Abstract
Senescence is characterized by an irreversible cell proliferation arrest. Specialized domains of facultative heterochromatin, called senescence-associated heterochromatin foci (SAHF), are thought to contribute to the irreversible cell cycle exit in many senescent cells by repressing the expression of proliferation-promoting genes such as cyclin A. SAHF contain known heterochromatin-forming proteins, such as heterochromatin protein 1 (HP1) and the histone H2A variant macroH2A, and other specialized chromatin proteins, such as HMGA proteins. Previously, we showed that a complex of histone chaperones, histone repressor A (HIRA) and antisilencing function 1a (ASF1a), plays a key role in the formation of SAHF. Here we have further dissected the series of events that contribute to SAHF formation. We show that each chromosome condenses into a single SAHF focus. Chromosome condensation depends on the ability of ASF1a to physically interact with its deposition substrate, histone H3, in addition to its cochaperone, HIRA. In cells entering senescence, HP1γ, but not the related proteins HP1α and HP1β, becomes phosphorylated on serine 93. This phosphorylation is required for efficient incorporation of HP1γ into SAHF. Remarkably, however, a dramatic reduction in the amount of chromatin-bound HP1 proteins does not detectably affect chromosome condensation into SAHF. Moreover, abundant HP1 proteins are not required for the accumulation in SAHF of histone H3 methylated on lysine 9, the recruitment of macroH2A proteins, nor other hallmarks of senescence, such as the expression of senescence-associated β-galactosidase activity and senescence-associated cell cycle exit. Based on our results, we propose a stepwise model for the formation of SAHF.
Senescence was initially described as a stable cell proliferation arrest resulting from the progression of primary human fibroblasts through a finite number of population doublings in vitro (35). However, activated oncogenes, oxidative stress, DNA damage, and drug-like inhibitors of specific enzymatic activities also induce senescence (14, 37, 82). In addition, senescence occurs in other cell types, such as primary human epithelial cells. In vivo, senescence is an important tumor suppression mechanism that restrains the proliferation of cells that harbor activated oncogenes (12, 16, 17, 51). Also, by limiting the self-renewal capacity of adult tissue stem cells, senescence is thought to contribute to tissue aging of many multicellular adult animals (38, 42, 53).
Senescent cells are typically characterized by a large flat morphology and the expression of a senescence-associated β-galactosidase (SA β-gal) activity of unknown function (16, 21). In the nucleus of senescent cells, the chromatin undergoes dramatic remodeling through the formation of domains of facultative heterochromatin called senescence-associated heterochromatin foci (SAHF) (56, 57, 86). Cytologically, SAHF appear as compacted punctate DAPI (4,6-diamidino-2-phenylindole)-stained foci of DNA in senescent cell nuclei. The formation of SAHF is also reflected in a general increase in the resistance of nuclear chromatin to digestion by nucleases (57). SAHF contain modifications and associated proteins characteristic of transcriptionally silent heterochromatin, such as methylated lysine 9 of histone H3 (H3K9Me), heterochromatin protein 1 (HP1), and the histone H2A variant macroH2A. In addition, Narita et al. recently showed that high-mobility group A (HMGA) proteins, a family of abundant non-histone chromatin proteins, are essential structural components of SAHF (56). Proliferation-promoting genes, such as E2F target genes (e.g., cyclin A), are recruited into SAHF, dependent on the pRB tumor suppressor protein, thereby irreversibly silencing expression of those genes.
Recently, we showed that two chromatin regulators, histone repressor A (HIRA) and antisilencing function 1a (ASF1a), drive the formation of SAHF in human cells (86). HIRA and ASF1a are the human orthologs of proteins known to create transcriptionally silent heterochromatin in yeasts, flies, and plants (9, 29, 39, 54, 63, 70-73, 78). In Saccharomyces cerevisiae, Hir1 and Hir2 are required for heterochromatin-mediated silencing of histone genes, telomeres, and mating loci, and the formation of pericentromeric chromatin structure (39, 70-73). Likewise, yeast Asf1p is required for heterochromatin-mediated silencing of telomeres, mating loci, and histone genes (40, 50, 70, 73, 75, 78) but also mediates nucleosome disassembly (2, 3, 68). Both HIRA and ASF1a bind to histones and exhibit histone chaperone activity in vitro (28, 64, 70, 78, 79). The HIRA/ASF1a-containing complex preferentially deposits the histone variant histone H3.3 into nucleosomes (46, 65, 76). Canonical human histone H3.1 and histone H3.3 differ in their primary amino acid sequences by only five amino acids. However, histone H3.1 is expressed periodically in the S phase of the cell cycle and is incorporated into chromatin during replication-coupled chromatin assembly (5, 36, 76). In contrast, histone H3.3 is expressed throughout the cell cycle and is incorporated into chromatin by the HIRA/ASF1a complex in a DNA replication- and repair-independent manner (5, 36, 76). Consistent with their partially overlapping biological and biochemical properties, yeast Asf1p and Hir proteins physically interact, and this interaction is necessary for telomeric silencing (19, 70). Likewise, the formation of SAHF in human cells by HIRA and ASF1a depends upon a physical interaction between these two proteins (76, 77, 86).
A previous careful kinetic analysis of SAHF formation from our laboratory indicated that formation of SAHF is likely a multistep process (87). In the earliest defined step, the histone chaperone proteins HIRA and HP1 are both recruited to a specific subnuclear organelle, the acute promyelocytic leukemia (PML) nuclear body (10, 67). Most human cells contain 20 to 30 PML nuclear bodies, which are typically 0.1 to 1 μm in diameter and are enriched in the protein PML, as well as many other nuclear regulatory proteins (10, 67). PML bodies have been previously implicated in various cellular processes, including tumor suppression and cellular senescence (20, 23, 61). At a molecular level, they have been proposed as sites of assembly of macromolecular regulatory complexes and protein modification (24, 31, 61). After HIRA's translocation into PML bodies, chromatin condensation occurs, as defined by the appearance of DAPI-stained foci. Finally, H3K9Me accumulates, and HP1 and macroH2A proteins are recruited to SAHF.
In this study, we set out to understand the series of events that contribute to the formation of SAHF in more detail and, in particular, to identify the molecular requirements for the different steps that were previously temporally defined. Here we report that during SAHF formation, each chromosome condenses into a single DAPI focus. Chromosome condensation mediated by the histone chaperone ASF1a depends on its binding to histone H3, as well as HIRA. Interestingly, HP1γ, but not HP1α and HP1β, is phosphorylated on serine 93 in senescent cells. This phosphorylation is not required for the protein's localization to PML bodies, but is required for its binding to SAHF. Remarkably, a large reduction in the amount of chromatin-bound HP1 proteins does not affect chromosome condensation, recruitment of histone variant macroH2A to SAHF, expression of SA β-gal, or senescence-associated cell cycle exit. Based on these data, we propose a multistep model of dependent and independent steps that culminate in the formation of mature SAHF.
MATERIALS AND METHODS
Cell culture and retroviral infection.
Cells were cultured according to ATCC. Retrovirus infection of WI38 cells was performed as described previously using phoenix packaging cells to make infectious virus (86). The following plasmids were used to produce viruses: pBabe-neo-H-RasV12 (a gift of Bill Hahn and Bob Weinberg), pQCXIP-HA-HP1γ and its mutants, pQCXIP-HA-ASF1a and its mutants, pQCXIN-myc-ASF1a and its mutants, pQCXIP-myc-HP1β (103-185), and pQCXIP-HA-HP1β (103-185).
Immunofluorescence, antibodies, SAHF, and SA β-gal staining.
Two color indirect immunofluorescence assays were performed as described previously (86). Antihemagglutinin (anti-HA) (Y11) (Santa Cruz), anti-myc (9E10) (Santa Cruz), anti-HP1γ (Chemicon), anti-histone H3 (Abcam), anti-glutathione S-transferase (anti-GST) (Santa Cruz), and anti-PML (AB1370) (Chemicon) were from the indicated suppliers. Anti-macroH2A and anti-HIRA antibodies were described previously (18, 33). Additional antibodies were raised to the macrodomain of macroH2A1.2 fused to GST, following a protocol described previously (34). Anticentromere antibody (ACA) was a gift from J. B. Rattner, University of Calgary. DAPI staining for SAHF and SA β-gal staining in senescent cells were performed essentially as described previously (86).
Chromosome painting and fluorescence in situ hybridization.
The protocol was adapted from that of Mahy and coworkers (48). Growing or senescent WI38 cells were cultured on coverslips, washed twice with phosphate-buffered saline (PBS), and incubated in 0.075 M KCl at room temperature for 20 min. The slides were first fixed in a 3:1 solution of methanol:acetic acid for 10 min at room temperature, followed by overnight fixation in 3:1 methanol:acetic acid at −20°C. The slides can be kept at −20°C for up to 1 week. After overnight fixation, the slides were washed three times in fresh 3:1 methanol:acetic acid and dried by steaming. Steaming was immediately stopped once the slides had dried. The slides were then treated with RNase (100 μg/ml in 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) at 37°C for 1 h, followed by pepsin treatment (0.1 mg/ml in 0.01 M HCl) for 3 min at 37°C. After pepsin treatment, the slides were washed with PBS and postfixed in 1% paraformaldehyde in PBS with 50 mM MgCl2 for 10 min at room temperature. After washing in PBS, the slides were dehydrated for 2 min each in 70%, 80%, and 100% ethanol at room temperature. The slides were dried completely at 37°C and then denatured in 70% formamide in 2× SSC in 73°C for 3 min. Immediately after denaturation, the slides were dehydrated with 70%, 80%, and 100% ethanol at −20°C for 2 min each. The slides were dried completely again and hybridized overnight in chromosome paint hybridization buffer with chromosome paint probes directly labeled with fluorescein isothiocyanate (whole chromosome paint probes were from Vysis) or together with a biotin-labeled probe for the cyclin A gene at 37°C. Biotin labeling of the cyclin A bacterial artificial chromosome probe CTD-2217D23 (Invitrogen) was performed using a Bioprimer DNA-labeling kit from Invitrogen. Hybridized biotin-labeled cyclin A probe was detected by the binding of Texas Red-avidin DCS (Vector Laboratories) and amplified by the binding of biotinylated anti-avidin D9 (Vector Laboratories), followed by another layer of binding of Texas Red-avidin DCS. The slides were counterstained for SAHF using 0.125 μg/ml DAPI for 5 min at room temperature before being visualized by epifluorescence.
GST pulldown and coimmunoprecipitation assays.
GST or GST-tagged wild-type ASF1a or its mutants were prebound to glutathione-Sepharose resin (Amersham Biosciences) and incubated with 1 μg histone H3 (a gift of Takashi Sekiya and Kenneth Zaret) in binding buffer (25 mM HEPES-NaOH [pH 7.5], 200 mM KCl, 13 mM MgCl2, 10% glycerol, 0.1% NP-40, and 0.3% β-mercaptoethanol) at 4°C for 2 h. After incubation, the resin was washed five times with binding buffer, and bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Coimmunoprecipitation was performed as described previously (1, 33).
RESULTS
Single chromosomes condense into a single SAHF.
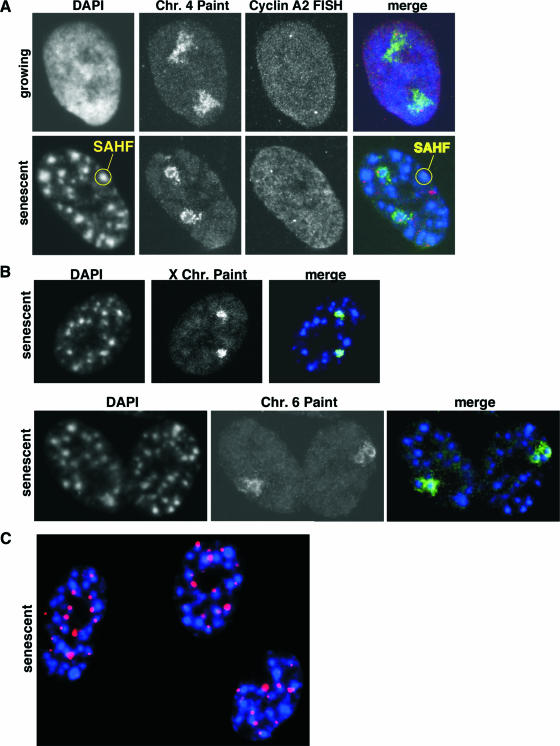
We reasoned that an individual chromosome might condense to form a single SAHF focus. Alternatively, a single chromosome might contribute to more than one SAHF focus. To distinguish between these two possibilities, we performed chromosome painting of specific chromosomes in WI38 primary human fibroblasts induced to undergo senescence by expression of an activated Ras oncogene. In growing cells, each copy of chromosome 4 occupied a dispersed nuclear territory, as described previously (27, 60) (Fig. 1A). In striking contrast, in senescent WI38 cells, each copy of these chromosomes was condensed into a single SAHF focus. Chromosome painting showed that the interior of the SAHF focus did not hybridize efficiently to the probe, presumably because the dense heterochromatin is inaccessible to the probe. When chromosome 4 painting was combined with cyclin A2 gene fluorescence in situ hybridization, this gene occupied the diffuse territory of chromosome 4 in growing cells. However, in senescent WI38 cells, the cyclin A2 gene either was localized at the periphery of the dense heterochromatin domain or was undetectable, presumably because it was localized to the nonhybridizing interior of the focus (Fig. 1A and data not shown). Similarly, in senescent cells, both copies of chromosome X and chromosome 6 were each condensed into a single SAHF focus (Fig. 1B). Recently, Funayama and coworkers also reported that SAHF result from the condensation of single chromosomes, based in part on chromosome painting with a chromosome 12-specific probe (25). Interestingly, as originally reported by Narita and coworkers (57), we found that telomeric and centromeric chromatin is located predominantly at the periphery of SAHF. In our experiments, telomeric DNA was detected with a telomeric DNA probe (84), and centromeric DNA localization was found by staining with ACA which recognize centromere-bound kinetochore proteins (Fig. 1C) (22). Funayama and coworkers also found centromeric chromatin to be peripheral to SAHF (25). Together, the data indicate that during SAHF formation, individual chromosomes condense to form a single SAHF focus. Domains of constitutive heterochromatin, such as pericentromeric and telomeric heterochromatin, do not appear to be integral to SAHF.
FIG. 1.
Individual chromosomes condense into a single SAHF focus. (A) Chromosome (Chr.) 4 painting combined with cyclin A2 gene fluorescence in situ hybridization (FISH) in growing and senescent WI38 cells. DNA was stained using DAPI. (B) Chromosome 6 and X chromosome painting and DAPI staining of senescent WI38 cells. Images were collected by confocal microscopy. (C) Senescent WI38 cells were stained with DAPI to visualize DNA (blue) and with ACA to visualize kinetochore proteins bound to centromeric DNA (red).
Formation of SAHF requires ASF1a histone H3 binding activity.
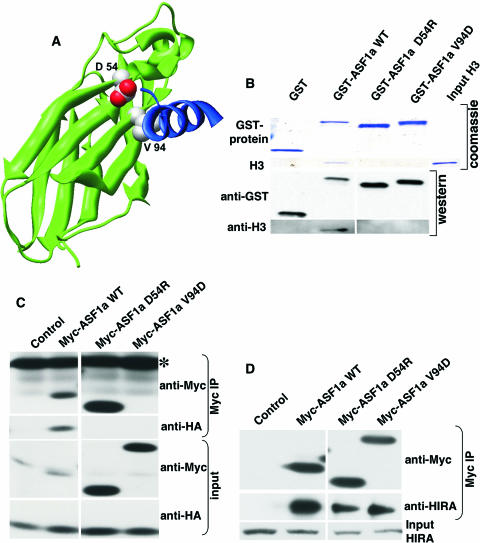
ASF1a binds to the histone chaperone HIRA and also to histone H3 (55, 70, 76, 77, 86). We previously showed that the formation of SAHF by ASF1a depends on its ability to bind to HIRA (86). To assess whether the histone H3 binding activity of ASF1a is also required for formation of SAHF, we introduced point mutations onto the surface of ASF1a used for histone H3 binding, based on the published nuclear magnetic resonance structure of human ASF1a bound to a histone H3-derived peptide (55) (Fig. 2A). We also made use of our recently solved X-ray crystal structure of ASF1a bound to a fragment of HIRA, to ensure that these mutants leave the HIRA interaction surface intact (77). Specifically, we generated ASF1a(D54R) and ASF1a(V94D). We tested the binding of these ASF1a mutants to histone H3.1. Although the HIRA/ASF1a complex preferentially uses histone H3.3 as a substrate (46, 76), ASF1a binds to both isoforms and makes key contacts with a peptide that is 100% conserved between histone H3.1 and H3.3 (55, 76). First, we tested the binding of the mutant proteins to histone H3.1 by GST pulldown assay, using purified recombinant histone H3.1 and GST-tagged wild-type ASF1a or its mutants. As predicted, both of the ASF1a mutants failed to bind to histone H3.1 in vitro, whereas wild-type ASF1a efficiently bound to histone H3.1 under identical conditions (Fig. 2B). To confirm these results, we tested the binding of wild-type ASF1a and its mutants to histone H3.1 in vivo by coexpressing myc epitope-tagged wild-type ASF1a or its mutants together with HA-tagged histone H3.1 in WI38 primary human fibroblasts and then testing for a physical interaction between the epitope-tagged proteins by immunoprecipitation and Western blotting analysis (Fig. 2C). Consistent with the in vitro binding results, both of the ASF1a mutants failed to bind to HA-histone H3.1, while wild-type ASF1a efficiently bound HA-histone H3.1. To eliminate the possibility that the ASF1a mutants failed to bind to histone H3.1 due to a gross disruption of protein folding, we tested the binding of wild-type ASF1a and the mutants to endogenous HIRA in WI38 cells, also by immunoprecipitation analysis (Fig. 2D). We found that, as intended, both of the ASF1a mutants still bound efficiently to HIRA, showing that the targeted mutations primarily disrupt the histone H3-binding surface and not the HIRA-binding surface.
FIG. 2.
Design of ASF1a mutants deficient in histone H3 binding. (A) Mutated residues (space fill, CPK coloring) on a modeled human ASF1a/H3 structure. Residues 1 to 156 of ASF1a are shown as the green ribbon. An ASF1a-binding H3 peptide (residues 122 to 135) is shown as the blue ribbon (data are taken from reference 55; see also reference 77). (B) GST pulldown assay using GST alone or GST-tagged wild-type (WT) ASF1a and its mutants and purified recombinant histone H3. After pulldown, the washed proteins bound to resin were separated on SDS-PAGE gels and visualized either by Coomassie blue staining or by Western blotting using the indicated antibodies. (C) Myc-tagged wild-type ASF1a or its mutants were coexpressed with HA-tagged histone H3 in WI38 cells, immunoprecipitated with anti-myc antibody (9E10), and Western blotted with the indicated antibodies. Note that Myc-ASF1aV94D comigrates with the light chain of the antibody used for immunoprecipitation (IP; marked with *) and that there is a background band in the control lane of the anti-Myc input. (D) Myc-tagged wild-type ASF1a or its mutants were expressed in WI38 cells, immunoprecipitated with the anti-myc antibody (9E10), and Western blotted with the indicated antibodies.
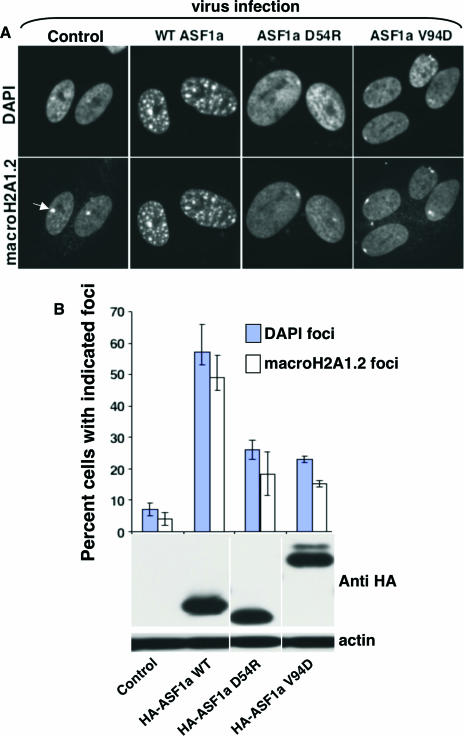
Using these ASF1a mutants, we tested whether the formation of SAHF by ASF1a requires its histone H3-binding activity, by ectopically expressing wild-type ASF1a or the mutants deficient in histone H3 binding in WI38 cells, using retroviruses. Compared to wild-type ASF1a, both ASF1a mutants deficient in histone H3 binding were grossly impaired in formation of SAHF, as judged by chromatin condensation using DAPI staining and the deposition of macroH2A proteins (Fig. 3A). The expression level of each of the HA-tagged ASF1a mutants was comparable to that of wild-type ASF1a, indicating that impaired SAHF formation by the mutants was not due to their underexpression (Fig. 3B). We conclude that formation of SAHF driven by ASF1a depends on its histone H3 binding activity.
FIG. 3.
Formation of SAHF by ASF1a depends on histone H3-binding activity. (A) HA-tagged wild-type (WT) ASF1a or its mutants deficient in histone H3 binding were expressed in WI38 cells by retrovirus infection and drug selected with 3 μg/ml puromycin. Ten days postinfection, drug-selected cells were stained with DAPI and antibodies to macroH2A1.2. The white arrow marks the inactive X chromosome. (B) One hundred cells shown in panel A were scored for the indicated nuclear foci. The inactive X chromosome was excluded when scoring cells with macroH2A foci. The bottom panel shows whole-cell lysates Western blotted with anti-HA and antiactin antibodies.
Phosphorylation of HP1γ is required for its deposition in SAHF but not its localization to PML bodies.
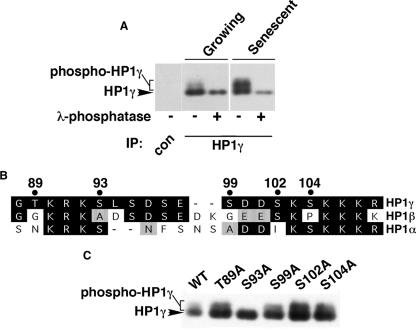
Next, in an attempt to understand how HP1 proteins are targeted to SAHF, we set out to identify posttranslational modifications of HP1 proteins that are regulated between young and senescent WI38 cells. Indicative of such a modification, we found by SDS-PAGE that HP1γ exhibited reduced mobility in senescent cells compared to that in growing cells (Fig. 4A). This apparent posttranslational modification was not observed for HP1α and HP1β (data not shown). To test whether this modification of HP1γ is due to phosphorylation, HP1γ was immunoprecipitated from growing or senescent WI38 cells and treated with or without λ-phosphatase. We found that phosphatase treatment completely abolished the modified form of the protein, confirming that HP1γ is phosphorylated in senescent cells (Fig. 4A).
FIG. 4.
HP1γ is phosphorylated on serine 93 in cells approaching senescence. (A) Control or HP1γ immunoprecipitates (IP) from growing (control-infected) or senescent (activated Ras-infected) WI38 cells were treated with or without λ-phosphatase, separated by SDS-PAGE, and Western blotted. (B) Alignment of regions of HP1α, -β, and -γ. Residues shaded gray are nonidentical but conserved; residues shaded black are identical. The numbers across the top indicate the residue numbers of candidate phosphoacceptor sites in HP1γ which are not conserved in both HP1α and -β. Residue numbers are taken from sequences in the Swiss-Prot database. (C) HA-tagged wild-type HP1γ and its mutants were ectopically expressed in WI38 cells, together with an activated Ras oncogene, and then Western blotted with anti-HA antibody.
Next, we wanted to identify the residue of HP1γ that is phosphorylated. Based on the observation that only HP1γ, and not HP1α or -β, becomes phosphorylated in senescent cells, each of the nonconserved serine or threonine residues in HP1γ was mutated to alanine to generate HP1γ(T89A), HP1γ(S93A), HP1γ(S99A), HP1γ(S102A), and HP1γ(S104A) (Fig. 4B and C). HA-tagged wild-type HP1γ or the mutants were expressed in WI38 cells, together with activated Ras to induce senescence. Western blotting analysis of the ectopically expressed HP1γ mutants showed that phosphorylation of HP1γ was completely abolished by the HP1γ(S93A) mutant (Fig. 4C). In contrast, there was no effect for any of the other mutations. We conclude that HP1γ is phosphorylated on serine 93 in senescent cells.
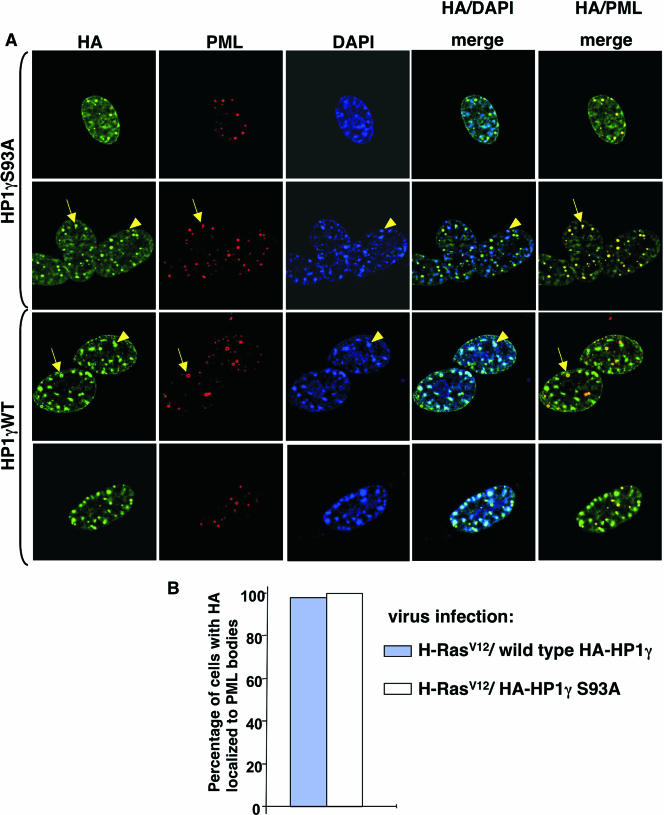
To test whether HP1γ phosphorylation is required for localization of HP1γ into PML bodies and/or SAHF, HA-tagged wild-type HP1γ and HP1γ(S93A) were coexpressed with activated Ras in WI38 cells. We found that both wild-type HP1γ and HP1γ(S93A) localized equivalently to PML bodies (Fig. 5A and B, yellow arrows). However, compared to wild-type HP1γ, the HP1γ(S93A) mutant was impaired in its deposition in SAHF (Fig. 5A, yellow arrowheads). We conclude that HP1γ is phosphorylated on serine 93 in senescent cells and that this phosphorylation is required for its efficient deposition in SAHF but not for its localization to PML bodies.
FIG. 5.
Phosphorylation of HP1γ is required for recruitment of HP1γ to SAHF but not its localization to PML bodies. (A) HA-tagged wild-type (WT) HP1γ or HP1γ(S93A) was coexpressed with an activated oncogene Ras (H-RasV12) in WI38 cells by retrovirus infection and drug-selected with 3 μg/ml puromycin and 500 μg/ml G418. Eights days postinfection, drug-selected cells were stained with the indicated antibodies and DAPI. Yellow arrows mark HP1γ wild type and HA-HP1γ(S93A) in a PML body, and yellow arrowheads mark HP1γ wild type in SAHF. The latter is largely absent from SAHF. Images were collected by confocal microscopy. (B) One hundred cells shown in panel A were scored for the colocalization of PML and HP1γ wild type or HP1γ(S93A).
Abundant HP1 proteins are not required for the formation of SAHF or senescence-associated cell cycle exit.
Given that all three HP1 family members are localized to SAHF in senescent cells (57, 86), we next asked whether these proteins are required for formation of SAHF and other features of the cellular senescence program. To do so, we took advantage of a dominant-negative HP1β mutant that we had developed previously (85). This dominant-negative mutant was created based on the prediction made by Lechner and coworkers that the deletion of the N-terminal chromodomain of HP1 proteins should prevent their binding to chromatin but not affect their ability to homo- or heterodimerize, thereby sequestering endogenous HP1 proteins from chromatin (43). Previously we showed that the ectopic expression of this mutant HP1β(103-185) (designated HP1βΔN) in WI38 cells does indeed deplete all three endogenous chromatin-bound HP1 subtypes by 70 to 80% (85) (see Fig. S1 in the supplemental material). Remarkably, ectopic expression of HP1βΔN in WI38 cells had no discernible effect upon cell viability or proliferation (85), so we were able to generate a polyclonal population of WI38 cells stably expressing HP1βΔN and markedly deficient in chromatin-bound HP1 proteins, or the appropriate empty vector-infected and drug-selected cells as a control.
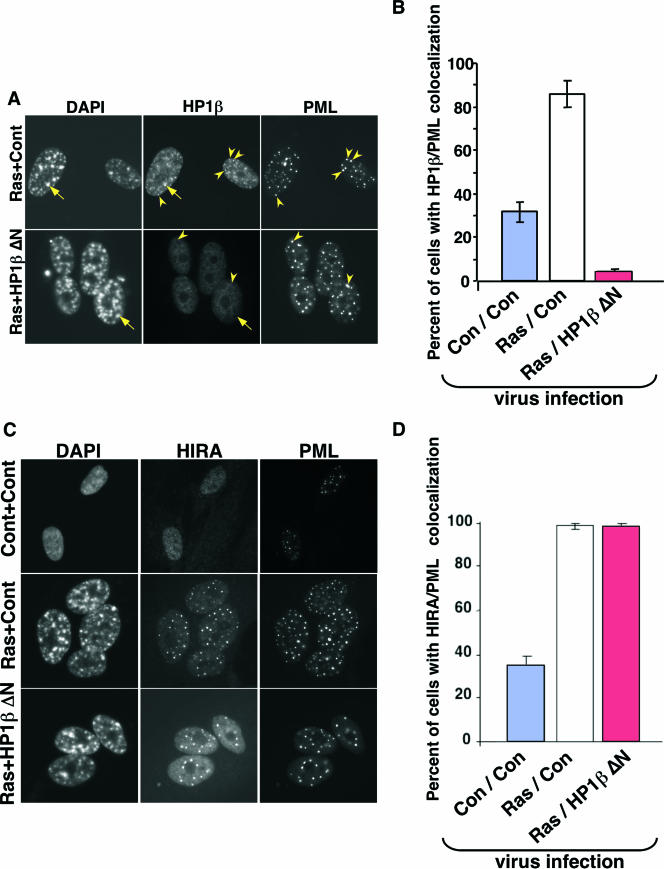
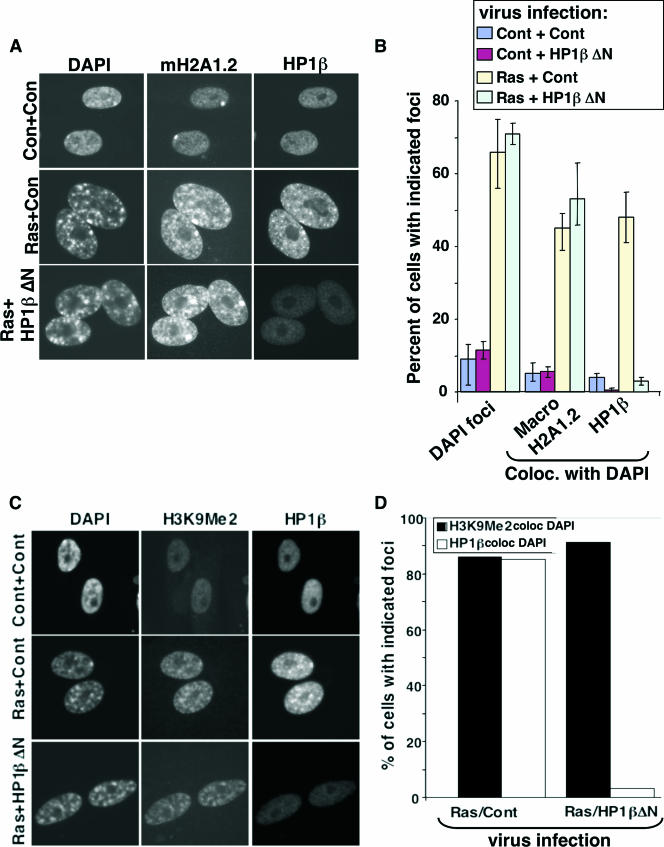
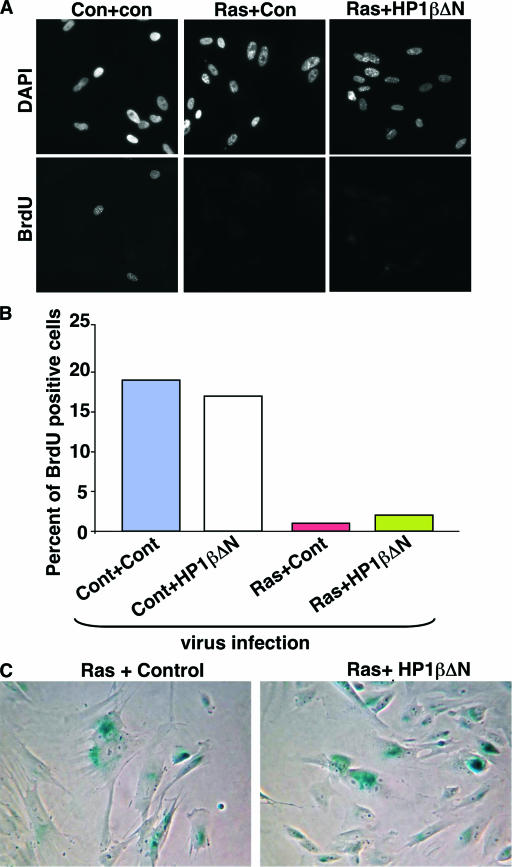
To test the requirement for chromatin-bound HP1 proteins for formation of SAHF and other aspects of the senescence program, we infected both control and HP1βΔN-expressing cells with a retrovirus encoding activated Ras and scored the effect on SAHF and other features of senescence. As expected, HP1βΔN efficiently removed the bulk of all three endogenous HP1 proteins from chromatin in WI38 cells coexpressing activated Ras. This is apparent from the almost complete absence of HP1 proteins from SAHF (Fig. 6A; also see Fig. S2 in the supplemental material). Interestingly, HP1βΔN also blocked the recruitment of endogenous HP1 proteins to PML bodies, a further indication of the ability of this mutant to disrupt the function of endogenous HP1 proteins (Fig. 6A and B, yellow arrowheads). However, HP1βΔN did not affect Ras-induced relocalization of HIRA to PML bodies (Fig. 6C and D). We previously showed that localization of HIRA to PML bodies is an early step in the formation of SAHF (86). Therefore, HP1βΔN did not impair early signaling events activated by oncogenic Ras. Remarkably, however, depletion of chromatin-bound HP1 proteins had no effect on chromosome condensation, as visualized by DAPI staining and the accumulation of H3K9Me2 and the macroH2A histone variant in SAHF (Fig. 6A, yellow arrows, and Fig. 7A to D; also see Fig. S2 in the supplemental material). No effect of HP1βΔN was observed on the rate of formation of SAHF (data not shown). We conclude that visible enrichment of HP1 proteins is not required for chromosome condensation and formation of SAHF.
FIG. 6.
HP1βΔN blocks the relocalization of HP1 proteins into PML bodies but has no effect on recruitment of HIRA into PML bodies. (A) WI38 cells were infected with vector control (Cont or Con) or HP1βΔN-expressing virus, together with a virus containing an activated Ras oncogene. The infected cells were selected with 3 μg/ml puromycin and 500 μg/ml G418 cells. Eight days postinfection, the cells were stained with antibodies to HP1β and PML. Yellow arrowheads indicate colocalizing HP1β and PML. Yellow arrows indicate HP1β in SAHF. (B) One hundred cells shown in panel A were scored for colocalizing HP1β and PML. (C) As in panel A but stained with antibodies to HIRA and PML. (D) One hundred cells shown in panel C were scored for colocalizing HIRA and PML.
FIG. 7.
HP1βΔN blocks deposition of HP1 proteins in SAHF, but has no effect on chromosome condensation, deposition of macroH2A protein, or accumulation of H3K9Me in SAHF. (A) WI38 cells were infected with vector control (Cont or Con) or HP1βΔN-expressing virus, together with a virus containing an activated Ras oncogene, as indicated. The infected cells were selected with 3 μg/ml puromycin and 500 μg/ml G418. Eight days postinfection, the cells were stained with antibodies to HP1β and macroH2A1.2. (B) One hundred cells shown in panel A were scored for chromosome condensation (DAPI foci) and deposition of macroH2A1.2 and HP1β in SAHF (colocalization of macroH2A1.2 and HP1β, respectively, with DAPI). Cells with macroH2A foci do not include cells with a visible inactive X chromosome only. (C) As in panel A but stained with antibodies to H3K9Me2 and HP1β. (D) One hundred cells shown in panel C were scored for incorporation of H3K9Me2 and HP1β in SAHF (colocalization of H3K9Me2 and HP1β, respectively, with DAPI). coloc., colocalization.
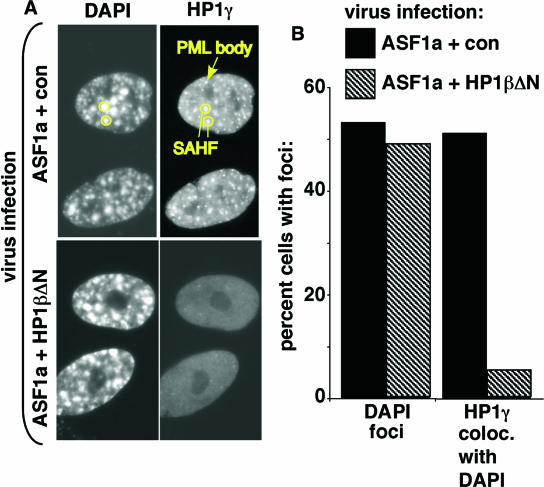
We previously showed that the histone chaperone ASF1a is an effector of Ras-mediated SAHF formation (86). We likewise found that the removal of chromatin-bound HP1 proteins by HP1βΔN did not affect chromosome condensation induced by ASF1a (Fig. 8A and B). Although we cannot exclude the possibility that the relatively small amount of remaining chromatin-bound HP1 proteins is sufficient for the observed changes in chromatin structure, we can conclude that a large depletion of chromatin-bound HP1 proteins does not obviously affect chromatin remodeling in senescent cells. Most notably, there is no effect on chromosome condensation to form morphological SAHF.
FIG. 8.
HP1βΔN has no effect on ASF1a-induced chromosome condensation. (A) WI38 cells were infected with vector control (con) or HP1βΔN-expressing virus, together with a virus encoding ASF1a, as indicated. The infected cells were selected with 3 μg/ml puromycin and 500 μg/ml G418. Ten days postinfection, the cells were stained with antibodies to HP1γ. The circles and arrow indicate HP1γ in SAHF and PML bodies, respectively. (B) One hundred cells shown in panel A were scored for chromosome condensation (DAPI foci) and HP1γ colocalized with SAHF (HP1γ coloc. with DAPI).
In light of this conclusion, we asked whether depletion of HP1 proteins affects other senescence phenotypes, specifically senescence-associated cell cycle exit and expression of SA β-gal activity. Remarkably, depletion of chromatin-bound HP1 proteins by HP1βΔN had no effect on either phenotype (Fig. 9A to C). We conclude that a large decrease in the binding of these proteins to chromatin and the resultant complete failure of these proteins to enrich in SAHF have no effect on any of the tested hallmarks of the senescence phenotype.
FIG. 9.
HP1βΔN does not affect senescence-associated cell cycle arrest and expression of SA β-gal. (A) WI38 cells stably infected with an HP1βΔN-expressing retrovirus or a control (Cont or Con) virus were infected with viruses encoding activated Ras or a control virus. The infected cells were selected with 3 μg/ml puromycin and 500 μg/ml G418. Eight days later, the cells were pulse-labeled with 5′-bromodeoxyuridine (BrdU) for 1 h and stained with antibodies to 5′-BrdU. (B) One hundred cells shown in panel A were scored as 5′-BrdU positive or negative. (C) Cells shown in panel A were stained to detect expression of SA β-gal (blue).
DISCUSSION
In this article, we report the following important findings regarding the mechanism of assembly of SAHF. First, SAHF results from condensation of individual chromosomes, each chromosome condensing into a single SAHF focus. Second, formation of SAHF by the histone chaperone ASF1a depends on its ability to bind to histone H3, in addition to the requirement for HIRA binding that we showed previously (86). Third, HP1γ is phosphorylated on serine 93 in senescent cells, and this modification is required for its deposition in SAHF but not for its localization to PML bodies. Fourth, high levels of chromatin-bound HP1 proteins are not required for chromosome condensation, deposition of macroH2A proteins in SAHF, or other hallmarks of the senescence program, such as expression of SA β-gal activity and senescence-associated cell cycle exit.
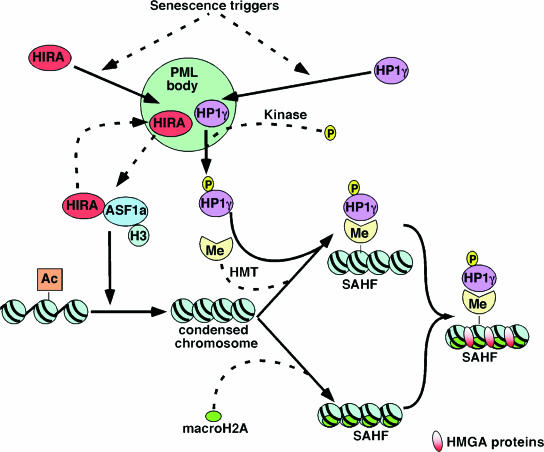
Based on these findings and others reported previously (57, 86), we propose the following stepwise model, comprised of dependent and independent steps, for the formation of SAHF (Fig. 10). Initially, histone chaperone protein HIRA and HP1 (HP1α, -β, and -γ) are recruited to PML nuclear bodies. Our previous kinetic analysis of SAHF formation showed that HIRA and HP1 proteins enter PML nuclear bodies prior to any detectable chromosome condensation or any other molecular marker of SAHF (86). Since HP1βΔN does not affect the localization of HIRA to PML bodies but does block recruitment of endogenous HP1 proteins to PML bodies and does not go to PML bodies itself (data not shown), we conclude that HIRA enters PML bodies independently of HP1 proteins.
FIG. 10.
A stepwise model for the formation of SAHF in senescent human cells. This model indicates the key steps in SAHF formation, after initiation by senescence triggers. Dashed lines are steps which are currently poorly defined (see text for details). Abbreviations: HMT, histone methyltransferase; Me, methylated lysine 9 histone H3; Ac, acetylated histone. HMGA proteins are shown in mature SAHF (56), but their point of entry is not known.
The role served by the translocation of HIRA and HP1 proteins to PML bodies is not known. However, PML bodies have been suggested as sites of assembly of macromolecular complexes and also as sites of protein modification (24, 31, 61). Significantly, in cells approaching senescence, HP1γ becomes phosphorylated on serine 93. Interestingly, we have not detected analogous phosphorylation of HP1α and -β. The nonphosphorylatable mutant HP1γ(S93A) efficiently enters PML bodies but does not efficiently localize to SAHF. Thus, HP1γ might be phosphorylated inside PML bodies and phosphorylation might target HP1γ to SAHF (86). Alternatively, HP1γ might be phosphorylated after the protein exits PML bodies en route to SAHF.
In the earliest discernible change in chromatin structure itself, individual chromosomes condense to form single SAHF. Previously, we showed that chromatin condensation depends on the histone chaperone ASF1a and is driven by a complex of ASF1a and its binding partner HIRA (86). Here we have extended this to show that chromosome condensation requires an interaction of ASF1a with histone H3 and HIRA. Since the HIRA/ASF1a complex serves as a chaperone for deposition of the histone H3/H4 complex into chromatin (65, 76), it seems likely that chromosome condensation by HIRA and ASF1a depends on their chaperone activity. Conceivably, chromosome condensation depends, in part, on increased nucleosome density due to HIRA/ASF1a-mediated nucleosome deposition. This is consistent with many previous reports that transcriptionally active chromatin is depleted of nucleosomes. This is true at both a genome-wide and a local chromatin level (2, 4, 6-8, 15, 44, 52, 87). Moreover, a previous study reported that the facultative heterochromatin of the inactive X chromosome has a higher nucleosome density than most other regions of the nucleus (62). Together, this suggests that whole chromosome condensation and gene silencing may result from increased nucleosome density throughout the chromosome.
The HIRA/ASF1a chaperone complex preferentially utilizes histone H3.3 as a deposition substrate (46, 76). Significantly, histone H3.3 accumulates in fibroblasts approaching senescence and in nondividing differentiated cells, in some cases to about 90% of the total histone H3, presumably with the majority being in inactive chromatin (11, 13, 30, 41, 59, 64, 66, 80, 83). Unfortunately, because histone H3.3 and canonical H3.1 differ only by five amino acids, they cannot presently be differentiated immunologically and there is no straightforward way to ask whether endogenous histone H3.3 is specifically enriched in SAHF. The idea that SAHF might contain histone H3.3 may initially seem unlikely, because deposition of histone H3.3 is typically linked to transcription activation (5, 49, 52, 69, 81), whereas SAHF is a form of transcriptionally silent facultative heterochromatin (56, 57, 86). However, the apparent inconsistency in this idea is merely an extension of an existing paradox. Specifically, HIRA and its orthologs in other species are typically involved in gene silencing and formation of heterochromatin (9, 29, 39, 40, 63, 70-72, 74), whereas HIRA's favored deposition substrate, histone H3.3, is linked to transcriptional activation (5, 49, 52, 69, 81). However, to our knowledge, histone H3.3 per se has not been shown to directly cause or contribute to transcription activation, and a proportion of histone H3.3 does carry posttranslational marks characteristic of transcriptionally silent chromatin (32, 47, 49). Therefore, histone H3.3 is unlikely to be exclusively linked to transcription activation. Instead, deposition of histone H3.3 may be associated with any major remodeling of chromatin, perhaps as a way to “reset” histone modifications. To express this idea, Ooi and coworkers have suggested that histone H3.3 is a chromatin “repair” variant (58). Concordant with this proposal, after egg fertilization in flies, dHIRA activity is required for the replacement of protamines by histone H3.3-containing nucleosomes in decondensing sperm chromatin (46). By this view, the HIRA/ASF1a complex might drive formation of SAHF by deposition of histone H3.3-containing nucleosomes.
In line with the idea that chromosome condensation to form SAHF results primarily from increased nucleosome density, chromosome condensation into SAHF does not require the accumulation of H3K9Me or the deposition of heterochromatic proteins HP1 and macroH2A. Our previous kinetic analysis showed that chromatin condensation occurs prior to the accumulation of H3K9Me and the deposition of HP1 and the histone variant macroH2A in chromatin (86). Here we have shown that chromosome condensation, triggered by an activated Ras oncogene or ectopic expression of ASF1a, efficiently occurs in the absence of high levels of stably bound HP1 proteins. Together, these results eliminate the possibility that H3K9Me, HP1, or macroH2A drives chromosome condensation. The finding that facultative heterochromatin can form in the absence of stably bound HP1 proteins is consistent with studies of facultative heterochromatin in nucleated vertebrate erythrocytes, which ordinarily forms without HP1 proteins (26). In sum, the HIRA/ASF1a complex appears to drive chromosome condensation by acting upstream of characteristic heterochromatin modifications and associated proteins, most likely by contributing to nucleosome assembly through the deposition of histone H3/H4 complexes.
The final steps of SAHF formation consist of recruitment of macroH2A and HP1 proteins to chromatin. These two steps are not separable, based on a temporal analysis alone (86). However, we have shown here that the recruitment of macroH2A occurs in the absence of stably bound HP1 proteins. At this time, we cannot exclude the possibility that recruitment of HP1 proteins to chromatin depends on prior loading of histone macroH2A. However, since we know of no evidence in support of this idea, we propose that HP1 and macroH2A proteins are independently loaded onto chromatin at approximately the same time. We find that phosphorylation of HP1γ on S93 is required for its efficient recruitment to heterochromatin. Interestingly, another study found that HP1γ phosphorylated on this residue is localized to euchromatin in immortal and transformed cells (45) (it should be noted that these authors numbered the processed form of HP1γ and so referred to the same residue as S83). Thus, phosphorylation of this site might target HP1γ to different chromatin sites depending on the physiological context.
Remarkably, loading of abundant HP1 proteins onto chromatin is not required for two hallmarks of the senescent phenotype: expression of SA β-gal and senescence-associated cell cycle exit. We obviously cannot rule out the possibility that the residual chromatin-bound HP1 proteins are sufficient to mediate HP1 functions that are required for these senescence phenotypes. However, these results raise the possibility that HP1 proteins do not contribute to the acute onset of the senescent phenotype. Instead, HP1 proteins might be required for the long-term maintenance of SAHF and the senescent state. Alternatively, HP1 proteins might secure the senescent state in the face of genetic alterations or cellular perturbations that compromise other aspects of the senescence program. These ideas remain to be tested.
In contrast to our results with HP1 proteins, Narita and coworkers found through shRNA knock-down experiments that the HMGA protein HMGA2 is required for the formation of SAHF (56). This might suggest that HMGA proteins are incorporated into SAHF quite early during SAHF assembly, perhaps at the time of chromosome condensation. However, until this is directly demonstrated, we have omitted HMGA's point of entry into SAHF from our model.
Although Fig. 10 provides a framework model for the formation of SAHF, many other questions remain. For example, we do not know the triggers responsible for localization of HIRA and HP1 proteins to PML bodies. We do not know the specific reason for HIRA's localization to PML bodies and the spatial and mechanistic relationships between its localization to PML bodies and the formation of SAHF. Finally, we do not know the identity of the kinase responsible for the phosphorylation of HP1γ, the histone methyltransferase that methylates lysine 9 of histone H3 to create H3K9Me, or the factors required for the deposition of macroH2A into SAHF. Studies to answer these questions are ongoing. Meanwhile, the model proposed in Fig. 10 provides a valuable conceptual framework for thinking about these questions, as well as summarizing a large body of existing knowledge.
Supplementary Material
Acknowledgments
We thank Ken Zaret and Takashi Sekiya for purified recombinant histone H3.1, John Pehrson for macroH2A antibodies, Bill Hahn and Bob Weinberg for pBABE-RasV12, and all members of the Adams laboratory for critical discussions.
This study was supported by NIH grant GM062281 and Leukemia and Lymphoma Society grant 1520-04 to P.D.A. and an AFAR grant to R.Z.
Footnotes
Published ahead of print on 22 January 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, P. D., W. R. Sellers, S. K. Sharma, A. D. Wu, C. M. Nalin, and W. G. Kaelin, Jr. 1996. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol. Cell. Biol. 16:6623-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 3.Adkins, M. W., and J. K. Tyler. 2004. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 279:52069-52074. [DOI] [PubMed] [Google Scholar]
- 4.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad, K., and S. Henikoff. 2002. The histone variant h3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 6.Angermayr, M., and W. Bandlow. 2003. Permanent nucleosome exclusion from the Gal4p-inducible yeast GCY1 promoter. J. Biol. Chem. 278:11026-11031. [DOI] [PubMed] [Google Scholar]
- 7.Angermayr, M., U. Oechsner, K. Gregor, G. P. Schroth, and W. Bandlow. 2002. Transcription initiation in vivo without classical transactivators: DNA kinks flanking the core promoter of the housekeeping yeast adenylate kinase gene, AKY2, position nucleosomes and constitutively activate transcription. Nucleic Acids Res. 30:4199-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 2004. Global nucleosome occupancy in yeast. Genome Biol. 5:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackwell, C., K. A. Martin, A. Greenall, A. Pidoux, R. C. Allshire, and S. K. Whitehall. 2004. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol. Cell. Biol. 24:4309-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosch, A., and P. Suau. 1995. Changes in core histone variant composition in differentiating neurons: the roles of differential turnover and synthesis rates. Eur. J. Cell Biol. 68:220-225. [PubMed] [Google Scholar]
- 12.Braig, M., S. Lee, C. Loddenkemper, C. Rudolph, A. H. Peters, B. Schlegelberger, H. Stein, B. Dorken, T. Jenuwein, and C. A. Schmitt. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436:660-665. [DOI] [PubMed] [Google Scholar]
- 13.Brown, D. T., S. E. Wellman, and D. B. Sittman. 1985. Changes in the levels of three different classes of histone mRNA during murine erythroleukemia cell differentiation. Mol. Cell. Biol. 5:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campisi, J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120:513-522. [DOI] [PubMed] [Google Scholar]
- 15.Chen, X., J. Wang, D. Woltring, S. Gerondakis, and M. F. Shannon. 2005. Histone dynamics on the interleukin-2 gene in response to T-cell activation. Mol. Cell. Biol. 25:3209-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, Z., L. C. Trotman, D. Shaffer, H. K. Lin, Z. A. Dotan, M. Niki, J. A. Koutcher, H. I. Scher, T. Ludwig, W. Gerald, C. Cordon-Cardo, and P. P. Pandolfi. 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436:725-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collado, M., J. Gil, A. Efeyan, C. Guerra, A. J. Schuhmacher, M. Barradas, A. Benguria, A. Zaballos, J. M. Flores, M. Barbacid, D. Beach, and M. Serrano. 2005. Tumour biology: senescence in premalignant tumours. Nature 436:642. [DOI] [PubMed] [Google Scholar]
- 18.Costanzi, C., and J. R. Pehrson. 2001. MACROH2A2, a new member of the MACROH2A core histone family. J. Biol. Chem. 276:21776-21784. [DOI] [PubMed] [Google Scholar]
- 19.Daganzo, S. M., J. P. Erzberger, W. M. Lam, E. Skordalakes, R. Zhang, A. A. Franco, S. J. Brill, P. D. Adams, J. M. Berger, and P. D. Kaufman. 2003. Structure and function of the conserved core of histone deposition protein Asf1. Curr. Biol. 13:2148-2158. [DOI] [PubMed] [Google Scholar]
- 20.de Stanchina, E., E. Querido, M. Narita, R. V. Davuluri, P. P. Pandolfi, G. Ferbeyre, and S. W. Lowe. 2004. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell 13:523-535. [DOI] [PubMed] [Google Scholar]
- 21.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Earnshaw, W., B. Bordwell, C. Marino, and N. Rothfield. 1986. Three human chromosomal autoantigens are recognized by sera from patients with anti-centromere antibodies. J. Clin. Investig. 77:426-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferbeyre, G., E. de Stanchina, E. Querido, N. Baptiste, C. Prives, and S. W. Lowe. 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 14:2015-2027. [PMC free article] [PubMed] [Google Scholar]
- 24.Fogal, V., M. Gostissa, P. Sandy, P. Zacchi, T. Sternsdorf, K. Jensen, P. P. Pandolfi, H. Will, C. Schneider, and G. Del Sal. 2000. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 19:6185-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Funayama, R., M. Saito, H. Tanobe, and F. Ishikawa. 2006. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 175:869-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert, N., S. Boyle, H. Sutherland, J. de Las Heras, J. Allan, T. Jenuwein, and W. A. Bickmore. 2003. Formation of facultative heterochromatin in the absence of HP1. EMBO J. 22:5540-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert, N., S. Gilchrist, and W. A. Bickmore. 2005. Chromatin organization in the mammalian nucleus. Int. Rev. Cytol. 242:283-336. [DOI] [PubMed] [Google Scholar]
- 28.Green, E. M., A. J. Antczak, A. O. Bailey, A. A. Franco, K. J. Wu, J. R. Yates III, and P. D. Kaufman. 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenall, A., E. S. Williams, K. A. Martin, J. M. Palmer, J. Gray, C. Liu, and S. K. Whitehall. 2006. Hip3 interacts with the HIRA proteins Hip1 and Slm9 and is required for transcriptional silencing and accurate chromosome segregation. J. Biol. Chem. 281:8732-8739. [DOI] [PubMed] [Google Scholar]
- 30.Grove, G. W., and A. Zweidler. 1984. Regulation of nucleosomal core histone variant levels in differentiating murine erythroleukemia cells. Biochemistry 23:4436-4443. [DOI] [PubMed] [Google Scholar]
- 31.Guo, A., P. Salomoni, J. Luo, A. Shih, S. Zhong, W. Gu, and P. P. Pandolfi. 2000. The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2:730-736. [DOI] [PubMed] [Google Scholar]
- 32.Hake, S. B., B. A. Garcia, E. M. Duncan, M. Kauer, G. Dellaire, J. Shabanowitz, D. P. Bazett-Jones, C. D. Allis, and D. F. Hunt. 2006. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 281:559-568. [DOI] [PubMed] [Google Scholar]
- 33.Hall, C., D. M. Nelson, X. Ye, K. Baker, J. A. DeCaprio, S. Seeholzer, M. Lipinski, and P. D. Adams. 2001. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol. Cell. Biol. 21:1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Hayflick, L. 1965. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 37:614-636. [DOI] [PubMed] [Google Scholar]
- 36.Henikoff, S., E. McKittrick, and K. Ahmad. 2004. Epigenetics, histone H3 variants, and the inheritance of chromatin states. Cold Spring Harbor Symp. Quant. Biol. 69:235-243. [DOI] [PubMed] [Google Scholar]
- 37.Herbig, U., and J. M. Sedivy. 2006. Regulation of growth arrest in senescence: telomere damage is not the end of the story. Mech. Ageing Dev. 127:16-24. [DOI] [PubMed] [Google Scholar]
- 38.Janzen, V., R. Forkert, H. E. Fleming, Y. Saito, M. T. Waring, D. M. Dombkowski, T. Cheng, R. A. Depinho, N. E. Sharpless, and D. T. Scadden. 2006. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16(INK4a). Nature 443:421-426. [DOI] [PubMed] [Google Scholar]
- 39.Kaufman, P. D., J. L. Cohen, and M. A. Osley. 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18:4793-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krawitz, D. C., T. Kama, and P. D. Kaufman. 2002. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 22:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krimer, D. B., G. Cheng, and A. I. Skoultchi. 1993. Induction of H3.3 replacement histone mRNAs during the precommitment period of murine erythroleukemia cell differentiation. Nucleic Acids Res. 21:2873-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnamurthy, J., M. R. Ramsey, K. L. Ligon, C. Torrice, A. Koh, S. Bonner-Weir, and N. E. Sharpless. 2006. p16(INK4a) induces an age-dependent decline in islet regenerative potential. Nature 443:453-457. [DOI] [PubMed] [Google Scholar]
- 43.Lechner, M. S., G. E. Begg, D. W. Speicher, and F. J. Rauscher III. 2000. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol. Cell. Biol. 20:6449-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 45.Lomberk, G., D. Bensi, M. E. Fernandez-Zapico, and R. Urrutia. 2006. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 8:407-415. [DOI] [PubMed] [Google Scholar]
- 46.Loppin, B., E. Bonnefoy, C. Anselme, A. Laurencon, T. L. Karr, and P. Couble. 2005. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437:1386-1390. [DOI] [PubMed] [Google Scholar]
- 47.Loyola, A., T. Bonaldi, D. Roche, A. Imhof, and G. Almouzni. 2006. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 24:309-316. [DOI] [PubMed] [Google Scholar]
- 48.Mahy, N. L., P. E. Perry, and W. A. Bickmore. 2002. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159:753-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKittrick, E., P. R. Gafken, K. Ahmad, and S. Henikoff. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 101:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meijsing, S. H., and A. E. Ehrenhofer-Murray. 2001. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 15:3169-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michaloglou, C., L. C. Vredeveld, M. S. Soengas, C. Denoyelle, T. Kuilman, C. M. van der Horst, D. M. Majoor, J. W. Shay, W. J. Mooi, and D. S. Peeper. 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436:720-724. [DOI] [PubMed] [Google Scholar]
- 52.Mito, Y., J. G. Henikoff, and S. Henikoff. 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37:1090-1097. [DOI] [PubMed] [Google Scholar]
- 53.Molofsky, A. V., S. G. Slutsky, N. M. Joseph, S. He, R. Pardal, J. Krishnamurthy, N. E. Sharpless, and S. J. Morrison. 2006. Increasing p16(INK4a) expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443:448-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moshkin, Y. M., J. A. Armstrong, R. K. Maeda, J. W. Tamkun, P. Verrijzer, J. A. Kennison, and F. Karch. 2002. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 16:2621-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mousson, F., A. Lautrette, J. Y. Thuret, M. Agez, R. Courbeyrette, B. Amigues, E. Becker, J. M. Neumann, R. Guerois, C. Mann, and F. Ochsenbein. 2005. Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc. Natl. Acad. Sci. USA 102:5975-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narita, M., M. Narita, V. Krizhanovsky, S. Nunez, A. Chicas, S. A. Hearn, M. P. Myers, and S. W. Lowe. 2006. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126:503-514. [DOI] [PubMed] [Google Scholar]
- 57.Narita, M., S. Nunez, E. Heard, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 58.Ooi, S. L., J. R. Priess, and S. Henikoff. 2006. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pantazis, P., and W. M. Bonner. 1984. Specific alterations in the pattern of histone-3 synthesis during conversion of human leukemic cells to terminally differentiated cells in culture. Differentiation 28:186-190. [DOI] [PubMed] [Google Scholar]
- 60.Parada, L., and T. Misteli. 2002. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 12:425-432. [DOI] [PubMed] [Google Scholar]
- 61.Pearson, M., R. Carbone, C. Sebastiani, M. Cioce, M. Fagioli, S. Saito, Y. Higashimoto, E. Appella, S. Minucci, P. P. Pandolfi, and P. G. Pelicci. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207-210. [DOI] [PubMed] [Google Scholar]
- 62.Perche, P. Y., C. Vourc'h, L. Konecny, C. Souchier, M. Robert-Nicoud, S. Dimitrov, and S. Khochbin. 2000. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 10:1531-1534. [DOI] [PubMed] [Google Scholar]
- 63.Phelps-Durr, T. L., J. Thomas, P. Vahab, and M. C. Timmermans. 2005. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell. 17:2886-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pina, B., and P. Suau. 1987. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol. 123:51-58. [DOI] [PubMed] [Google Scholar]
- 65.Ray-Gallet, D., J. P. Quivy, C. Scamps, E. M. Martini, M. Lipinski, and G. Almouzni. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9:1091-1100. [DOI] [PubMed] [Google Scholar]
- 66.Rogakou, E. P., and K. E. Sekeri-Pataryas. 1999. Histone variants of H2A and H3 families are regulated during in vitro aging in the same manner as during differentiation. Exp. Gerontol. 34:741-754. [DOI] [PubMed] [Google Scholar]
- 67.Salomoni, P., and P. P. Pandolfi. 2002. The role of PML in tumor suppression. Cell 108:165-170. [DOI] [PubMed] [Google Scholar]
- 68.Schwabish, M. A., and K. Struhl. 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22:415-422. [DOI] [PubMed] [Google Scholar]
- 69.Schwartz, B. E., and K. Ahmad. 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463-473. [DOI] [PubMed] [Google Scholar]
- 71.Sharp, J. A., A. A. Franco, M. A. Osley, P. D. Kaufman, D. C. Krawitz, T. Kama, E. T. Fouts, and J. L. Cohen. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sherwood, P. W., S. V.-M. Tsang, and M. A. Osley. 1993. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:28-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Mahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spector, M. S., A. Raff, H. DeSilva, K. Lee, and M. A. Osley. 1997. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 17:545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sutton, A., J. Bucaria, M. A. Osley, and R. Sternglanz. 2001. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 77.Tang, Y., M. V. Poustovoitov, K. Zhao, M. Garfinkel, A. Canutescu, R. Dunbrack, P. D. Adams, and R. Marmorstein. 2006. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 13:921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402:555-560. [DOI] [PubMed] [Google Scholar]
- 79.Tyler, J. K., K. A. Collins, J. Prasad-Sinha, E. Amiott, M. Bulger, P. J. Harte, R. Kobayashi, and J. T. Kadonaga. 2001. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 21:6574-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Urban, M. K., and A. Zweidler. 1983. Changes in nucleosomal core histone variants during chicken development and maturation. Dev. Biol. 95:421-428. [DOI] [PubMed] [Google Scholar]
- 81.Wirbelauer, C., O. Bell, and D. Schubeler. 2005. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 19:1761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wright, W. E., and J. W. Shay. 2002. Historical claims and current interpretations of replicative aging. Nat. Biotechnol. 20:682-688. [DOI] [PubMed] [Google Scholar]
- 83.Wunsch, A. M., and J. Lough. 1987. Modulation of histone H3 variant synthesis during the myoblast-myotube transition of chicken myogenesis. Dev. Biol. 119:94-99. [DOI] [PubMed] [Google Scholar]
- 84.Ye, X., B. Zerlanko, R. Zhang, N. Somaiah, M. Lipinski, and P. D. Adams. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci (SAHF). Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 85.Zhang, R., S.-T. Liu, W. Chen, B. Bonner, J. Pehrson, T. J. Yen, and P. D. Adams. 2007. HP1 proteins are essential for a dynamic nuclear response that rescues the function of perturbed heterochromatin in primary human cells. Mol. Cell. Biol. 27:949-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang, R., M. V. Poustovoitov, X. Ye, H. A. Santos, W. Chen, S. M. Daganzo, J. P. Erzberger, I. G. Serebriiskii, A. A. Canutescu, R. L. Dunbrack, J. R. Pehrson, J. M. Berger, P. D. Kaufman, and P. D. Adams. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8:19-30. [DOI] [PubMed] [Google Scholar]
- 87.Zhao, J., J. Herrera-Diaz, and D. S. Gross. 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 25:8985-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.