Abstract
Circadian rhythms control the temporal arrangement of molecular, physiological, and behavioral processes within an organism and also synchronize these processes with the external environment. A cell autonomous molecular oscillator, consisting of interlocking transcriptional/translational feedback loops, drives the approximately 24-hour duration of these rhythms. The cryptochrome protein (CRY) plays a central part in the negative feedback loop of the molecular clock by translocating to the nucleus and repressing CLOCK and BMAL1, two transcription factors that comprise the positive elements in this cycle. In order to gain insight into the inner workings of this feedback loop, we investigated the structure/function relationships of Xenopus laevis CRY1 (xCRY1) and xCRY2 in cultured cells. The C-terminal tails of both xCRY1 and xCRY2 are sufficient for their nuclear localization but achieve it by different mechanisms. Through the generation and characterization of xCRY/photolyase chimeras, we found that the second half of the photolyase homology region (PHR) of CRY is important for repression through facilitating interaction with BMAL1. Characterization of these functional domains in CRYs will help us to better understand the mechanism of the known roles of CRYs and to elucidate new intricacies of the molecular clock.
Organisms ranging from cyanobacteria to humans exhibit circadian rhythms in many processes, from gene expression to cell physiology and from hormone levels to locomotor activity. Circadian rhythms are approximately 24 hours in duration and persist in constant conditions. These oscillations do not accelerate or decelerate within a physiological range of temperatures and, importantly, can be reset by cues from the environment. Having an internal timekeeping mechanism allows an organism to temporally arrange physiological processes internally and also to anticipate changes in the external environment (reviewed in reference 1).
The clocks driving these rhythms are intracellular mechanisms composed of interlocking transcriptional/translational feedback loops (reviewed in reference 23). At the core of the vertebrate molecular oscillator is a negative feedback loop that is necessary for rhythmicity (19). Two positive elements, CLOCK and BMAL1, which are basic helix-loop-helix PAS transcription factors, heterodimerize and bind to E-box enhancer elements in the promoters of the Period (Per1, Per2, and Per3) and Cryptochrome (Cry1 and Cry2) genes, activating their transcription. The Per and Cry mRNAs are then translated, and the proteins accumulate in the cytoplasm. PER, CRY, and casein kinase Iɛ (CKIɛ) proteins form a complex in the cytoplasm, which translocates into the nucleus, where it represses CLOCK-BMAL1-mediated transcription. The repression complex is eventually degraded or dismantled, CLOCK-BMAL1 transcription is activated, and the cycle begins again (reviewed in reference 1).
Since the molecular clock is temporally precise and longer in duration than most intracellular feedback loops, many regulatory strategies may be required to maintain this unique oscillation. The detailed mechanisms that contribute to the cycle's robust rhythmicity and stable period are largely unknown. We hypothesize that one of these regulation steps may be to control the intracellular localization of the CRY proteins, in order to prevent repression before the appropriate time.
Both CRY proteins, CRY1 and CRY2, are components of the core molecular oscillator in vertebrates, and it has been shown that their repression of CLOCK-BMAL1-mediated transcription is essential for rhythmicity at both the molecular and behavioral levels (19, 21). The inhibition of the CLOCK-BMAL1 heterodimer is independent of light and is strong even at low doses of CRY (6, 11, 25). While the mechanism for how this repression occurs is not known, some ideas have been proposed. One hypothesis is that CRYs repress CLOCK-BMAL1 by reducing CLOCK-BMAL1's affinity for the E box (17). Another idea is that CRYs inhibit CLOCK-BMAL1 by interacting with the heterodimer and then recruiting histone deacetylases (16) or inhibiting histone acetylases (5).
In order to gain insight into these aspects of the molecular clock, we sought to identify and define functional domains in CRYs that are responsible for their subcellular localization and ability to repress CLOCK-BMAL1. We chose to examine CRYs from Xenopus laevis for identification of key regions of the protein that are involved in the circadian function of CRYs, because these animals, unlike mammals, possess both CRYs and also a closely related protein, Xenopus 6-4 photolyase (xPHOTO). Both Xenopus CRYs (xCRYs) and xPHOTO are members of the CRY/PHOTO family, which share significant sequence similarity over the main body (known as the photolyase homology region [PHR]) but have C-terminal tails that differ greatly in length and amino acid composition (reviewed in reference 2)). Despite the high degree of similarity in the PHR, xCRYs can repress CLOCK-BMAL1-mediated transcription, while xPHOTO, even when expressed at high levels, cannot (24). We have previously shown that the C-terminal tails of both CRY proteins are necessary for their nuclear localization in COS7 cells (24), while the PHRs are sufficient for repression. The combination of primary amino acid sequence similarity and functional diversity between xCRYs and xPHOTO provides an optimal paradigm for studying structure/function relationships.
Here, we report that the C-terminal tails of xCRY1 and xCRY2 (formerly known as xCRY2b) are sufficient for nuclear localization and appear to have distinct nuclear localization mechanisms. We also demonstrate, through the generation and characterization of xCRY/xPHOTO chimeras, that residues that are conserved in repressive CRYs, but not in xPHOTO, in the second half of the PHR are required for full repression of xCLOCK-xBMAL1 via facilitating interaction with xBMAL1. Interestingly, even with these residues replaced with residues from xPHOTO, the chimeras still exhibit some repression.
MATERIALS AND METHODS
Plasmids. (i) Cry C terminus-nocturnin constructs.
The C termini of the xCry1 and xCry2 genes (residues E550 to R596 for xCRY1 and residues G515 to R556 for xCRY2) were amplified by PCR and then subcloned in frame between enhanced green fluorescent protein (eGFP) and nocturnin (NOC) in the peGFP-C2 vector (BD Biosciences, Clontech). To make serial deletions of the xCRY1 C terminus in this construct, EcoRI sites were inserted using the QuikChange site-directed mutagenesis kit (Stratagene). The clones were then digested and religated with T4 ligase, deleting a portion of the region encoding the C terminus. Four deletion mutants were generated: pEGFP-xCry1CtermΔ 1-Noc, which is missing amino acids E550 to G564; pEGFP-xCry1CtermΔ 2-Noc, which is missing amino acids S565 to D574; pEGFP-xCry1CtermΔ 1-2-Noc, which is missing amino acids S550 to D574; and pEGFP-xCry1CtermΔ 2-3-Noc, which is missing amino acids S565 to R596. To introduce amino acid substitutions into the C terminus of xCRY2, site-directed mutagenesis was also used, to change either a single residue or a small cluster of residues.
(ii) xCry1/xPhoto chimeras.
Both xCry/xPhoto chimeras were cloned in the pCMV-Tag2b vector (Stratagene) in frame with the FLAG tag in the vector and verified by sequencing. Both xCry constructs (and the chimeras that were derived from them) contain a portion of the 5′ untranslated region following the N-terminal FLAG tag that is translated in the resulting fusion proteins. We verified that the resulting CRY proteins showed repression comparable to that of wild-type mammalian CRY1 (mCRY1) and mCRY2. To generate the xCry1/xPhoto chimeras, the region of xCry1 corresponding to amino acids 1 to 257 and the region of xPhoto corresponding to amino acids 258 to stop were amplified. A unique AflII site was added to the 3′ end of the xCry1 piece and to the 5′ end of the xPhoto piece to facilitate joining of the fragments but was designed to prevent amino acid substitution. To generate the xCry2/xPhoto chimera, the region of xPhoto corresponding to amino acids 262 to stop was amplified with a SalI site added at the 5′ end and an ApaI site added at the 3′ end. pCMV-xCry2 was digested with SalI and ApaI, and the xPhoto fragment was inserted. To create the nuclear localization signal (NLS) chimera constructs, the xCry/xPhoto chimeras were subcloned into a pCMV vector which had a heterologous NLS (PPKKKRKVEGEF) cloned in frame with the FLAG tag (24).
(iii) xCry1/Photo chimeras with further xCry1 substitutions.
The xCry1/Photo chimeras with further xCry1 substitutions were made by an overlap extension cloning method (20). The xCRY1/P1,C2,P3-5 chimera consists of xCRY1 residues 1 to 257, xPHOTO residues 258 to 284, xCRY1 residues 285 to 315, and xPHOTO residues 315 to stop. xCRY1/P1-2,C3,P4-5 consists of xCRY1 amino acids 1 to 257, xPHOTO amino acids 258 to 314, xCRY1 amino acids 316 to 348, and xPHOTO amino acids 347 to stop. xCRY1/P1-3,C4,P5 consists of xCRY1 residues 1 to 257, xPHOTO residues 258 to 346, xCRY1 residues 349 to 496, and xPHOTO residues 494 to stop. All constructs were verified by sequencing.
Cell culture, transient transfection, and Western blotting analysis.
COS7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1 mM sodium pyruvate. COS7 cells were transfected with expression plasmids, using Fugene (Roche), according to the manufacturer's instructions. For Western blotting analysis, lysates were harvested using 1× passive lysis buffer from the dual luciferase reporter assay system (Promega). Total protein levels were measured using a Bradford-based protein assay kit (Bio-Rad), and 20 μg of total protein from each sample was run on a 10% sodium dodecyl sulfate-polyacrylamide gel and then transferred onto a polyvinylidene difluoride membrane (Bio-Rad). The membrane was then blocked with BLOTTO solution (0.1% Tween 20 and 5% dry nonfat milk powder in Tris-buffered saline [TBS], pH 7.4) overnight at 4°C. The membrane was treated for 1 h at room temperature (RT) with the primary antibody, monoclonal mouse anti-FLAG M2 antibody (Sigma), which was diluted 1:1,000 in BLOTTO. The membrane was then washed twice for 10 min at RT with BLOTTO and then twice more for 10 min at RT with TBS-Tween (0.1% Tween 20 in 1× TBS, pH 7.4), followed by incubation for 30 min at RT with the secondary antibody, anti-mouse immunoglobulin G-peroxidase (Chemicon), which was diluted 1:1,500 in BLOTTO. The membrane was washed four times with TBS-Tween for 15 min at RT. The bound antibody was visualized using a chemiluminescence Western blotting kit (Roche).
Immunocytochemistry.
COS7 cells were seeded on coverslips in six-well cell culture dishes, transfected 24 h later with a total of 1 μg DNA as described above, and then allowed to incubate for 24 h before fixation. Cells expressing eGFP constructs were rinsed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 15 min, washed two more times with PBS, and then mounted onto slides, using Fluoromount G (Electron Microscopy Sciences). Cells expressing FLAG-tagged constructs were fixed with ice-cold methanol for 15 min and were put in blocking solution (0.3% Triton, 1% protease-free bovine serum albumin, 0.25% l-carageenan, 0.1% sodium azide in 1× TBS, pH 7.6) overnight at 4°C. These slides were then treated with monoclonal mouse anti-FLAG (Sigma), diluted 1:1,000 in blocking solution for 1.5 h at RT, washed with PBS three times, and then treated with digoxigenin (DIG)-conjugated sheep anti-mouse secondary antibody (Chemicon), diluted 1:1,000 in blocking solution, for 1 h at RT. The coverslips were then washed with PBS, treated with 1:1,000 rhodamine-conjugated rabbit anti-DIG tertiary antibody (Boehringer) for 1 h at RT, washed, stained for 5 min with Hoechst's stain (Sigma), and mounted as described above. The cells were viewed using an Olympus inverted epifluorescence microscope (IX-70). A blind count of 200 cells for each sample was done, and the percentage of cells in each cell compartment (nuclear, cytoplasmic, or both) was calculated.
Immunoprecipitation.
COS7 cells, which were 60% confluent in 10-cm dishes, were transfected in duplicate with 150 ng of null-Renilla luc, 2 μg of per-luciferase reporter gene, 1.5 μg of xClock, 1.5 μg of xBmal1, and either 1.5 μg of wild-type Cry or 3.0 μg of xCry/xPhoto in 600 μl Opti-MEM (Invitrogen) and 22.5 μl Fugene (Roche). After 24 h, the cells were washed with sterile PBS, harvested by trypsinization, and solublized in 0.7 ml TGED buffer (50 mM Tris [pH 7.4], 100 mM NaCl, 1 mM EDTA, 5% glycerol, 0.5 mM dithiothreitol) with protease inhibitors (Sigma) and 0.5% Triton. The duplicate lysates were pooled, nutated at 4°C for 10 min, and centrifuged for 20 min at 13,000 rpm. Total protein levels were measured using a Bradford-based protein assay kit (Bio-Rad). Lysates were analyzed by Western blotting to confirm protein expression levels. Equal amounts of total protein, 1.2 mg, were loaded on anti-FLAG-conjugated resin (Sigma), which was prepared according to the manufacturer's instructions. The lysate-resin mix was nutated overnight at 4°C. Bound proteins were washed three times with wash buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl) and then were eluted with 0.25 μg/μl 3XFLAG peptide (Sigma) in wash buffer for 1 h at 4°C. The eluted protein was resolved on a sodium dodecyl sulfate-polyacrylamide gel, transferred, and then blotted with monoclonal mouse anti-V5 (Invitrogen) or monoclonal mouse anti-FLAG M2 (Sigma) antibody to detect xBMAL1-V5 or FLAG-xCRY protein, respectively.
Luciferase repression assay.
The luciferase reporter assay was carried out as previously described (24). COS7 cells were transfected with 200 ng of per-luciferase reporter gene and 15 ng of null-Renilla luc (14), which was used as an internal control, and 150 ng of Clock, Bmal1, wild-type xCry, and/or chimera plasmid was added as indicated below each graph in the figures. The total amount of DNA transfected was kept constant at 1 μg by supplementation with empty pCMV-Tag2b vector (Stratagene). Transcriptional activity was assessed with the dual-luciferase reporter assay system (Promega) by measuring the ratio of firefly luciferase activity to Renilla luciferase activity in each cellular lysate.
xCRY1 PHR structure model.
A homology model of the xCRY1 PHR was generated and evaluated using tools available at the SWISS-MODEL Workspace (http://swissmodel.expasy.org/workspace/). The model was constructed by the SWISS-MODEL protein homology modeling server (7) using the sequence of the xCRY1 PHR and the coordinates of an Anacystis nidulans cyclobutane pyrimidine dimer photolyase structure (9) (Protein Data Bank [PDB] code 1OWL). The stereochemical characteristics of the model, analyzed using PROCHECK (12), were similar to those of the template structure, with 88.5% and 9.3% of the residues in the core and allowed regions, respectively, of the Ramachandran plot (compared to 91.9 and 7.8% in the template) and an overall G-factor of 0.05 (compared to 0.42). This model was then visualized using PyMOL (4) (http://www.pymol.org).
RESULTS
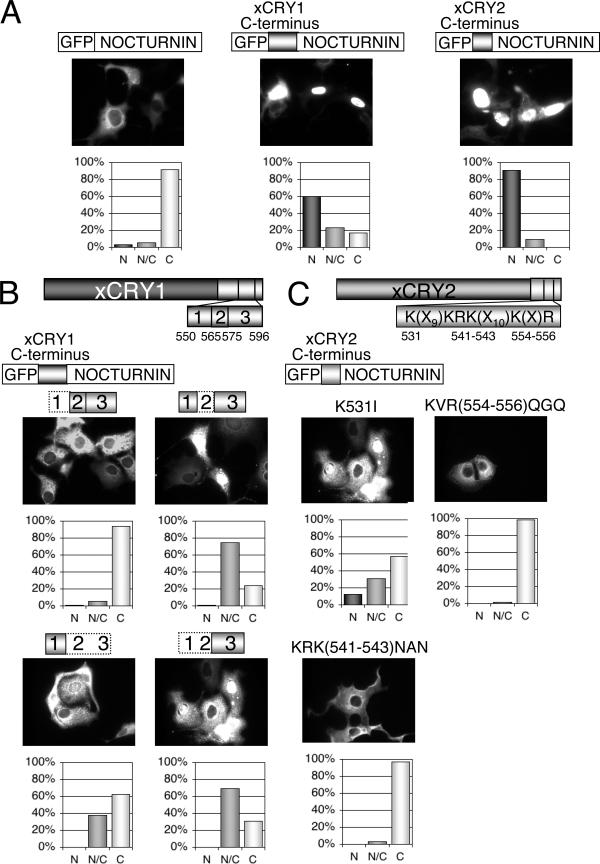
In order to gain insight into the role of CRYs in the molecular circadian clock, we sought to identify and further define functional domains in the CRY proteins of Xenopus laevis. We have previously shown that the C-terminal tails of both xCRY proteins are necessary for each protein's nuclear localization in vitro (24). To determine whether these regions are sufficient for nuclear localization, we fused these portions of the C-terminal tail of each xCRY protein in frame with eGFP and NOC, a cytoplasmic protein, and assayed these constructs for subcellular localization (Fig. 1). While eGFP-NOC was localized in the cytoplasm as expected, both eGFP-xCRY1Cterm-NOC and eGFP-xCRY2Cterm-NOC were found predominantly in the nucleus (Fig. 1A). Therefore, residues E550 to R596 of xCRY1 and residues G515 to R556 of xCRY2 are not only necessary but also sufficient for nuclear localization in COS7 cells.
FIG. 1.
The C termini of xCRY1 and xCRY2 are sufficient for nuclear localization of the CRY protein when expressed in COS7 cells. (A) Visualization of NOC, when cloned in frame with the C terminus of either xCRY1 (residues 550 to 596) or xCRY2 (residues 515 to 556). All constructs were cloned in the pEGFP-C2 vector and therefore were tagged with eGFP on their N terminus, as indicated in each schematic. One microgram of each plasmid was transfected into COS7 cells. N, N/C, and C, nuclear, nuclear and cytoplasmic, and cytoplasmic localization, respectively. (B) Effect of serial deletions of the C terminus of xCRY1 on the localization of GFP-CRYCterm-NOC. The C terminus was divided into three regions: 1, residues 550 to 564; 2, residues 565 to 574; and 3, residues 575 to 596. Deletions were made as indicated in each schematic, and then each construct was tested for localization, as described for panel A. (C) Effect of amino acid substitutions in the putative canonical bipartite NLS in the C terminus of xCRY2 on the localization of a cytoplasmic protein. Substitutions were made as indicated in each schematic, and then the localization of each mutant was tested using immunocytochemistry as described for panel A.
Next, we set out to define which residues within these domains are important for nuclear localization. In the C-terminal tail of xCRY1, residues 595 to 612 comprise a sequence that weakly resembles a bipartite NLS. This sequence is highly conserved in mCRY1 and has been shown to be functional in that protein. To determine if this sequence is sufficient for localization of xCRY1, we cloned it in frame with eGFP and NOC and assayed for subcellular localization. We found that there was only a small increase of this NOC fusion protein in the nucleus or in both the nucleus and the cytoplasm compared to eGFP-NOC without the putative NLS (37% versus 22%, respectively). It is for this reason that we did not include this putative NLS in our eGFP-xCry1Cterm-NOC constructs.
Since we found that the putative NLS in the C-terminal portion of xCRY1 was not sufficient for nuclear localization, we then investigated the area comprised of residues 550 to 596 into three regions, i.e., residues 550 to 564, residues 565 to 574, and residues 575 to 596, and then made various deletions in the xCRY1 C-terminal portion of eGFP-xCRY1Cterm-NOC. Any region or combination of regions that we deleted within the xCRY1 C terminus in the eGFP-xCRY1Cterm-NOC fusion protein led to partial or complete loss of nuclear localization (Fig. 1B). Deletion of region 1 from the xCRY1 C-terminal region had the most severe effect on NOC localization, but when it alone was fused to NOC, it was not sufficient to bring NOC into the nucleus. While we have shown that the xCRY1 C-terminal tail, residues 550 to 596, is sufficient for nuclear localization in these cells, it seems that this function is not confined to a single small domain.
In contrast, the C-terminal tail of xCRY2 contains a putative canonical nuclear NLS, which consists of six positively charged residues in a K(X9)KRK(X10)K(X)R pattern (15). In order to test the functionality of this sequence, we replaced each cluster of basic residues with noncharged residues in the xCRY2Cterm-NOC fusion protein. When tested for subcellular localization, all of the mutated fusion proteins exhibited decreased nuclear localization; in fact, mutation of KRK541-543 or KVR554-556 resulted in complete loss of nuclear localization (Fig. 1C). Mutation of these same residues in full-length xCRY2 also resulted in complete loss of nuclear localization (data not shown). Therefore, xCRY2 has a functional bipartite NLS in its C-terminal tail.
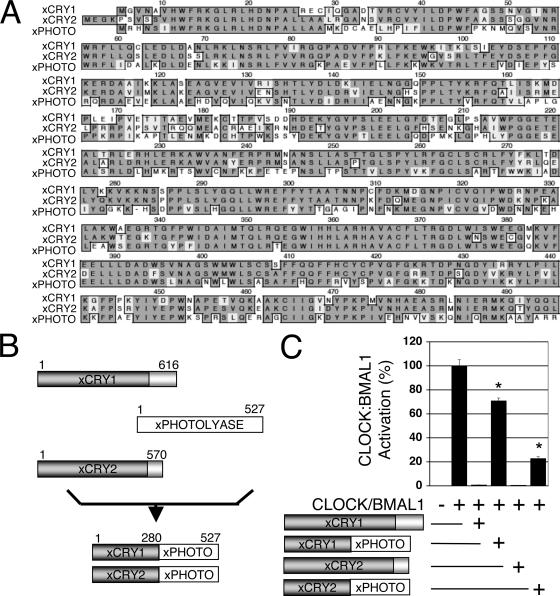
Despite the fact that the C-terminal tails of xCRYs seem to be necessary and sufficient for the proteins' nuclear localization, the PHR of xCRY1 and xCRY2 is sufficient for repression of CLOCK-BMAL1-mediated activation when it is localized to the nucleus by a heterologous NLS (24). Although the primary amino acid sequences of xCRY1, xCRY2, and xPHOTO are 85% similar in this region (Fig. 2A), the PHRs of xCRYs can repress CLOCK-BMAL1-mediated activation, but xPHOTO cannot (24). The fact that these proteins share a high degree of sequence similarity yet have distinct functions allowed us to use a chimeric protein approach to characterize the function of the PHR of xCRY in the repression mechanism of CRYs and also to define functional domains within this region, while preserving the protein's structural integrity.
FIG. 2.
xCRY/PHOTO chimeras show reduced repression of CLOCK-BMAL1-mediated activation. (A) Protein sequence alignment of the PHRs of xCRY1, xCRY2, and xPHOTO. The xCRY sequences are very similar to those of xPHOTO in the PHR, which includes all of the protein except the highly variable C terminus. Conserved amino acid residues are indicated by gray shading, while the nonconserved amino acids are not shaded. (B) Generation of xCry/xPhoto chimeras. Both chimeras were tagged with a FLAG tag on their N terminus. xCRY1/xPHOTO consists of xCRY1 amino acids 1 to 257 and xPHOTO amino acids 258 to stop. xCRY2/xPHOTO consists of xCRY2 amino acids 1 to 261 and xPHOTO amino acids 262 to stop. Both constructs were fused in a conserved region to avoid amino acid substitution at the fusion site. (C) Luciferase repression assay. In this experiment, COS7 cells were transfected with the Per-luciferase reporter gene, and 150 ng of Clock, Bmal1, wild-type xCry, and/or chimera plasmids were added as indicated. Each data point is averaged from six replicates, with the error bars representing the standard error of the mean. Both chimeras, xCRY1/xPHOTO and xCRY2/xPHOTO, exhibit decreased repression significantly different from each wild-type CRY (P < 0. 01).
xCRY/xPHOTO chimeras which had the first half of either xCRY1 or xCRY2 and the second half of PHOTO were generated (Fig. 2B). Since xCRYs and xPHOTO are so similar, this was equivalent to making 46 amino substitutions in the second half of the PHR of xCRY. These chimeras were then tested for their ability to repress CLOCK-BMAL1-mediated transcription in a luciferase repression assay in COS7 cells. Both chimeras were still able to repress CLOCK-BMAL1, but they exhibited a significant decrease in repression compared to wild-type xCRY (P < 0.01) (Fig. 2C).
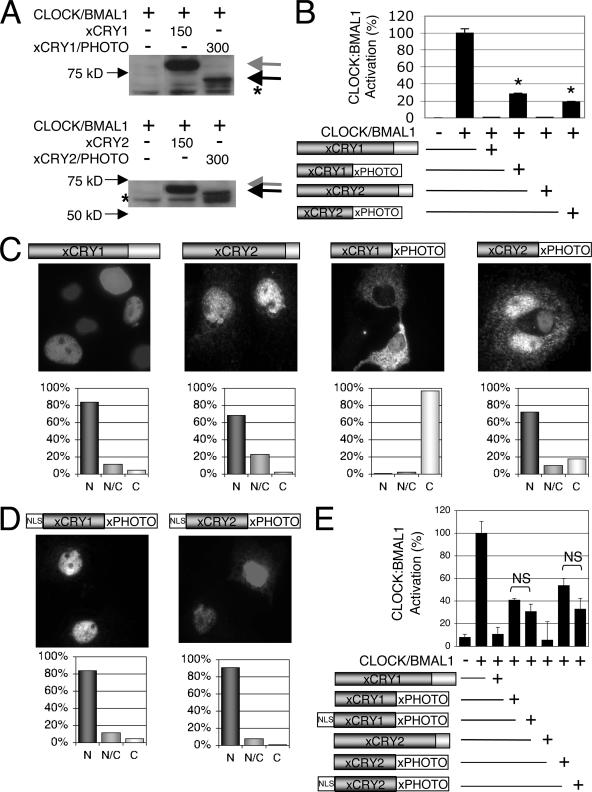
The decreased repression shown by the chimeras in the luciferase assay could be for many reasons; some examples include decreased protein stability, improper subcellular localization, or decreased interaction with CLOCK-BMAL1. To see if the chimeras' decreased repression is due to decreased protein expression or stability, the protein levels of the chimeras versus wild-type xCRYs were examined. These experiments demonstrated that the chimeric proteins were present at somewhat reduced levels compared to their wild-type xCRY counterparts: xCRY1/xPHOTO expression was reduced approximately twofold, and xCRY2/xPHOTO expression was reduced approximately fourfold (Fig. 3A). When the luciferase repression assay was repeated using doses of plasmid adjusted to yield comparable protein expression levels, the chimeras' abilities to repress CLOCK-BMAL1 improved slightly but still were significantly lower than that of wild-type xCRY (P < 0.01) (Fig. 3B). Therefore, the decrease in repression shown by the chimeras cannot be fully explained by decreased protein expression or stability.
FIG. 3.
Decreased repression exhibited by chimeras is not due to decreased protein expression or improper subcellular localization. (A) Protein expression levels of xCRY and chimera proteins. COS7 cells were transfected with the indicated plasmids. A DNA dose of 150 ng was used for each clock plasmid, except for the chimeras, of which 300 ng was added to compensate for decreased expression levels. Twenty micrograms of total protein lysate was run, and both membranes were blotted with anti-FLAG (1:1,000; Sigma) and anti-mouse-POD (1:1,500; Chemicon). The gray arrowheads denote the wild-type CRY band, while the black arrowhead denotes the chimera band. Nonspecific bands are indicated with asterisks. (B) Luciferase repression assay with protein normalization. Instead of using the same DNA dose for each construct, these data were normalized to protein level. The experiment was done as described for Fig. 2, except for the following changes. The black bars indicate activation seen when chimera plasmids or wild-type Cry DNA (300 ng and 150 ng, respectively) was added as indicated. These DNA doses lead to equal protein levels of wild-type CRYs and the chimeras, as verified by Western blotting with anti-FLAG antibody. Both chimeras, xCRY1/xPHOTO and xCRY2/xPHOTO, still exhibit decreased repression significantly different from both wild-type CRYs (P < 0.01). (C) Immunocytochemistry of wild-type xCRYs and chimeras. One microgram of each plasmid was transfected separately into COS7 cells. These cells were then fixed and incubated with monoclonal mouse anti-FLAG M2 primary antibody, DIG-conjugated sheep anti-mouse secondary antibody, rhodamine-conjugated goat anti-DIG tertiary antibody, and Hoechst's stain. The florescence shown here is the rhodamine signal. The graphs below each immunocytochemistry picture denote the percentage of cells showing nuclear (N), nuclear and cytoplasmic (N/C), or cytoplasmic (C) localization of the expressed protein. (D) Schematic and immunocytochemistry of NLS-chimera proteins. The heterologous NLS was cloned between the FLAG-tag and the beginning of the Xenopus chimera coding sequence. Immunocytochemistry experiments were done as described for panel C, revealing that both chimeras reside only in the nucleus. (E) Luciferase repression assay, comparing chimeric CRYs to NLS-chimeric CRYs. There was no significant difference in the repression ability of either chimera versus its NLS-fused counterpart.
Next, the subcellular localization of each chimera was investigated to see if the decrease in repression is due to mislocalized protein. While wild-type xCRY1, xCRY2, and xCRY2/xPHOTO were found predominantly in the nucleus, xCRY1/xPHOTO was localized mostly in the cytoplasm (Fig. 3C). Coexpression of CLOCK and BMAL1 with the xCRY1 chimera did not alter its localization (data not shown). To further elucidate whether the decrease in repression exhibited by xCRY1/PHOTO was due to its cytoplasmic localization, both Xenopus chimeras were cloned in frame with a heterologous NLS. Although both NLS-xCRY/xPHOTO proteins were localized to the nucleus (Fig. 3D), neither protein's repressive ability improved significantly (Fig. 3E), suggesting that improper localization of the chimera cannot explain its decreased repression.
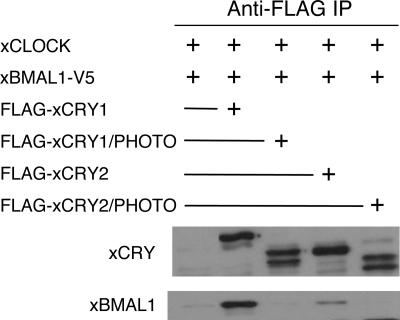
Previous studies have shown that CRY binds to the CLOCK-BMAL1 heterodimer (6, 11, 13) and that this binding is required for repression (19). In light of these data, we hypothesized that that the disruption of repression seen with the xCRY chimeras may be due to an inability to bind the CLOCK-BMAL1 heterodimer. To test this hypothesis, we transfected COS7 cells with xCLOCK, xBMAL1-V5, and either FLAG-xCRY1, FLAG-xCRY2, FLAG-xCRY1/PHOTO, or FLAG-xCRY2/PHOTO. We then examined whether xBMAL1 immunoprecipitated with each CRY protein. While wild-type xCRY1 and xCRY2 exhibit a strong interaction with xBMAL1, neither chimera showed detectable interaction with xBMAL1 (Fig. 4). This suggests that reduced interaction with theBMAL1 explains the decrease in repression by the chimeras.
FIG. 4.
Chimeras do not interact with xBMAL1, unlike wild-type CRY proteins. IP, immunoprecipitation. COS7 cells were transfected with xCLOCK, xBMAL1-V5, and FLAG-xCRY constructs as indicated. CRY and chimera proteins were pulled down, using anti-FLAG-conjugated resin (Sigma). Copurified proteins were detected by Western blotting, using mouse anti-V5 (Invitrogen) or monoclonal mouse anti-FLAG M2 (Sigma) antibodies.
We could not determine whether CLOCK can bind to the chimeras since CLOCK was present at very low levels in our immunoprecipitation lysates and did not coimmunoprecipitate even with wild-type xCRY1 (data not shown). This is not surprising, since it has been previously shown that when CLOCK and BMAL are coexpressed in cell culture, CLOCK is rapidly degraded (8).
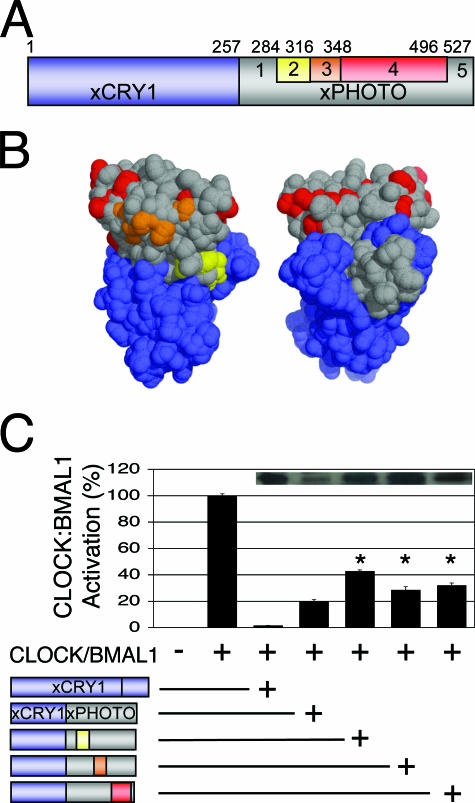
The characterization of the xCRY/xPHOTO chimeras indicates that the second half of the xCRY PHR is required for full repression of CLOCK-BMAL1. To further define functionally important regions of this second half, we replaced portions of the xPHOTO moiety of the xCRY1/xPHOTO chimera with the equivalent portions of xCRY1 to see if restoration of xCRY1 residues would rescue repression of CLOCK-BMAL1. These substitutions corresponded to clusters of residues that are conserved in repressive-type vertebrate CRYs but not in xPHOTO (Fig. 2A and 5A and B). When tested in the luciferase repression assay at a subsaturating dose (150 ng), these chimeras were expressed at levels comparable to those of wild-type xCRY1 and higher than those of the original xCRY1/xPHOTO chimera (Fig. 5C). However, the substitutions in these chimeras did not improve their repression of CLOCK-BMAL transcriptional activity, and in fact, the chimeras with further substitutions were less effective (Fig. 5C). From these data, we can conclude that restoration of any single cluster of xPHOTO residues to xCRY1 residues in the second half of the PHR was insufficient for full repression.
FIG. 5.
Replacing portions of the xPHOTO portion of xCRY1/xPHOTO with xCRY1 residues does not rescue repression. (A) Schematic diagram of the primary structure of the xCRY1/xPHOTO chimera (xCRY1 moiety, blue; xPHOTO moiety, gray), highlighting portions of the xPHOTO moiety that were replaced with the equivalent portions of xCRY1 to generate further chimeras: xCRY1/P1,C2,P3-5; xCRY1/P1-2,C3,P4-5; and xCRY1/P1-3,C4,P5 (yellow, red, and orange, respectively). (B) Homology model of the xCRY1 PHR generated using the SWISS-MODEL protein homology-modeling server (7) based on the structure of A. nidulans photolyase (PDB code 1OWL) (9) and visualized as a solvent-accessible surface using PyMOL (http://www.pymol.org) (4). This model displays residues that are from xCRY1 (blue) and xPHOTO (gray) in all xCRY/xPHOTO chimeras and also shows xCRY1 residues in the rescue chimeras that are conserved in vertebrate repressive-type CRYs but not xPHOTO (yellow, orange, and red). (C) Luciferase assay, carried out as described for Fig. 2. Full-length xCRY1 or xCRY1/xPHOTO chimeras were added as indicated (represented schematically). Repression by rescue chimeras was significantly worse than that by the original chimera (P < 0.01). Inset, Western blotting of FLAG-tagged xCRY1 and xCRY/xPHOTO chimeras, performed as described for Fig. 3. Rescue chimeras were expressed at levels similar to those of full-length xCRY1 and higher than those of the original chimera.
DISCUSSION
Although both CRY1 and CRY2 are potent repressors of CLOCK-BMAL1-mediated transcription, mice lacking either protein individually have opposing circadian locomotor phenotypes (21). Knocking out CRY1 makes the clock run fast, while knocking out CRY2 makes the clock run slow. In this paper, we have shown that the C-terminal tails of both xCRY1 and xCRY2 are sufficient for nuclear localization of the protein in cell culture, but by different mechanisms. Differential regulation of nuclear translocation is one potential mechanism by which CRYs could share a common function but affect the molecular circadian clock in opposite ways.
We report that in xCRY1, the entire region of amino acids 550 to 596 is both necessary and sufficient for localization when expressed in COS7 cells. Although in the C-terminal tail of xCRY1, residues 595 to 612 comprise a sequence that weakly resembles a bipartite NLS that has shown to be functional in mCRY1 (3), we show that this sequence is not sufficient for NLS on its own. In addition, previous studies from our lab showed that an xCRY1 truncation mutant missing a large portion of this sequence did show partial loss of nuclear localization. These data suggest that this sequence may have a role in determining the localization of xCRY1 in the context of the entire protein (24).
Interestingly, Chaves et al. also have reported a second mechanism by which mCRY1 can gain entry into the nucleus via interaction with BMAL1 though mCRY1's coiled-coil region, located at amino acids 471 to 493 (3). At first glance, it appears that this mechanism may not be conserved in Xenopus, since we have previously reported that xCRY1 with an intact coiled-coil region but lacking a C-terminal tail exhibits cytoplasmic localization (24). In that localization experiment, however, the xCRY1 mutant was not coexpressed with xBMAL1, leaving the possibility open that xBMAL1 can affect the subcellular localization of xCRY1. In addition, we have previously shown that xCRY1 with a coiled-coil domain but without a C-terminal tail exhibited partial repression of CLOCK-BMAL1 (24), providing further evidence that this second mechanism for nuclear localization may be conserved in Xenopus. The C-terminal tail of xCRY2 contains a canonical bipartite NLS, which is necessary and sufficient for localization in COS7 cells, while the region most important for localization in xCRY1 appears to be noncanonical. This bipartite NLS is conserved in mouse CRY2 and has been shown to mediate nuclear localization through active nuclear import via importin alpha/beta (18). Similar sequences in other proteins are tightly regulated through posttranslational modifications, such as phosphorylation (10).
In this paper, we have characterized two functional domains that are important for nuclear localization of the CRY proteins when expressed in COS7 cells. Many groups have reported that in cell culture CRY proteins are only observed in the nucleus, but curiously, in vivo there is a constant pool of CRY1 and CRY2 in the cytoplasm and only a portion of the CRY protein population rhythmically translocates to the nucleus (13). This suggests that nuclear translocation of CRY protein is tightly regulated in cells with a functional circadian clock. We hypothesize that nuclear translocation of CRY in the molecular clock may be important for establishing period length. Furthermore, the fact that nuclear localization of CRY1 and CRY2 is achieved by different mechanisms offers a potential way for the two CRY proteins to have different effects on period length.
One potential mechanism for the regulation of the subcellular localization of CRYs is through changing their binding partners. It is known that CRY translocates to the nucleus in a complex with PERs and CKIɛ (13). Perhaps there is some other mechanism by which CRYs are retained in the cytoplasm until PER protein levels trigger nuclear localization. It is important to mention that other researchers have shown that CRY affects the localization of PER and that PER can also influence the localization of CRY (11, 13, 22). Although it is clear that in vivo CRYs, PERs, and CKIɛ translocate to the nucleus as a complex (13), it is not clear whether CRY, being the only nuclear protein when expressed alone in cell culture, is the driving force for the movement of the entire complex. The contribution of each protein, and each localization domain within each protein, to the localization of the complex as a whole has been and continues to be debated in the literature.
Our laboratory has previously shown that the PHR of xCRY, when attached to an NLS, is sufficient for repression of CLOCK-BMAL1. In this report, we showed that when the second half of the PHR of xCRY is replaced with the homologous region of xPHOTO, repression is significantly decreased. This cannot be explained by decreased protein expression or mislocalization. While the xCRY1 chimera is localized predominantly to the cytoplasm, this does not explain its decreased repression, since when it is pulled into the nucleus by a heterologous NLS, its repressive ability does not improve. This suggests that even though xCRY1/PHOTO appears to be cytoplasmic in our immunocytochemistry assay, enough protein enters the nucleus to repress CLOCK-BMAL1.
We did, however, observe that the chimeras do not interact with BMAL1, which we believe to play a central role in their decreased repressive ability. These data are consistent with data reported by Chaves et al., showing that residues 471 to 493 comprise the BMAL interaction domain in mCRY1 (3). Interestingly, replacement of these residues from xPHOTO back to xCRY1 was not sufficient to restore the repressive ability of our xCRY1 chimera. These data imply that CRY's interaction with BMAL1 may depend on more than just the coiled-coil domain or that the decreased interaction with BMAL1 may not fully explain their decreased abilities. Although it is clear that interaction with BMAL1 is required for full repression by CRY, the significance of this binding is for repression is not clear. It may be that CRY binding decreases the affinity of the CLOCK-BMAL1 heterodimer for the E box, allows recruitment of corepressors, or inhibits coactivators. Our experiments do not distinguish between these possibilities.
While the chimeras exhibit significantly decreased repression of CLOCK-BMAL1, it is important to emphasize that the chimeras still repress significantly. One interpretation of these data is that the first half of the PHR contains a region that is sufficient for some repression. An alternative interpretation could be that it is the amino acids that are conserved between xPHOTO and CRYs in the second half of the PHR that are important for repression. In this scenario, residues in the PHR that are not conserved between CRYs and xPHOTO and residues in the C-terminal tail confer specificity, facilitating interaction with BMAL1. This interaction allows CRYs to repress CLOCK-BMAL1, while xPHOTO cannot. This idea is consistent with the publication by Chaves et al., which reported that when PHOTO is fused to the last 100 amino acids of the PHR, coil-coil domain, and C-terminal tail of mCRY1, it can repress CLOCK-BMAL1 (3). The fact that the chimeras still repress well is especially interesting since we have shown that the chimeras do not bind xBMAL1.
In summary, our data suggest that the C-terminal tails of both xCRY proteins are sufficient for nuclear localization of the proteins but act through different mechanisms. We also have identified regions in the PHR of CRYs that are important for interaction with BMAL1 and also repression of the CLOCK-BMAL1 heterodimer. While these data give insight into the structure/function relationships of CRYs, the true value of these insights will emerge as the resulting hypotheses are tested in the context of a functioning molecular clock.
Acknowledgments
This work was supported by grants from the National Institute of Mental Health (MH-61461) and the Jack Kent Cooke Foundation.
Footnotes
Published ahead of print on 8 January 2007.
REFERENCES
- 1.Bell-Pedersen, D., V. M. Cassone, D. J. Earnest, S. S. Golden, P. E. Hardin, T. L. Thomas, and M. J. Zoran. 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6:544-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cashmore, A. R., J. A. Jarillo, Y. J. Wu, and D. Liu. 1999. Cryptochromes: blue light receptors for plants and animals. Science 284:760-765. [DOI] [PubMed] [Google Scholar]
- 3.Chaves, I., K. Yagita, S. Barnhoorn, H. Okamura, G. T. van der Horst, and F. Tamanini. 2006. Functional evolution of the photolyase/cryptochrome protein family: importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol. Cell. Biol. 26:1743-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLano, W. L. 2002. The PyMOL user's manual. DeLano Scientific, San Carlos, CA.
- 5.Etchegaray, J. P., C. Lee, P. A. Wade, and S. M. Reppert. 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421:177-182. [DOI] [PubMed] [Google Scholar]
- 6.Griffin, E. A., Jr., D. Staknis, and C. J. Weitz. 1999. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286:768-771. [DOI] [PubMed] [Google Scholar]
- 7.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 8.Kondratov, R. V., M. V. Chernov, A. A. Kondratova, V. Y. Gorbacheva, A. V. Gudkov, and M. P. Antoch. 2003. BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 17:1921-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kort, R., H. Komori, S. Adachi, K. Miki, and A. Eker. 2004. DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction. Acta Crystallogr. D 60:1205-1213. [DOI] [PubMed] [Google Scholar]
- 10.Kumagai, A., and W. G. Dunphy. 1999. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 13:1067-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kume, K., M. J. Zylka, S. Sriram, L. P. Shearman, D. R. Weaver, X. Jin, E. S. Maywood, M. H. Hastings, and S. M. Reppert. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193-205. [DOI] [PubMed] [Google Scholar]
- 12.Laskowski, R. A., M. M. W, D. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 13.Lee, C., J. P. Etchegaray, F. R. Cagampang, A. S. Loudon, and S. M. Reppert. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107:855-867. [DOI] [PubMed] [Google Scholar]
- 14.Liu, X., and C. B. Green. 2002. Circadian regulation of nocturnin transcription by phosphorylated CREB in Xenopus retinal photoreceptor cells. Mol. Cell. Biol. 22:7501-7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naruse, Y., K. Oh-hashi, N. Iijima, M. Naruse, H. Yoshioka, and M. Tanaka. 2004. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 24:6278-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ripperger, J. A., and U. Schibler. 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38:369-374. [DOI] [PubMed] [Google Scholar]
- 18.Sakakida, Y., Y. Miyamoto, E. Nagoshi, M. Akashi, T. J. Nakamura, T. Mamine, M. Kasahara, Y. Minami, Y. Yoneda, and T. Takumi. 2005. Importin alpha/beta mediates nuclear transport of a mammalian circadian clock component, mCRY2, together with mPER2, through a bipartite nuclear localization signal. J. Biol. Chem. 280:13272-13278. [DOI] [PubMed] [Google Scholar]
- 19.Sato, T. K., R. G. Yamada, H. Ukai, J. E. Baggs, L. J. Miraglia, T. J. Kobayashi, D. K. Welsh, S. A. Kay, H. R. Ueda, and J. B. Hogenesch. 2006. Feedback repression is required for mammalian circadian clock function. Nat. Genet. 38:312-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shevchuk, N. A., A. V. Bryksin, Y. A. Nusinovich, F. C. Cabello, M. Sutherland, and S. Ladisch. 2004. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 32:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Horst, G. T., M. Muijtjens, K. Kobayashi, R. Takano, S. Kanno, M. Takao, J. de Wit, A. Verkerk, A. P. Eker, D. van Leenen, R. Buijs, D. Bootsma, J. H. Hoeijmakers, and A. Yasui. 1999. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398:627-630. [DOI] [PubMed] [Google Scholar]
- 22.Yagita, K., F. Tamanini, M. Yasuda, J. H. Hoeijmakers, G. T. van der Horst, and H. Okamura. 2002. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 21:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young, M. W., and S. A. Kay. 2001. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2:702-715. [DOI] [PubMed] [Google Scholar]
- 24.Zhu, H., F. Conte, and C. B. Green. 2003. Nuclear localization and transcriptional repression are confined to separable domains in the circadian protein CRYPTOCHROME. Curr. Biol. 13:1653-1658. [DOI] [PubMed] [Google Scholar]
- 25.Zhu, H., and C. B. Green. 2001. Three cryptochromes are rhythmically expressed in Xenopus laevis retinal photoreceptors. Mol. Vis. 7:210-215. [PubMed] [Google Scholar]