assessed whether the poor growth and premature senescence shown by Hus1neo/Δ1 MEFs was associated with genomic instability. Metaphase spreads were prepared from MEFs of each genotype in the allelic series at passages one through three, and gross chromosomal abnormalities were scored (Fig. 4A). At passage one, an increased percentage of Hus1neo/Δ1 metaphases had chromosomal aberrations compared to all other genotypes. The increase in chromosomal abnormalities in Hus1neo/Δ1 cells was even greater at passage two, and by passage three 48% of Hus1neo/Δ1 MEFs had at least one chromosomal abnormality and 32% had multiple abnormalities. By contrast, 84% of Hus1+/+ metaphases were normal at passage three, and only 9% had multiple abnormalities. Hus1neo/neo MEFs showed an intermediate level of chromosomal instability which was not apparent until later passages. By passage three, 35% of Hus1neo/neo metaphases contained at least one chromosomal abnormality and 13% contained multiple abnormalities. As reported previously for primary Hus1-null MEFs (63), the chromosomal abnormalities in Hus1neo/Δ1 MEFs were primarily chromatid gaps and breaks, and the relative frequency for different types of chromosomal lesions was similar for all genotypes (data not shown; see also Table 1). Representative metaphases are shown in Fig. 4B. Together, these data indicate that a partial reduction in Hus1 expression leads to spontaneous chromosomal abnormalities that accumulate over time and are associated with premature cellular senescence.
FIG. 4.
A partial reduction in Hus1 expression results in chromosomal instability. (A) Summary of the frequency of chromosomal abnormalities in MEFs with reduced Hus1 expression. A total of 60 to 93 metaphases, prepared from at least three independent MEF cultures, were scored for each genotype at each passage. (B) Representative metaphase spreads from primary cultures expressing reduced levels of Hus1. Metaphase spreads were prepared from Hus1+/+, Hus1neo/neo, and Hus1neo/Δ1 MEFs at passage three and stained with Giemsa. Arrows highlight some of the chromosomal abnormalities.
TABLE 1.
Aphidicolin-induced chromosomal abnormalities in cells with reduced Hus1 expressiona
| Hus1 genotype | Aphidicolin (μM) | Total no. of metaphases | No. of breaks/gaps
|
No. of chromatid interchanges
|
No. with extensive damageb | ||||
|---|---|---|---|---|---|---|---|---|---|
| Chromosome type | Chromatid type | Total | Avg | Total | Avg | ||||
| +/neo | Untreated | 19 | 0 | 3 | 3 | 0.16 | 0 | 0 | 0 |
| 0.05 | 20 | 3 | 2 | 5 | 0.25 | 0 | 0 | 0 | |
| 0.1 | 20 | 1 | 11 | 12 | 0.6 | 0 | 0 | 0 | |
| neo/neo | Untreated | 20 | 0 | 9 | 9 | 0.45 | 3 | 0.15 | 0 |
| 0.05 | 20 | 0 | 9 | 9 | 0.45 | 10 | 0.5 | 1 | |
| 0.1 | 20 | 2 | 19 | 20 | 0.95 | 14 | 0.7 | 1 | |
| neo/Δ1 | Untreated | 19 | 0 | 6 | 6 | 0.32 | 0 | 0 | 0 |
| 0.05 | 19 | 2 | 22 | 24 | 1.26 | 15 | 0.79 | 0 | |
| 0.1 | 19 | 0 | 19 | 19 | 1.00 | 14 | 0.74 | 3 | |
MEFs of the indicated Hus1 genotype were left untreated (control) or treated with the indicated dose of aphidicolin for 24 h. Metaphase spreads were then prepared, and chromosomal abnormalities were scored.
Severely damaged metaphases with too many abnormalities to count.
Cells with partial impairment of Hus1 expression are hypersensitive to DNA damage and replication stress.
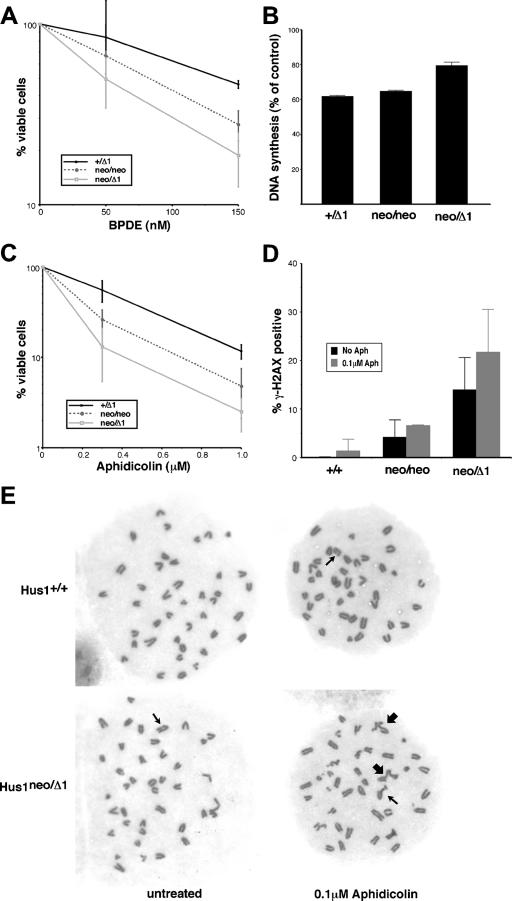
Because the level of Hus1 expressed from Hus1neo was insufficient for genomic stability under normal culture conditions, it was also of interest to test how partial Hus1 impairment would impact cellular responses to DNA damage or replication blockage. Sensitivity to the bulky DNA lesion-inducing agent BPDE, a genotoxin to which Hus1-null cells are hypersensitive (64), was tested in short-term viability assays (Fig. 5A). These assays employed MEFs at passage one, a passage at which untreated cells of all genotypes proliferated similarly (Fig. 3C and data not shown). Hus1neo/Δ1 MEFs showed a significant increase in BPDE sensitivity. At 72 h after treatment with 150 nM BPDE, only 18.8% ± 6.2% of Hus1neo/Δ1 cells remained viable, compared to 46.3% ± 2.3% of control Hus1+/Δ1 cells. Although untreated Hus1neo/neo MEFs proliferated normally, it was of interest to test whether the level of Hus1 expressed in these cells would allow cells to cope with a greater level of genome damage. Hus1neo/neo MEFs showed an intermediate sensitivity to BPDE relative to Hus1+/Δ1 and Hus1neo/Δ1 MEFs, with 27.8% ± 5.2% of cells of this genotype surviving treatment with 150 nM BPDE. Additional control experiments indicated that Hus1+/neo and Hus1+/Δ1 MEFs were as sensitive to BPDE and all other genotoxins tested as Hus1+/+ MEFs (data not shown). The BPDE hypersensitivity of cells with a partial reduction in Hus1 expression was associated with defects in an intra-S DNA damage checkpoint that inhibits DNA synthesis in response to genome damage (Fig. 5B). At 1 h after treatment with 100 nM BPDE, the level of DNA synthesis in Hus1+/Δ1 MEFs was reduced to 61.9% ± 0.5% of that for untreated control cultures. Hus1neo/neo MEFs showed a slightly greater level of DNA synthesis following BPDE treatment (64.7% ± 0.7% of untreated control levels), but this was not significantly different from the value for Hus1+/Δ1 cells. However, in BPDE-treated Hus1neo/Δ1 MEFs, DNA synthesis was maintained at 79.4% ± 2.0% of that for untreated control cultures. Impaired proliferation in untreated Hus1neo/Δ1 MEFs could lead to an overestimation of the extent of the S-phase checkpoint defect, although similar results were obtained with cells at passage one, when cells of all genotypes proliferate similarly (data not shown). Thus, a partial reduction in Hus1 expression leads to defective S-phase checkpoint function and increased sensitivity to DNA damage. The results additionally identify a level of Hus1 in Hus1neo/neo MEFs that is sufficient under normal growth conditions but is sublimiting for the response to extrinsic genotoxic stresses.
FIG. 5.
Defective responses to DNA damage and replication blockage in cells expressing reduced levels of Hus1. (A) Cell viability following BPDE treatment. Hus1+/Δ1, Hus1neo/neo, and Hus1neo/Δ1 MEFs at passage one were treated with BPDE, and cell viability was measured 72 h posttreatment. The percentage of viable cells, relative to mock-treated control cultures, is plotted. Values are the mean of triplicate samples, with error bars representing the standard deviation. (B) DNA synthesis following BPDE treatment. Hus1+/Δ1, Hus1neo/neo, and Hus1neo/Δ1 MEFs at passage two were treated with BPDE and then assayed for DNA synthesis 1 h later by measurement of radiolabeled thymidine incorporation. The percentage of DNA synthesis, relative to that for matched mock-treated control cells, is shown. Values are the mean of triplicate samples, with error bars representing the standard deviation. (C) Cell viability following aphidicolin treatment. Hus1+/Δ1, Hus1neo/neo, and Hus1neo/Δ1 MEFs at passage one were treated with aphidicolin for 24 h, and cell viability was measured 72 h posttreatment. The percentage of viable cells, relative to mock-treated control cultures, is plotted. Values are the mean of triplicate samples, with error bars representing the standard deviation. (D) γ-H2AX accumulation following aphidicolin treatment. Hus1+/+, Hus1neo/neo, and Hus1neo/Δ1 cells at passage three were treated with 0.1 μM aphidicolin for 24 h, and the percentage of γ-H2AX-positive cells was then determined by indirect immunofluorescence. Values are the mean for three independent 40× microscope fields, with error bars representing the standard deviation. (E) Representative metaphase spreads from aphidicolin-treated MEFs. Hus1+/+ and Hus1neo/Δ1 MEFs at passage two were mock treated or treated with 0.1 μM aphidicolin for 24 h. Metaphase spreads were then prepared, stained with Giemsa, and imaged with a 100× objective lens. Thin arrows indicate chromosome gaps and breaks, while thick arrows indicate chromatid interchanges.
The response of cells with reduced Hus1 expression to replication stress was also examined, and Hus1 expression from Hus1neo was found to be sublimiting for cellular responses to the replication inhibitor aphidicolin (Fig. 5C). In short-term viability assays, Hus1neo/Δ1 MEFs at passage one showed an approximately fivefold increase in sensitivity to aphidicolin relative to Hus1+/Δ1 MEFs. Hus1neo/neo MEFs showed intermediate aphidicolin sensitivity. A similar pattern of hypersensitivity was observed when MEFs of the allelic series were treated with hydroxyurea, another inhibitor of DNA synthesis (data not shown).
To gain insights into the basis for increased sensitivity to replication inhibitors in cells with reduced Hus1 expression, we examined the accumulation of double-strand breaks (DSB) following blockage of DNA synthesis in these cells. For this purpose, we performed immunofluorescence assays to detect γ-H2AX, the phosphorylated form of histone H2AX that marks sites of DSB formation (53). Although Atr is responsible for H2AX phosphorylation in response to replication stress, Hus1 is dispensable for this process (62). As shown in Fig. 5D, reduced Hus1 expression resulted in an increased frequency of γ-H2AX-positive cells, even in the absence of extrinsic genotoxin. Treatment of these cells with aphidicolin resulted in a substantial increase in γ-H2AX staining. Following low-dose (0.1 μM) aphidicolin treatment, only 1.4% of Hus1+/+ MEFs were γ-H2AX positive, while 6.6% and 21.7% of Hus1neo/neo and Hus1neo/Δ1 MEFs, respectively, were γ-H2AX positive after aphidicolin exposure. Similar results were obtained when γ-H2AX accumulation was analyzed by immunoblotting (data not shown). These results suggest that reduced Hus1 expression results in accumulation of DSB under conditions of replication stress. To further assess the genomic consequences of blockage of DNA synthesis when Hus1 levels are sublimiting, we produced metaphase spreads from MEFs of the allelic series after low-dose aphidicolin treatment. As expected, treatment of control Hus1+/neo MEFs with aphidicolin resulted in an increase in the frequency of chromatid-type and chromosome-type gaps and breaks (Table 1). In untreated Hus1neo/neo and Hus1neo/Δ1 MEFs, there were greater numbers of chromatid gaps and breaks than in Hus1+/neo MEFs, and following aphidicolin treatment the frequency of these lesions, as well as chromosome-type gaps and breaks, increased significantly. Interestingly, the frequency of chromatid interchanges also increased dramatically following aphidicolin treatment in both Hus1neo/neo and Hus1neo/Δ1 MEFs, but not in control Hus1+/neo MEFs. Representative metaphase spreads are shown in Fig. 5E. These findings indicate that the level of Hus1 expressed from Hus1neo is inadequate for responding to replication stress, leading to the formation of DSB and aberrant chromosomal structures.
Mice with reduced Hus1 expression are grossly normal despite increased genomic instability.
Because MEFs with reduced Hus1 expression showed premature senescence, spontaneous chromosomal abnormalities, and impaired DNA damage responses, we expected that mice of the corresponding genotypes also would display severe phenotypes. Surprisingly, mice of all genotypes in the allelic series were born at the expected frequency. Intercrosses between Hus1+/neo mice produced Hus1neo/neo animals in an approximately one-in-four ratio (41 Hus1+/+/86 Hus1+/neo/49 Hus1neo/neo). Likewise, approximately one-quarter of the offspring from crosses between Hus1+/neo and Hus1+/Δ1 mice were of the Hus1neo/Δ1 genotype (61 Hus1+/+/69 Hus1+/neo/74 Hus1+/Δ1/65 Hus1neo/Δ1). Hus1neo/neo and Hus1neo/Δ1 mice were of normal size and appeared indistinguishable from control littermates. Litters of typical size were produced from three independent matings of adult Hus1neo/Δ1 mice, indicating that mice of this genotype were fertile.
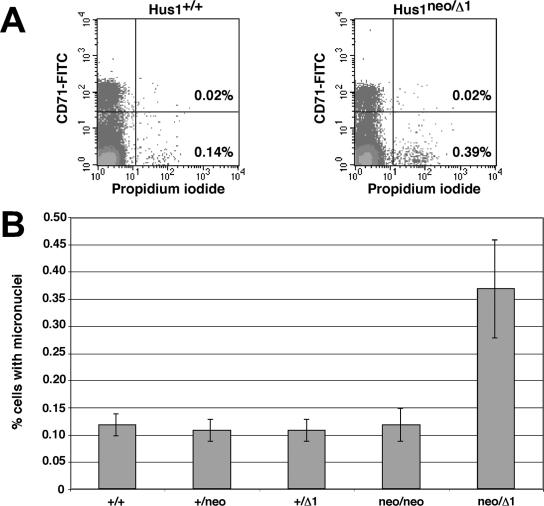
To test whether reduced Hus1 expression resulted in genomic instability in vivo, we quantitated the frequency of micronucleus formation in peripheral blood cells from mice of the allelic series. Micronuclei arise from chromosomes that mis-segregate during mitosis or from acentric fragments that are not incorporated into the nucleus (22). Micronucleus formation is a proven indicator of genomic instability and can be induced by genotoxin exposure or by genetic defects, such as mutation of the Atm checkpoint gene (47). A flow cytometric assay has been developed to quantify micronucleus formation in peripheral blood cells, based on the fact that maturing reticulocytes expel their main nucleus but not micronuclei (15). Peripheral blood was drawn from mice of the allelic series and stained with anti-CD71 antibody, to distinguish reticulocytes from mature normochromatic erythrocytes, and with propidium iodide, to identify cells with micronuclei (Fig. 6A). In peripheral blood from Hus1+/+ mice, the fraction of erythrocytes with micronuclei was 0.12% ± 0.02% on average (Fig. 6B). A similarly low level of micronucleus formation was observed for Hus1+/neo, Hus1+/Δ1, and Hus1neo/neo mice. Notably, a significantly increased fraction of erythrocytes with micronuclei (0.37% ± 0.09%) was observed in peripheral blood from Hus1neo/Δ1 mice. For clarity, results are shown only for female mice because micronucleus formation is generally lower in female mice than in male mice. A similar proportional increase in micronucleus formation in Hus1neo/Δ1 mice also was observed in male animals (data not shown). These data indicate that reduced Hus1 expression results in genomic instability in vivo.
FIG. 6.
Increased micronucleus formation in peripheral blood cells from mice with reduced Hus1 expression. (A) Representative FACS plots. Peripheral blood was drawn from mice of the indicated genotypes, stained with anti-CD71 antibody and propidium iodide, and analyzed by flow cytometry. The lower right quadrant contains mature normochromatic erythrocytes harboring micronuclei. The upper right quadrant contains reticulocytes with micronuclei. (B) Summary of micronucleus formation in peripheral blood from mice of the Hus1 allelic series. The mean frequency of micronucleus-containing erythrocytes is graphed for Hus1+/+ (n = 5), Hus1+/neo (n = 6), Hus1+/Δ1 (n = 6), Hus1neo/neo (n = 3), or Hus1neo/Δ1 (n = 10) female mice, with error bars representing the standard deviation.
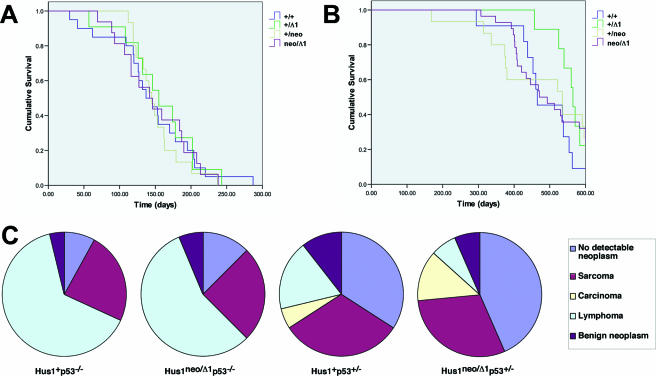
To determine whether increased genomic instability would induce tumor development in mice with a partial Hus1 impairment, we aged and monitored mice of all genotypes in the allelic series for 14 to 22 months. Overall survival was similar for all genotypes, and no increase in spontaneous tumor development was observed for mice with reduced Hus1 expression. We therefore tested whether partial Hus1 inactivation would promote tumorigenesis in combination with a defined oncogenic stimulus. For this purpose, we produced the Hus1 allelic series in p53−/− and p53+/− genetic backgrounds and examined whether the kinetics of tumor development or the spectrum of tumor types in p53-deficient mice would be affected by a partial Hus1 defect. Consistent with published results (16, 17, 25), Hus1+/+p53−/− mice rapidly lost viability due to tumor development, demonstrating a median survival of 146 days. Reduced Hus1 gene dosage did not alter the kinetics of tumor development in p53−/− mice, with median survival times of 152, 157, and 149 days observed for Hus1+/neop53−/−, Hus1+/Δ1p53−/−, and Hus1neo/Δ1p53−/− mice, respectively (Fig. 7A) (P = 0.951 [log rank test]). Partial Hus1 impairment also did not affect the spectrum of tumors arising in p53−/− mice (Fig. 7C) (P = 0.913 [chi-square test]). Regardless of Hus1 genotype, the majority of p53-null mice developed lymphomas, with most remaining mice developing sarcomas.
FIG. 7.
Partial Hus1 impairment does not accelerate tumorigenesis or alter the tumor spectrum in p53+/− or p53−/− mice. Survival curves for mice of the indicated Hus1 genotypes in p53−/− (A) or p53+/− (B) backgrounds. The following number of animals was analyzed for each genotype: Hus1+/+p53−/−, 20; Hus1+/neop53−/−, 15; Hus1+/Δ1p53−/−, 11; Hus1neo/Δ1p53−/−, 16; Hus1+/+p53+/−, 11; Hus1+/neop53+/−, 15; Hus1+/Δ1p53+/−, 9; Hus1neo/Δ1p53+/−, 28. (C) Pie charts showing the tumor spectrum for the mice described above at the conclusion of the experiment. Hus1+p53−/− and Hus1+p53+/− charts include the combined results for Hus1+/+, Hus1+/neo, and Hus1+/Δ1 mice. Hematoxylin- and eosin-stained tissue sections prepared from necropsied mice were analyzed blind with respect to genotype, and tumors were categorized as sarcoma, carcinoma, lymphoma and other hematopoietic neoplasms, or benign neoplasms. “No detectable neoplasm” refers to mice that remained healthy at the completion of the experiment or for which no neoplasm was identified at necropsy.
Because the strong tumor predisposition in p53−/− mice might mask the impact of partial Hus1 impairment, we also generated the Hus1 allelic series in a p53+/− background. Relative to p53−/− animals, p53+/− mice develop tumors with significantly delayed kinetics and typically display a more diverse tumor spectrum (16, 17, 25). Similar to the results with p53−/− mice, reduced Hus1 gene dosage did not significantly alter tumor development in p53+/− mice (Fig. 7B). The kinetics of tumor development were actually slightly delayed in mice with reduced Hus1 expression (median survival of 490, 504, 571, and 508 days for Hus1+/+p53−/−, Hus1+/neop53−/−, Hus1+/Δ1p53−/−, and Hus1neo/Δ1p53−/− mice, respectively), but these differences were not statistically significant (P = 0.531 [log rank test]). Likewise, the distribution of tumor types in p53+/− mice did not vary significantly by Hus1 genotype (P = 0.456 [chi-square test]). Taken together, the analysis of mice with reduced Hus1 expression suggests that partial Hus1 impairment, alone or in combination with targeted p53 inactivation, does not promote tumor development.
DISCUSSION
A full understanding of the physiological functions of the mammalian cell cycle checkpoint pathway involving Atr, Chk1, and the 9-1-1 complex has been elusive in part because complete inactivation of Hus1 or other components of this pathway in mice causes embryonic lethality. In this report, we describe a system for assessing the functional consequences of partial impairment of the Hus1-dependent cell cycle checkpoint pathway. This new mouse model centers on a hypomorphic allele, Hus1neo, that expresses wild-type Hus1 transcripts at a reduced level as well as aberrant transcripts with minimal coding capacity. A Hus1 allelic series was generated by combining this and other Hus1 alleles. Analysis of the allelic series in cells and mice indicated that Hus1 gene dosage has a significant impact on genomic integrity during normal growth conditions and in response to DNA damage and further identified an essential role for Hus1 in the maintenance of genomic stability in adult mice. Interestingly, however, mice with reduced Hus1 expression developed normally and were not tumor prone.
Hus1neo/Δ1 MEFs underwent premature senescence, whereas cells with a slightly greater level of Hus1 expression (Hus1neo/neo) were capable of normal proliferation. The senescence of cultured mouse cells is believed to reflect a DNA damage response to stressful culture conditions and to be independent of telomere shortening (24). That Hus1neo/Δ1 cells underwent premature senescence in culture while mice of the same genotype were born at the expected frequency suggests that some aspect of in vitro culture creates a requirement for an intact checkpoint apparatus. The MEFs described here were cultured under atmospheric oxygen levels (∼20% oxygen), and others have found that this high oxygen level causes DNA damage that contributes to the senescence of wild-type MEFs (38). During embryonic development, on the other hand, Hus1neo/Δ1 cells experience lower, physiological oxygen levels and under these conditions the level of Hus1 expressed from Hus1neo is sufficient for apparently normal proliferation and differentiation. Thus, the partial Hus1 impairment in Hus1neo/Δ1 MEFs might trigger premature senescence by sensitizing cells to oxidative stress. Consistent with this possibility, treatment of Hus1neo/Δ1 cultures with the antioxidant NAC largely suppressed the premature senescence phenotype. The improved proliferation of Hus1neo/Δ1 cells following antioxidant treatment was associated with a significant reduction in the frequency of spontaneous chromosomal abnormalities (data not shown). It is worth noting that the 9-1-1 checkpoint complex associates with a number of proteins required for base excision repair, the DNA repair pathway that mediates many of the responses to oxidative DNA damage (10, 20, 48, 55, 58, 59). These findings suggest that an essential function of the 9-1-1 complex during an unperturbed cell cycle is responding to spontaneous oxidative DNA lesions.
Analysis of the Hus1 allelic series also revealed how incremental reductions in Hus1 expression affect responses to extrinsic genotoxins. Hus1neo/Δ1 MEFs were found to be hypersensitive to both DNA damaging agents and replication inhibitors. An intermediate level of genotoxin sensitivity was observed for Hus1neo/neo MEFs, even though untreated cells of this genotype were fully capable of normal proliferation. This suggests that the level of Hus1 in Hus1neo/neo MEFs is sufficient for the response to intrinsic cellular stresses but is sublimiting upon additional genome damage. Importantly, increasing Hus1 expression via a Hus1-expressing retrovirus restored normal genotoxin sensitivity to both Hus1neo/neo and Hus1neo/Δ1 MEFs (data not shown). Consistent with our previous results (64), the sensitivity of cells with reduced Hus1 expression to the DNA adducting agent BPDE roughly correlated with the functioning of an intra-S cell cycle checkpoint mechanism that represses DNA synthesis following DNA damage. Sensitivity to the replication inhibitor aphidicolin was associated with increased DSB formation, as indicated by increased H2AX phosphorylation, as well as increased formation of chromosomal abnormalities. These findings are in accord with current models suggesting that replication stress in checkpoint-defective cells results in replication fork collapse (5). Interestingly, while Hus1+/+ cells showed a slight increase in chromosomal gaps and breaks following low-dose aphidicolin treatment, Hus1neo/neo and Hus1neo/Δ1 cells accumulated not only gaps and breaks but also chromatid interchanges at high frequency. This phenotype is somewhat reminiscent of that observed for Fanconi anemia cells following treatment with mitomycin C, which also involves high-frequency radial chromosome formation (13). Genotoxin-induced chromatid interchanges are believed to reflect a failure of DNA repair by homologous recombination and increased use of error-prone pathways such as nonhomologous end joining and single-strand annealing. A similar shift in repair pathway usage might account for the occurrence of certain chromosome aberrations in cells with reduced Hus1 expression, as mammalian Hus1, Rad9, Rad17, and Chk1 proteins are required for homologous recombinational repair (7, 37, 49, 61) and the yeast 9-1-1 complex has been implicated in translesion DNA synthesis (26, 42).
Consistent with the observed increase in chromosomal abnormalities in Hus1neo/Δ1 MEFs, peripheral blood cells from Hus1neo/Δ1 mice showed increased micronucleus formation. It is important to note that the micronucleus frequency measurements gauge genomic instability specifically in red blood cell precursors, and the extent of genomic instability in other tissues of Hus1neo/Δ1 mice remains unknown. Nevertheless, despite this indicator of genomic instability, Hus1neo/Δ1 mice were not predisposed to spontaneous tumor development. Furthermore, reduced Hus1 expression did not affect the kinetics of tumor development in p53+/− or p53−/− mice or significantly change the spectrum of tumors arising in these animals. Several other mouse models featuring defects in cell cycle checkpoints and/or DNA repair similarly do not show increased cancer incidence (1, 13, 47, 52, 66). These data contribute to the emerging picture that a particular type and level of genomic instability may be critical to tumor development (9). In the case of Hus1neo/Δ1 mice, the frequency of cancer-inducing chromosomal abnormalities may be too low or, alternatively, genetically unstable preneoplastic cells with reduced Hus1 expression may be incapable of extensive proliferation, as suggested by their propensity to undergo premature senescence in culture. One interesting possibility is that the low level of genome damage in Hus1neo/Δ1 mice could accumulate over multiple generations. We have interbred Hus1neo/Δ1 mice for over five generations and continue to obtain viable, fertile Hus1neo/Δ1 offspring. Whether these animals will develop late-onset phenotypes remains to be determined.
While cell cycle checkpoints in general are key tumor suppressor mechanisms (3, 21, 27), the available evidence suggests that defects in the Atr-dependent checkpoint mechanism may not strongly promote tumorigenesis. In mammals, the components of this pathway are essential, and their complete inactivation may not be compatible with tumor cell proliferation. Chk1+/− mice are not prone to spontaneous tumor development (31), and only a slight tumor predisposition is seen for Atr+/− mice (6). Consistent with these animal studies, there are only limited reports of ATR and CHK1 mutations in human cancers (32, 56) and it is uncertain whether these mutations are causative. Seckel syndrome, caused by a hypomorphic ATR mutation, is primarily a developmental disorder that has not been associated with increased tumor predisposition, although relatively few patients have been analyzed (35). However, heterozygosity for Chk1 does accelerate Wnt1-induced mammary tumorigenesis to a limited extent (31), while Atr heterozygosity increases tumor frequency in mismatch repair-defective mice (18). Thus, it may be that only very specific hypomorphic mutations affecting this pathway, in particular genetic or environmental contexts, will stimulate oncogenesis. Further analysis of the mice described here should provide additional insights into possible tumor suppressor functions for Hus1 and further clarify the impact of checkpoint defects and genomic instability on cancer initiation and progression.
Acknowledgments
We thank Naoko Shima for advice on micronucleus formation assays; Francoise Vermeylen, of the Cornell Statistical Consulting Unit, for advice on statistical analyses; Young Lu and Rinti Mukherjee for assistance with data processing; Fei Sun for performing preliminary antioxidant treatment experiments; the staff of the Cornell Center for Animal Resources and Education (CARE) and Lab Animal Services for excellent animal care; and Cyrus Vaziri, Tom Wolkow, and members of the Weiss lab for helpful discussion and comments on the manuscript.
This work was supported by NIH grant R01 CA108773 (R.S.W.). A.C. was supported in part through the Cornell University Veterinary Investigator Program, and J.D. was a participant in the Lansing High School Authentic Scientific Research Program.
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Baker, D. J., K. B. Jeganathan, J. D. Cameron, M. Thompson, S. Juneja, A. Kopecka, R. Kumar, R. B. Jenkins, P. C. de Groen, P. Roche, and J. M. van Deursen. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744-749. [DOI] [PubMed] [Google Scholar]
- 2.Bao, S., T. Lu, X. Wang, H. Zheng, L. E. Wang, Q. Wei, W. N. Hittelman, and L. Li. 2004. Disruption of the Rad9/Rad1/Hus1 (9-1-1) complex leads to checkpoint signaling and replication defects. Oncogene 23:5586-5593. [DOI] [PubMed] [Google Scholar]
- 3.Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864-870. [DOI] [PubMed] [Google Scholar]
- 4.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 5.Branzei, D., and M. Foiani. 2005. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17:568-575. [DOI] [PubMed] [Google Scholar]
- 6.Brown, E. J., and D. Baltimore. 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14:397-402. [PMC free article] [PubMed] [Google Scholar]
- 7.Budzowska, M., I. Jaspers, J. Essers, H. De Waard, E. Van Drunen, K. Hanada, B. Beverloo, R. W. Hendriks, A. De Klein, R. Kanaar, J. H. Hoeijmakers, and A. Maas. 2004. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 23:3548-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burtelow, M. A., S. H. Kaufmann, and L. M. Karnitz. 2000. Retention of the human Rad9 checkpoint complex in extraction-resistant nuclear complexes after DNA damage. J. Biol. Chem. 275:26343-26348. [DOI] [PubMed] [Google Scholar]
- 9.Cahill, D. P., K. W. Kinzler, B. Vogelstein, and C. Lengauer. 1999. Genetic instability and Darwinian selection in tumours. Trends Cell Biol. 9:M57-60. [PubMed] [Google Scholar]
- 10.Chang, D. Y., and A. L. Lu. 2005. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 280:408-417. [DOI] [PubMed] [Google Scholar]
- 11.Cimprich, K. A. 2003. Fragile sites: breaking up over a slowdown. Curr. Biol. 13:R231-R233. [DOI] [PubMed] [Google Scholar]
- 12.Cortez, D. 2005. Unwind and slow down: checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 19:1007-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Andrea, A. D. 2003. The Fanconi road to cancer. Genes Dev. 17:1933-1936. [DOI] [PubMed] [Google Scholar]
- 14.de Klein, A., M. Muijtjens, R. van Os, Y. Verhoeven, B. Smit, A. M. Carr, A. R. Lehmann, and J. H. Hoeijmakers. 2000. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr. Biol. 10:479-482. [DOI] [PubMed] [Google Scholar]
- 15.Dertinger, S. D., D. K. Torous, and K. R. Tometsko. 1996. Simple and reliable enumeration of micronucleated reticulocytes with a single-laser flow cytometer. Mutat. Res. 371:283-292. [DOI] [PubMed] [Google Scholar]
- 16.Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. J. Montgomery, J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215-221. [DOI] [PubMed] [Google Scholar]
- 17.Donehower, L. A., M. Harvey, H. Vogel, M. J. McArthur, C. A. Montgomery, Jr., S. H. Park, T. Thompson, R. J. Ford, and A. Bradley. 1995. Effects of genetic background on tumorigenesis in p53-deficient mice. Mol. Carcinog. 14:16-22. [DOI] [PubMed] [Google Scholar]
- 18.Fang, Y., C. C. Tsao, B. K. Goodman, R. Furumai, C. A. Tirado, R. T. Abraham, and X. F. Wang. 2004. ATR functions as a gene dosage-dependent tumor suppressor on a mismatch repair-deficient background. EMBO J. 23:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francia, S., R. S. Weiss, M. P. Hande, R. Freire, and F. d'Adda di Fagagna. 2006. Telomere and telomerase modulation by the mammalian Rad9/Rad1/Hus1 DNA-damage-checkpoint complex. Curr. Biol. 16:1551-1558. [DOI] [PubMed] [Google Scholar]
- 20.Friedrich-Heineken, E., M. Toueille, B. Tannler, C. Burki, E. Ferrari, M. O. Hottiger, and U. Hubscher. 2005. The two DNA clamps Rad9/Rad1/Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J. Mol. Biol. 353:980-989. [DOI] [PubMed] [Google Scholar]
- 21.Gorgoulis, V. G., L. V. Vassiliou, P. Karakaidos, P. Zacharatos, A. Kotsinas, T. Liloglou, M. Venere, R. A. Ditullio, Jr., N. G. Kastrinakis, B. Levy, D. Kletsas, A. Yoneta, M. Herlyn, C. Kittas, and T. D. Halazonetis. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907-913. [DOI] [PubMed] [Google Scholar]
- 22.Heddle, J. A., M. C. Cimino, M. Hayashi, F. Romagna, M. D. Shelby, J. D. Tucker, P. Vanparys, and J. T. MacGregor. 1991. Micronuclei as an index of cytogenetic damage: past, present, and future. Environ. Mol. Mutagen. 18:277-291. [DOI] [PubMed] [Google Scholar]
- 23.Hopkins, K. M., W. Auerbach, X. Y. Wang, M. P. Hande, H. Hang, D. J. Wolgemuth, A. L. Joyner, and H. B. Lieberman. 2004. Deletion of mouse Rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol. Cell. Biol. 24:7235-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itahana, K., J. Campisi, and G. P. Dimri. 2004. Mechanisms of cellular senescence in human and mouse cells. Biogerontology 5:1-10. [DOI] [PubMed] [Google Scholar]
- 25.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 26.Kai, M., and T. S. Wang. 2003. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 17:64-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kastan, M. B., and J. Bartek. 2004. Cell-cycle checkpoints and cancer. Nature 432:316-323. [DOI] [PubMed] [Google Scholar]
- 28.Kinzel, B., J. Hall, F. Natt, J. Weiler, and D. Cohen. 2002. Downregulation of Hus1 by antisense oligonucleotides enhances the sensitivity of human lung carcinoma cells to cisplatin. Cancer 94:1808-1814. [DOI] [PubMed] [Google Scholar]
- 29.Kwan, K. M. 2002. Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genesis 32:49-62. [DOI] [PubMed] [Google Scholar]
- 30.Levitt, P. S., H. Liu, C. Manning, and R. S. Weiss. 2005. Conditional inactivation of the mouse Hus1 cell cycle checkpoint gene. Genomics 86:212-224. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Q., S. Guntuku, X. S. Cui, S. Matsuoka, D. Cortez, K. Tamai, G. Luo, S. Carattini-Rivera, F. DeMayo, A. Bradley, L. A. Donehower, and S. J. Elledge. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14:1448-1459. [PMC free article] [PubMed] [Google Scholar]
- 32.Menoyo, A., H. Alazzouzi, E. Espin, M. Armengol, H. Yamamoto, and S. Schwartz, Jr. 2001. Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 61:7727-7730. [PubMed] [Google Scholar]
- 33.Meyers, E. N., M. Lewandoski, and G. R. Martin. 1998. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet. 18:136-141. [DOI] [PubMed] [Google Scholar]
- 34.Mitelman, F. (ed.). 1995. ISCN (1995): an international system for human cytogenetic nomenclature. S. Karger, Basel, Switzerland.
- 35.O'Driscoll, M., A. R. Gennery, J. Seidel, P. Concannon, and P. A. Jeggo. 2004. An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair 3:1227-1235. [DOI] [PubMed] [Google Scholar]
- 36.O'Driscoll, M., V. L. Ruiz-Perez, C. G. Woods, P. A. Jeggo, and J. A. Goodship. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 33:497-501. [DOI] [PubMed] [Google Scholar]
- 37.Pandita, R. K., G. G. Sharma, A. Laszlo, K. M. Hopkins, S. Davey, M. Chakhparonian, A. Gupta, R. J. Wellinger, J. Zhang, S. N. Powell, J. L. Roti Roti, H. B. Lieberman, and T. K. Pandita. 2006. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol. Cell. Biol. 26:1850-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parrinello, S., E. Samper, A. Krtolica, J. Goldstein, S. Melov, and J. Campisi. 2003. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reinholdt, L., T. Ashley, J. Schimenti, and N. Shima. 2004. Forward genetic screens for meiotic and mitotic recombination-defective mutants in mice. Methods Mol. Biol. 262:87-107. [DOI] [PubMed] [Google Scholar]
- 40.Roos-Mattjus, P., K. M. Hopkins, A. J. Oestreich, B. T. Vroman, K. L. Johnson, S. Naylor, H. B. Lieberman, and L. M. Karnitz. 2003. Phosphorylation of human Rad9 is required for genotoxin-activated checkpoint signaling. J. Biol. Chem. 278:24428-24437. [DOI] [PubMed] [Google Scholar]
- 41.Roos-Mattjus, P., B. T. Vroman, M. A. Burtelow, M. Rauen, A. K. Eapen, and L. M. Karnitz. 2002. Genotoxin-induced Rad9-Hus1-Rad1 (9-1-1) chromatin association is an early checkpoint signaling event. J. Biol. Chem. 277:43809-43812. [DOI] [PubMed] [Google Scholar]
- 42.Sabbioneda, S., B. K. Minesinger, M. Giannattasio, P. Plevani, M. Muzi-Falconi, and S. Jinks-Robertson. 2005. The 9-1-1 checkpoint clamp physically interacts with polζ and is partially required for spontaneous polζ-dependent mutagenesis in Saccharomyces cerevisiae. J. Biol. Chem. 280:38657-38665. [DOI] [PubMed] [Google Scholar]
- 43.Savage, J. R. 1976. Classification and relationships of induced chromosomal structural changes. J. Med. Genet. 13:103-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shechter, D., V. Costanzo, and J. Gautier. 2004. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair 3:901-908. [DOI] [PubMed] [Google Scholar]
- 45.Shechter, D., and J. Gautier. 2005. ATM and ATR check in on origins: a dynamic model for origin selection and activation. Cell Cycle 4:e74-e77. [PubMed] [Google Scholar]
- 46.Shi, G., D. Y. Chang, C. C. Cheng, X. Guan, C. Venclovas, and A. L. Lu. 2006. Physical and functional interactions between MutY homolog (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem. J. 400:53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shima, N., R. J. Munroe, and J. C. Schimenti. 2004. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol. Cell. Biol. 24:10381-10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smirnova, E., M. Toueille, E. Markkanen, and U. Hubscher. 2005. The human checkpoint sensor and alternative DNA clamp Rad9-Rad1-Hus1 modulates the activity of DNA ligase I, a component of the long-patch base excision repair machinery. Biochem. J. 389:13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorensen, C. S., L. T. Hansen, J. Dziegielewski, R. G. Syljuasen, C. Lundin, J. Bartek, and T. Helleday. 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 7:195-201. [DOI] [PubMed] [Google Scholar]
- 50.Sorensen, C. S., R. G. Syljuasen, J. Lukas, and J. Bartek. 2004. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle 3:941-945. [PubMed] [Google Scholar]
- 51.Takai, H., K. Tominaga, N. Motoyama, Y. A. Minamishima, H. Nagahama, T. Tsukiyama, K. Ikeda, K. Nakayama, M. Nakanishi, and K. Nakayama. 2000. Aberrant cell cycle checkpoint function and early embryonic death in Chk1−/− mice. Genes Dev. 14:1439-1447. [PMC free article] [PubMed] [Google Scholar]
- 52.Theunissen, J. W., M. I. Kaplan, P. A. Hunt, B. R. Williams, D. O. Ferguson, F. W. Alt, and J. H. Petrini. 2003. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol. Cell 12:1511-1523. [DOI] [PubMed] [Google Scholar]
- 53.Thiriet, C., and J. J. Hayes. 2005. Chromatin in need of a fix: phosphorylation of H2AX connects chromatin to DNA repair. Mol. Cell 18:617-622. [DOI] [PubMed] [Google Scholar]
- 54.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toueille, M., N. El-Andaloussi, I. Frouin, R. Freire, D. Funk, I. Shevelev, E. Friedrich-Heineken, G. Villani, M. O. Hottiger, and U. Hubscher. 2004. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 32:3316-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vassileva, V., A. Millar, L. Briollais, W. Chapman, and B. Bapat. 2002. Genes involved in DNA repair are mutational targets in endometrial cancers with microsatellite instability. Cancer Res. 62:4095-4099. [PubMed] [Google Scholar]
- 57.Venclovas, C., and M. P. Thelen. 2000. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 28:2481-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, W., P. Brandt, M. L. Rossi, L. Lindsey-Boltz, V. Podust, E. Fanning, A. Sancar, and R. A. Bambara. 2004. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl. Acad. Sci. USA 101:16762-16767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, W., L. A. Lindsey-Boltz, A. Sancar, and R. A. Bambara. 2006. Mechanism of stimulation of human DNA ligase I by the Rad9-Rad1-Hus1 checkpoint complex. J. Biol. Chem. 281:20865-20872. [DOI] [PubMed] [Google Scholar]
- 60.Wang, X., J. Guan, B. Hu, R. S. Weiss, G. Iliakis, and Y. Wang. 2004. Involvement of Hus1 in the chain elongation step of DNA replication after exposure to camptothecin or ionizing radiation. Nucleic Acids Res. 32:767-775. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Wang, X., B. Hu, R. S. Weiss, and Y. Wang. 2006. The effect of Hus1 on ionizing radiation sensitivity is associated with homologous recombination repair but is independent of nonhomologous end-joining. Oncogene 25:1980-1983. [DOI] [PubMed] [Google Scholar]
- 62.Ward, I. M., and J. Chen. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759-47762. [DOI] [PubMed] [Google Scholar]
- 63.Weiss, R. S., T. Enoch, and P. Leder. 2000. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 14:1886-1898. [PMC free article] [PubMed] [Google Scholar]
- 64.Weiss, R. S., P. Leder, and C. Vaziri. 2003. Critical role for mouse Hus1 in an S-phase DNA damage cell cycle checkpoint. Mol. Cell. Biol. 23:791-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiss, R. S., S. Matsuoka, S. J. Elledge, and P. Leder. 2002. Hus1 acts upstream of Chk1 in a mammalian DNA damage response pathway. Curr. Biol. 12:73-77. [DOI] [PubMed] [Google Scholar]
- 66.Williams, B. R., O. K. Mirzoeva, W. F. Morgan, J. Lin, W. Dunnick, and J. H. Petrini. 2002. A murine model of Nijmegen breakage syndrome. Curr. Biol. 12:648-653. [DOI] [PubMed] [Google Scholar]
- 67.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]