Abstract
Porphyria cutanea tarda (PCT), the most common form of porphyria in humans, is due to reduced activity of uroporphyrinogen decarboxylase (URO-D) in the liver. Previous studies have demonstrated that protein levels of URO-D do not change when catalytic activity is reduced, suggesting that an inhibitor of URO-D is generated in hepatocytes. Here, we describe the identification and characterization of an inhibitor of URO-D in liver cytosolic extracts from two murine models of PCT: wild-type mice treated with iron, δ-aminolevulinic acid, and polychlorinated biphenyls; and mice with one null allele of Uro-d and two null alleles of the hemochromatosis gene (Uro-d+/−, Hfe−/−) that develop PCT with no treatments. In both models, we identified an inhibitor of recombinant human URO-D (rhURO-D). The inhibitor was characterized by solid-phase extraction, chromatography, UV-visible spectroscopy, and mass spectroscopy and proved to be uroporphomethene, a compound in which one bridge carbon in the uroporphyrinogen macrocycle is oxidized. We synthesized uroporphomethene by photooxidation of enzymatically generated uroporphyrinogen I or III. Both uroporphomethenes inhibited rhURO-D, but the III isomer porphomethene was a more potent inhibitor. Finally, we detected an inhibitor of rhURO-D in cytosolic extracts of liver biopsy samples of patients with PCT. These studies define the mechanism underlying clinical expression of the PCT phenotype, namely oxidation of uroporphyrinogen to uroporphomethene, a competitive inhibitor of URO-D. The oxidation reaction is iron-dependent.
Keywords: heme, porphyrins, iron, oxidation
Porphyria cutanea tarda (PCT) results from subnormal activity of uroporphyrinogen decarboxylase (URO-D) in the liver. The URO-D reaction converts uroporphyrinogen to coproporphyrinogen in the pathway ultimately leading to heme (1). The disease phenotype results from hepatic accumulation of uroporphyrin, the oxidized substrate of URO-D, which circulates in plasma and is finally excreted in the urine. The fluorescent property of uroporphyrin is responsible for the photodermatitis that occurs on sun-exposed areas. Porphyric disorders are usually due to mutations affecting enzymes in the heme biosynthetic pathway, but only approximately one-third of patients with PCT are heterozygous for a mutant allele of URO-D (designated familial PCT, or F-PCT) (1, 2). Most heterozygotes for URO-D mutations do not express a phenotype unless additional genetic or environmental factors are present. These include homozygosity for mutant hemochromatosis alleles, hepatitis C, alcohol abuse, and the use of medicinal estrogens (2). No mutations in the URO-D gene have been identified in the remaining two-thirds of patients (designated sporadic PCT, or S-PCT). In both types of PCT, the clinical and biochemical phenotypes are identical. Hepatic iron overload is an almost universal finding, and depletion of hepatic iron stores by repetitive phlebotomy corrects the clinical phenotype (3, 4). One or more of the same factors associated with expression of F-PCT are present in almost all sporadic cases.
Immunoreactive URO-D in the liver of patients with S-PCT is normal, whereas in F-PCT, protein levels are approximately half the normal level (5). In contrast, hepatic URO-D enzyme activity is ≈25% of the normal level in both types of PCT, suggesting that the specific activity of hepatic URO-D has been affected (6, 7). Immunologic and catalytic URO-D assays have been repeated in patients with S-PCT after phlebotomy-induced clinical remission when hepatic porphyrins were normal (5). Catalytic activity had returned to normal, suggesting that an iron-dependent process is responsible for the reduction in the specific activity of hepatic URO-D (5).
We, and others, have generated mouse models of S-PCT by administering iron and inducers of the aryl-hydrocarbon receptor battery of cytochrome P450s (8–13). Early reports indicated the presence of an inhibitor of URO-D activity in heat-denatured liver cytosols from porphyric animals (14–16), a finding we confirmed (13). We also created a mouse model of F-PCT by disrupting one allele of Uro-d and crossing Uro-d+/− mice with mice homozygous for null alleles of the hemochromatosis gene (Hfe) (17). A porphyric phenotype occurred at 14 weeks of age, when hepatic URO-D activity was further reduced but immunoreactive URO-D did not change (17). The reduction in URO-D activity is comparable to values reported in the models of S-PCT referred to above.
Collectively, these data suggest that a common mechanism is responsible for expression of the porphyric phenotype, namely a reduction in the specific activity of hepatic URO-D through the generation of an inhibitor of URO-D by an iron-dependent mechanism. Here, we identify the inhibitor of URO-D as a porphomethene in which a single bridge carbon between adjacent pyrrole rings of uroporphyrinogen has been oxidized.
Results
A URO-D Inhibitor in Liver Homogenates.
Wild-type C57BL/6J and Uro-d+/− animals were treated with iron dextran and Aroclor 1254, and given δ-aminolevulinic acid (ALA)-supplemented drinking water as described in ref. 8. Animals were killed after 3 weeks and liver homogenates were prepared. Liver porphyrin content, mainly uroporphyrin I, was dramatically increased in both groups, but was slightly higher in Uro-d+/− animals (Table 1). The accumulation of uroporphyrin I was not due to diminished uroporphyrinogen III synthase (U3S) activity, because U3S assays revealed no abnormalities (data not shown).
Table 1.
Hepatic porphyrin content, URO-D activity, and inhibitor activity in treated and untreated mice
| Genotype | Treatment (n) | Hepatic porphyrins* | Hepatic URO-D activity† | rhURO-D inhibition, % |
|---|---|---|---|---|
| Uro-d+/+ | None (6) | 0.5 ± 0.2 | 1.07 ± 0.08 | 0.5 ± 1.4 |
| Uro-d+/+ | Fe/ALA/PCB‡ (7) | 940.1 ± 95.8 | 0.22 ± 0.03 | 54.2 ± 2.0 |
| Uro-d+/− | None (13) | 0.4 ± 0.1 | 0.50 ± 0.04 | 3.1 ± 0.9 |
| Uro-d+/− | Fe/ALA/PCB‡ (13) | 1,165.1 ± 62.0 | 0.13 ± 0.02 | 49.5 ± 3.8 |
Numbers in parentheses indicate number of animals per group.
*nmol/g of liver, wet weight.
†nmol of decarboxylated products per h per mg of protein.
‡Fe/ALA/PCB, iron dextran/δ -aminolevulinic acid/polychlorinated biphenyl (Aroclor 1254).
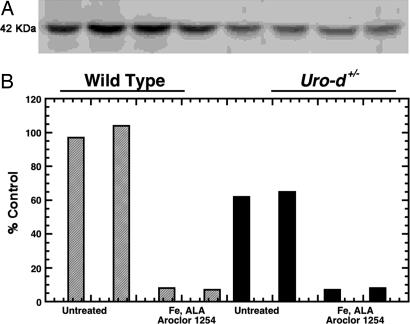
URO-D activity in untreated Uro-d+/− animals was approximately half (47%) of that seen in wild-type animals. When treatment-induced PCT was present, URO-D activity in animals of both genotypes was approximately equal (Table 1). URO-D protein was measured by Western blotting in two animals from each group. URO-D protein was not changed when URO-D activity was markedly reduced, indicating that the specific activity of URO-D had been diminished (Fig. 1). Cytosolic extracts of the liver homogenates were denatured and clarified by centrifugation, and aliquots of the supernatant were tested for inhibitory activity against recombinant human URO-D (rhURO-D) (see Materials and Methods). The mean inhibitory activity from treated wild type and Uro-d+/− was ≈50% (Table 1). To exclude the possibility that the URO-D protein had been modified as the porphyric phenotype developed, we performed isoelectric focusing and ran 2D gels of URO-D in liver homogenates. No differences were seen when the properties of URO-D from treated animals were compared with those of controls (data not shown).
Fig. 1.
The specific activity of URO-D is reduced in cytosolic extracts of porphyric mouse liver. Both wild-type (gray bars) and Uro-d+/− (black bars) mice were treated with iron dextran (Fe), drinking water supplemented with ALA, and a mixture of polychlorinated biphenyls (Aroclor 1254) and killed after 21 days, when porphyria was fully developed. (A) Western blotting of 30 μg of total cytosolic protein from each animal by using a 1:200 dilution of a polyclonal rabbit anti-human URO-D antiserum as a primary antibody and a 1:2,000 dilution of a horseradish-peroxidase-labeled goat anti-rabbit IgG as a secondary antibody. The blot was developed by using ECL reagents (Amersham Biosciences/GE Healthcare, Piscataway, NJ). URO-D protein in Uro-d+/− animals was approximately half of the wild-type value. Protein content did not change when URO-D catalytic activity fell in animals of either genotype. (B) URO-D catalytic activity before and after treatment. Two animals of each genotype were analyzed per group. URO-D activity for each animal is aligned below the corresponding band on the Western blot.
Characterization of the Inhibitor.
To further define the inhibitory activity in heat-denatured homogenates, the clarified supernatant was acidified by the addition of 2% acetic acid and applied to a hydrophilic–lipophilic balanced resin (HLB) column. The column was eluted with increasing concentrations of methanol in a stepwise fashion with 20% increments, and fractions were evaporated to dryness under argon, resuspended in 50 mM Tris buffer (pH 6.8), and again tested for inhibitory activity against rhURO-D. The column retained most of the porphyrins present in the clarified supernatant. Inhibitory activity was assayed in all fractions. Inhibitory activity, ranging from 40% to 60%, was found only in the 80–100% methanol eluate. To exclude the possibility that inhibitory activity was generated from Aroclor 1254, we used the Uro-d+/−, Hfe−/− mouse, which develops a porphyric phenotype without any manipulations (17). Liver homogenates from these animals, prepared and processed identically, also contained inhibitory activity (mean inhibition 38%), as previously reported (13).
Inhibitory activity of the 100% methanol HLB eluate was rapidly lost on exposure to ambient light and air. Processing all materials under red light preserved inhibitory activity and tubes were purged with argon until inhibition assays were performed. Size-exclusion membranes were used to estimate the molecular mass of the inhibitor. The inhibitor passed through the 2,000-Da cut-off membrane.
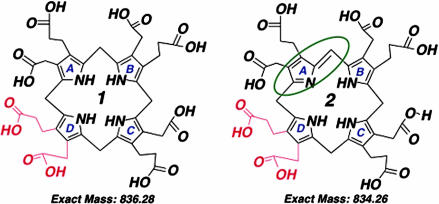
The optical spectra of the HLB column eluates with inhibitory activity displayed complex profiles that included a peak at ≈500 nm (data not shown), an absorption characteristic of uroporphomethene (18). Aliquots of 100% methanol eluates, prepared from 10 ml of porphyric cytosol, were evaporated to dryness as above and dissolved in 5% acetonitrile with 0.2% formic acid and analyzed by mass spectroscopy. A dominant peak with a protonated molecular mass of 835 Da was noted [see supporting information (SI) Fig. 6]. These data indicate that a porphomethene, in which one of the four bridge carbons of uroporphyrinogen (protonated molecular mass 837 Da) has been oxidized (Fig. 2), was present in the inhibitor fraction.
Fig. 2.
Structures of the I isomers of uroporphyrinogen and uroporphomethene. (Left) Uroporphyrinogen, a fully reduced substrate of URO-D. (Right) Uroporphomethene, the partially oxidized inhibitor of URO-D, has lost two hydrogens and gained a double bond on one bridge carbon, as indicated in the green oval. The molecular mass of each compound is indicated. Switching of the acetate and propionate groups (shown in red) on the D ring produces the III isomer compounds. Uroporphyrin, the fully oxidized fluorescent compound, is not a substrate of URO-D.
Synthesis of a URO-D Inhibitor from Uroporphyrinogen.
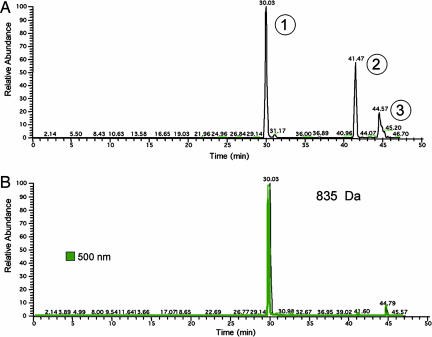
To verify that a porphomethene derived from uroporphyrinogen could inhibit rhURO-D, we generated uroporphyrinogen I from porphobilinogen (PBG) incubated with PBG-deaminase (PBG-D) (19). The porphyrinogen generated was oxidized with UV light in air for various times. Optimal inhibitory activity was generated after 8 min of UV exposure (data not shown). The inhibitor was passed over HLB to remove any fully oxidized uroporphyrin generated by the UV exposure. As before, inhibitory activity was eluted in 100% methanol. The inhibitor was further purified by HPLC with a C18 column and an aqueous/acetonitrile gradient (see Materials and Methods). Fractions were collected at 1-min intervals. Inhibitory activity was eluted at 30 min. This liquid chromatography system was then coupled to the mass spectrometer. Three peaks were detected (Fig. 3A). The peak eluting at 30 min had an absorbance maximum at 500 nm and contained a compound with a protonated molecular mass of 835 Da (Fig. 3), indicating that this peak contained uroporphomethene. The molecular mass of peak 2 was consistent with uroporphyrinogen (protonated molecular mass 837 Da) and peak 3 was consistent with uroporphyrin (protonated molecular mass 831 Da, absorbance maximum at 400 nm).
Fig. 3.
Visible absorbance and HPLC/MS base peak chromatogram overlays obtained from the analysis of partially purified inhibitor. (A) Base peak HPLC/MS chromatogram of substances detected between m/z 200 and 1,200. The base peak ions for peaks 1, 2, and 3 were m/z 835 (uroporphomethene), 837 (uroporphyrinogen), and 831 (uroporphyrin, visible absorbance at 400 nm), respectively. (B) Base peak HPLC/MS chromatogram representing ions of m/z 835 ± 1.0 and visible absorbance at 500 nm.
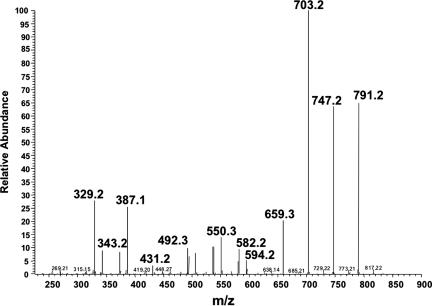
The 30-min peak containing the uroporphomethene was analyzed by tandem mass spectrometry. Product ions were generated with molecular masses differing by 44 Da, representing hepta-, hexa-, penta-, and tetracarboxylic porphomethenes (Fig. 4), verifying that the parent compound was an octacarboxylic tetrapyrrole. Additional product ions with lower protonated molecular masses were also identified. These products were consistent with decomposition of the porphomethene (m/z 835 Da) to tri-, di-, and monopyrroles. Tandem mass spectrometry of the 835-Da peak identified in the 100% methanol HLB eluate of porphyric mouse cytosol revealed an identical profile of product ions, verifying that the inhibitor identified in the mouse is identical to the uroporphomethene generated in vitro.
Fig. 4.
Tandem mass analysis of uroporphomethene I. The 835-Da base peak at 30 min was further analyzed by tandem mass spectroscopy (SI Methods). Product ions with masses of 791, 747, 703, and 659 Da represent the loss of one or more carboxyl groups from the octacarboxylic uroporphomethene. The smaller product ions represent additional breakdown products of the tetrapyrrole macrocycle that are consistent with tri- and dipyrroles. Neutral loss of 197 or 209 Da from m/z 791 via elimination of one pyrrole ring by cleavage of bonds at one of two positions yields m/z 594 and 582 (tripyrroles) with additional breakdown via loss of CO2. Neutral loss of 195 or 209 from m/z 582 yields m/z 387 and 373, which also decompose by loss of CO2. Products of heterolytic cleavage of m/z 835 are also detectable at m/z 419, 431, and 448/387.
The dominant porphyrins in the urine of patients with PCT are uroporphyrin (mainly isomer I) and heptacarboxyl porphyrin (mainly isomer III) (20). Similar isomer profiles were found in urine samples from our porphyric mice (data not shown). To determine whether porphomethenes generated from the I isomer or the III isomer of uroporphyrinogen were equivalent inhibitors of URO-D, inhibition assays were performed with either the I or the III isomer porphomethenes and the I or the III isomer porphyrinogen substrates (SI Table 2). To ensure that approximately equal amounts of the I and III isomer porphomethene were used in the inhibition assays, aliquots of the HLB eluates were analyzed by HPLC, using the C18 column and diode array detector system described (see Materials and Methods). The area under the curve at 500 nm was approximately the same for both isomers. The III isomer porphomethene appeared to be a more effective (two- to threefold) inhibitor against either substrate. Kinetic assays using dilutions of the HLB eluate prepared from porphyric mouse liver cytosol were performed with 1.5–10 μM uroporphyrinogen I or III as substrates and 0.4 μg of rhURO-D. Lineweaver–Burk plots were consistent with a competitive mechanism of inhibition (SI Fig. 7).
The interaction of the inhibitor and the enzyme was determined by soaking crystals of rhURO-D, prepared as previously described (21), with either the I or the III isomer porphomethene for 2 h. The porphomethenes were generated by photooxidization of enzymatically synthesized porphyrinogens and purified by solid-phase extraction followed by separation by HPLC on a C18 column as described above. The 30-min peak was collected, dried under argon, resuspended in crystallization buffer, and added to rhURO-D crystals in an anaerobic chamber. Data collected from these crystals indicated that the porphomethene was bound in the active site of URO-D. The electron density analysis was somewhat ambiguous but was consistent with a tetrapyrrole macrocycle in which two of the pyrrole rings were in a planar configuration (data not shown).
Human Studies.
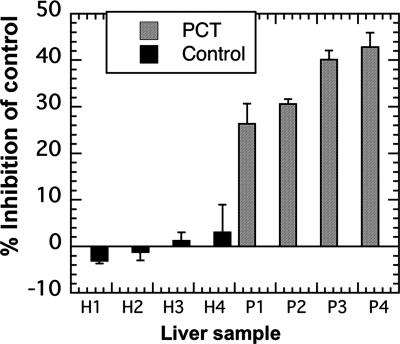
To determine whether the results of the animal studies had relevance to humans with PCT, we sought evidence for an identical inhibitor of URO-D in liver homogenates prepared from patients with PCT. Controls for these studies were untreated patients with hereditary hemochromatosis who did not have PCT. Liver biopsies were performed on four patients with PCT (three with S-PCT and one with F-PCT) and on four patients with hemochromatosis. All biopsy samples from patients with PCT fluoresced under UV illumination. The portion of the sample illuminated was used for histologic examination. The samples used for the URO-D inhibition assay ranged from 8 to 22 mg wet weight and were never illuminated. Inhibitory activity ranging from 25% to 42% was found in the PCT samples (Fig. 5). No inhibitory activity was found in the controls.
Fig. 5.
Inhibition of rhURO-D by denatured human cytosolic extracts. Biopsy samples ranging in weight from 8 to 22 mg were processed as described (see Materials and Methods) and assayed for inhibition of rhURO-D activity. Samples H1–H4 were obtained from untreated patients with hemochromatosis. Samples P1–P4 were obtained from untreated patients with PCT. Sample P2 was from a patient with F-PCT. All other porphyric samples were from patients with S-PCT.
Discussion
The finding that porphyrins are present in the livers of patients with PCT, hereditary coproporphyria, and porphyria variegata led Heikel et al. (22) to suggest that the mechanism responsible for photosensitizing porphyrias involved oxidation of porphyrinogen substrates to the corresponding porphyrins. Once formed, the porphyrins accumulated and were excreted as useless but harmful products. The “substrate oxidation hypothesis” was widely accepted until 1976, when an inherited defect in URO-D activity was demonstrated in patients with F-PCT (6), and subsequently an acquired defect was shown in hepatic URO-D activity in S-PCT (7, 23). The mechanism responsible for the reduction in URO-D activity was unclear, but the possibility of an inhibitor-mediated mechanism was suggested by the demonstration that the specific activity of URO-D was reduced in liver samples of patients with PCT (5).
In Turkey, an epidemic of PCT induced by hexachlorobenzene (24, 25) led to the development of animal models of PCT induced by polyhalogenated aromatic hydrocarbons. These models generated reports of a heat-stable inhibitor of URO-D (14, 15, 26–28). Later studies suggested that halogenated hydrocarbon induction of cytochrome P4501A2 (CYP1A2), a member of the aryl-hydrocarbon receptor battery of P450s, led to the oxidation of uroporphyrinogen to uroporphyrin by an iron-dependent mechanism and that oxidation reactions might generate an inhibitor of URO-D (29, 30). Here, we demonstrate that uroporphomethene, a partially oxidized porphyrinogen (Fig. 2), is the inhibitor of URO-D in experimental PCT in mice treated with PCB, iron, and drinking water supplemented with ALA. The optical properties, the molecular mass, and the MS/MS profile of the inhibitor all support this conclusion. Uroporphomethene was also identified in liver homogenates of the Uro-d+/−, Hfe−/− mouse, an animal that develops PCT without any manipulation. The porphomethene generated by partial photooxidation of uroporphyrinogen in vitro also proved to be an inhibitor of URO-D, and the physical properties of this porphomethene were identical to the inhibitor identified in porphyric mouse cytosol. A heat-stable inhibitor of URO-D was also identified in liver homogenates of patients with both F-PCT and S-PCT. The finding of an inhibitor of URO-D in widely differing mouse models and in humans with and without URO-D mutations provides a common mechanism responsible for the PCT phenotype.
Uroporphyrin is not a substrate for URO-D because oxidation of all of the bridge carbons yields a planar, nonflexible macrocycle. We have demonstrated that the flexible, fully reduced porphyrinogen substrate is accommodated in the active site of URO-D when the porphyrinogen adopts a domed configuration. This permits all four pyrrole nitrogens to form hydrogen bonds with Asp-86, a residue critical for enzymatic activity deep within the active site (21). The uroporphomethene, with three reduced bridge carbons (Fig. 2), is predicted to have sufficient flexibility to adapt to the spatial requirement of the active site, although the two pyrrole rings flanking the oxidized bridge carbon would be in the same plane. The mechanism of the decarboxylation catalyzed by URO-D has not been defined but this single oxidized bridge carbon variance from the structure of uroporphyrinogen is predicted to prevent the inhibitor from serving as a substrate.
The URO-D reaction with the III isomer substrate is less affected than the reaction with the I isomer substrate by either the I or the III isomer porphomethene inhibitor (SI Table 2). Our kinetic data with the inhibitor from porphyric mouse cytosol (SI Fig. 7) reflect this and support the view that the mechanism of inhibition is competitive. Kinetic studies with bovine and human URO-D indicate a lower Km and a higher Vmax for the III isomer uroporphyrinogen substrate compared with the I isomer substrate (31, 32). The structural basis of this observation is the position of the propionates of the substrate and the position of the arginine side chains (Arg-37 and Arg-41) in the active site. The III isomer reaction product (coproporphyrinogen) forms a more extensive network of hydrogen bonds than the I isomer product (11 hydrogen bonds vs. 6 hydrogen bonds) (21). These additional hydrogen bonds provide a higher binding energy to the III isomer product and a similar hydrogen bonding pattern is predicted for both the III isomer substrate (uroporphyrinogen) and the III isomer uroporphomethene. The more extensive network of hydrogen bonding predicted for the III isomer uroporphomethene is a probable explanation for our finding that the I isomer uroporphomethene is a less effective inhibitor.
In humans, uroporphyrinogen I is normally produced and excreted (as uroporphyrin I) in very small amounts. Total urinary uroporphyrin is normally <50 μg/d, almost all of which is uroporphyrin III (33). In patients with PCT, total urinary porphyrin excretion generally exceeds 1,000 μg/d and may be as high as 10,000 μg/d (1). The isomer composition of the excess urinary uroporphyrin is ≈80% isomer I and 20% isomer III (33), indicating that both isomers of uroporphyrin are excreted in excess. In our mouse models, most hepatic uroporphyrin was isomer I despite the fact that U3S activity was normal. Our studies with uroporphomethene inhibitors offer a plausible explanation for these observations. The accumulation of uroporphyrinogen I (and some uroporphyrinogen III), because of inhibited URO-D, would accelerate generation of porphomethene inhibitors by an iron-dependent oxidation reaction in which unmetabolized porphyrinogen substrates are converted to inhibitors. Our previous studies support this view because generation of a small amount of an inhibitor is followed by a rapid increase in inhibitory activity (34). Most of the small amount of uroporphyrinogen I generated daily would not be decarboxylated and would be partially oxidized to the porphomethene or fully oxidized and excreted as uroporphyrin I. In contrast, most (but not all) of the uroporphyrinogen III generated would be decarboxylated and ultimately converted to heme. This scenario is supported by the observation that hepatic heme production is not deficient in PCT or in animal models of PCT as functional hemoproteins, such as cytochrome P450s, can be induced (34, 35).
The mechanism responsible for generating the porphomethene inhibitor in vivo has not been defined, but CYP1A2 likely plays a central role. Sinclair et al. (9) have demonstrated that the Cyp1A2−/− mouse is resistant to regimens that produce experimental PCT. Our results in the Uro-d+/−, Hfe−/− mouse demonstrate that induction of CYP1A2 is not required for generation of the inhibitor, suggesting that constitutive levels of activity are sufficient. This interpretation is supported by our findings in humans. We demonstrated that the amount of immunoreactive CYP1A2 in liver biopsy samples from patients with PCT varies widely, as does the activity of CYP1A2 measured in vivo by disposition of caffeine (35). Immunoreactive CYP1A2 and CYP1A2 activity also varies widely in nonporphyric humans (36, 37), but values well below average values in controls were found in some patients with PCT (35).
Iron also plays a central role in the development of PCT. Hepatic siderosis is found in most cases. Iron depletion through phlebotomy therapy is almost universally effective and replacement of the iron removed through phlebotomy therapy produces a prompt relapse (38). In many mouse models of PCT, iron depletion either prevents or dramatically attenuates the porphyric phenotype, whereas iron excess accelerates development of the porphyric phenotype (39, 40). Approximately 20% of patients with PCT are homozygous for the Cys-282→Tyr mutation of the HFE gene and an additional 15% are heterozygous for this mutation (2). In two-thirds of cases, however, there is no apparent explanation for the hepatic siderosis. A key consideration concerning liver iron and PCT is the finding that very few patients with hemochromatosis or other diseases associated with iron overload develop PCT. Increased liver iron stores alone, therefore, are not sufficient to produce the porphyric phenotype (2). This is true in mice (17) and in humans (2). Other genes influencing iron disposition probably play a role in the majority of patients with PCT. This appears to be the situation in syngeneic strains of mice that are either susceptible or resistant to experimentally induced porphyria (41). The environmental risk factors associated with PCT (hepatitis C, excess alcohol use, and medicinal estrogens) are far more common than the observed frequency of PCT, which is estimated to occur in 1 in 5,000 to 1 in 25,000 people (2). Genetic variations influencing the production of the porphomethene inhibitor of URO-D on exposure to these environmental risk factors also probably play a role in patients who develop PCT.
Here, we have shown that uroporphyrinogen can be partially oxidized to generate a porphomethene and that the porphomethene is an inhibitor of URO-D. Collectively, the published data suggest that in vivo CYP1A2 converts uroporphyrinogen to uroporphomethene in an oxidative, iron-dependent reaction. The chemistry of this reaction and the precise interactions of genetic and environmental factors remain to be determined.
Materials and Methods
Animals.
Mice (C57BL/6J) with the Uro-d+/− genotype were generated as described previously (17). Mice were crossed with mice homozygous for a null mutation of the Hfe locus (Hfe−/−) on a C57BL/6J background (17). Wild-type C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All animals used were female. All were housed in the University of Utah Animal Resource Center. All studies were approved by the University of Utah's Institutional Animal Care and Use Committee in accordance with National Institutes of Health guidelines. Animals were maintained on a 12 h light/dark cycle with free access to food (Teklad 3080; Harlan Teklad, Madison, WI) and water. The porphyria-inducing regimen has been previously described (8, 42). Briefly, drinking water was supplemented with ALA (Frontier Scientific, Logan, UT) (2 g/liter), neutralized to pH 7.0. On day 0, an i.p. injection of Aroclor 1254 (a mixture of polychlorinated biphenyls; Monsanto, St. Louis, MO; 4 mg in corn oil) and an i.p. injection of iron dextran (Sigma, St. Louis, MO; 10 mg of iron and 11 mg of dextran) were administered.
Animals were killed on day 21, and their livers were analyzed for total porphyrin content as described in refs. 8 and 17. The activity of URO-D was measured as described below. Cytosolic URO-D protein was estimated by separating 30 μg of cytosolic protein on a 12% polyacrylamide gel followed by immunoblotting with a rabbit anti-human URO-D polyclonal antiserum as previously described (43). Protein concentrations were measured by using the BCA reagent (Pierce, Rockford, IL).
URO-D Activity Assays.
Activity of URO-D present in cytosolic extracts prepared from 1.5 g of mouse liver homogenized in 4 vol of 0.25 M sucrose was assayed as previously described (8, 19). Processing of cytosolic extracts was carried out under minimal illumination by using darkroom safelights. Hepatic cytosol (250 μl) was denatured by heating (100°C, 30 min) and clarified by centrifugation (14,000 × g) for 10 min. Inhibitory activity of the processed extract was assayed in a 200-μl mixture containing 70 μl of processed extract, 0.4 μg of rhURO-D, and 6.0–6.5 nmol of uroporphyrinogen I (8, 13). Hydroxymethylbilane was enzymatically generated from 276 μM PBG (Frontier Scientific) with 3–4 μg of purified PBG-D in the presence of 7.5 mM DTT and 0.1 M Tris, pH 7.65 (total volume 100 μl), for 30 min (19). The uroporphyrinogen I macrocycle is rapidly formed by nonenzymatic closure of the linear hydroxymethylbilane. Uroporphyrinogen III was generated by addition of 2–3 μg of purified recombinant human U3S (44) to the above reaction mixture. Each substrate-generating mixture was incubated for 30 min at 37°C, neutralized with 20 μl of 0.15 M potassium phosphate, and added to the processed extract and rhURO-D. The rhURO-D activity assay mixture was then incubated for 30 min at 37°C. The processed extract was replaced with buffer as a control for uninhibited rhURO-D activity. Porphyrins present in the processed extract were measured by replacing the substrate-generating mixture and rhURO-D with buffer. Reactions were stopped by adding an equal volume (200 μl) of 3 M HCl and then illuminated under UV light for 30 min to convert all reaction products to porphyrins. Porphyrins were then separated and quantified by HPLC as previously described (19).
U3S Assays.
U3S assays were performed by using our previously described method (44), using aliquots of cytosolic extract in place of recombinant human U3S.
Isolation of the Inhibitor.
The inhibitor in processed extracts was partially purified by solid-phase extraction by using an Oasis HLB cartridge (Waters, Milford, MA) preconditioned with methanol followed by 2% (vol/vol) acetic acid. The extract was acidified to pH 4.0 by adding an equal volume of 2% acetic acid and was then applied to the prepared column. Columns were then washed with 2% acetic acid. Inhibitor activity was eluted with 100% methanol, dried under argon in the dark, and resuspended in 50 mM Tris, pH 6.8, and tested for inhibition as described above. Aliquots were also analyzed by mass spectroscopy (see below).
Uroporphomethene was prepared by photooxidizing enzymatically generated uroporphyrinogen I or III as described above except that the incubation time was doubled to 70 min. The reaction mixture, at pH 7.0, was then exposed to longwave UV light (Model B-100 A; Ultra-Violet Products, San Gabriel, CA) at a distance of 15 cm for 1–30 min at 4°C. The partially oxidized product was acidified to approximately pH 4.0 with the addition of glacial acetic acid (2% by volume) and passed through an Oasis HLB column as described above. The 100% methanol eluate was dried under argon and resuspended in buffer for direct rhURO-D inhibition assays (see above). Heating (100°C, 30 min) had no effect on inhibitor activity. Aliquots of the dried methanol eluates were resuspended in 0.2% formic acid for additional purification by HPLC and analysis by HPLC/MS.
Mass Spectroscopy Analysis.
All analyses were performed by using a Thermo LCQ Advantage MAX HPLC–MSn system (Thermo, San Jose, CA). Dried HLB eluates, from 10 ml of liver extract, and HPLC fractions were resuspended in 200 μl of a mobile phase of 95% (vol/vol) 0.2% aqueous formic acid and 5% (vol/vol) acetonitrile. A loop injection of 20 μl was applied on a C18 column and separated by HPLC, programmed to run at a flow rate of 0.05 ml/min with an isocratic mobile phase consisting of 95% acetonitrile/5% 0.2% aqueous formic acid, and the effluent was directed into the mass spectrometer. The mass spectrometer was operated in positive electrospray ionization mode as described (SI Methods).
Other Methods.
Isoelectric focusing of URO-D in liver homogenates was performed by using the Protean IEF system (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instructions. Strips from the Protean IEF system were run in a second dimension by using a Bio-Rad miniProtean 2D electrophoresis cell on a precast 12% polyacrylamide system.
Studies in Humans.
Percutaneous needle biopsies of the liver were performed at the University of Utah's General Clinical Research Center. Biopsy samples were divided into three portions. One was analyzed by atomic absorption spectroscopy to determine iron content. Another was fixed in formalin for histologic examination. The third, weighing from 8 to 22 mg, was snap-frozen in liquid nitrogen, and later thawed and homogenized in 250 mM sucrose, and the proteins were heat-denatured as above. The denatured homogenate was clarified by centrifugation and the supernatant was analyzed for inhibitory activity against rhURO-D as above. Approval for the use of human liver biopsy material was obtained from the University of Utah's Institutional Review Board. All experiments were conducted according to the Declaration of Helsinki.
Supplementary Material
Acknowledgments
We thank Dr. Kevin Smith for his advice on the physical properties of porphomethenes and Dr. Chris Hill for his help in interpreting the crystal structure of inhibitor-bound URO-D. This work was supported in part by National Institutes of Health Grants R01 DK20503, M01 RR00064, P30 DK072437 (to J.P.K.), and R01 GM856775 (to Chris Hill, University of Utah).
Abbreviations
- URO-D
uroporphyrinogen decarboxylase
- PCT
porphyria cutanea tarda
- F-PCT
familial porphyria cutanea tarda
- S-PCT
sporadic porphyria cutanea tarda
- ALA
δ-aminolevulinic acid
- HLB
hydrophilic–lipophilic balanced resin
- U3S
uroporphyrinogen III synthase
- PBG
porphobilinogen
- PBG-D
porphobilinogen deaminase.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700547104/DC1.
References
- 1.Anderson KE, Sassa S, Bishop DF, Desnick RJ. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. Vol 2. New York: McGraw–Hill; 2001. pp. 2991–3062. [Google Scholar]
- 2.Bulaj ZJ, Phillips JD, Ajioka RS, Franklin MR, Griffen LM, Guinee DJ, Edwards CQ, Kushner JP. Blood. 2000;95:1565–1571. [PubMed] [Google Scholar]
- 3.Felsher BF, Kushner JP. Semin Hematol. 1977;14:243–251. [PubMed] [Google Scholar]
- 4.Lundvall O. Acta Med Scand. 1971;189:33–49. doi: 10.1111/j.0954-6820.1971.tb04337.x. [DOI] [PubMed] [Google Scholar]
- 5.Elder GH, Urquhart AJ, De Salamanca RE, Munzo JJ, Bonkovsky HL. Lancet. 1985;ii:229–232. doi: 10.1016/s0140-6736(85)90287-9. [DOI] [PubMed] [Google Scholar]
- 6.Kushner JP, Barbuto AJ, Lee GR. J Clin Invest. 1976;58:1089–1097. doi: 10.1172/JCI108560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elder GH, Lee GB, Tovey JA. N Engl J Med. 1978;299:274–278. doi: 10.1056/NEJM197808102990603. [DOI] [PubMed] [Google Scholar]
- 8.Franklin MR, Phillips JD, Kushner JP. Toxicol Appl Pharmacol. 1997;147:289–299. doi: 10.1006/taap.1997.8282. [DOI] [PubMed] [Google Scholar]
- 9.Sinclair PR, Gorman N, Dalton T, Walton HS, Bement WJ, Sinclair JF, Smith AG, Nebert DW. Biochem J. 1998;330:149–153. doi: 10.1042/bj3300149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith AG, Clothier B, Robinson S, Scullion MJ, Carthew P, Edwards R, Luo J, Lim CK, Toledano M. Mol Pharmacol. 1998;53:52–61. doi: 10.1124/mol.53.1.52. [DOI] [PubMed] [Google Scholar]
- 11.Smith AG, Francis JE. Biochem J. 1993;291:29–35. doi: 10.1042/bj2910029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urquhart AJ, Elder GH, Roberts AG, Lambrecht RW, Sinclair PR, Bement WJ, Gorman N, Sinclair JA. Biochem J. 1988;253:357–362. doi: 10.1042/bj2530357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin MR, Phillips JD, Kushner JP. Hepatology. 2002;36:805–811. doi: 10.1053/jhep.2002.35621. [DOI] [PubMed] [Google Scholar]
- 14.Smith AG, Francis JE. Biochem J. 1987;246:221–226. doi: 10.1042/bj2460221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Billi de Catabbi S, Rios de Molina MC, San Martin de Viale LC. Int J Biochem. 1991;23:675–679. doi: 10.1016/0020-711x(91)90037-n. [DOI] [PubMed] [Google Scholar]
- 16.Cantoni L, Graziani A, Rizzardini M, Saletti MC. IARC Sci Publ. 1986;77:449–456. [PubMed] [Google Scholar]
- 17.Phillips JD, Jackson LK, Bunting M, Franklin MR, Thomas KR, Levy JE, Andrews NC, Kushner JP. Proc Natl Acad Sci USA. 2001;98:259–264. doi: 10.1073/pnas.011481398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mauzerall D. J Am Chem Soc. 1962;84:2437–2445. [Google Scholar]
- 19.Phillips JD, Kushner JP. In: Current Protocols in Toxicology. Maines MD, Costa LG, Reed DJ, Sassa S, Sipes IG, editors. New York: Wiley; 1999. pp. 8.4.1–8.4.13. [Google Scholar]
- 20.Nacht S, San Martin de Viale LC, Grinstein M. Clin Chim Acta. 1970;27:445–452. doi: 10.1016/0009-8981(70)90297-4. [DOI] [PubMed] [Google Scholar]
- 21.Phillips JD, Whitby FG, Kushner JP, Hill CP. EMBO J. 2003;22:6225–6233. doi: 10.1093/emboj/cdg606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heikel T, Lockwood WH, Rimington C. Nature. 1958;182:313. doi: 10.1038/182313a0. [DOI] [PubMed] [Google Scholar]
- 23.Felsher BF, Carpio NM, Engleking DW, Nunn AT. N Engl J Med. 1982;306:766–769. doi: 10.1056/NEJM198204013061302. [DOI] [PubMed] [Google Scholar]
- 24.Cam C, Nigogosyan G. J Am Med Assoc. 1963;183:90–93. [Google Scholar]
- 25.Schmid R. N Engl J Med. 1960;263:397–398. doi: 10.1056/NEJM196008252630807. [DOI] [PubMed] [Google Scholar]
- 26.Rios de Molina MC, Wainstok de Calmanovici R, San Martin de Viale LC. Int J Biochem. 1980;12:1027–1032. doi: 10.1016/0020-711x(80)90206-2. [DOI] [PubMed] [Google Scholar]
- 27.Cantoni L, dal Fiume D, Rizzardini M, Ruggieri R. Toxicol Lett. 1984;20:211–217. doi: 10.1016/0378-4274(84)90149-8. [DOI] [PubMed] [Google Scholar]
- 28.Sinclair PR, Bement WJ, Lambrecht RW, Gorman N, Sinclair JF. Arch Biochem Biophys. 1990;281:225–232. doi: 10.1016/0003-9861(90)90436-3. [DOI] [PubMed] [Google Scholar]
- 29.Sinclair PR, Gorman N, Walton HS, Bement WJ, Jacobs JM, Sinclair JF. Arch Biochem Biophys. 1993;304:464–470. doi: 10.1006/abbi.1993.1376. [DOI] [PubMed] [Google Scholar]
- 30.Gorman N, Ross KL, Walton HS, Bement WJ, Szakacs JG, Gerhard GS, Dalton TP, Nebert DW, Eisenstein RS, Sinclair JF, Sinclair PR. Hepatology. 2002;35:912–921. doi: 10.1053/jhep.2002.32487. [DOI] [PubMed] [Google Scholar]
- 31.Straka JG, Kushner JP. Biochemistry. 1983;22:4664–4672. doi: 10.1021/bi00289a009. [DOI] [PubMed] [Google Scholar]
- 32.de Verneuil H, Sassa S, Kappas A. J Biol Chem. 1983;258:2454–2460. [PubMed] [Google Scholar]
- 33.Smith SG, Rao KR, Jackson AH. Int J Biochem. 1980;12:1081–1084. doi: 10.1016/0020-711x(80)90216-5. [DOI] [PubMed] [Google Scholar]
- 34.Franklin MR, Phillips JD, Kushner JP. J Biochem Mol Toxicol. 2001;15:287–293. doi: 10.1002/jbt.10000. [DOI] [PubMed] [Google Scholar]
- 35.Bulaj ZJ, Franklin MR, Phillips JD, Miller KL, Bergonia HA, Ajioka RS, Griffen LM, Guinee DJ, Edwards CQ, Kushner JP. J Lab Clin Med. 2000;136:482–488. doi: 10.1067/mlc.2000.111024. [DOI] [PubMed] [Google Scholar]
- 36.Kalow W, Tang BK. Clin Pharmacol Ther. 1991;50:508–519. doi: 10.1038/clpt.1991.176. [DOI] [PubMed] [Google Scholar]
- 37.Kalow W, Tang BK. Clin Pharmacol Ther. 1993;53:503–514. doi: 10.1038/clpt.1993.63. [DOI] [PubMed] [Google Scholar]
- 38.Felsher BF, Jones ML, Redeker AG. J Am Med Assoc. 1973;226:663–665. [PubMed] [Google Scholar]
- 39.Franklin MR, Phillips JD, Kushner JP. Environ Toxicol Pharmacol. 2005;20:417–423. doi: 10.1016/j.etap.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Sweeny GD, Jones KG, Cole FM, Basford D, Krestynski F. Science. 1979;204:332–335. doi: 10.1126/science.432648. [DOI] [PubMed] [Google Scholar]
- 41.Smith AG, Francis JE, Walters DG, Lake BG. Biochem Pharmacol. 1990;40:2564–2568. doi: 10.1016/0006-2952(90)90102-q. [DOI] [PubMed] [Google Scholar]
- 42.Franklin MR, Phillips JD, Kushner JP. Biochem Pharmacol. 2000;60:1325–1331. doi: 10.1016/s0006-2952(00)00449-4. [DOI] [PubMed] [Google Scholar]
- 43.Hansen JL, Pryor MA, Kennedy JB, Kushner JP. Am J Hum Genet. 1988;42:847–853. [PMC free article] [PubMed] [Google Scholar]
- 44.Mathews MA, Schubert HL, Whitby FG, Alexander KJ, Schadick K, Bergonia HA, Phillips JD, Hill CP. EMBO J. 2001;20:5832–5839. doi: 10.1093/emboj/20.21.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.