Abstract
Amplification of large chromosomal regions (gene amplification) is a common somatic alteration in human cancer cells and often is associated with advanced disease. A critical event initiating gene amplification is a DNA double-strand break (DSB), which is immediately followed by the formation of a large DNA palindrome. Large DNA palindromes are frequent and nonrandomly distributed in the genomes of cancer cells and facilitate a further increase in copy number. Although the importance of the formation of large DNA palindromes as a very early event in gene amplification is widely recognized, it is not known how a DSB is resolved to form a large DNA palindrome and whether any local DNA structure determines the location of large DNA palindromes. We show here that intrastrand annealing following a DNA double-strand break leads to the formation of large DNA palindromes and that DNA inverted repeats in the genome determine the efficiency of this event. Furthermore, in human Colo320DM cancer cells, a DNA inverted repeat in the genome marks the border between amplified and nonamplified DNA. Therefore, an early step of gene amplification is a regulated process that is facilitated by DNA inverted repeats in the genome.
Structural aberrations of chromosomes are a hallmark of cancer and result in either a gain or a loss of chromosomal regions (1, 15, 21). Studies of gene amplification in cancer cells often focus on the increased dosage of cellular oncogenes and its association with advanced stages of diseases, whereas in most cases the “gene” amplification involves large chromosomal regions that span several megabases of DNA (32, 38). Recent advances in the technology for genomewide copy-number surveys have identified a number of chromosomal regions in different cancer types as candidates for loci driving tumor progression. However, the molecular mechanisms and DNA contexts that initiate regional amplification and establish boundaries of amplicons remain largely unknown.
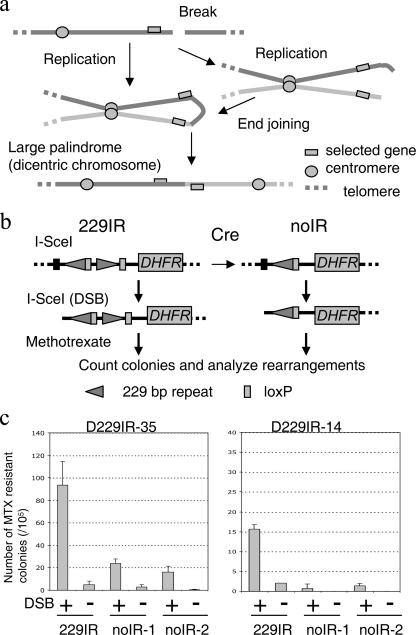
Mammalian cells treated with either radiation, DNA replication inhibitors, or other DNA-damaging agents show increased frequencies of gene amplification, suggesting chromosome breaks as initiating lesions (4, 12, 24, 39). In fact, either a chromosome break at a fragile site or a site-specific chromosomal DNA double-strand break (DSB) induced by I-SceI endonuclease leads to gene amplification by initiating the formation of large palindromic chromosomes (9, 37). Large palindromic dicentric chromosomes generate further chromosome breaks during chromosome segregation (see Fig. 1a). These breaks are again resolved into large palindromic chromosomes (breakage-fusion-bridge [BFB] cycle), establishing intrachromosomal gene amplification (9, 19, 29, 40). Thus, the formation of large palindromes occurs at a very early stage of gene amplification as a consequence of illegitimate repair of DSBs. How a DSB is processed to form a large palindrome remains unknown. A potential mechanism includes the joining of broken DNA ends of sister chromatids by nonhomologous end joining (NHEJ) (see Fig. 1a, right), a mechanism of chromosome end fusion in critically short telomeres (30). However, palindromic gene amplification still occurs in mice deficient in NHEJ (44), suggesting another, yet-unknown mechanism for large DNA palindrome formation in mammalian cells (see Fig. 1a, left).
FIG. 1.
DNA double-strand break and palindrome formation. a. A chromosome break is resolved into a large DNA palindrome and initiates gene amplification (model). A chromosome break leads to a large DNA palindrome (a dicentric chromosome) either by NHEJ of broken sister chromatids (right) or a mechanism independent of NHEJ (left). Subsequent chromosome segregation generates another chromosome break that can be further resolved into a large DNA palindrome (breakage-fusion-bridge cycle). Solid lines represent chromosomes. b. A system for studying palindrome formation in the presence or absence of DNA IR at the same integration site. A Cre-loxP-mediated system generated transformants with or without 229IR at the same genomic locus. Both 229IR and noIR transformants were transfected with the I-SceI expression vector to induce a chromosomal DSB at the I-SceI site, and clones with elevated DHFR activity were selected with MTX. c. The number of MTX-resistant colonies (per 105 cells plated) with an induction of DSB (+) or without an induction of DSB (−). The numbers shown here are the averages from three independent experiments. Error bars indicate standard deviations. For both original 229IR transformants (229IR), two independent noIR subtransformants were used for this experiment.
Gene amplification in simple eukaryotes also involves the formation of large palindromes at a very early stage. In Tetrahymena, amplification of the rRNA (rDNA) gene is initiated by the formation of large palindromic chromosomes with the developmentally regulated induction of a site-specific DSB next to a short DNA inverted repeat (DNA IR) in the genome (42). DSBs initiate intramolecular recombination (intrastrand annealing) at the site of the short DNA IR, creating a large hairpin molecule prior to DNA replication. This hairpin molecule is resolved into a large palindrome following DNA replication, with the original DNA IR at a boundary between amplified and nonamplified DNA (6, 43). A short DNA IR is required for this process in Tetrahymena, as well as in Saccharomyces cerevisiae (5). Thus, a DNA IR in the genome is a critical cis-acting element for the formation of large palindromes by an intramolecular recombination pathway and sets a boundary of the amplicons in lower eukaryotes.
Large palindromes are widespread in cancer cells and provide a platform for subsequent gene amplification (36). In a prior study, we showed that a short DNA IR adjacent to a DSB could catalyze the formation of a large palindrome and subsequent gene amplification in mammalian cells (37). Thus, gene amplification in cancer cells might proceed through the same intramolecular pathway as the developmentally regulated gene amplification in single-cell eukaryotes. To test this emerging model of mammalian gene amplification, it is necessary to show the following: (i) that a DNA IR in the genome facilitates large palindrome formation through a process consistent with intrastrand annealing of broken ends and (ii) that a DNA IR in the genome marks the boundary between amplified and nonamplified DNA in spontaneous palindrome-associated gene amplification in human cancer cells.
We have now developed a system for measuring the efficiency of large palindrome formation after a DSB in the presence or absence of a short DNA IR at the same integration site in mammalian cells. Analysis of multiple amplicons at very early stages of amplification indicates that large palindrome formation after DSB is mediated by intrastrand annealing and that a DNA IR next to a DSB significantly facilitates this process. We further determine that in human colon cancer cells, the center of the large palindrome sets the boundary between amplified and nonamplified DNA, and a 26-kb DNA IR preexisting in the human genome marks the region of recombination at the center of the palindrome. Therefore, a newly discovered mechanism of mammalian gene amplification occurs through a process similar to the intramolecular recombination used by simple eukaryotes, resulting in an amplicon bordered by a DNA IR in the genome.
MATERIALS AND METHODS
Constructs.
To construct pD229IRScelox2, primer pairs, either XbaIIR and XholoxIR or XhoCenPt and PstloxIR, were used to amplify each one of the two parts of the 229-base-pair inverted repeat (229IR) with pD229IRSce as a template (37). PCR products were cloned into pCRII-TOPO (Invitrogen) using the TOPO TA cloning kit to generate pTOPOXbaIR and pTOPOPstIR. An XbaI-XhoI fragment of pTOPOXbaIR was ligated into XbaI and XhoI sites of pTOPOPstIR to generate pTOPO229IRlox2. An XbaI-PstI fragment of pTOPO229IRlox2 replaced an XbaI-PstI fragment of pD79IRSce to generate pD229IRlox2Sce. DNA sequences of the PCR primers are available upon request.
Cell cultures, transfections, and DNA analysis.
Dihydrofolate reductase (DHFR)-deficient CHO cells were transformed with pD229IRlox2Sce as described previously (37). Single-copy transformants were determined by Southern analysis and were subjected to transfection with 5 μg of pCMV3xnls-I-SceI (16) for DSB induction using Superfect (QIAGEN, Chatsworth, CA). As a control, pCMVnoSce, a vector without the I-SceI coding sequence, was transfected. Forty-eight hours after transfection, 1 × 105 cells were plated in the medium with 0.2 μM methotrexate (MTX). Resistant colonies were scored after 10 to 12 days. Colonies were picked and expanded in 24-well plates for further genomic analysis. Genomic DNA extraction and Southern analysis were done as described previously (37). Briefly, 2 μg of high-molecular-weight genomic DNA was digested with a restriction enzyme, run on either a 0.8% or a 0.6% agarose gel, and blotted to a nylon membrane. Snap-back DNA was prepared as follows: 2 μg of genomic DNA in 50 μl water with 100 mM NaCl was boiled for 7 min and immediately transferred on ice. DNA was precipitated by ethanol and digested with a restriction enzyme. A 2.5-kb molecular ruler (Bio-Rad), a 1-kb DNA ladder (New England Biolabs), or a 200-bp DNA stepladder (Promega) was used as a size marker. To generate a probe for Southern analysis, human genomic DNA was amplified by PCR, and the fragment was cloned by using the TOPO TA cloning kit (Invitrogen). For sequence analysis of the centers of palindromes, either bisulphite-treated DNA or genomic DNA from each MTX-resistant clone was amplified using nested PCR. Bisulfite treatment of genomic DNA was done following published procedures (23). DNA sequences of the PCR primers are available upon request.
GAPF.
The genome-wide analysis of palindrome formation (GAPF) procedure was performed as described previously (36) with modifications. After snap-back treatment, 6 μl of S1 nuclease buffer, 4 μl of 3 M NaCl, and 100 U of S1 nuclease (Invitrogen) were added to the DNA and incubated at 37°C for 1 h. S1 nuclease was inactivated by addition of 10 mM EDTA and phenol-chloroform extraction. DNA was precipitated with ethanol, dissolved in water, and digested with 40 U of MspI, TacI, or MseI (New England Biolab) for 16 h. DNA was precipitated, dissolved in 21 μl of water, and ligated to a MspI-, TacI- or MseI-specific linker by adding 5 μl of a 20 mM linker, 3 μl of T4 DNA ligase buffer, and 400 U of T4 DNA ligase (New England Biolabs) at 16°C for 16 h. DNA was precipitated and dissolved in 200 μl Tris-EDTA, followed by application on a Microcon YM-50 spin column (Millipore) to remove excess linker. DNA was recovered in 20 μl H2O. Thus, for each Colo320DM and normal human foreskin fibroblast (HFF2) cell culture, templates with three different linkers were prepared. For PCR, 2 μl of DNA, 0.5 μl of Faststart Taq DNA polymerase (Roche), 2.5 μl of 2 mM deoxynucleoside triphosphate, 5 μl of 10× PCR buffer with MgCl2 (for Faststart Taq DNA polymerase; Roche), and 2 μM of a linker-specific primer were mixed with H2O for a total reaction volume of 50 μl. For MspI-linker-ligated DNA, 5 μl of GC-rich solution (for Faststart Taq DNA polymerase; Roche) was also included in the reaction mixture. PCR was performed at 96°C for 6 min followed by 25 cycles of 96°C for 30 s, 55°C for 30 s, and 72°C for 30 s on a GeneAmp PCR system 9700 thermocycler (Perkin-Elmer). PCRs using the same template with three different linker-specific primers were concentrated using a Microcon YM-50 spin column. DNA was recovered in 25 μl H2O and fragmented using DNase I (New England Biolabs). After heat inactivation of DNase I, DNA was labeled with biotin-11-dATP using terminal transferase (Roche). The procedure was performed in triplicate to produce three independent preparations of DNA for hybridization on GeneChip Human Genome U133A arrays (Affymetrix). After hybridization, the microarrays were washed and then scanned with GENECHIP software (Affymetrix, Santa Clara, CA). Statistical analysis was done as described previously (8). The perfect-match probe intensities were corrected by robust multichip analysis, normalized by quantile normalization, and summarized by Tukey's median polish procedure using the Affy package of Bioconductor. The comparison of GAPF profiles between Colo320DM and normal human foreskin fibroblasts was done using the LIMMA package of Bioconductor. The differentially expressed genes were determined to be significant according to a false discovery rate (FDR) (34) of <0.05.
Microarray data accession number.
All microarray data have been deposited in the Gene Expression Omnibase database at NCBI (accession no. GSE6274).
RESULTS
Significant increase of MTX-resistant clones in response to DSBs.
To determine whether the formation of large palindromes is facilitated by the presence of DNA IRs in mammalian cells, we created a system to measure the efficiency of large palindrome formation in the presence or absence of a 229IR at the same integration site. In the construct, pD229IRlox2Sce, one of the two parts of the repeat is flanked by a pair of loxP sites (Fig. 1b). This construct was randomly integrated into the genome of dhfr-deficient Chinese hamster ovary cells (CHO dhfr−/−). For each single-copy transformant (229IRlox2-35 and -14 229IR transformants), we generated two independent subtransformants in which one repeat element was excised by Cre recombinase (noIR subtransformants) (see Fig. S1 in the supplemental material). Transformants were transfected with either pCMV3xnlsI-SceI (16) for DSB induction at the I-SceI site or pCMVnoSce, a control vector without I-SceI coding sequence, followed by MTX selection (0.2 μM) for 10 to 12 days before the number of colonies was determined. Compared to the 229IR transformants, the numbers of MTX-resistant colonies induced after DSBs were significantly decreased for the noIR subtransformants, indicating that the 229IR promotes the generation of MTX-resistant colonies (Fig. 1c). The IR-mediated increase in colony formation was 3.9-fold for D229IRlox2-35 (93.6 ± 21.0 for 229IR and 24.0 ± 4.0 for noIR-1) and 12-fold for D229IRlox2-14 (15.7 ± 1.2 for 229IR and 1.3 ± 0.6 for noIR-2). It should be noted, however, that I-SceI-induced DSBs generated a significant increase in MTX-resistant colonies for both noIR subtransformants of D229IRlox2-35 (24.0 ± 4.0 with DSB and 3.0 ± 1.7 without DSB for noIR-1; 16.3 ± 4.9 with DSB and 0.3 ± 0.6 without DSB for noIR-2 [Fig. 1c, left]).
Intrastrand annealing for palindrome formation is facilitated by a 229IR.
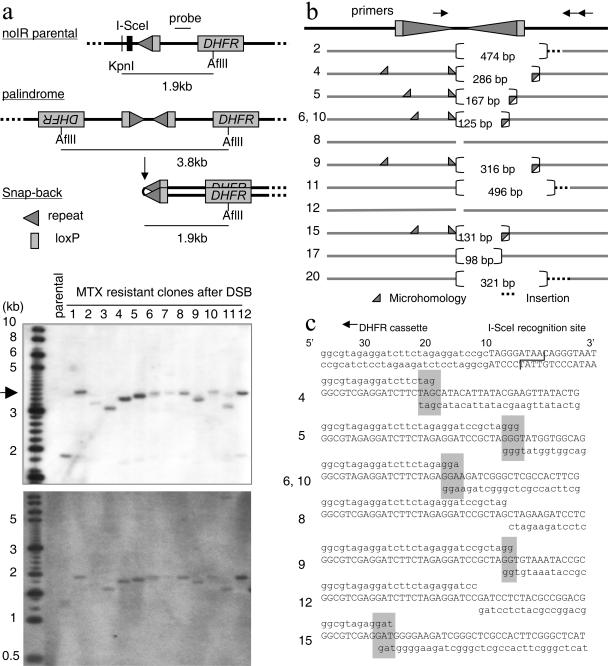
Southern analysis was used to analyze DSB-induced DNA rearrangements in MTX-resistant clones from the noIR subtransformants. In contrast to a 1.9-kb KpnI-AflII fragment in parental cells, MTX-resistant clones gave fragments of about 3.8 kb or a little smaller (Fig. 2a). The fragment in each MTX-resistant clone was converted to half-size after denaturing and rapid cooling (snap-back) (36), indicating that a DSB alone, without an adjacent DNA IR, is sufficient to generate large palindromes. Heterogeneous sizes of palindromic fragments suggest variable processing of I-SceI-induced DSBs. The DNA sequences at the centers of large palindromes hold a key to identifying molecular mechanisms. To determine the DNA sequences at the centers of palindromes, bisulphate modification of genomic DNA was applied (25, 37). Bisulphite treatment converted cytosines to uracils, which disrupted the palindromes to allow the centers of palindromes to be amplified by PCR for subsequent sequencing analysis. Two clones (8 and 12) had an almost perfect palindrome with very small deletions (less than 25 bp) from the I-SceI cutting site on both arms (Fig. 2b and c). This very small loss of sequences is similar to the end of an I-SceI-induced DSB repaired by NHEJ (16, 17). Thus, these large palindromes were probably generated by an intermolecular mechanism in which two sister chromatids became joined head to head by NHEJ. However, in the other clones, the sequences at the junction showed biased processing of DSB ends: one end of the palindrome arm had very small deletions (<25 bp), while the other end had deletions of more than 100 nucleotides from the I-SceI cutting site. To confirm the sequencing results obtained by bisulfite sequencing and further facilitate the analysis of the centers of palindromes, we developed a PCR assay using genomic DNA as templates. The centers of palindromes are difficult to amplify by regular PCR. We designed one PCR primer very close to the I-SceI cutting site with the other primer further downstream, which minimized palindromic sequence in the PCR products. In two clones, deletions on both arms are very small (5 and 14 bp in clone 8 and 11 and 20 bp in clone 12). In other clones, deletion on one arm is much larger, between 496 and 98 bp. There was microhomology of several nucleotides at the junction in five clones (clones 4, 5, 6, 9, and 15), and insertions of 1, 22, 64, or 372 bp were seen at the junction in the other four clones. In total, PCR products were obtained with 12 out of 24 clones, among which 10 clones showed biased processing of DSB ends. Biased processing of the ends of DSBs with the microhomology at the junction strongly suggests intrastrand annealing; one end of the DSB underwent a 5′-3′ resection of one strand, followed by intrastrand base pairing by microhomology and DNA replication to form large palindromes (Fig. 3b). Nucleotide insertions in four clones could have occurred by an addition of nucleotides to the free end, followed by a 5′-to-3′ resection (27) and intrastrand annealing.
FIG. 2.
Biased processing of the DSB ends at the junctions of palindromes without a short IR. a. Southern analysis for the genomic DNA (top) and snap-back genomic DNA (bottom) was performed using the probe indicated. A DSB at the I-SceI site and palindrome formation eliminated the KpnI site that was located upstream of the I-SceI site. If two DHFR cassettes are joined to generate a palindrome after I-SceI-induced DSB, a 3.8-kb AflII fragment is expected. Most of the MTX-resistant clones showed a fragment that is a little smaller than 3.8 kb (arrowhead), and palindromes in these fragments were confirmed by Southern analysis using snap-back DNA (bottom). A 1-kb DNA ladder (NEB) mixed with a 200-bp DNA stepladder (Promega) is shown as a size marker. b. Schematic drawing of the centers of palindromes from MTX-resistant clones. Solid gray bars represents DNA sequences present in the palindromes, and dotted bars represent insertions of foreign sequences at the junctions. Each numbers in brackets represents the number of nucleotides missing at the junction. Gray triangles indicate the sites of microhomology. The locations of primers used for PCR are shown on the top. c. Sequence analysis of the junctions of palindromes. A part of the I-SceI recognition sequence (actual cutting site indicated with a line) and surrounding sequence are shown at the top. The sequence on the top line is the end sequence of the left arm of a palindrome (aligned with the sequence shown on the top), sequence in the middle is the actual sequence at the junction, and sequence on the bottom line is the sequence of the right arm of the palindrome. Microhomology at the junctions is indicated by gray rectangles.
FIG. 3.
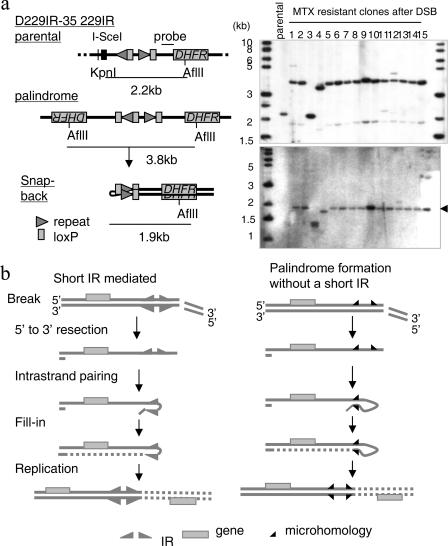
A 229-bp DNA IR facilitates palindrome formation a. Palindrome formation mediated by a 229IR. Southern analysis for the genomic DNA (right, top) and snap-back genomic DNA (right, bottom) was performed using the probe indicated (left). A DSB at the I-SceI site and palindrome formation eliminated the KpnI site that was located upstream of the I-SceI site and generated a 3.8-kb fragment in most of the clones. A 1.9-kb fragment of lower intensity was also seen, which might represent a half-sized fragment formed in vitro by intramolecular pairing of a large 3.8-kb palindrome (20, 36). Palindrome formation was confirmed by the snap-back procedure on genomic DNA, which generated a single 1.9-kb fragment (indicated by an arrowhead) in most of the clones. A 1-kb DNA ladder (NEB) is shown as a size marker. b. Intramolecular recombination model. After 5′-to-3′ resection of the DSB end, intrastrand pairing occurs either between short IR (left) (5, 6, 37) or bymicrohomology (right). This initially generates a hairpin molecule, and a large DNA palindrome forms after DNA replication. Microhomology serves as a very small version of IR.
In contrast, MTX-resistant clones from D229IR transformants showed homogeneous sizes of DNA fragments (Fig. 3a). A 2.2-kb KpnI-AflII fragment corresponding to the original construct was replaced with a 3.8-kb fragment. This 3.8-kb fragment was converted into a half-sized fragment (1.9-kb) after snap-back, indicating large palindromes nucleated by the 229IR. In summary, the majority of MTX-resistant clones (15/23 clones from 229IRlox2-35 and 8/14 clones from 229IRlox2-14) had a palindromes nucleated by the 229IR (Fig. 3a; also data not shown). The increased frequency of large palindrome formation in D229IR transformants strongly suggests that the length of homology determines the frequency by facilitating intrastrand annealing. The average copy number of the DHFR transgene in four MTX-resistant clones analyzed from 229IRlox2-35 remained low, between 3.3 and 4.7 (data not shown). The fact that we recovered clones that had a single palindromic fragment with low-copy-number amplification strongly supports the idea that large palindrome formation is an initial rearrangement triggered by the I-SceI-induced DSB. DNA sequence upstream of the I-SceI site was not amplified (data not shown), indicating that the 229IR formed the boundary of the amplicon.
A boundary of gene amplification in cancer cells is defined by the center of a palindrome.
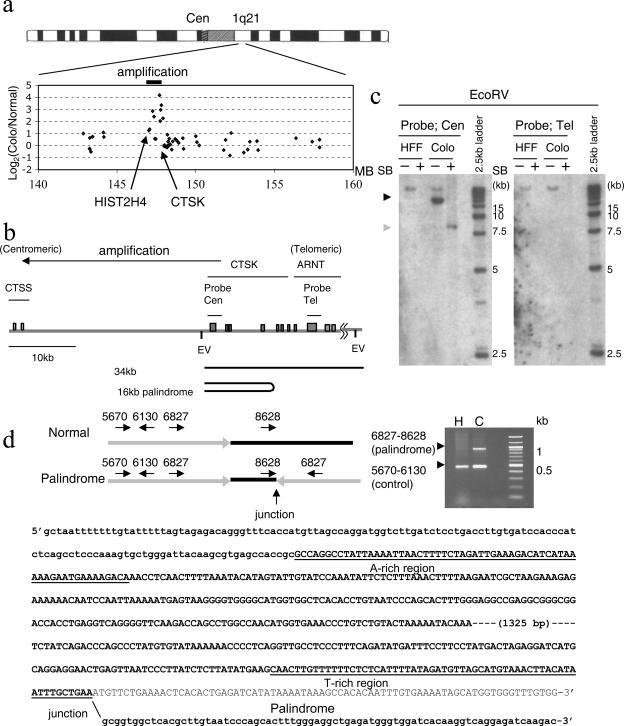
In order to investigate whether intrastrand annealing and recombination sets a boundary of amplicons in cancer cells, GAPF was used (22, 36). GAPF identifies regions of the genome at or near centers of DNA palindromes. Genomic DNA from Colo320DM and HFF2 (a human foreskin fibroblast cell strain) cells were processed to enrich for DNA palindromes and hybridized to the GeneChip Human Genome U133A array. Among the 42 features that were significantly increased in Colo320DM DNA (FDR [34] of <0.05) (see Table S1 in the supplemental material), four genes were located at 1q21, which is an amplified locusin Colo320DM DNA (13, 36) (Fig. 4a). CTSK was among the most significantly increased genes (FDR = 0.00889). Digesting Colo320DM DNA with EcoRV and hybridizing with a probe covering the CTSK gene (Fig. 4b) yielded an amplified 16-kb fragment that was distinct from the normal fragment of 34 kb, indicating a DNA rearrangement and gene amplification (Fig. 4c). This 16-kb fragment was converted to a half-size (8-kb) fragment after snap-back, confirming its palindromic structure. The palindrome extended in the direction of the centromere (see Fig. S2 in the supplemental material). DNA sequence analysis of the center of the palindrome indicates that the palindrome has a small asymmetry of 1.8 kb (Fig. 4d). Identical stretches of sequence homology were not found. However, the genomic sequences at the proximal and distal ends of the nonpalindromic region are highly AT rich (72% and 73%, respectively). The distribution of adenine and thymine has a strong strand bias (Fig. 4d), with 47% (28/60) adenine in the proximal end of the nonpalindromic spacer (“A-rich” region) and 45% (27/60) thymine in the distal end (“T-rich” region). The AT richness and strand bias distribution of nucleotides might have promoted intrastrand annealing during the formation of this large palindrome. The region covering the ARNT gene was not amplified (Fig. 4c), indicating that the center of the palindrome marks the telomeric boundary between amplified and nonamplified regions.
FIG. 4.
The center of a palindrome marks the boundary of the amplicon at 1q21 in the Colo320DM cell line. a. Array-CGH analysis identified the regional amplification at 1q21 in Colo320DM cells (36). The log2 ratio of CGH signal intensity of Colo320DM relative to that of human foreskin fibroblast (HFF2) cells on genes within the 20-Mb region at 1q21 is shown. A spotted human cDNA array was used for this array-CGH analysis. Genes with increased intensity in Colo320DM cluster within a 1-Mb region (solid bar). Note that the CTSK and HIST2H4 genes are located near the telomeric and centromeric boundaries, respectively, of this amplification. MB, million base pairs. b. Schematic drawing of the 50-kb region at 1q21, covering three genes (CTSS, CTSK, and ARNT). Exons of these genes are represented by gray rectangles. Probes used for Southern analysis (Cen and Tel) are indicated. Restriction sites for EcoRV (EV) are also shown. c. Palindrome formation at the boundary of the amplicon. EcoRV-digested DNA is hybridized by the probes indicated in panel b. A 16-kb amplified fragment was seen in Colo320DM (Colo) cells using the Cen probe (black arrowhead), and this fragment was converted into a half-sized one after snap-back (gray arrowhead), indicating palindromic organization of this fragment. A probe in the ARNT gene (Tel) hybridized to a single fragment of similar intensity in both human foreskin fibroblast (HFF) and Colo cells, and this fragment was also seen in Colo cells using the probe Cen, implying a normal allele was present in Colo320DM cells. SB, snap-back. A 2.5-kb molecular ruler (Bio-Rad) is shown as a DNA size marker. d. DNA sequence at the center of the palindrome. Multiplex PCR was performed using four primers shown on the left. Primer pairs 5670 and 6130 generated a 460-bp fragment from both normal DNA from human foreskin fibroblast (H) cells and DNA from Colo320 DNA (C) cells. Primers 6827 and 8628 were normally on the same strand and did not generate a PCR product on the normal allele but amplified the 900-bp region including the novel junction (left/right) in the palindrome. The gray line represents the symmetric region in the palindrome, and the black line represents the asymmetric sequence at the center. The DNA sequence at the nonpalindromic center and flanking palindromic regions is shown with bold letters, and normal genomic sequence downstream of the junction is shown with normal letters. Uppercase letters represent the DNA sequence at the asymmetric center, and lower-case letters represent DNA sequences in the palindrome. DNA sequences for both A-rich and T-rich regions are shown underlined.
A naturally existing DNA inverted repeat forms the centromeric boundary of the amplicon.
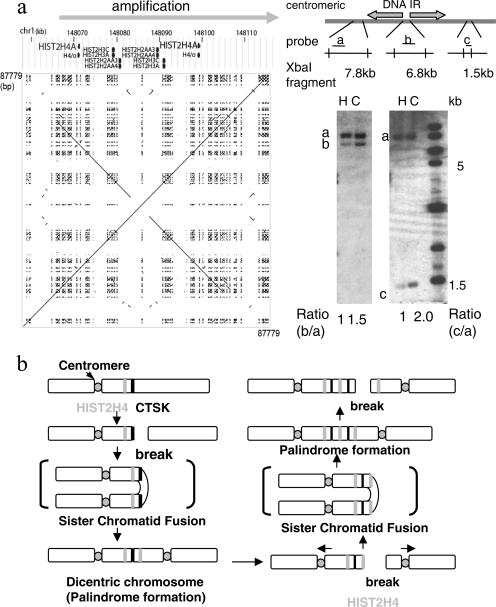
Another GAPF-positive gene at 1q21 in Colo320DM DNA was the HIST2H4 gene, which is in a naturally existing 26.1-kb large DNA IR (Fig. 5a). The sequence identity of the arms is 99.7%, and there is a 6.7-kb spacer between arms (41). To determine whether this DNA IR is involved in gene amplification, DNA copy numbers were compared in the regions centromeric to, within, and telomeric to the DNA IR. Although the DNA region 7 kb centromeric to the IR showed similar copy number intensities (2N) between HFF2 and Colo320DM DNA (probe a), the regions within and telomeric to the IR (probes b and c) showed amplification. However, the degree of amplification was different between the nonpalindromic spacer (probe b) and the telomeric region (probe c): the telomeric region showed a 4N copy number (an increase of 2N), whereas the nonpalindromic spacer showed a 3N copy number (an increase of 1N). This twofold difference in copy number increase is consistent with palindromic amplification of the entire region, with the nonpalindromic spacer at the center, in which the palindromic arms are amplified twice as many times as the nonpalindromic spacer. Thus, a naturally existing IR in the genome sets the centromeric boundary of the amplicon (Fig. 5b). Therefore, the large palindrome formed in cancer cells seems to have a structure consistent with those generated by DSB-induced intrastrand annealing in our model system of DHFR amplification.
FIG. 5.
A naturally existing IR marks the centromeric border of a palindromic amplification. a. Schematic drawing and a dot plot of the 87.8-kb region at 1q21, covering HIST2H4, shows a 27-kb, naturally existing IR. A dot plot for the DNA sequence in this region (chromosome 1, 148,043,413 to 148,131,191) is shown. Southern analysis for XbaI-digested DNA of human foreskin fibroblast (H) and Colo320DM (C) cells indicated that the region centromeric to this IR (probe a for a 7.8-kb XbaI fragment) was not amplified. In contrast, the regions within (probe b for a 6.8-kb fragment) and telomeric to (probe c for a 1.5-kb fragment) the IR were amplified. Note the different degree of amplification between the nonpalindromic spacer (probe b) and telomeric region (probe c). The DNA sequence was obtained from the UCSC genome browser (http://genome.ucsc.edu/), and the dot plot was drawn using the Pipmaker (http://pipmaker.bx.psu.edu/pipmaker/). b. A model for the initiation of regional amplification at 1q21 by the BFB cycle. An initial break at the CTSK gene (black bar) led to palindrome formation and the dicentric chromosome. Subsequent BFB cycles likely resulted in another chromosome break near the 27-kb IR at the HIST2H4 locus (gray bar), generating a large DNA palindrome with the IR at the center for this regional amplification. In both cases, the centers of the palindrome set the boundary of the amplified region.
DISCUSSION
We show here that intrastrand annealing is a prevalent mechanism for palindromic gene amplification in mammalian cells and that a DNA IR significantly facilitates this process. Furthermore, the boundaries of genomic amplification in cancer cells are consistent with palindrome formation by intrastrand annealing, and a DNA IR in the human genome marks the boundary between amplified and nonamplified DNA. These findings suggest that human gene amplification might occur through a conserved process also used for developmental gene amplification in Tetrahymena. A DNA IR in the genome is involved in this process, also indicating that local sequence homologies, at least in some cases, determine the regions most susceptible to amplification.
In our model system of DHFR amplification, a 229-bp IR significantly increases the efficiency of palindrome formation when inserted at a chromosomal locus. Furthermore, in Colo320DM cells, a 26-kb IR at 1q21 marks the boundary of an amplicon. Thus, some regions of genomic amplification are likely to be determined by the local DNA homology. The extent of the IR homology facilitates palindrome formation, which in turn determines the location of the amplicon boundaries. Several lines of evidence suggest that IR-mediated palindrome formation is a major pathway for generating DNA palindromes in cancer cells. First, large IRs with very high identity between the arms preexist in the human genome. A study by Warburton et al. identified a number of DNA IRs in the human genome with more than 8 kb in each arm length and 99% identity between the arms (41). In addition, Alu sequences often form DNA IRs with a high degree of homology (>80%) and substantial alignment (>275 bp) when present next to each other in the genome (33). Second, a DNA IR also serves either as an “at-risk-motif” or a “fragile site” to regulate large palindrome formation in S. cerevisiae when DNA replication, repair, and checkpoint function are compromised (14, 18). Furthermore, IR sequences preexisting in the genome are found at the centers of palindromes in Leishmania and Schizosaccharomyces pombe (2, 11). This DNA IR-facilitated model of gene amplification predicts that palindrome formation is in part regulated by the presence of DNA IRs and occurs nonrandomly in the genome, which may provide a mechanistic basis for the nonrandom distribution of DNA palindromes in human cancer cells (36).
It is also important to note that a DSB alone is sufficient to initiate large palindrome formation in mammalian cells. Even in these cases, however, many clones show biased processing of the ends of the I-SceI-induced DSB, which is likely to be derived from intrastrand annealing. Excision (5′ to 3′) of an I-SceI-induced DSB end generates a 3′ single-strand overhang that forms a hairpin molecule prior to DNA replication through intrastrand base pairing, resulting in a large palindrome after DNA replication (Fig. 3b). This model is analogous to the intramolecular mechanism of palindrome formation mediated by the short DNA IR in Tetrahymena and to the small IR-assisted intramolecular mechanism for palindrome formation in yeast (Fig. 3b, left) (6, 20, 26). Microhomology represents a very short DNA IR (Fig. 3b, right), and the length of homology determines the efficiency of intrastrand annealing. This process is likely to be independent of NHEJ, since an intramolecular mechanism using a short stretch of homology can occur in the absence of RAD50, YKU70, and LIG4 in S. cerevisiae (20, 26).
The amplicon at 1q21 in Colo320DM DNA covers a 1-Mb genomic region, and the copy number increase at some loci in this region is more than fourfold (36). Regional amplification of 1q21 is often seen in primary cancer tissues, including those of osteosarcoma and ovarian cancer (3, 35), suggesting an involvement of 1q21 amplification in tumor progression. The CTSK gene is located at the telomeric end of this regional amplification. An amplicon extending centromeric to the CTSK gene suggests that a DSB at this region initiated palindrome formation and generated a dicentric chromosome (Fig. 5b). Subsequent BFB cycles probably resulted in another chromosome break near the 27-kb DNA IR at the HIST2H4 locus, again generating a large palindrome with the DNA IR at the center for this regional amplification. The presence of one large palindrome along with the duplication of thenormal allele generates a copy number profile that is consistent with the copy number increases presented here. In both cases, the center of the large palindrome sets the boundary of an amplicon.
Using both a model experimental system with CHO cells and DNA analysis with cancer cells, we show here an important molecular process that regulates the development and progression of cancer. An initial chromosome break followed by palindrome formation triggers BFB cycles, which further generate genetic diversity in premalignant and cancer cell populations (10, 28). With a genetic deficiency, such as the loss of p53 function, which leads to permissivity for gene amplification (31), local genomic sequences, such as DNA IRs, might determine the incidence and location of gene amplification. IRs might not be accurately represented in the current human genome sequences, since DNA IR sequences are well known to be unstable upon cloning in Escherichia coli. DNA IRs may also exist as a form of genetic polymorphism among individuals (7). Further mapping of the epicenters of large palindromes should determine genomic and genetic attributes that lead to initial palindrome formation in cancer cells.
Supplementary Material
Acknowledgments
We thank J. Delrow and the FHCRC Genomics facility for helpful advice on the array studies and M. Marotta for technical assistance. We thank G. Stark, B. Trask, D. Gottschling, R. Monnat, M. Thayer, and members of the Yao lab for suggestions and comments on the manuscript.
This work was supported by National Institutes of Health grants R01GM 26210 (M.-C.Y.) and R01AR 045113, R01AR 045113, R01AR 045203, and POINS 46788 (S.J.T) and by grants from the Avon Breast Cancer Crusade Opportunity Fund (to H.T.). H.T. is a recipient of an Interdisciplinary Dual Mentor Fellowship from the Fred Hutchinson Cancer Research Center.
Footnotes
Published ahead of print on 22 January 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Albertson, D. G., C. Collins, F. McCormick, and J. W. Gray. 2003. Chromosome aberrations in solid tumors. Nat. Genet. 34:369-376. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht, E. B., A. B. Hunyady, G. R. Stark, and T. E. Patterson. 2000. Mechanisms of sod2 gene amplification in Schizosaccharomyces pombe. Mol. Biol. Cell 11:873-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atiye, J., M. Wolf, S. Kaur, O. Monni, T. Bohling, A. Kivioja, E. Tas, M. Serra, M. Tarkkanen, and S. Knuutila. 2005. Gene amplifications in osteosarcoma-CGH microarray analysis. Genes Chromosomes Cancer 42:158-163. [DOI] [PubMed] [Google Scholar]
- 4.Brown, P. C., T. D. Tlsty, and R. T. Schimke. 1983. Enhancement of methotrexate resistance and dihydrofolate reductase gene amplification by treatment of mouse 3T6 cells with hydroxyurea. Mol. Cell. Biol. 3:1097-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler, D. K., L. E. Yasuda, and M. C. Yao. 1996. Induction of large DNA palindrome formation in yeast: implications for gene amplification and genome stability in eukaryotes. Cell 87:1115-1122. [DOI] [PubMed] [Google Scholar]
- 6.Butler, D. K., L. E. Yasuda, and M. C. Yao. 1995. An intramolecular recombination mechanism for the formation of the rRNA gene palindrome of Tetrahymena thermophila. Mol. Cell. Biol. 15:7117-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caburet, S., C. Conti, C. Schurra, R. Lebofsky, S. J. Edelstein, and A. Bensimon. 2005. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 15:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, Y., R. M. Kumar, B. H. Penn, C. A. Berkes, C. Kooperberg, L. A. Boyer, R. A. Young, and S. J. Tapscott. 2006. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 25:502-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coquelle, A., E. Pipiras, F. Toledo, G. Buttin, and M. Debatisse. 1997. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell 89:215-225. [DOI] [PubMed] [Google Scholar]
- 10.Gisselsson, D., L. Pettersson, M. Hoglund, M. Heidenblad, L. Gorunova, J. Wiegant, F. Mertens, P. Dal Cin, F. Mitelman, and N. Mandahl. 2000. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc. Natl. Acad. Sci. USA 97:5357-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grondin, K., G. Roy, and M. Ouellette. 1996. Formation of extrachromosomal circular amplicons with direct or inverted duplications in drug-resistant Leishmania tarentolae. Mol. Cell. Biol. 16:3587-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn, P., B. Nevaldine, and W. F. Morgan. 1990. X-ray induction of methotrexate resistance due to dhfr gene amplification. Somat. Cell Mol. Genet. 16:413-423. [DOI] [PubMed] [Google Scholar]
- 13.Ishkanian, A. S., C. A. Malloff, S. K. Watson, R. J. DeLeeuw, B. Chi, B. P. Coe, A. Snijders, D. G. Albertson, D. Pinkel, M. A. Marra, V. Ling, C. MacAulay, and W. L. Lam. 2004. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 36:299-303. [DOI] [PubMed] [Google Scholar]
- 14.Lemoine, F. J., N. P. Degtyareva, K. Lobachev, and T. D. Petes. 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120:587-598. [DOI] [PubMed] [Google Scholar]
- 15.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 16.Liang, F., M. Han, P. J. Romanienko, and M. Jasin. 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 95:5172-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, Y., T. Lukacsovich, and A. S. Waldman. 1999. Multiple pathways for repair of DNA double-strand breaks in mammalian chromosomes. Mol. Cell. Biol. 19:8353-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183-193. [DOI] [PubMed] [Google Scholar]
- 19.Ma, C., S. Martin, B. Trask, and J. L. Hamlin. 1993. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev. 7:605-620. [DOI] [PubMed] [Google Scholar]
- 20.Maringele, L., and D. Lydall. 2004. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 18:2663-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murnane, J. P., and L. Sabatier. 2004. Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. Bioessays 26:1164-1174. [DOI] [PubMed] [Google Scholar]
- 22.Neiman, P. E., R. Kimmel, A. Icreverzi, K. Elsaesser, S. J. Bowers, J. Burnside, and J. Delrow.2006. Genomic instability during Myc-induced lymphomagenesis in the bursa of Fabricius. Oncogene 25:6325-6335. [DOI] [PubMed] [Google Scholar]
- 23.Paulin, R., G. W. Grigg, M. W. Davey, and A. A. Piper. 1998. Urea improves efficiency of bisulphite-mediated sequencing of 5′-methylcytosine in genomic DNA. Nucleic Acids Res. 26:5009-5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulson, T. G., A. Almasan, L. L. Brody, and G. M. Wahl. 1998. Gene amplification in a p53-deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol. Cell. Biol. 18:3089-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rattray, A. J. 2004. A method for cloning and sequencing long palindromic DNA junctions. Nucleic Acids Res. 32:e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rattray, A. J., B. K. Shafer, B. Neelam, and J. N. Strathern. 2005. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 19:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson, C., and M. Jasin. 2000. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol. Cell. Biol. 20:9068-9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudolph, K. L., M. Millard, M. W. Bosenberg, and R. A. DePinho. 2001. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat. Genet. 28:155-159. [DOI] [PubMed] [Google Scholar]
- 29.Smith, K. A., M. B. Stark, P. A. Gorman, and G. R. Stark. 1992. Fusions near telomeres occur very early in the amplification of CAD genes in Syrian hamster cells. Proc. Natl. Acad. Sci. USA 89:5427-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smogorzewska, A., J. Karlseder, H. Holtgreve-Grez, A. Jauch, and T. de Lange. 2002. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 12:1635-1644. [DOI] [PubMed] [Google Scholar]
- 31.Stark, G. R. 1993. Regulation and mechanisms of mammalian gene amplification. Adv. Cancer Res. 61:87-113. [DOI] [PubMed] [Google Scholar]
- 32.Stark, G. R., M. Debatisse, E. Giulotto, and G. M. Wahl. 1989. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell 57:901-908. [DOI] [PubMed] [Google Scholar]
- 33.Stenger, J. E., K. S. Lobachev, D. Gordenin, T. A. Darden, J. Jurka, and M. A. Resnick. 2001. Biased distribution of inverted and direct Alus in the human genome: implications for insertion, exclusion, and genome stability. Genome Res. 11:12-27. [DOI] [PubMed] [Google Scholar]
- 34.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100:9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takano, M., K. Fujii, T. Kita, Y. Kikuchi, and K. Uchida. 2004. Amplicon profiling reveals cytoplasmic overexpression of MUC1 protein as an indicator of resistance to platinum-based chemotherapy in patients with ovarian cancer. Oncol. Rep. 12:1177-1182. [PubMed] [Google Scholar]
- 36.Tanaka, H., D. A. Bergstrom, M. C. Yao, and S. J. Tapscott. 2005. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat. Genet. 37:320-327. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka, H., S. J. Tapscott, B. J. Trask, and M. C. Yao. 2002. Short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. Proc. Natl. Acad. Sci. USA 99:8772-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tlsty, T. D. 1997. Genomic instability and its role in neoplasia. Curr. Top. Microbiol. Immunol. 221:37-46. [DOI] [PubMed] [Google Scholar]
- 39.Tlsty, T. D., P. C. Brown, and R. T. Schimke. 1984. UV radiation facilitates methotrexate resistance and amplification of the dihydrofolate reductase gene in cultured 3T6 mouse cells. Mol. Cell. Biol. 4:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toledo, F., D. Le Roscouet, G. Buttin, and M. Debatisse. 1992. Co-amplified markers alternate in megabase long chromosomal inverted repeats and cluster independently in interphase nuclei at early steps of mammalian gene amplification. EMBO J. 11:2665-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warburton, P. E., J. Giordano, F. Cheung, Y. Gelfand, and G. Benson. 2004. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 14:1861-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao, M. C., C. H. Yao, and B. Monks. 1990. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell 63:763-772. [DOI] [PubMed] [Google Scholar]
- 43.Yasuda, L. F., and M. C. Yao. 1991. Short inverted repeats at a free end signal large palindromic DNA formation in Tetrahymena. Cell 67:505-516. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, C., K. D. Mills, D. O. Ferguson, C. Lee, J. Manis, J. Fleming, Y. Gao, C. C. Morton, and F. W. Alt. 2002. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109:811-821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.