Abstract
The core histone tail domains play a central role in chromatin structure and epigenetic processes controlling gene expression. Although little is known regarding the molecular details of tail interactions, it is likely that they participate in both short-range and long-range interactions between nucleosomes. Previously, we demonstrated that the H3 tail domain participates in internucleosome interactions during MgCl2-dependent condensation of model nucleosome arrays. However, these studies did not distinguish whether these internucleosome interactions represented short-range intra-array or longer-range interarray interactions. To better understand the complex interactions of the H3 tail domain during chromatin condensation, we have developed a new site-directed cross-linking method to identify and quantify interarray interactions mediated by histone tail domains. Interarray cross-linking was undetectable under salt conditions that induced only local folding, but was detected concomitant with salt-dependent interarray oligomerization at higher MgCl2 concentrations. Interestingly, lysine-to-glutamine mutations in the H3 tail domain to mimic acetylation resulted in little or no reduction in interarray cross-linking. In contrast, binding of a linker histone caused a much greater enhancement of interarray interactions for unmodified H3 tails compared to “acetylated” H3 tails. Collectively these results indicate that H3 tail domain performs multiple functions during chromatin condensation via distinct molecular interactions that can be differentially regulated by acetylation or binding of linker histones.
The genomic DNA of eukaryotic cells is assembled with core and linker histones and other proteins into extensive arrays of nucleosomes, the primary repeating subunit of chromatin (27). Native nucleosome arrays spontaneously assemble under physiological conditions into secondary chromatin structures such as the 25- to 30-nm-diameter chromatin fiber (25) and higher-order tertiary structures such as the 300- to 400-nm “chromonema” fibers observed by Belmont and colleagues (4). Importantly, many essential elements of the primary, secondary, and tertiary levels of chromatin packaging and compaction can be recapitulated with model nucleosome arrays containing DNA and just the four core histone proteins (13, 14, 29). In vitro hydrodynamic studies have shown that model nucleosome arrays undergo MgCl2-dependent folding into secondary structures resembling the 30-nm-diameter chromatin fiber, as well as reversible self-association into higher-order tertiary structures, a process that mimics fiber-fiber association observed in native chromatin (4, 13, 29).
Besides their structural role in organizing the genome within the confines of the nucleus, secondary and tertiary chromatin structures play essential roles in the epigenetic regulation of gene expression by providing heritable repressive or permissive environments for transcription. Importantly, the N-terminal tail domains of the core histones are critical components of this regulation. The tail domains are essential for condensation of nucleosome arrays into secondary and tertiary chromatin structures (1, 10, 21) and interact with multiple protein and DNA targets in chromatin (11, 13, 33). Moreover, specific posttranslational modifications within the tail domains are associated with gene activation and repression and likely elicit distinct chromatin states by either directly or indirectly altering tail interactions (16, 18, 33). For example, acetylation alters the ability of nucleosome arrays to undergo salt-dependent folding and oligomerization, presumably by directly modulating tail interactions with protein or DNA targets (9, 10, 26). In one case it is know that a portion of the H4 tail interacts with a surface of the H2A/H2B dimer of an adjoining nucleosome (7, 8) and that acetylation of lysine 16 within the H4 tail inhibits salt-dependent condensation of model nucleosome arrays based on tandem repeats of the 601 nucleosome positioning sequence (22). However, all four tails contribute to chromatin condensation (11, 13). Thus, despite the central role of the core histone tail domains in defining multiple levels of chromatin structure and function, little is known regarding the complicated molecular interactions of these domains.
In order to elucidate the mechanisms by which the tails direct chromatin condensation, in a previous work we investigated internucleosome interactions of the H3 N-terminal tail during MgCl2-dependent condensation of a model nucleosome array (34). We found that the H3 tail makes primarily intranucleosome interactions in extended arrays but rearranges to primarily internucleosome interactions upon salt-induced folding of the array (34). Importantly, both chromatin folding and oligomerization take place in >2 mM MgCl2 and the H3 tail could contribute to either or both of these processes. To define the interactions and function of the H3 tail during chromatin condensation, we developed a system to document interarray interactions of a core histone tail during formation of tertiary chromatin structures. Here we provide evidence that the H3 tail domain partitions between long-range, bridging interactions between nucleosome arrays and shorter-range interactions between nucleosomes of the same array. Moreover we show that association of linker histone but not mutants with mutations modeling lysine acetylation substantially enhances interarray interactions.
MATERIALS AND METHODS
Expression of 3H-labeled H3 proteins.
Recombinant H3T6C, H3A24C and H3V35C, which contain a single cysteine substituted for amino acids 6, 24, and 35, respectively, were prepared based on a Xenopus major H3 as previously described (34). In order to simulate acetylation of lysine residues, H3 proteins containing 2, 4 or 6 lysine→glutamine substitutions at amino acid residues 9 and 14, 14, 18, 23, and 27 or 4, 9, 14, 18, 23, and 27 were generated by using the QIAGEN Quikchange mutagenesis kit. pET3a vectors containing the recombinant H3 sequences were transformed into BL21(DE3) by standard methods. Transformed Escherichia coli cells were grown in minimal medium (30). At an optical density at 600 nm of ∼0.6, [3H]lysine was added to minimal medium following induction by isopropyl-β-d-thiogalactopyranoside (IPTG) and the cells were grown for another 3 h. Incorporation of [3H]lysine into recombinant H3 proteins was examined by autoradiography.
Purification of core histone proteins and linker histone.
Recombinant H3 and H4 were prepared and tetramers purified by acid extraction and Biorex 70 ion-exchange chromatography as described previously (24). Briefly, after 3 h of induction, cells were resuspended in 10 mM EDTA- 0.5% Triton X-100 and placed on ice for 30 min. HCl was added to a final concentration of 0.4 M followed by a 30-min incubation on ice, the mixture was centrifuged, and then pellets were resuspended in 8 M urea. A second acid precipitation was performed, and H3 was found in the soluble fraction. The solution was neutralized to pH 7 with NaOH. H3/H4 tetramers were formed by mixing acid-extracted H3 with H4 in 1:1 ratio and dialyzing against 2 M NaCl-Tris-EDTA (TE) overnight. Refolded tetramers were diluted three times with TE and applied to a Biorex 70 ion-exchange column, washed extensively with 0.6 M NaCl-TE, and eluted with 2 M NaCl-TE. Purified H3/H4 tetramers were modified directly in storage buffer with a twofold excess of 4-azidophenacylbromide (APB) in room temperature for 1 h in the dark as described previously (17, 32). The efficacy of APB was periodically tested by UV irradiation of modified H3/H4 tetramers, which produces intratetramer protein-protein cross-links (unpublished results). All manipulations with APB-modified proteins and reconstitutes were carried out in the dark. H2A/H2B dimers and linker histone H1 were expressed and purified as described previously (17, 32).
Reconstitution and analysis of nucleosome arrays.
Nucleosome array DNA templates, containing 12 tandem repeats of a 208-bp 5S rRNA gene sequences from Lytechinus variegates (23), were reconstituted with core histone proteins by serial salt dialysis as described previously (20, 26). Core histone proteins were mixed with DNA in 1.2:1 ratio in 2 M NaCl-TE, followed by serial dialysis against 1 M NaCl-TE for 4 h, 0.75 M NaCl-TE for another 3 h, and finally 10 mM Tris, 0.1 mM EDTA, and 0.1 mM EGTA overnight. All procedures were performed in the dark. Reconstitution quality was examined by EcoRI digestion and analysis of nucleosome and free DNA bands on nucleoprotein gels as described previously (26). The tendency of nucleosome arrays to self-associate into oligomeric structures in the presence of divalent cations was also used to gauge the extent of nucleosome occupancy, as self-association is dependent on the level of saturation (20). The 35-mer nucleosome array was reconstituted by a similar method with a DNA fragment containing 35 tandem 5S rRNA gene repeats and core histone proteins purified from chicken erythrocytes as described previously (20).
UV-induced cross-linking and identification of interarray cross-linking.
The 35-mer nucleosome array (8.1 μl, 0.2 mg/ml) was mixed with 8.1 μl of 12-mer nucleosome array (0.04 mg/ml) reconstituted with 3H-labeled and APB-modified recombinant H3 proteins, followed by addition of 1.8 μl of 10× concentrated MgCl2 solutions containing 10 mM Tris to reach a final MgCl2 concentration of 0 to 8 mM. The sample was placed inside a Falcon 5-ml polystyrene tube and enclosed in a 15-ml Pyrex 9820 glass tube. The sample was irradiated using a VMR LM-20E light box for 1 min at 365 nm. The irradiated samples were then adjusted to 0.01% sodium dodecyl sulfate (SDS) and 0.2 μg/ml ethidium bromide (EtBr), and the samples were separated on a 0.7% agarose gel (1/2× Tris-borate-EDTA, 0.01% SDS, 0.2 μg/ml EtBr). After UV photography, the gels were soaked in 45% methanol-10% acetic acid and then 1 M sodium salicylate and dried. The gels were exposed to Kodak MR film at −70°C for 1 to 2 weeks.
Linker histone binding assay.
Purified recombinant Xenopus H1° (15) was mixed with 8.1 μl of 35-mer nucleosome array (0.2 mg/ml) in an approximately 1:1 protein/nucleosome ratio and 8.1 μl of 12-mer nucleosome array (0.04 mg/ml) in the presence of 50 mM NaCl on ice for 3 h. The exact amount of H1 required to saturate the nucleosome arrays was empirically determined by centrifugation of H1-bound arrays at 13,000 rpm for 15 min in a microcentrifuge in the presence of 1.3 mM MgCl2 to precipitate oversaturated arrays. We determined that an apparent H1/nucleosome ratio of ∼1.3 causes saturation of the arrays. The stoichiometries of H1 bound 12-mer and 35-mer arrays were further confirmed by SDS-polyacrylamide gel electrophoresis (PAGE) and comparison to native chromatin samples.
RESULTS
We previously established that the H3 tail domain participates in primarily internucleosome interactions with DNA when model oligonucleosome arrays are extensively condensed and oligomerized in solutions containing elevated levels of MgCl2 (34). These internucleosome interactions could play an important role in formation of condensed secondary chromatin structures via short-range “intra-array” interactions, and/or they may contribute to array oligomerization via long-range “interarray” contacts. To further define how the H3 tail contributes to both processes, we designed a novel experimental system to differentiate between intra- (short range) and interarray (long range) histone tail-DNA interactions (Fig. 1). This system employs a 35-mer nucleosome array containing native unlabeled core histone proteins and a 12-mer nucleosome array reconstituted with native core histones H2A, H2B and H4 and 3H-labeled H3 (*H3) that has been site-specifically modified with a photoactivateable cross-linking agent, APB. We confirmed the quality of the reconstitutions by two methods. Incubation of the reconstituted arrays with a restriction enzyme that cleaves in the putative “linker” regions between nucleosome cores liberated <5% of the repeats as naked DNA (Fig. 2A) (5). Moreover, the reconstituted arrays, either individually (not shown) or when mixed together (Fig. 2B), undergo MgCl2-induced self-association to form large oligomeric structures at the MgCl2 concentration expected for arrays nearly saturated with nucleosomes (13, 20). Specifically, oligomerization occurs cooperatively in the range of 3 to 4 mM MgCl2 and is essentially 100% in >5 mM MgCl2. These data indicated that the arrays were appropriately and equally saturated with histones and the APB modification did not significantly alter MgCl2-dependent self-association of the arrays.
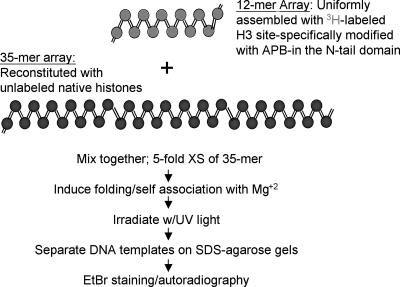
FIG. 1.
Experimental strategy for detecting interarray H3 tail-DNA interactions. The 12-mer nucleosome arrays reconstituted with 3H-labeled and APB-modified H3 are mixed with a fivefold (wt/wt) excess (XS) of 35-mer nucleosome arrays reconstituted with native, unlabeled core histones. Self-association of the arrays is induced by the addition of various amounts of MgCl2, and then the sample is subjected to UV irradiation to induce cross-linking. Cross-linking is detected by separation of the templates on SDS-agarose gels, as described in Materials and Methods, followed by staining with EtBr and subsequent fluorography.
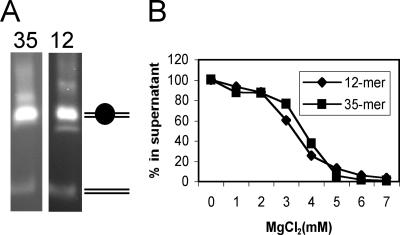
FIG. 2.
Nucleosome arrays are equally saturated with nucleosomes and self-associate in identical levels of MgCl2. (A) The 12-mer and 35-mer nucleosome arrays were prepared as described in Materials and Methods, and the level of nucleosome saturation was determined by EcoRI restriction enzyme digestion assays. Digestion yields about 95% mononucleosomes on agarose nucleoprotein gels, as expected for saturated templates (26). (B) The 12-mer and 35-mer arrays self-associate at identical MgCl2 concentrations.
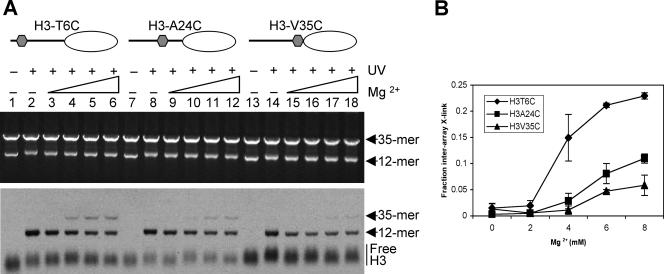
To determine whether internucleosome interactions of the H3 tail involved intra- or interarray interactions or both, 12-mer arrays containing *H3 site-specifically modified with APB at the sixth amino acid residue within the H3 tail domain (Fig. 3A; H3T6C), were combined with a fivefold (wt/wt) excess of 35-mer arrays reconstituted with native histones. The arrays were mixed with increasing concentrations of MgCl2 to induce various extents of oligomerization, and then the sample was UV irradiated to induce cross-linking. The samples were then analyzed by SDS-agarose gel electrophoresis, which separates the 12-mer- and 35-mer DNA templates. In samples not subjected to UV irradiation, the SDS stripped all proteins from the DNAs and the free *H3 appeared as a single fast-moving diffuse band on the autoradiograph that did not comigrate with either DNA template (Fig. 3A, lane 1). However, after UV irradiation, some of the radiolabeled H3 comigrated with the 12-mer DNA in the presence of SDS, indicating that the *H3 was covalently cross-linked to the template (Fig. 3A, lanes 2 to 6). Importantly, irradiation in the absence of divalent cations, where no self-association of the templates takes place, results in no detectable *H3 comigrating with the 35-mer DNA band in the gel (Fig. 3A, lane 2). The addition of lower levels of MgCl2 (2 mM) induces folding into secondary chromatin structures (13) but had no discernible effect on the cross-linking pattern (lane 3). However, when the MgCl2 concentration was raised above the threshold that induces array oligomerization, some *H3 was found cross-linked to the 35-mer DNA template, indicating interarray cross-linking (Fig. 3A, lanes 4 to 6). About 15% of all cross-linking of *H3 occurred to the 35-mer DNA template in the presence of 4 mM MgCl2 (Fig. 3B), which induces 70 to 80% array oligomerization (Fig. 2B). Interarray cross-linking increased to ∼20% and ∼22% of total cross-linking when the MgCl2 concentration was increased to 6 and 8 mM, respectively (Fig. 3B), levels of salt that induce 100% array oligomerization (Fig. 2B). Thus, interarray cross-linking reaches a maximum of about 20% of total cross-links when the nucleosome arrays are completely condensed and oligomerized.
FIG. 3.
Detection of interarray H3 tail-DNA interactions at three positions within the H3 tail domain. (A). The 12-mer nucleosome arrays reconstituted with *H3 modified with APB at the 6th, 24th, or 35th residue (H3T6C, lanes 1 to 6; H3A24C, lanes 7 to 12; or H3V35C, lanes 13 to 18, respectively) were mixed with 35-mer arrays, and the extent of MgCl2-dependent interarray H3 tail-DNA cross-linking was determined. Cross-linking reactions were carried out in 0, 2, 4, 6, and 8 mM MgCl2 (lanes 2 to 6, 8 to 12, and 14 to 18, respectively) as indicated. Samples in lanes 1, 7, and 13 were incubated in 10 mM Tris without UV irradiation. (Upper panel) EtBr-stained gel. (Lower panel) Autoradiograph of the same gel. (B). Interarray cross-linking (X-link) was quantified and plotted as the fraction of total cross-linking at each MgCl2 concentration.
We next asked whether other locations within the H3 tail domain have the same propensity to mediate interarray interactions by moving the cross-linker from the N-terminal position (6th amino acid) to either the middle of the tail domain (24th amino acid) or a position near the histone fold domain (35th amino acid) (Fig. 3A). The *H3 proteins with the cross-linker attached to each of these positions were reconstituted into 12-mer nucleosome arrays, and the extent of interarray histone-DNA cross-linking at various levels of MgCl2 was determined. All three proteins exhibited very similar extents of intra-array cross-link formation to the 12-mer template (Fig. 3A, compare lanes 2 to 6, 8 to 12, and 14 to 18). However, H3T6C-APB clearly exhibited the highest proportion of interarray cross-linking among the three. When *H3A24C-APB was incorporated into the 12-mer array, only about 10% of the cross-links were to the 35-mer array at 8 mM MgCl2 (Fig. 3A, lanes 9 to 12, and B). Similarly, *H3V35C-APB exhibited robust cross-linking to the 12-mer array DNA template but cross-linking to the 35-mer template was barely detectable even at the highest MgCl2 concentrations investigated (Fig. 3A, lane 18, and B), consistent with previous results indicating that this position does not participate in internucleosome interactions (34).
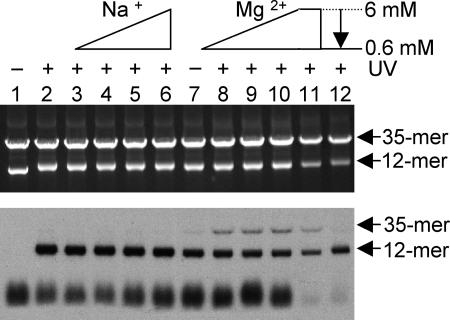
The covalent attachment of *H3 to the 35-mer DNA template indicates that the H3 tails within the 12-mer array must be in very close proximity (∼10 Å) to the 35-mer DNA after self-association. However, it is possible that the presence of salts such as MgCl2, could induce a small amount of exchange of histones between the two arrays, thus allowing some *H3 to be incorporated into the 35-mer arrays. In this case, cross-linking to the 35-mer DNA would represent intra-array rather than interarray interactions. In order to investigate this possibility, we designed two control experiments to determine if exchange takes place. Monovalent salts such as NaCl are unable to induce array oligomerization (20), and histone exchange can take place in solutions containing elevated NaCl. We therefore used NaCl to generate similar ionic strengths to the MgCl2 used in our experimental system, but without array oligomerization. However, no cross-linking of *H3 to the 35-mer template was detected at any of the NaCl concentrations tested (Fig. 4, lanes 3 to 6). We next tested the possibility that the exchange may occur specifically in MgCl2. Arrays were incubated in the presence of 6 mM MgCl2, and then the salt concentration was reduced to 0.6 mM by dilution before UV irradiation (Fig. 4, lane 12). Control arrays were diluted in the same manner except without reduction of the MgCl2 concentration (Fig. 4, lane 11). We find that dilution of the MgCl2 results in almost complete loss of cross-linking to the 35-mer array (Fig. 4, compare lane 11 with lane 12), indicating that the cross-linking was not due to histone exchange between templates in elevated MgCl2. Finally, a comparison of cross-links obtained with H3T6C-APB and H3V35C-APB (Fig. 3) also argues against exchange as octamers containing either of these H3s would be expected to exchange between arrays with equal efficiency.
FIG. 4.
Cross-linking of *H3-APB to the 35-mer array is not due to histone exchange. Samples were prepared as described above (see Fig. 3, lanes 1 to 6) and then incubated with 0, 10, 15, 20, or 25 mM NaCl (lanes 2 to 6) or 0, 2, 4, and 6 mM MgCl2 (lanes 7 to 10), and then the extent of interarray cross-linking was determined. Samples in lanes 11 and 12 were treated identically to the sample in lane 10, except that before UV irradiation the samples were diluted 10-fold with either 6 mM MgCl2 (lane 11) or 10 mM Tris (lane 12). (Top panel) EtBr-stained gel. (Bottom panel) Autoradiograph of the same gel.
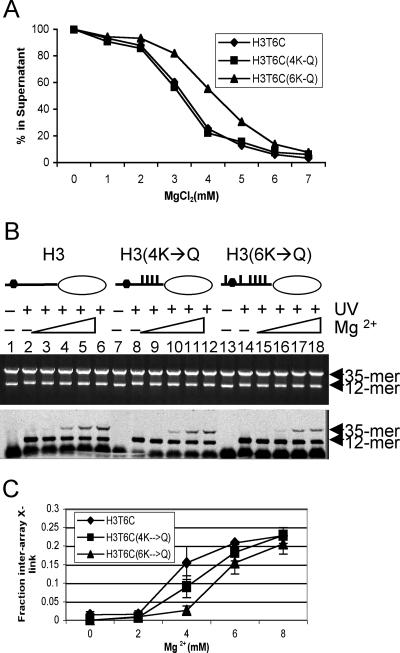
Lysine acetylation within the core histone tail domains reduces the ability of nucleosome arrays to fold into secondary structures and to self-associate into tertiary structures, probably by directly altering tail domain interactions (3, 10, 26). However, it is unclear which acetylation events among the core histones are involved in either of these processes. We thus investigated the effect of mutations modeling lysine acetylation on interarray interactions of the H3 tail. To simulate lysine acetylation within the H3 tail domain, we generated specific lysine-to-glutamine (K→Q) point mutations (27, 31). Mutant H3 proteins with two, four, and six K→Q mutations (2K→Q, 4K→Q, and 6K→Q, respectively), were constructed based on the protein sequence of H3T6C (Fig. 5A). Arrays containing H3s with two or four K→Q mutations undergo self-association in a manner identical to native arrays, while those containing six K→Q mutations self-associated at slightly higher MgCl2 concentrations (Fig. 5A) (results not shown). These results suggest that the functions mediated by the H3 tail during oligomerization are not strongly sensitive to acetylation. However, since all four tails contribute to array self-association (11), effects targeted to the H3 tail may not be apparent in our assays. Thus, it was important to test these arrays for interarray cross-linking of the H3 tail domain. Like the native arrays, mutant arrays did not exhibit interarray cross-linking in 0 or 2 mM MgCl2 (Fig. 5B). Interestingly, at 4 mM MgCl2 arrays containing mutant H3s with two K→Q substitutions were unchanged in their ability to undergo interarray cross-linking compared to controls (not shown) while arrays containing four or six K→Q mutations exhibited a reduced ability to cross-link to the 35-mer template. However, these differences disappeared at higher levels of MgCl2 (Fig. 5B and C).
FIG. 5.
Effect of mutations modeling acetylation on interarray H3 tail-DNA interactions. The 12-mer arrays were prepared with *H3 containing four or six K→Q substitutions in the tail domain, and interarray cross-linking was analyzed. (A) Analysis of interarray cross-linking for 12-mer arrays containing H3T6C (lanes 1 to 6), H3T6C(4K→Q) (lanes 7 to 12), and H3T6C(6K→Q) (lanes 13 to 18). The experimental conditions were as described in the legend to Fig. 3. (B) Interarray cross-linking of native and mutant *H3s was quantified and plotted as in Fig. 3B. (C) Graphic representation of interarray cross-linking (X-link) for H3T6C, H3T6C(4K→Q), and H3T6C(6K→Q).
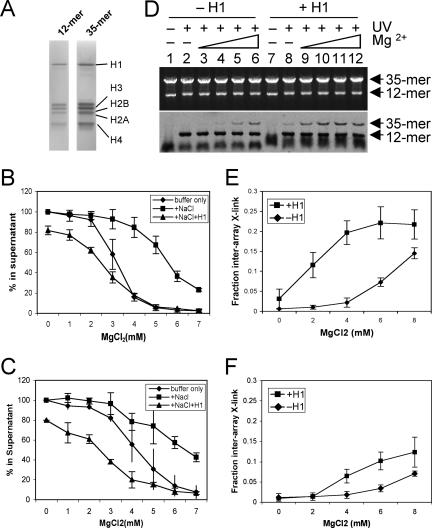
Linker histones stabilize both secondary and tertiary chromatin structures (5, 25), and the binding of H1 alters specific tail-DNA interactions within mononucleosomes (12, 17). We thus examined whether the presence of linker histones affects interarray interactions of the H3 tail domain. Arrays were incubated in 50 mM NaCl with stoichiometric amounts of H1, based on masses of the protein and arrays (Fig. 6A) (results not shown), and H1 induced oligomerization of both arrays in the presence of 1.3 mM MgCl2 (Fig. 6B) (5). Note that the H1 binding buffer contains 50 mM NaCl, which competes with MgCl2 for binding to chromatin (20, 28). Accordingly, we observed that MgCl2-induced self-association is reduced in the presence of 50 mM NaCl, such that 50% self-association occurs in >5 mM MgCl2 compared to ∼3 mM MgCl2 (Fig. 6B), as expected: the inclusion of NaCl simply alters the level of MgCl2 required to induced self-association (21). Likewise interarray cross-linking in 50 mM NaCl was significantly reduced compared to cross-linking in buffers lacking NaCl and did not appear to reach a plateau at the highest levels of MgCl2 tested (Fig. 6D, lanes 2 to 6, and compare Fig. 6E to Fig. 3B). Interestingly, H1 binding greatly stimulated both array self-association (Fig. 6B) (5) and interarray cross-linking (Fig. 6D, lanes 8 to 12). Notably, while no interarray cross-linking was detected for arrays incubated in the absence of MgCl2, interarray cross-linking was detectable for H1-bound arrays in the presence of only 50 mM NaCl (Fig. 6D, compare lanes 2 and 8). Furthermore, while interarray cross-linking in the absence of H1 did not appear to reach a plateau even in 7 to 8 mM MgCl2, this plateau was attained in 4 mM MgCl2 in the presence of H1 (Fig. 6E). Thus, although 50 mM NaCl significantly reduced interarray interactions, the binding of H1 restored these interactions to levels observed in the absence of NaCl.
FIG. 6.
H1 association enhances interarray interactions by unmodified but not mutated H3 tail domains. (A) H1 was incubated with 12-mer-35-mer array mixtures, and then proteins were analyzed by SDS-PAGE. The effects of 50 mM NaCl and H1 on MgCl2-induced self-association of arrays containing native histones and H3 6K→Q are shown in panels B and C, respectively. Self-association was assayed as in Fig. 2B, in standard low-salt buffer, buffer plus 50 mM NaCl, and buffer plus NaCl and H1, and is plotted as diamonds, squares, and triangles, respectively, versus MgCl2 concentration. (D) Effect of H1 on interarray cross-linking (X-link). Arrays containing native histones were incubated in the absence (lanes 1 to 6) or presence (lanes 7 to 12) of H1 and assayed for interarray H3 tail-DNA cross-linking. Experimental conditions and data analysis were as described in the legend to Fig. 3, except all samples contained 50 mM NaCl to facilitate H1 binding. (E and F) Quantification of interarray cross-linking. Cross-linking data as in panel D were quantified and plotted for native arrays (E) and arrays containing H3 6K→Q (F) versus MgCl2 concentration in the absence (diamonds) and presence (squares) of H1.
Interestingly, stimulation of interarray interactions by H1 is significantly attenuated by mutations modeling hyperacetylation within the H3 tail domain. Arrays containing the H3 6K→Q mutant exhibited NaCl-dependent changes in self-association similar to that of arrays containing native histones. Further, upon the addition of H1 the midpoint of self-association occurred at approximately the same lower MgCl2 concentration for both arrays (Fig. 6B and C). However, while the addition of 50 mM NaCl significantly reduced interarray cross-linking for the H3 6K→C mutant, similar to native arrays (Fig. 6E and F), the addition of H1 did not restore cross-linking within H3 6K→Q arrays to the levels observed for arrays in the absence of NaCl (Fig. 5), in contrast to the effect of H1 on cross-linking within native arrays (see above).
DISCUSSION
The core histone tail domains define multiple levels of chromatin structure via a complicated set of molecular interactions with both DNA and protein targets. In this work, we demonstrate that the H3 tail mediates both short-range and long-range internucleosome interactions that likely play important roles in formation of secondary and tertiary chromatin structures. We find that under maximally folded and condensed conditions, about 20% of the H3 tails provide bridging interactions in trans between arrays, while about 80% interact in cis with DNA of other nucleosomes within the same array. Note that these fractions assume that all interarray interactions mediated by H3 tail domains emanating from the 12-mer array result in cross-links to the 35-mer template, and thus our method may underestimate the fraction of interarray interactions. Nonetheless, this suggests that the mechanism of folding and oligomerization involves both intra-array and interarray nucleosome-nucleosome interactions and is consistent with the idea that each of the core histone tail domains fulfills multiple distinct functions during chromatin condensation (13). In previous work, we documented that the H3 tail also interacts with the DNA of its own nucleosome when nucleosome arrays are unfolded, rearranging to internucleosome interactions upon MgCl2-dependent folding (34). Thus, the H3 tail is mobile and participates in numerous distinct interactions during formation of secondary and tertiary chromatin structures. These data highlight the multiple, surprisingly plastic interactions of the tail domains.
The exact mechanisms by which the H3 tail domains contribute to folding and oligomerization remain to be elucidated. As mentioned above, in previous work we found that nucleosomal arrays are largely folded but not self-associated in 2 mM MgCl2 and that the H3 tail makes primarily intranucleosomal interactions at this salt concentration (34). The H3 tails rearrange to mainly internucleosomal interactions in higher concentrations of MgCl2, concomitant with array oligomerization (34). This suggests that internucleosome interactions of the H3 tail do not contribute to folding of oligonucleosome arrays but rather primarily play a role in array-array self-association. However, in the present work, we find that in elevated MgCl2 only about 20% of the internucleosomal interactions of the H3 tails actually participate in interarray bridging interactions, presumably directly facilitating array-array self-association. Thus, this leaves open the question of the function of the remaining 80% of the H3 tails in the fully condensed and self-associated arrays, which participate in internucleosomal but intra-array interactions. One possibility is that additional folding of the array occurs beyond the compaction observed in 2 mM MgCl2 and that the H3 internucleosome interactions are important for this process. Alternatively, the majority of the H3 tails may mediate an intra-array conformational change that facilitates oligomerization, with a subset directly mediating the self-association process via interarray interactions. These non-mutually exclusive possibilities are currently under investigation.
The addition of linker histones to the nucleosome arrays greatly stimulates interarray interactions of the H3 tail domain at low MgCl2 concentrations. We detected interarray tail-DNA interactions between H1-bound arrays in salt concentrations as low as 2 mM MgCl2, reaching a maximum in 4 mM MgCl2, compared to 8 mM MgCl2 for arrays lacking linker histones. However, binding of linker histone did not increase the fraction of interarray cross-linking observed at high levels of MgCl2 (∼20% of total cross-linking). Thus, linker histone stabilizes array self-association, perhaps by promoting interarray interactions by the H3 tail, but does not appear to substantially alter the structure of the self-associated complex. Early microscopic studies as well as recent hydrodynamic studies show that linker histone-bound nucleosome arrays are capable of forming oligomeric structures in the presence of NaCl or low MgCl2, conditions normally insufficient for array oligomerization in the absence of linker histone (5, 25). Thus, the stimulation of interarray interactions in the presence of linker histone may be due to a greater stability of an array conformation that has increased propensity to undergo self-association. Alternatively, previous work has shown that linker DNA between nucleosomes is a primary binding site of the core histone tails as well as a major target of linker histones (2, 6) and cross-linking experiments demonstrate that H1 binding to nucleosomes can induce rearrangement of core histone tail-DNA interactions (12, 17). This raises the possibility that linker histones directly compete with the tails for binding linker DNA and force greater extents of interarray interactions. It is also interesting to note that binding of linker histone did not increase the fraction of total cross-links that were interarray, but only promoted their formation at lower MgCl2 concentrations. These results suggest that H1 binding stabilizes self-associated structures but does not significantly alter the configuration or structure of the oligomerized complex.
Interestingly, mutations modeling moderate levels of H3 lysine acetylation have little or no effect on interarray cross-linking in solutions containing no NaCl and high MgCl2 concentrations (Fig. 5). However, at intermediate levels of MgCl2, H3 tails bearing four or six lysine-to-glutamine substitutions—representing a physiologically high level of acetylation—did exhibit significant differences in interarray interactions at intermediate (4 mM) MgCl2 concentrations. These experiments predict that lower levels of acetylation would have little effect on the extent of interarray interactions mediated by the H3 tail domain but that the extent of such interactions for tails with high (≥4) acetylation events may depend on the microenvironment experienced by the chromatin fibers. Importantly, differences between “acetylated” and “unacetylated” tails appear to be enhanced in the presence of NaCl. While H1 binding “rescues” interarray interactions for the unmodified H3 tail domains in 50 mM NaCl, such interactions are not restored to the low-salt levels when the H3 tails contain mutations modeling acetylation. These data, coupled with data indicating that acetylation does not result in a loss of tail-DNA interactions (19), suggest that acetylation may override the effect of H1 in stabilizing higher-order chromatin structures by redirecting tail interactions, resulting in the observed reduction in MgCl2-induced self-association of arrays containing acetylated histones (26). Moreover, some lysine acetylation events serve to create specific binding surfaces for ancillary proteins, which may in turn alter interarray interactions of the H3 tail. It will be interesting to test this possibility using the methods developed in this work.
Acknowledgments
We thank Tamara Caterino for the generous gift of Xenopus H1.
This work was supported by NIH grants GM52426 (J.J.H.) and GM45916 (J.C.H.).
Footnotes
Published ahead of print on 22 January 2007.
REFERENCES
- 1.Allan, J., N. Harborne, D. C. Rau, and H. Gould. 1982. Participation of the core histone tails in the stabilization of the chromatin solenoid. J. Cell Biol. 93:285-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelov, D., J. M. Vitolo, V. Mutskov, S. Dimitrov, and J. J. Hayes. 2001. Preferential interaction of the core histone tail domains with linker DNA. Proc. Natl. Acad. Sci. USA 98:6599-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annunziato, A. A., L.-L. Y. Frado, R. L. Seale, and C. L. F. Woodcock. 1988. Treatment with sodium butyrate inhibits the complete condensation of interphase chromatin. Chromosoma 96:132-138. [DOI] [PubMed] [Google Scholar]
- 4.Belmont, A. S., and K. Bruce. 1994. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 127:287-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carruthers, L. M., J. Bednar, C. L. Woodcock, and J. C. Hansen. 1998. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry 37:14776-14787. [DOI] [PubMed] [Google Scholar]
- 6.Clark, D. J., and T. Kimura. 1990. Electrostatic mechanism of chromatin folding. J. Mol. Biol. 211:883-896. [DOI] [PubMed] [Google Scholar]
- 7.Dorigo, B., T. Schalch, K. Bystricky, and T. J. Richmond. 2003. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327:85-96. [DOI] [PubMed] [Google Scholar]
- 8.Dorigo, B., T. Schalch, A. Kulangara, S. Duda, R. R. Schroeder, and T. J. Richmond. 2004. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 306:1571-1573. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher, T. M., and J. C. Hansen. 1995. Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. J. Biol. Chem. 270:25359-25362. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Ramirez, M., C. Rocchini, and J. Ausio. 1995. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 270:17923-17928. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, F., K. Luger, and J. C. Hansen. 2005. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J. Biol. Chem. 280:33701-33706. [DOI] [PubMed] [Google Scholar]
- 12.Guschin, D., S. Chandler, and A. P. Wolffe. 1998. Asymmetric linker histone association directs the asymmetric rearrangement of core histone interactions in a positioned nucleosome containing a thyroid hormone response element. Biochemistry 37:8629-8636. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, J. C. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31:361-392. [DOI] [PubMed] [Google Scholar]
- 14.Hansen, J. C., J. Ausio, V. H. Stanik, and K. E. van Holde. 1989. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry 28:9129-9136. [DOI] [PubMed] [Google Scholar]
- 15.Hayes, J. J., and K. M. Lee. 1997. In vitro reconstitution and analysis of mononucleosomes containing defined DNAs and proteins. Methods 12:2-9. [DOI] [PubMed] [Google Scholar]
- 16.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 17.Lee, K. M., and J. J. Hayes. 1998. Linker DNA and H1-dependent reorganization of histone-DNA interactions within the nucleosome. Biochemistry 37:8622-8628. [DOI] [PubMed] [Google Scholar]
- 18.Luger, K., and J. C. Hansen. 2005. Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 15:188-196. [DOI] [PubMed] [Google Scholar]
- 19.Mutskov, V., D. Gerber, D. Angelov, J. Ausio, J. Workman, and S. Dimitrov. 1998. Persistent interactions of core histone tails with nucleosomal DNA following acetylation and transcription factor binding. Mol. Cell. Biol. 18:6293-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz, P. M., A. Felthauser, T. M. Fletcher, and J. C. Hansen. 1996. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry 35:4009-4015. [DOI] [PubMed] [Google Scholar]
- 21.Schwarz, P. M., and J. C. Hansen. 1994. Formation and stability of higher order chromatin structures. Contributions of the histone octamer. J. Biol. Chem. 269:16284-16289. [PubMed] [Google Scholar]
- 22.Shogren-Knaak, M., H. Ishii, J. M. Sun, M. J. Pazin, J. R. Davie, and C. L. Peterson. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311:844-847. [DOI] [PubMed] [Google Scholar]
- 23.Simpson, R. T., F. Thoma, and J. M. Brubaker. 1985. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell 42:799-808. [DOI] [PubMed] [Google Scholar]
- 24.Thiriet, C. 2004. Analysis of chromatin assembled in vivo using exogenous histones in Physarum polycephalum. Methods 33:86-92. [DOI] [PubMed] [Google Scholar]
- 25.Thoma, F., T. Koller, and A. Klug. 1979. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 83:403-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tse, C., T. Sera, A. P. Wolffe, and J. C. Hansen. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18:4629-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Holde, K. E. 1989. Chromatin. Springer Verlag, New York, NY.
- 28.Widom, J. 1986. Physicochemical studies of the folding of the 100 A nucleosome filament into the 300 A filament. Cation dependence. J. Mol. Biol. 190:411-424. [DOI] [PubMed] [Google Scholar]
- 29.Woodcock, C. L., and S. Dimitrov. 2001. Higher-order structure of chromatin and chromosomes. Curr. Opin. Genet. Dev. 11:130-135. [DOI] [PubMed] [Google Scholar]
- 30.Yang, Z., C. Zheng, C. Thiriet, and J. J. Hayes. 2005. The core histone N-terminal tail domains negatively regulate binding of transcription factor IIIA to a nucleosome containing a 5S RNA gene via a novel mechanism. Mol. Cell. Biol. 25:241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, K., H. Tang, L. Huang, J. W. Blankenship, P. R. Jones, F. Xiang, P. M. Yau, and A. L. Burlingame. 2002. Identification of acetylation and methylation sites of histone H3 from chicken erythrocytes by high-accuracy matrix-assisted laser desorption ionization-time-of-flight, matrix-assisted laser desorption ionization-postsource decay, and nanoelectrospray ionization tandem mass spectrometry. Anal. Biochem. 306:259-269. [DOI] [PubMed] [Google Scholar]
- 32.Zheng, C., and J. J. Hayes. 2003. Intra- and inter-nucleosomal protein-DNA interactions of the core histone tail domains in a model system. J. Biol. Chem. 278:24217-24224. [DOI] [PubMed] [Google Scholar]
- 33.Zheng, C., and J. J. Hayes. 2003. Structures and interactions of the core histone tail domains. Biopolymers 68:539-546. [DOI] [PubMed] [Google Scholar]
- 34.Zheng, C., X. Lu, J. C. Hansen, and J. J. Hayes. 2005. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J. Biol. Chem. 280:33552-33557. [DOI] [PubMed] [Google Scholar]