Abstract
RNA interference with one of the eight Caenorhabditis elegans linker histone genes triggers desilencing of a repetitive transgene and developmental defects in the hermaphrodite germ line. These characteristics are similar to the phenotype of the C. elegans Polycomb group genes mes-2, mes-3, mes-4, and mes-6 (M. A. Jedrusik and E. Schulze, Development 128:1069-1080, 2001; I. Korf, Y. Fan, and S. Strome, Development 125:2469-2478, 1998). These Polycomb group proteins contribute to germ line-specific chromatin modifications. Using a his-24 deletion mutant and an isoform-specific antibody, we characterized the role of his-24 in C. elegans germ line development. We describe an unexpected cytoplasmic retention of HIS-24 in peculiar granular structures. This phenomenon is confined to the developing germ lines of both sexes. It is strictly dependent on the activities of the chromatin-modifying genes mes-2, mes-3, mes-4, and mes-6, as well as on the C. elegans sirtuin gene sir-2.1. A temperature shift experiment with a mes-3(ts) mutant revealed that mes gene activity is required in a time window ranging from L3 to the early L4 stage before the onset of meiosis. We find that the his-24(ok1024) mutant germ line is characterized by an increased level of the activating H3K4 methylation mark concomitant with a decrease of the repressive H3K9 methylation. In the germ line of his-24(ok1024) mes-3(bn35) double mutant animals, the repressive H3K27 methylation is more reduced than in the respective mes single mutant. These observations distinguish his-24 as an unusual element in the developmental regulation of germ line chromatin structure in C. elegans.
Linker histones are highly abundant chromatin proteins that bind to the elementary structural unit of the chromatin, the nucleosome. Our previous work (9) characterized the Caenorhabditis elegans linker histone variant gene his-24 (H1.1) as a gene involved in the control of hermaphrodite germ line development. Using RNA interference (RNAi), desilencing of a repetitive transgene in the germ lines of both sexes had been shown. Additionally a low-penetrant cytological gonad phenotype occurred, where the germ line substantially lacked proliferation and differentiation. This cytological phenotype was observed in hermaphrodites but not in males. The combination of both observations related his-24 to the phenotype of the C. elegans Polycomb group genes, mes-2, mes-3, mes-4, and mes-6 (18; for reviews, see references 25 and 27). The precise gonadal expression pattern of HIS-24, its general mode of action, and its specific functional relationship to the mes genes remained unclear. No germ line phenotype of a linker histone mutant has been reported for mammals so far, although the mouse linker histone complement has recently been recognized as essential for embryogenesis (6). The present view on linker histones in general describes them as highly dynamic chromatin components. They are considered to be dispensable in single-cell eukaryotes (4, 16).
The C. elegans SET domain histone methyl transferase MES-2 forms a complex with MES-3 and MES-6 that is responsible for H3K27 di- and trimethylation in the adult germ line and in the embryo. The target loci in the germ line are concentrated on the X chromosome (2). A germ line-specific methylation of H3 at lysine 9 of the X chromosome had been shown previously (14, 23). Both modifications are expected to participate in repression of specific target genes. The mammalian homolog of MES-2, the Enhancer of Zeste (EZH2), methylates mammalian linker histones (19). This raises the question of whether this is a valid model for the function of his-24 in the C. elegans germ line.
To address this question, we used an isoform-specific anti-HIS-24 antibody and a his-24 deletion mutant. Surprisingly, we identified his-24 as a germ line-specific cytoplasmic element that supports germ line chromatin modification and hermaphrodite germ line development. The germ line-specific cytoplasmic presence of HIS-24 is controlled by all four mes genes and by the putative histone deacetylase SIR-2.1, a protein type known to synergize with EZH2-dependent methylation of linker histones in mammals.
MATERIALS AND METHODS
Strains and alleles.
Strains were maintained following standard procedures (3). C. elegans N2 variety Bristol is the wild-type reference strain. Strains with the following genotypes were obtained from the Caenorhabditis Genetics Center (CGC), which is funded by the NIH National Center for Research Resources (NCRR): mes-2(bn11) unc-4(e120)/mnC1 dpy-10(e128) unc-52(e444) II, mes-3(bn21)I, mes-3(bn35) dpy-5(e61) I, sDp2(I;f), dpy-11(e224) mes-4(bn23) unc-76(e911) V/nT1[unc-?(n754) let-?] (IV;V), mes-6(bn66) dpy-20(e1282) IV/nT1[unc-?(n754) let-?] (IV;V), sir-2.1(ok434). Strain PD7271 [pha-1(e2123) Ex 412.5/8 (ccEx7271)] carrying the repetitive extrachromosomal array of the plasmids pBK48.1 and pC1 (let-858::gfp and pha-1 rescue) was kindly provided by W. G. Kelly. Strain EC602 [unc-119(ed3)III; eels602[unc-119(+) his-24::gfp] was generated by biolistic transformation of C. elegans (22). The his-24(ok1024) deletion mutant was prepared by the C. elegans Gene Knockout Consortium with the mutagen UV/TMP. It was outcrossed five times and used as strain EC109. The sequence flanking the deletion is GCAGCTCAAGGACCGCAAAG/CACTTCTAACTACTGTACGA, and the size of the deletion is 2,548 bp.
Genetics.
The his-24(ok1024) mes-3 double mutant strains were generated by crossing. Double-homozygous animals were selected from the F2 self-progeny. A PCR-based analysis was used for ok1024, a test for temperature sensitivity was used for bn21, and the single-nucleotide substitution bn35 was detected by PCR amplification followed by AlwI restriction enzyme cleavage. Germ line desilencing was analyzed by crossing his-24(ok1024) strain EC109 with the let-858::gfp reporter strain PD7271 (13). In subsequent generations, a PCR-based analysis was used to identify his-24(ok1024).
RNA interference experiments.
Linker histone RNAi was accomplished by a combination of double-stranded-RNA injection and feeding as described earlier (9, 12). For the analysis of the his-24(ok1024) mutant, two additional double-stranded RNAs were used. his-24-N spans the 300 bp of the his-24 coding region maintained in this mutant, whereas his-24-D spans an additional 600 bp downstream, to target the his-24(ok1024)-specific mRNA.
Generation of a specific anti-HIS-24 antibody.
The synthetic peptide HIS-24-CT (N-CAAKKAAKPAAKA-C) was chemically synthesized and coupled with sulfo-maleimidobenzoyl-N-hydroxysuccinimide ester (Pierce) to bovine serum albumin. Five hundred milligrams of antigen was used for rabbit immunization in a series of three injections. Antigen injections and resulting antiserum collections were performed by BioScience (Göttingen). Anti-HIS-24 antibody was affinity purified with a SulfoLink (Pierce) column using 1 mg of the synthetic peptide.
Western blot analysis.
A C. elegans lysate was prepared by boiling worms in sodium dodecyl sulfate sample buffer. The samples were subsequently separated on a 12% sodium dodecyl sulfate-polyacrylamide gel. After transfer onto a nitrocellulose membrane, unspecific binding sites were blocked for 1 h at room temperature with 0.1% Tween 20 and 5% dry milk powder in TBS (150 mM NaCl, 10 mM KCl, 10 mM Tris-HCl, pH 7.6). The membrane was washed with TBS, incubated with 0.01 to 0.03 μg/ml anti-HIS-24 in TBS overnight at 4°C, and washed with 0.1% Tween 20 in TBS at room temperature. An anti-acetyl-H3 antibody (0.01 μg/ml) (Upstate; catalog no. 06-599) was used as a loading control. The membrane was then incubated for an additional hour with an anti-rabbit horseradish peroxidase-conjugated antibody diluted 1:5,000. After extensive rinsing with Tween 20-TBS the immunoblots were developed using chemiluminescence detection kits from New England Biolabs or Bio-Rad according to the manufacturer's instructions. Luminescence was recorded on Kodak blue X-Omat XB-1 film.
Immunofluorescence staining.
We use the nomenclature of Turner (28) for the modified histone epitopes. The following primary antibodies were used: polyclonal rabbit anti-HIS-24 in a final concentration of 2.7 μg/ml; two monoclonal mouse antibodies directed against unknown P-granule epitopes (K76 and OIC1D4 (26), obtained from the Developmental Studies Hybridoma Bank, (University of Iowa) and diluted 1:10 from a cell culture supernatant prior to use; rabbit anti-H3K9me2 diluted 1:100 (Upstate Biotechnology); rabbit anti-H3K4me2 (Lys4) diluted 1:1,000 (Upstate Biotechnology); rabbit anti-H3K27me3 diluted 1:1,000 (kindly provided by T. Jenuwein, Vienna); and mouse anti-green fluorescent protein (anti-GFP) diluted 1:1,000 (Chemicon). Secondary antibodies purchased from Molecular Probes were Alexa 488-goat anti-rabbit (1:500), Alexa 488-goat anti-mouse (1:500), Alexa 555 goat anti-rabbit (1:500), and Cy3-anti-mouse (1:500) antibodies from Jackson ImmunoResearch Laboratories diluted in PBST (10 ml 10× phosphate-buffered saline, 0.5 ml Triton X-100 [Sigma], 0.2 ml 0.5 M EDTA [pH 8], 87.3 ml double-distilled water).
Adult hermaphrodites and males were cut directly posterior of the pharynx or, alternatively, at the distal tail to release the gonad. Gonads and embryos were fixed in 1% paraformaldehyde in 1× sperm salts [50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 7.0), 25 mM KCl, 1 mM MgSO4, 45 mM NaCl, 2 mM CaCl2] on a positively charged glass slide (SuperFrost, Menzel, Germany). The slides were incubated for 5 min and then frozen in liquid nitrogen. The coverslips were quickly removed with a blade, and the slides were immediately placed in 95% cold ethanol for 1 min. The slides were incubated three times for 10 min in PBST. The primary antibody was diluted in PBST with 0.5 mg/ml bovine serum albumin and 10% goat serum and added after the washing step. The slides were then placed in a humid chamber and incubated overnight at 4°C. After this incubation the slides were washed three times for 10 min in PBST, and 100 μl of secondary antibody dilution was added. The slides were placed in a humid chamber and incubated overnight at 4°C. The washing step in PBST was repeated. The slides were mounted with 20 μl of SlowFade Light antifade reagent with DAPI (4′,6′-diamidino-2-phenylindole) (1.5 μg/ml) (Molecular Probes).
Temperature shift experiments.
The temperature shift experiments with mes-3(bn21)ts (30) were performed as follows. For upshift experiments (16°C to 25°C), hermaphrodites, embryos, and L1 to L4 stage larvae were raised at the permissive temperature (16°C). The embryos, L1 to L4 stage larvae, or young hermaphrodites were isolated and transferred to 25°C for 16 h. To determine the localization of HIS-24, the animals were fixed and immunostained. For downshift experiments, gravid adults were transferred from 16°C to 25°C. The L1 to L4 stage larvae and young adults (F1) of the subsequent generation were then transferred to 16°C. After 16 h at 16°C, the worms were fixed and immunostained.
Microscopy.
Microscopy was performed with a Zeiss Axioplan 2 microscope equipped with a Zeiss LSM 510 confocal laser-scanning module as described previously (9).
RESULTS
Cytoplasmic retention of linker histone variant HIS-24 distinguishes the developing germ line from the soma.
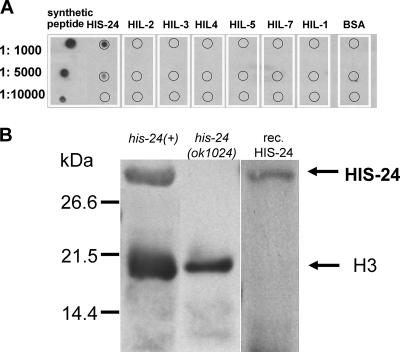
An affinity-purified polyclonal antibody was obtained to allow specific detection of HIS-24. A dot blot experiment with a set of seven recombinant C. elegans linker histones was used to identify an animal serum with this property (Fig. 1). The specificity of the purified antibody was characterized in Western blot experiments with lysates of wild-type and his-24 mutant C. elegans. A single band with the appropriate electrophoretic mobility was recognized in the wild type but not in the mutant (Fig. 1B).
FIG. 1.
Specificity of the HIS-24 antibody. (A) The specific polyclonal antibody detects recombinant HIS-24, but none of six other recombinant C. elegans linker histone variants, in a dot blot experiment. One hundred nanograms of recombinant protein or 1 ng of peptide was used for each dot. BSA, bovine serum albumin. (B) In a Western blot experiment with C. elegans lysates, the purified antibody recognizes a single band with an apparent molecular mass of about 31 kDa in a his-24(+) lysate but no band in a lysate from the his-24(ok1024) deletion mutant. One hundred nanograms of recombinant HIS-24 protein was used as a positive control, and an anti-acetyl-H3 antibody was added to provide a loading control. Actyl-H3 is detected at an apparent molecular mass of about 17 kDa. The designations HIS-24, HIL-2-HIL-5, HIL-7, and HIL-1 correspond to H1.1 to H1.5, H1.Q, and H1.X (9), respectively. Expression and purification of the recombinant proteins were described previously (9).
Indirect immunofluorescence analysis of wild-type animals revealed the expression pattern and the subcellular localization of HIS-24 (Fig. 2). In most, if not all, somatic cells, HIS-24 is expressed and exclusively associated with chromatin. It is, however, absent from the primordial germ cells Z2 and Z3, which are born in the embryo and rest during the first larval stage (L1). Germ line HIS-24 expression starts in the late L3 stage concomitant with gonad development and continues during adulthood. The protein, however, does not translocate into the germ nuclei but is associated with specific cytoplasmic granular structures surrounding the nuclei. Costaining with P-granule-specific antibodies revealed that the structures containing the cytoplasmic HIS-24 protein are not the P-granules. Cytoplasmic HIS-24 is characteristic for most developmental stages of the germ line, it is found in the mitotic region of the gonad, in the transition zone, in the meiotic region, and during all stages of oogenesis (Fig. 3B). SYTO-RNA-select staining of hermaphrodite gonads identified the well-known RNA component of the P granules but showed no staining of the HIS-24-containing granular structures (data not shown). Only during the late pachytene stage is a small fraction of HIS-24 associated with chromatin (Fig. 3A). In the male gonad the germ line expression level of HIS-24 is considerably lower than that in hermaphrodites, but the protein is accordingly localized in the cytoplasm (Fig. 2E).
FIG. 2.
Antibody staining of HIS-24 in a zygote, in embryos, and in gonads of adult wild-type C. elegans (HIS-24, green; DAPI, blue; P granules, red). In adult gonads of both sexes (A to H and S) HIS-24 is detected in cytoplasmic granules that surround the germ nuclei but not inside the nuclei. An enlarged part of panel D is presented in panel S. It shows that the structures stained with the HIS-24 antibody are distinct from the P granules. In both sexes HIS-24 is prominently expressed in the somatic distal tip cells of the gonads (A to H, arrows). In the zygote (P to R) cytoplasmic HIS-24 translocates to the pronuclei and polar bodies (arrows) shortly after fertilization. During the time of embryonic founder cell divisions (M to O) HIS-24 is localized in the nuclei of all cells, including P4 (arrow). Later in embryonic development (I to L) HIS-24 is not detected in the nuclei of the primordial germ cells Z2 and Z3, but it is present in all somatic nuclei. Bars, 20 μm.
FIG. 3.
Immunofluorescence detection of HIS-24 in the gonads of wild-type (A to C), fertile (M+Z−) mes-3(bn35) (D to F), and sterile (M−Z−) mes-3(bn35) (G and H) animals (HIS-24, green; DAPI, blue). In fertile mes-3 mutant animals that had heterozygous mothers (M+Z−), the mislocalization of HIS-24 to the pachytene nuclei (D) and to the chromosomes during diakinesis (E) is clearly detectable but is incomplete. In sterile mes-3 mutant animals (M−Z−), HIS-24 is almost completely mislocalized to the nuclear compartment (G and I). Interestingly, a comparable situation was also found in the sir-2.1(ok434) mutant (H). Straight arrows point to a nuclear compartment; the curved arrow (B) points to a somatic nucleus. Cartoon symbols: large white circles, germ nuclei; triangles, sperm nuclei; hexagons, oocyte nuclei; stars, nuclei of the somatic gonad. Small cyan-filled circles represent small granular structures labeled with the HIS-24 antibody. In the corresponding oocytes, HIS-24 associates in granular structures with the condensed chromosomes of the diakinesis stage of meiosis (E). Bars, 5 μm. DTC, distal tip cell.
As a specificity control for the immunofluorescence experiment, the his-24(ok1024) mutant was also stained with the HIS-24 antibody. No signal was observed in these experiments (data not shown). Moreover, the HIS-24 staining results were reproduced with an anti-GFP antibody and an integrated his-24::gfp transgenic animal line (data not shown).
Fertilization triggers fast nuclear translocation of HIS-24, and P4 cell cleavage induces HIS-24 degradation in primordial germ cells.
Immediately after sperm entry, HIS-24 starts to associate with chromatin (Fig. 2P to R). During the pronucleus stage, HIS-24 is found in the chromatin of the polar bodies and in both pronuclei. Only a small residual fraction remains transiently in the cytoplasm. During the cleavage stage of embryogenesis, HIS-24 segregates to all somatic nuclei and also to the nuclei of the P lineage (P0 to P4) (Fig. 2 M to O). HIS-24, however, is absent from the primordial germ cells Z2 and Z3, which are the daughters of P4 (Fig. 2I to K). A faint staining of Z2 and Z3 was occasionally observed shortly after P4 cleavage. This indicates a transcriptional shutdown of his-24, coinciding with a successive germ line-specific degradation of HIS-24 after P4 cleavage.
Germ line-specific cytoplasmic retention and degradation of HIS-24 are lost in the sterile mes-2, -3, -4, and -6 mutants and in the C. elegans sir-2.1(ok434) sirtuin mutant.
In previous studies we identified a relationship of HIS-24 with the epigenetic silencing system formed by the four maternal-effect sterile genes mes-2, mes-3, mes-4, and mes-6. These mutants are fertile if a maternal contribution of wild-type mRNA is available for a homozygous mutant zygote (M+Z−) but are sterile if they originated from homozygous mes mutant mothers (M−Z−). The sterility in mes mutant animals is caused by a failure of germ cell proliferation and differentiation. We tested the expression and the subcellular localization of HIS-24 in mes mutant animals. In sterile (M−Z−) mutants of any of these four genes, HIS-24 was expressed in the remaining hermaphrodite germ line (Fig. 4). This protein, however, was chromatin associated, as it normally is, only in somatic cells. We analyzed mes-2(bn11), mes-3(bn21)ts (at 25°C), mes-3(bn35), mes-4(bn23), and mes-6(bn66) mutants. A mislocalization to the germ nuclei was found in all strains. The HIS-24 degradation subsequent to P4 cell division that occurs typically did not happen in the mes-3(bn35) (M−Z−) mutant embryos, which develop to sterile adults (Fig. 4D). A comparable observation was made for mes-4(bn23). mes-2 and mes-6 mutants were not tested for this effect. This observation suggests that the subcellular mislocalization and ectopic expression of his-24 in the Z2 and Z3 cells is indicative for mes phenotypic primordial germ cells that later fail to proliferate and to differentiate. Interestingly, we identified a comparable mislocalization of HIS-24 to the germ nuclei in the C. elegans sir-2.1(ok434) sirtuin mutant (Fig. 3H). In contrast to the mes mutants, the sir-2.1(ok434) mutant has no obvious germ line phenotype; however, we have shown earlier that sir-2.1 contributes to the silencing of the germ line (11).
FIG. 4.
Immunofluorescence detection of HIS-24 in mes-3(bn35) mutant hermaphrodites (HIS-24, green; DAPI, blue; P granules, red). (A to D) Pregastrulation stage embryo that originated from a mes-3(bn35) homozygous mother. HIS-24 is misexpressed in the primordial germ cells Z2 and Z3 (arrows). Interestingly, the HIS-24 signal in the Z2 and Z3 cells is surrounded by the cytoplasmic P granules and colocalizes with the DAPI staining (D), indicating that HIS-24 has a nuclear localization. In the germ line of the first homozygous mes-3(bn35) generation (fertile M+Z−), HIS-24 is partially mislocalized to the nuclear compartment (E to G). In the subsequent homozygous mes-3(bn35) generation (sterile M−Z−), HIS-24 is completely mislocalized to the nuclear compartment (H to J). Bars, 20 μm.
First generation homozygous (M+Z−) fertile mes-3(bn35) mutants show a partial mislocalization of HIS-24 to germ nuclei.
To investigate whether the mislocalization of HIS-24 in sterile (M−Z−) mes mutant animals is a consequence merely of the strong sterile gonad phenotype, the situation in fertile (M+Z−) mes-3(bn35) mutant animals was additionally analyzed. Interestingly, it was noted that a considerable fraction of HIS-24 associates with the hermaphrodite germ line chromatin in peculiar granular structures in these animals, whereas the remaining quantity of this protein was located in the cytoplasm. This intermediary situation was seen in mitotic and meiotic germ nuclei as well as in oocytes (Fig. 3).
The germ line-specific subcellular localization of HIS-24 is determined during early gonadogenesis.
Using temperature shift experiments with the temperature-sensitive mes-3(bn21) allele, we investigated the time period during which the mes gene function is required for normal subcellular localization of HIS-24 in the hermaphrodite germ line (Table 1). It is known from previous work (30) that MES-3 function is essentially required in the pregastrulation embryo to achieve fertility in the adult stage. However, MES-3 function is not required for this in later stages of embryonic and larval development. When mes-3(bn21) mutant animals were raised at the restrictive temperature of 25°C, the resulting hermaphrodites showed a complete mislocalization of HIS-24 to the chromatin of the germ nuclei (Fig. 5).
TABLE 1.
HIS-24 mislocalization in a mes-3(bn21)ts temperature shift experiment
| Developmental stage | % of animals with mislocalization (no. with mislocalization/total) after shift froma:
|
|
|---|---|---|
| 16°C to 25°C | 25°C to 16°C | |
| L3 | 83 (25/30) | 10 (3/40) |
| Young L4 | 60 (12/20) | 40 (8/20) |
| L4 | 10 (2/20) | 90 (18/20) |
| Young hermaphrodite | 0 (0/10) | 100 (20/20) |
| Older hermaphrodite | 0 (0/10) | 100 (20/20) |
The temperature shift experiment was done as indicated in Materials and Methods. Numbers represent the percentage of animals that had a mislocalization of HIS-24 to the germ nuclei 16 h after the shift.
FIG. 5.
Indirect immunofluorescence detection of HIS-24 in temperature shift experiments performed with mes-3(bn21)ts mutant animals. Animals raised at the restrictive temperature of 25°C show a complete nuclear mislocalization of HIS-24 (A to C). A temperature downshift of L3 stage larvae and also of young L4 stage larvae to the permissive temperature of 16°C is sufficient to produce a wild-type-like localization of HIS-24 (D to F). Conversely, animals raised at the permissive temperature of 16°C show wild-type-like cytoplasmic localization of HIS-24 (G to I). A temperature upshift of L3 stage larvae and also of young L4 stage larvae to the restrictive temperature of 25°C is sufficient to produce a mes phenotypic nuclear mislocalization of HIS-24 (J to L). The animals were analyzed 16 h after the temperature shift. Bars, 20 μm.
When the animals were shifted to the permissive temperature in the stage of young L4 larvae (or in even younger stages of development), HIS-24 was found in the cytoplasm surrounding the germ nuclei of the resulting adult animals. A small fraction of these animals developed to fertile adults. When the temperature shift was done at later developmental stages than young L4 larvae, HIS-24 remained in the nuclei and the animals were sterile.
Conversely, when mes-3(bn21) mutant animals were raised at the permissive temperature of 16°C, the resulting hermaphrodites showed a wild-type-like localization of HIS-24 in the cytoplasm of the germ line. When we shifted young mes-3(bn21) L4 larvae (or even younger stages of development) to the restrictive temperature and analyzed them as young adults, HIS-24 was found in the germ nuclei. Again this was not observed when the temperature shift was done at later developmental stages than young L4. Because the gonads of adult hermaphrodites contain more than 1,000 germ nuclei and mitosis and meiosis continue during adulthood, we conclude that it is not the time of germ nucleus division during which mes-3 activity is required for a normal germ line-specific cytoplasmic localization of HIS-24. Instead, our results indicate that mes-3 activity is required for HIS-24 distribution just transiently during a very specific developmental situation when the young L4 stage gonads mature.
The deletion allele his-24(ok1024) produces defects in germ line proliferation, gametogenesis, and transgene silencing.
Homozygous his-24(ok1024) animals are vital, have a normal appearance, and grow in a way comparable to wild-type C. elegans. However, a more detailed analysis of his-24(ok1024) revealed a cytological gonad phenotype similar to the phenotype reported in our earlier his-24 RNAi studies (9). Nine percent of the adult hermaphrodite animals lacked sperm and oocytes in one of the two gonad arms (Fig. 6). Moreover, the corresponding gonad arms also contained a reduced number of undifferentiated germ nuclei. These germ nuclei had an unusual doughnut-like morphology. Sterility of both gonad arms occurs rarely in his-24(ok1024) animals. Three of 651 hermaphrodites analyzed (0.5%) were completely sterile.
FIG. 6.
(A) Cytological appearance of a his-24(ok1024) mutant hermaphrodite. One of the two gonad arms (left side) is sterile. It contains a reduced number of germ nuclei, no oocytes, and no sperm. Some germ nuclei are doughnut shaped (left arrow). The other gonad arm (right side) produces oocytes (right arrow) and is fertile, as indicated by the presence of a zygote (triangle). The inset shows an enlarged view of swollen, doughnut-shaped germ nuclei from the gonad of a different animal. (B) When a silenced repetitive let-858::gfp transgene is crossed into a his-24(ok1024) background, germ line expression of the transgene is activated in undifferentiated germ nuclei and oocytes (arrows). (C) Corresponding Nomarski picture. Bars, 20 μm.
Transgene silencing in the germ line of his-24(ok1024) animals in crosses with a strain carrying a repetitive extrachromosomal let-858::gfp transgene was also analyzed. This indicator transgene is typically active in the soma but silenced in the germ line. The heterozygous first generation [his-24(ok1024)/+] showed an activation of transgene expression in all animals (Fig. 6). This was not observed in his-24 wild-type control animals. The germ line expression of the transgene was activated in his-24(ok1024)/+ animals which had a maternal contribution of his-24(+) as well as in animals that had a paternal contribution of his-24(+). We analyzed the germ line transgene expression in his-24(ok1024) homozygous animals in the subsequent generations (Table 2). All animals of the first homozygous generation showed activation of the transgene, whereas in the subsequent two generations germ line expression of the transgene was successively lost. In the fourth homozygous his-24(ok1024) generation, transgene expression was completely lost in the germ lines of all animals.
TABLE 2.
Germ line desilencing of a repetitive transgene in the his-24(ok1024) mutanta
| Cross | Generation (his-24 genotype) | % of hermaphrodites with desilencing effectb |
|---|---|---|
| PD7271 × his-24(ok1024) (male) | F1 (ok1024/+) | 100 (87/87) |
| F2 (ok1024/ok1024) | 100 (10/10) | |
| F3 (ok1024/ok1024) | 80 (177/142) | |
| F4 (ok1024/ok1024) | 20 (100/20) | |
| F5 (ok1024/ok1024) | 0 (100/0) | |
| PD7271 (male) × his-24(ok1024) | F1 (ok1024/+) | 100 (121/121) |
| F2 (ok1024/ok1024) | 100 (10/10) | |
| F3 (ok1024/ok1024) | 70 (141/99) | |
| F4 (ok1024/ok1024) | 10 (100/10) | |
| F5 (ok1024/ok1024) | 0 (100/0) | |
| Controls | ||
| PD7271 × N2 (male) | F1 (+/+) | 0 (531/0) |
| F2 (+/+) | 0 (101/0) | |
| F3 (+/+) | 0 (378/0) | |
| PD7271 (male) × N2 | F1 (+/+) | 0 (341/0) |
| F2 (+/+) | 0 (110/0) | |
| F3 (+/+) | 0 (211/0) |
The repetitive extrachromosomal transgene let-858::gfp was crossed from a his-24(+) source strain (PD7271) to the his-24(ok1024) mutant. Germ line transgene expression (Fig. 6) in ok1024 heterozygous F1 animals and in subsequent generations of his-24(ok1024) homozygous animals was analyzed.
The numbers in parentheses are the number of animals analyzed followed by the number of animals with LET-858::GFP fluorescence in the germ line.
his-24 RNAi and his-24 deletion promote transgene methylation of histone H3 at lysine 4 and antagonize transgene methylation of histone H3 at lysine 9.
The germ line expression of silenced let-858::gfp transgenes was activated either by his-24 RNAi as shown before (9) or by crossing into a his-24(ok1024) background. The methylation status of histone H3 at lysine residues 4 and 9 was determined using indirect immunofluorescence. Antibody staining of histone H3 methylated at lysine 4 showed a gain of this modification on desilenced transgenes in pachytene stage germ nuclei and in oocytes, whereas this modification was absent from silenced transgenes in control animals (Fig. 7).
FIG. 7.
Analysis of H3K9me2 and H3K4me2 in the context of the desilencing of a repetitive extrachromosomal transgene in his-24(ok1024) mutant hermaphrodites (A to D and M to P) and in his-24 wild-type animals. Pachytene stage germ nuclei are shown. The transgenes were detected as brightly DAPI-stained spot-like structures (arrows). Desilencing of the let-858::gfp transgene (B and N) is detected only in the his-24(ok1024) mutant. This correlates with a loss of the typically very intense H3-K9me2 modification at the transgenes (C) as well as with a spreading of the H3K4me2 to the transgene (O). This modification is normally not detected at the transgene (K). Bars, 5 μm.
In contrast, antibody staining of histone H3K9me2 showed a loss of H3K9 methylation in the desilenced extrachromosomal transgene chromatin in mature oocytes and also in pachytene stage germ nuclei (Fig. 7). In control animals, which had a silenced transgene, prominent H3K9me2 staining was detected. Both experiments were done with the his-24(ok1024) mutant and in addition with his-24 RNAi. In all cases a strict correlation of the H3 modification status of the extrachromosomal transgene with let-858::gfp expression was observed.
The his-24(ok1024) mutation is a putative null allele.
Sequencing of the his-24 deletion allele ok1024 revealed a 2,548-bp deletion eliminating most of the coding sequence and all of the 3′ untranslated region of his-24. The remaining coding sequence represents the N-terminal first 57 amino acid residues of his-24 wild-type protein, which has a total length of 208 amino acids. The deletion preserves the N-terminal domain but eliminates the complete C-terminal domain and the globular domain after the first alpha helix, as well as the single his-24 intron. Because reverse transcription-PCR experiments detected an mRNA that originated from the his-24(ok1024) allele, we tested with an allele-specific RNAi experiment whether the his-24(ok1024) phenotype could be modulated. Because no phenotypic changes were detected, we assumed that the remaining N-terminal domain of his-24(ok1024) does not confer any function. Because the locus-specific RNAi does not change the phenotype and two out of three functional domains of HIS-24 are deleted, it is highly probable that ok1024 represents a null allele.
The his-24 deletion allele his-24(ok1024) enhances the cytological defects of mes-3 mutants.
In order to investigate the functional relation between his-24 and the mes genes, his-24(ok1024) mes-3 double mutants were created. We used the temperature-sensitive allele mes-3(bn21)ts as well as the non-temperature-sensitive allele mes-3(bn35). The mes-3(bn21)ts his-24(ok1024) double mutant reproduced the general phenotypic characteristics of the mes-3(bn21) mutant, which is fertile at 16°C and sterile at 25°C. Similarly the mes-3(bn35) his-24(ok1024) double mutant reproduced the general phenotypic effects of mes-3(bn35), such as fertility of the mes-3 M+Z− generation and sterility of the M−Z− animals. No parental his-24(+) contribution was provided in these experiments. In both double mutants the sterility is caused by a failure of germ cell proliferation and differentiation, as is expected for the mes-3 mutants. A detailed analysis of the sterile gonads of double mutant adult animals was done by DAPI staining. The data presented in Table 3 revealed that, compared to the mes-3 mutants alone, the mes-3 his-24(ok1024) double mutants possess a further reduced number of germ nuclei. The synthetic phenotype was also studied for H3K27 methylation in fertile (M+Z−) mes-3(bn35) his-24(ok1024) mutant animals. The reaction conditions used do not allow one to distinguish between mono-, di-, and trimethylation, although the antibody used has a strong preference for H3K27me3. In germ nuclei of wild-type adult hermaphrodites, methylation of H3 at lysine K27 is readily detectible in all stages of germ line development. The M+Z− mes-3(bn35) mutant animals show a strongly reduced, although detectable, H3K27me staining, whereas the methylation is completely lost in the germ line of sterile (M−Z−) mes-3(bn35) animals (2). In M+Z− mes-3(bn35) his-24(ok1024) double mutant animals, no H3K27me was detectable (Fig. 8). The intensity of the anti-H3K27me staining of somatic nuclei was comparable in all experiments. Animals that were his-24 mutant but mes wild type showed an H3K27me status undistinguishable from the wild type. These observations show that his-24(ok1024) specifically enhances the deficiency of H3 methylation at K27 in the germ line of fertile (M+Z−) mes-3(bn35) mutant animals.
TABLE 3.
Numbers of germ nuclei in mes-3 his-24 double mutantsa
| Strain | No. of germ nuclei per gonad (mean ± SD)b |
|---|---|
| N2 | 1,140 ± 120 |
| mes-3(bn21)ts | 38 ± 10 |
| mes-3(bn21)ts; his-24(ok1024) | 12 ± 20 |
| mes-3(bn35) | 172 ± 80 |
| mes-3(bn35); his-24(ok1024) | 81 ± 34 |
mes-3(bn21) animals were raised at 25°C. mes-3(bn35) mutant hermaphrodites were derived from homozygous mutant mothers and raised at 20°C.
Values were determined from counting DAPI-stained germ nuclei in both gonad arms of at least 20 hermaphrodites.
FIG. 8.
Analysis of H3K27me in a mes-3(bn35) his-24(ok1024) double mutant. Fertile (M+Z−) mes-3(bn35) animals originating from mes heterozygous mothers are shown. The mes-3(bn35) mutant shows a residual H3K27 methylation in the germ nuclei (B and I) (2). H3K27me is detectable in somatic nuclei (arrows) and in germ nuclei from late pachytene to oogenesis (arrowheads). H3K27me is enhanced on the X chromosome (stars) in pachytene nuclei (B and I). In the double mutant, no H3K27me is detectable in germ nuclei (C). The H3K27me epitope is detected only in somatic nuclei (arrows). In wild-type animals H3K27me is present in all nuclei of the gonad (A). Bars, 20 μm.
DISCUSSION
The linker histone HIS-24 promotes core histone modifications in the hermaphrodite germ line.
Our first observations using HIS-24 RNAi indicated a function of this protein in the germ line (9). This was concluded from the decrease of the repeat-dependent silencing of transgenes in both sexes and also from the occurrence of a hermaphrodite-specific low-penetrant sterile phenotype. Both observations were similar to observations for the mes gene phenotype. Our results here present the corresponding analysis of chromatin modifications for RNAi experiments and for the deletion mutant his-24(ok1024). HIS-24 RNAi or the introduction of a silenced extrachromosomal array in the his-24(ok1024) background led to transgene activation and to a specific loss of the repressive H3K9me2 modification on the transgene. Concomitantly, the activating chromatin modification H3K4me2, which is typically enriched on germ line autosomes, spread to the transgene. In both types of experiments a persistent permanent activation of the transgene that would last for more than one generation (RNAi) or a few generations [his-24(ok1024)] could not be achieved. This suggests that his-24 promotes the establishment and maintenance of germ line-specific chromatin modifications in a partially redundant way so that the germ cells can compensate effectively for the loss of HIS-24 after a few generations.
The linker histone mutation his-24(ok1024) enhances the mes gene phenotype.
The C. elegans mes genes mes-2, mes-3, mes-4, and mes-6 are required to establish a distinct silenced chromatin state of the X chromosome during hermaphrodite germ line development. This process is essential for hermaphrodite germ line development and fertility. Besides this, the mes genes also function in the silencing of repetitive transgenes. The MES complex is a histone methyl transferase and consists of MES-2, MES-3, and MES-6 (15). It is responsible for H3K27 di- and trimethylation in the adult germ line and in embryos. The target loci in the germ line are concentrated on the X chromosome (2). We show here that the his-24(ok1024) mutation enhances the severity of the mes-3 phenotype at the cytological level and also at the level of histone H3K27 methylation. This poses the question of how a possible synergistic interaction between his-24 and the mes genes could happen.
The mes genes and sir-2.1 control a germ line-specific cytoplasmic retention of HIS-24.
Surprisingly, we identified HIS-24 in a granular cytoplasmic structure that is characteristic for most developmental stages of the germ line but is absent in the soma. In contrast to the case for the later germ line, HIS-24 is localized to the nuclei of the germ line precursor cells from P0 to P4. Histones, including linker histones, translocate very efficiently to the nuclear compartment (1, 8). Therefore, the cytoplasmic pool of histones is typically very low. Only in a few cases has an extracellular (7) or a cytoplasmic localization of linker histones or linker histone-like proteins been described (10, 17, 21). Because this observation is very unexpected, careful validation is necessary. Different observations support our claim. (i) The characteristic granular signal is absent from the his-24(ok1024) deletion mutant, which has lost the sequence that encodes the peptide used for immunization. (ii) Independent antibody staining using an anti-GFP antibody and his-24::gfp transgenic animals reproduced the initial observation. (iii) The anti-HIS-24 staining of the wild-type gonad contains a significant internal control: nuclei of somatic cells are stained very intensely, whereas the adjacent germ nuclei are not stained. (iv) The characteristic cytoplasmic enrichment seen in the adult hermaphrodite germ line was not detected in the mes mutants. (v) A polyclonal antibody raised against H1.4, which is cross-reactive with H1.1 to H1.5 and H1.Q, labels the germ line chromatin at all stages (9). Taken together, these observations allow exclusion of the possibility of unspecific antibody cross-reactivity of the primary or the secondary antibody as well as insufficient permeabilization of the specimen.
A mutation in a single one of the four mes genes mes-2, mes-3, mes-4, and mes-6 or in sir-2.1 caused a loss of the cytoplasmic retention of HIS-24. This indicates that either the direct function of the mes genes or a later consequence of their activity causes the very unusual cytoplasmic retention of the linker histone HIS-24. The mislocalization of HIS-24 in fertile M+Z− mes mutant animals points to a more direct role of the mes genes in the nuclear exclusion of HIS-24. This observation also allowed us to speculate that nuclear HIS-24 could be a causative element in the sterile mes phenotype. Our analysis of the his-24(ok1024) mes-3(bn35) double mutant, however, showed clearly that this is not the case.
Models for the developmental function of HIS-24.
The primary protein structure of HIS-24 is that of a typical linker histone with no particular variations. The nuclear localization of HIS-24 in the C. elegans soma as well as in transgenic Saccharomyces cerevisiae (11) suggests the nucleus as the default subcellular target compartment. We conclude that our observations can be explained only by a germ line-specific inhibition of nuclear translocation. This could be caused by a posttranslational modification of HIS-24, by the binding of specific RNA or protein factors to this protein, or by a combination of both. Because the default target compartment for HIS-24 is the nucleus, this process could also involve a primary nuclear import of HIS-24, followed by HIS-24 modification and a subsequent nuclear reexport. The modified protein then would be stably retained in the cytoplasm. Such a model would also imply that the nuclear export of HIS-24 is a continuous process in the germ line. Our present data suggest that the best candidates for HIS-24-modifying enzymes are the MES complex and the sirtuin SIR-2.1. Both function in the nucleus, and both contribute to germ line silencing in C. elegans (11, 13). This model is supported by data from mammalian homologs, i.e., the Polycomb repressive complexes and SirT1. SirT1 demethylates mammalian histone H1b at lysine 26 and interacts with the Polycomb repressive complex PRC4, which preferentially methylates this lysine residue (19, 20, 29). Mammalian SirT1 is the putative ortholog of the C. elegans sirtuin SIR-2.1. It has been shown that another histone methyltransferase (Set9) controls the protein stability of p53 (5). Interestingly, HIS-24 is degraded in the germ nuclei shortly after the primordial germ cells Z2 and Z3 are born. This coincides with the activation of the MES-2 histone methyltransferase and the establishment of germ line-specific chromatin structures (23, 24, 25, 27).
Additionally, our data suggest a function of HIS-24 in germ line development. The his-24 mutant shows germ line defects and enhances the reduced germ cell proliferation in mes-3 mutants. HIS-24 is also implicated in germ line-specific H3 methylation at three different lysine residues and therefore contributes to a process which maintains autosome activity and reduces the activity of X chromosomes and repetitive transgenes. Considering these observations, we conclude that HIS-24 supports the establishment of germ line-specific chromatin structures. We proved that the germ line-specific cytoplasmic localization of HIS-24 is an unusual consequence of the function of germ line-specific chromatin-modifying systems. Therefore, we may speculate that HIS-24 is part of a positive regulative feedback loop that establishes and maintains germ line chromatin structure. Although our data suggest a non-chromatin-based function for HIS-24, we cannot completely exclude a function of HIS-24 in the chromatin because a small fraction of HIS-24 is associated with germ line chromatin during the late pachytene stage. Moreover, our suggestion of a continuous nuclear reexport of HIS-24 implies a constant source of a small amount of HIS-24 in the germ nuclei. Possible non-chromatin-associated functions of HIS-24 could be the nuclear coexport of RNAs or proteins bound to it, the translational control of mRNAs in the cytoplasm, or the prevention of the nuclear translocation of RNAs or proteins in germ cells. Nonchromatin functions of linker histones have been recognized in a few cases (7, 10). Because in the his-24(ok1024) mutant transgenes are first reactivated and then efficiently silenced after a few generations, we believe that chromatin structure is controlled by multiple redundantly functioning regulatory systems, which can compensate for the loss of his-24. This would also explain the relatively mild phenotype of the his-24(ok1024) mutant.
Acknowledgments
We thank Sabine Pitzel and Angelika Schäfer for their excellent technical assistance, Bettina Schulze and Krzysztof Drabikowski for critically reading the manuscript, and Ralf Baumeister (Freiburg, Germany) and Ernst Wimmer (Göttingen, Germany) for their support of this work in their respective labs. We gratefully received the excellent anti-H3K27me3 antibody from T. Jenuwein (Vienna, Austria) and the LET-858::GFP reporter strain PD7271 from W. G. Kelly (Atlanta, GA).
Some C. elegans strains used were obtained from the Caenorhabditis Genetics Center (CGC), which is funded by the NIH National Center for Research Resources (NCRR). The his-24(ok1024) deletion mutant was provided by the C. elegans Gene Knockout Consortium, which is publicly funded. This work was supported by German National Funding Agency (DFG) grants SCHU 1033/3-4 to E. Schulze and JE 505/1-1 to M. Jedrusik.
Footnotes
Published ahead of print on 8 January 2007.
REFERENCES
- 1.Baake, M., M. Bauerle, D. Doenecke, and W. Albig. 2001. Core histones and linker histones are imported into the nucleus by different pathways. Eur. J. Cell Biol. 80:669-677. [DOI] [PubMed] [Google Scholar]
- 2.Bender, L. B., R. Cao, Y. Zhang, and S. Strome. 2004. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr. Biol. 14:1639-1643. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustin, M., F. Catez, and J. H. Lim. 2005. The dynamics of histone H1 function in chromatin. Mol. Cell 17:617-620. [DOI] [PubMed] [Google Scholar]
- 5.Chuikov, S., J. K. Kurash, J. R. Wilson, B. Xiao, N. Justin, G. S. Ivanov, K. McKinney, P. Tempst, C. Prives, S. J. Gamblin, N. A. Barlev, and D. Reinberg. 2004. Regulation of p53 activity through lysine methylation. Nature 432:353-360. [DOI] [PubMed] [Google Scholar]
- 6.Fan, Y., T. Nikitina, E. M. Morin-Kensicki, J. Zhao, T. R. Magnuson, C. L. Woodcock, and A. I. Skoultchi. 2003. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 23:4559-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henriquez, J. P., J. C. Casar, L. Fuentealba, D. J. Carey, and E. Brandan. 2002. Extracellular matrix histone H1 binds to perlecan, is present in regenerating skeletal muscle and stimulates myoblast proliferation. J. Cell Sci. 115:2041-2051. [DOI] [PubMed] [Google Scholar]
- 8.Jakel, S., W. Albig, U. Kutay, F. R. Bischoff, K. Schwamborn, D. Doenecke, and D. Gorlich. 1999. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 18:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jedrusik, M. A., and E. Schulze. 2001. A single histone H1 isoform (H1.1) is essential for chromatin silencing and germline development in Caenorhabditis elegans. Development 128:1069-1080. [DOI] [PubMed] [Google Scholar]
- 10.Jedrusik, M. A., S. Vogt, P. Claus, and E. Schulze. 2002. A novel linker histone-like protein is associated with cytoplasmic filaments in Caenorhabditis elegans. J. Cell Sci. 115:2881-2891. [DOI] [PubMed] [Google Scholar]
- 11.Jedrusik, M. A., and E. Schulze. 2003. Telomeric position effect variegation in Saccharomyces cerevisiae by Caenorhabditis elegans linker histones suggests a mechanistic connection between germ line and telomeric silencing. Mol. Cell. Biol. 23:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jedrusik, M. A., and E. Schulze. 2004. Analysis of germline chromatin silencing by double-stranded RNA-mediated interference (RNAi) in Caenorhabditis elegans. Methods Mol. Biol. 254:35-48. [DOI] [PubMed] [Google Scholar]
- 13.Kelly, W. G., and A. Fire. 1998. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125:2451-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly, W. G., C. E. Schaner, A. F. Dernburg, M. H. Lee, S. K. Kim, A. M. Villeneuve, and V. Reinke. 2002. X-chromosome silencing in the germline of C. elegans. Development 129:479-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ketel, C. S., E. F. Andersen, M. L. Vargas, J. Suh, S. Strome, and J. A. Simon. 2005. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol. Cell. Biol. 25:6857-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khochbin, S. 2001. Histone H1 diversity: bridging regulatory signals to linker histone function. Gene 271:1-12. [DOI] [PubMed] [Google Scholar]
- 17.Konishi, A., S. Shimizu, J. Hirota, T. Takao, Y. Fan, Y. Matsuoka, L. Zhang, Y. Yoneda, Y. Fujii, A. I. Skoultchi, and Y. Tsujimoto. 2003. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell 114:673-688. [DOI] [PubMed] [Google Scholar]
- 18.Korf, I., Y. Fan, and S. Strome. 1998. The Polycomb group in Caenorhabditis elegans and maternal control of germline development. Development 125:2469-2478. [DOI] [PubMed] [Google Scholar]
- 19.Kuzmichev, A., T. Jenuwein, P. Tempst, and D. Reinberg. 2004. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell 14:183-193. [DOI] [PubMed] [Google Scholar]
- 20.Kuzmichev, A., R. Margueron, A. Vaquero, T. S. Preissner, M. Scher, A. Kirmizis, X. Ouyang, N. Brockdorff, C. Abate-Shen, P. Farnham, and D. Reinberg. 2005. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc. Natl. Acad. Sci. USA 102:1859-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni, J. Q., L. P. Liu, D. Hess, J. Rietdorf, and F. L. Sun. 2006. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 20:1959-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Praitis, V., E. Casey, D. Collar, and J. Austin. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157:1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuben, M., and R. Lin. 2002. Germline X chromosomes exhibit contrasting patterns of histone H3 methylation in Caenorhabditis elegans. Dev. Biol. 245:71-82. [DOI] [PubMed] [Google Scholar]
- 24.Schaner, C. E., G. Deshpande, P. D. Schedl, and W. G. Kelly. 2003. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev. Cell 5:747-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaner, C. E., and W. G. Kelly. 24 January 2006, posting date. Chapter 1.73.1. Germline chromatin. In WormBook. doi: 10.1895/wormbook.1.73.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 26.Strome, S., and W. B. Wood. 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35:15-25. [DOI] [PubMed] [Google Scholar]
- 27.Strome, S. 28 July 2005, posting date. Chapter 1.9.1. Specification of the germ line. In WormBook. doi: 10.1895/wormbook.1.9.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 28.Turner, B. M. 2005. Reading signals on the nucleosome with a new nomenclature for modified histones. Nat. Struct. Mol. Biol. 12:110-112. [DOI] [PubMed] [Google Scholar]
- 29.Vaquero, A., M. Scher, D. Lee, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16:93-105. [DOI] [PubMed] [Google Scholar]
- 30.Xu, L., J. Paulsen, Y. Yoo, E. B. Goodwin, and S. Strome. 2001. Caenorhabditis elegans MES-3 is a target of GLD-1 and functions epigenetically in germline development. Genetics 159:1007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]