Abstract
Nuclear NAD+ metabolism constitutes a major component of signaling pathways. It includes NAD+-dependent protein deacetylation by members of the Sir2 family and protein modification by poly(ADP-ribose) polymerase 1 (PARP-1). PARP-1 has emerged as an important mediator of processes involving DNA rearrangements. High-affinity binding to breaks in DNA activates PARP-1, which attaches poly(ADP-ribose) (PAR) to target proteins. NMN adenylyl transferases (NMNATs) catalyze the final step of NAD+ biosynthesis. We report here that the nuclear isoform NMNAT-1 stimulates PARP-1 activity and binds to PAR. Its overexpression in HeLa cells promotes the relocation of apoptosis-inducing factor from the mitochondria to the nucleus, a process known to depend on poly(ADP-ribosyl)ation. Moreover, NMNAT-1 is subject to phosphorylation by protein kinase C, resulting in reduced binding to PAR. Mimicking phosphorylation, substitution of the target serine residue by aspartate precludes PAR binding and stimulation of PARP-1. We conclude that, depending on its state of phosphorylation, NMNAT-1 binds to activated, automodifying PARP-1 and thereby amplifies poly(ADP-ribosyl)ation.
Keywords: apoptosis, NAD biosynthesis, poly(ADP-ribosyl)ation, protein phosphorylation
NMN adenylyl transferase (NMNAT) is an essential enzyme because it catalyzes the final step of NAD+ biosynthesis (1). There are three isoforms in humans that exhibit tissue- and organelle-specific expression (2). Because the biological role of NAD+ and NADH (collectively termed NAD) has long been thought to be confined to their function as electron carriers, the nuclear localization of the major isoforms, human NMNAT-1 (3) and yeast Nma2 (4), was somewhat puzzling. However, recent research has revealed many important regulatory functions of the pyridine nucleotides, some of which take place within the nucleus (5).
NAD+ serves as substrate for covalent protein modifications such as mono(ADP-ribosyl)ation (6), poly(ADP-ribosyl)ation (7, 8), and protein deacetylation by sirtuins, proteins of the silent information regulator 2 (Sir2) family (9). Pyridine nucleotides are also direct precursors of two calcium-mobilizing messengers, cyclic ADP ribose and nicotinic acid adenine dinucleotide phosphate (NAADP+) (10–13). The predominant NAD+-consuming activity, poly(ADP-ribosyl)ation by poly(ADP-ribose) polymerase 1 (PARP-1), occurs within the nucleus, as does NAD+-dependent protein deacetylation by sirtuins. Consequently, substrate supply by NMNAT-1 may directly influence these processes. Overexpression of NMNAT-1 extends the lifespan of eukaryotic cells, which has been attributed to augmented NAD+-dependent histone deacetylation catalyzed by Sir2p or its homologs (4, 14–18). Increased NMNAT activity is also required for the axon-sparing activity of the chimeric slow Wallerian degeneration (Wlds) protein, which is composed of 19 amino acids of the ubiquitin assembly protein Ufd2a and full-length NMNAT-1 (19).
Although suggested previously (3, 20, 21), a functional interaction between NMNAT-1 and PARP-1 has not been demonstrated. PARP-1 is an abundant nuclear enzyme that binds to DNA single-strand breaks. This binding triggers its catalytic activity, the synthesis of ADP-ribose polymers, and their attachment to acceptor proteins including itself (7, 8, 22–25), thereby initiating such events as recruitment of DNA repair proteins (7, 8, 26–28) and modulation of p53- and NF-κB-dependent signaling pathways (29–33) or chromatin structure (34).
Nuclear poly(ADP-ribosyl)ation seems to be the major NAD+-consuming process. In the absence of DNA strand breaks, poly(ADP-ribosyl)ation is low, but it increases >100-fold upon DNA damage. Under these conditions, ≈90% of poly(ADP-ribose) (PAR) is synthesized by PARP-1 (35). Overactivation of the enzyme may almost completely deplete cellular NAD+ stores (36, 37). Poly(ADP-ribosyl)ation also triggers cell death by releasing apoptosis-inducing factor (AIF) from the mitochondria (38–40). Such a broad range of functions requires mechanisms controlling the catalytic activity of PARP-1 to assure the balanced activation of individual pathways.
Here we show that NMNAT-1 associates with PARP-1, which undergoes automodification, thereby increasing the extent of poly(ADP-ribosyl)ation. Moreover, specific phosphorylation of NMNAT-1 by protein kinase C (PKC) precludes the NMNAT-1-mediated activation of PARP-1. Therefore, NMNAT-1 not only provides the substrate but also regulates PARP-1 activity, depending on its state of phosphorylation.
Results
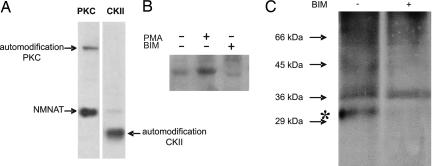
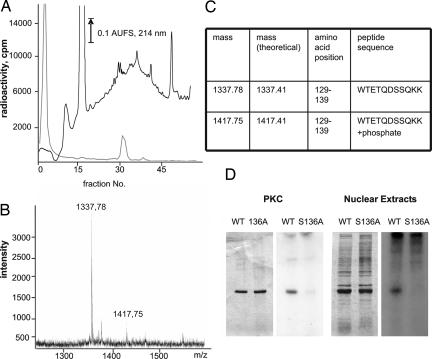
According to a previous study (3), purified NMNAT-1 has been phosphorylated in cell extracts; however, this modification was not further explored (3). Prediction programs (3) indicated several PKC- and casein kinase II (CKII)-specific phosphorylation sites. Indeed, commercial PKC phosphorylated NMNAT-1 (Fig. 1A), yielding up to 20% of 32P-labeled NMNAT-1 polypeptides as estimated by Cherenkov counting of the excised band. No significant phosphorylation of NMNAT-1 was detected by using CKII, only automodification of the kinase (Fig. 1A). In line with a PKC-specific modification, phosphorylation of NMNAT-1 by nuclear extracts was inhibited in the presence of bisindolylmaleimide (BIM) and activated by phorbol myristate acetate (PMA) (Fig. 1B). The relevance of these findings is supported by the detection of the phosphorylation of NMNAT-1 in living cells. Human fibroblasts were maintained for 8 h in phosphate-free medium supplemented with 32P-labeled orthophosphate. Subsequent immunoprecipitation by an NMNAT-1-specific antibody (3) revealed a radiolabeled protein corresponding to NMNAT-1 (Fig. 1C, left lane). As in nuclear extracts, no labeling of NMNAT-1 was detected when the PKC inhibitor BIM was added (Fig. 1C, right lane). To identify the site(s) of phosphorylation, NMNAT-1 was 32P-phosphorylated by PKC and subjected to tryptic digestion, and the peptides were separated by HPLC. A single radioactive eluate fraction was found (Fig. 2A). The same elution fraction contained the radioactive label when HeLa nuclear extract was used to 32P-phosphorylate NMNAT-1 (data not shown). MALDI-TOF mass spectrometry of the isolated fraction (Fig. 2B) revealed a peak (m/z = 1,337.78) corresponding to a peptide containing serine 136 that is predicted to be a PKC consensus phosphorylation site (Fig. 2C), as well as a mass peak accounting for this peptide plus the phosphate group (m/z = 1,417.75). Fragmentation of this species by postsource decay confirmed the peptide sequence and suggested serine 136 as the site of phosphorylation (data not shown).
Fig. 1.
PKC-mediated phosphorylation of NMNAT-1. (A) Recombinant NMNAT-1 (500 ng) was incubated with PKC or casein kinase II (CKII) (10 units) and [γ-32P]ATP. Proteins were then separated by SDS/PAGE. The autoradiograph of the gel is shown. (B) NMNAT-1 (500 ng) was incubated with nuclear extracts (1 μg) in the presence of [γ-32P]ATP and phorbol myristate acetate (PMA) or BIM as indicated. Proteins were separated by SDS/PAGE, and gels were analyzed by autoradiography. (C) Human fibroblasts were incubated with [32P]orthophosphate for 8 h. Where indicated, BIM (1 μM) was added 1 h before cell lysis. After immunoprecipitation of endogenous NMNAT-1, proteins were separated by SDS/PAGE. Labeled proteins were visualized by autoradiography. The numbers on the left indicate the mobility of marker proteins. The asterisk shows the position of NMNAT-1.
Fig. 2.
Identification of the NMNAT-1 phosphorylation site as serine 136. (A) Recombinant NMNAT-1 (25 μg) was 32P-phosphorylated by PKC and then subjected to tryptic digestion. The generated peptides were separated by HPLC. The elution profile is shown. The upper, black trace shows absorbance at 214 nm. The lower, gray trace shows radioactivity of the collected fractions. AUFS, Absorbance units full scale. (B) The fraction containing a radioactive peptide was analyzed by MALDI-TOF mass spectrometry. The relevant part of the mass spectrum is presented. (C) The obtained peptide masses and the corresponding stretches of the NMNAT-1 sequence are shown. (D) WT NMNAT-1 or the S136A mutant was incubated with PKC or HeLa nuclear extracts as indicated in the presence of [γ-32P]ATP. Proteins were separated by SDS/PAGE (PKC Left and Nuclear Extracts Left) and then subjected to autoradiography (PKC Right and Nuclear Extracts Right).
When we replaced serine 136 with alanine, phosphorylation of recombinant NMNAT-1 by either PKC or nuclear extract was essentially absent (Fig. 2D). To preclude alternative phosphorylation at the adjacent serine residue S135, we also generated a double mutant, S135/136A. As expected, the overexpressed S135/136A mutant was no more phosphorylated (data not shown). The catalytic properties of the S/A mutants were indistinguishable from those of the WT protein (data not shown).
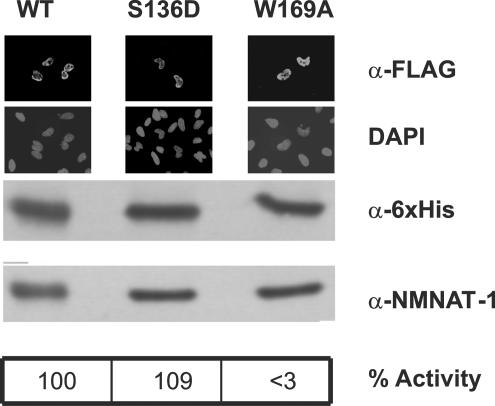
According to the crystal structure of NMNAT-1 (41), the identified phosphorylation site resides within an external loop that is well suited to mediate the interactions of NMNAT-1 with other proteins, e.g., the nuclear import machinery or PARP-1. Indeed, the nuclear localization sequence (amino acids 123–129) is also located within this loop, in close proximity to the identified phosphorylation site, S136. We tested whether the phosphorylation of NMNAT-1 influenced its nuclear localization. Treatment of HeLa or HEK 293 cells with phorbol myristate acetate (PMA) or BIM had no detectable influence on the nuclear localization of endogenous NMNAT-1 (data not shown). We generated a mutant, S136D, in which the negatively charged side chain of the aspartate residue was intended to mimic phosphorylation. The recombinant proteins were overexpressed in Escherichia coli and HeLa cells. The WT and the S136D and S135/136A mutants (data not shown) had similar catalytic activities and were localized to the nucleus (Fig. 3). In addition, a catalytically inactive W169A mutant was constructed that was similar to the W170A mutation of mouse NMNAT-1 (19). The overexpressed protein was inactive and located within the nucleus. When expressed in E. coli, all these proteins could be purified virtually to homogeneity on nickel nitriloacetic acid (Ni-NTA) resins, exhibited the same mobility during SDS/PAGE, and were recognized by antibodies directed against the N-terminal His6 tag or WT NMNAT-1 (Fig. 3).
Fig. 3.
Characterization of generated NMNAT-1 proteins. FLAG-tagged WT and mutant proteins were overexpressed in HEK 293 cells. The FLAG tag and nuclei were visualized by fluorescence microscopy using FLAG-specific antibodies and DAPI, respectively (as indicated). Western blots of the purified NMNAT-1 proteins overexpressed in E. coli (as His6-tagged proteins) were probed with antibodies specific for the His6 tag or against WT NMNAT-1 (as indicated). The catalytic activities of the purified recombinant proteins are given as the average value obtained from two independent protein preparations. The activity of the W169A mutant, if any, was below the detection level.
These observations indicated that neither the phosphorylation of NMNAT-1 at serine 136 nor the catalytic activity had any influence on the subcellular localization of the protein. The recombinantly expressed proteins were also similarly stable with regard to enzymatic activity and intactness of the protein. When transiently overexpressed in HeLa cells, no fragmentation or degradation was detected, and the time courses of expression were similar. Nor did we observe any differences after immunoprecipitation with regard to differential ubiquitination (data not shown). Given the absence of any detectable effect of the modification of S136 on enzymatic activity, subcellular localization, or protein stability, we decided to explore the possibility of a physical interaction between NMNAT-1 and PARP-1.
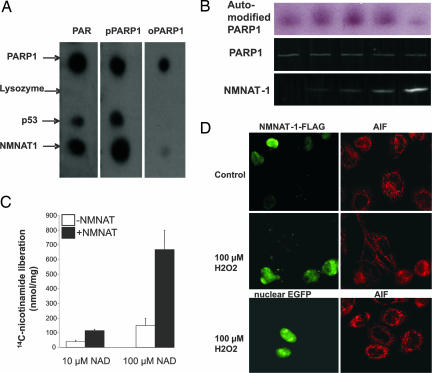
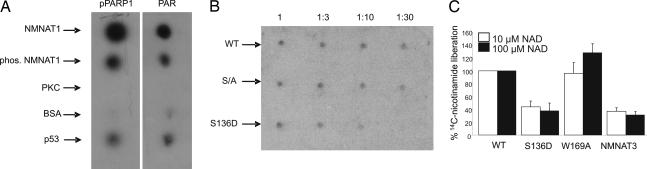
To visualize the possible affinity of NMNAT-1 for PARP-1, purified NMNAT-1 that had been immobilized on nitrocellulose was incubated with purified 32P-labeled poly(ADP-ribosyl)ated PARP-1, protein-free 32P-labeled PAR, or 32P-labeled oligo(ADP-ribosyl)ated PARP-1 (42). The latter was obtained by automodification of PARP-1 in the presence of ≈10 nM 32P-labeled NAD+, thereby attaching only short radiolabeled ADP-ribose oligomers and enabling the detection of PARP-1 binding (43). As positive controls, PARP-1 and p53, known to bind preferentially to PAR (44), were used. NMNAT-1 exhibited a strong affinity for free and PARP-1-bound PAR, but not for oligo(ADP-ribosyl)ated PARP-1 (Fig. 4A). Therefore, NMNAT-1 did not appear to interact directly with PARP-1, but rather via ADP-ribose polymers. This observation was somewhat surprising, because PAR did not influence the catalytic activity of NMNAT-1 (2). A possible influence of NMNAT-1 on the catalytic activity of PARP-1 was tested by assaying PARP-1 automodification in the presence of NMNAT-1 and a low concentration of 32P-labeled NAD+. The proteins were separated by SDS/PAGE, and automodification of PARP-1 was monitored by autoradiography. An excess of NMNAT-1 of up to 10-fold stimulated the automodification, whereas higher amounts of NMNAT-1 seemed to inhibit PARP-1 (Fig. 4B), presumably because of the increased binding of NAD+ to NMNAT-1 (2), which thereby limited the substrate for PARP-1. The gel-based assay is inapplicable at higher NAD+ concentrations, because the extensively modified protein does not migrate as a distinct band but rather as a smear. Therefore, we quantified the byproduct of the reaction, nicotinamide. The amount of [14C]nicotinamide released from [14C]NAD+ was determined by HPLC using radioisotope detection. At 10 μM NAD+, PARP-1 was stimulated by NMNAT-1 by ≈3-fold (Fig. 4C). When using 100 μM [14C]NAD+ as substrate, the stimulatory effect of NMNAT-1 on PARP-1 activity was even stronger (Fig. 4C). Stimulation of PARP-1 activity by NMNAT-1 was also detected by using 32P-NAD+ as substrate followed by precipitation of the automodified protein with trichloroacetic acid (data not shown). Consequently, the increased nicotinamide release during the HPLC-based assay was not due just to an increase of the glycohydrolase activity of PARP-1. To test whether stimulation by NMNAT-1 has an influence on PARP-1-mediated downstream processes, the translocation of AIF, which is known to be triggered by PAR (38–40), was studied. HeLa cells were transiently transfected with NMNAT-1 that was N-terminally endowed with a FLAG epitope. Control cells were transfected with enhanced green fluorescent protein (EGFP) directed to the nucleus. After treatment with 100 μM hydrogen peroxide, cells transfected with NMNAT-1 exhibited AIF translocation, whereas the mitochondrial localization of AIF was largely retained in the untransfected cells in the same experiment (Fig. 4D). As shown by the EGFP control, it was not the transfection per se that sensitized the NMNAT-1-expressing cells to H2O2.
Fig. 4.
NMNAT-1 associates with PAR and automodified PARP-1 and accelerates poly(ADP-ribosyl)ation and AIF translocation. (A) Specific interaction of NMNAT-1 with free or PARP-1-bound PAR was visualized by spotting the indicated proteins (0.5 μg) on a nitrocellulose membrane. The membrane was then incubated with 32P-labeled oligo(ADP-ribosyl)ated PARP-1 (oPARP-1, 0.25 μg), poly(ADP-ribosyl)ated PARP-1 (pPARP-1, 0.25 μg), or protein-free PAR (PAR, 5 μM). After washing, specific binding was visualized by autoradiography. (B) PARP-1 (25 ng) was incubated with 32P-NAD+ in the absence or presence of NMNAT-1 in a mass ratio (PARP-1/NMNAT-1) of 1:2, 1:4, 1:10, or 1:20. Thereafter, the proteins were separated by SDS/PAGE. (Top) The autoradiograph of the gel is shown. (Middle and Bottom) Proteins were visualized by the fluorescent SYPRO Ruby stain (Invitrogen, Carlsbad, CA). (C) PARP-1 (100 ng) was incubated with nicotinamide/[14C]NAD+ in the presence or absence of NMNAT-1 (mass ratio of 1:5). [14C]Nicotinamide released during automodification of PARP-1 was identified by HPLC using radioisotope detection. (D) PAR-mediated AIF translocation is enhanced by NMNAT-1 overexpression. FLAG-tagged NMNAT-1 was transiently overexpressed in HeLa cells. (Bottom Left) As a control, nuclear EGFP was overexpressed. The cells were treated with 100 μM H2O2 as indicated. FLAG-tagged NMNAT-1 and AIF were visualized by immunostaining.
Finally, we tested the possibility that the phosphorylation of NMNAT-1 might influence the catalytic activation of PARP-1. First, the association of NMNAT-1 phosphorylated in vitro (Fig. 5A) or the pseudophosphorylated S136D mutant (Fig. 5B) with automodified PARP-1 or PAR was assessed. Even though only 15–20% of the NMNAT-1 polypeptides were phosphorylated, a noticeable reduction of the affinity for PAR and automodified PARP-1 was detected (Fig. 5A). Serial dilutions of the WT and the S135/136A and S136D mutants confirmed that introducing a negative charge into position 136 of NMNAT-1 reduced the affinity for PARP-1-bound PAR (Fig. 5B).
Fig. 5.
Phosphorylated NMNAT-1 has less affinity for automodified PARP-1 and does not stimulate PARP-1 activity. (A) Recombinant NMNAT-1 was phosphorylated by PKC, as indicated. Proteins (0.5 μg) were spotted onto nitrocellulose. The membrane was then incubated with 32P-labeled poly(ADP-ribosyl)ated PARP-1 or free polymers. Thereafter, the membrane was washed, and specific binding was visualized by autoradiography. (B) Serial dilutions of the indicated recombinant NMNAT-1 proteins were spotted onto nitrocellulose. The membrane was then incubated with 32P-labeled poly(ADP-ribosyl)ated PARP-1 (0.25 μg/ml), washed, and subjected to autoradiography. S/A, serial mutant in which serines 135 and 136 were replaced by alanine residues. (C) PARP-1 was incubated with [14C]nicotinamide/NAD+ in the presence of the indicated NMNAT-1 proteins or NMNAT-3, as a control. [14C]Nicotinamide released during automodification of PARP-1 was analyzed by HPLC using radioisotope detection.
Given the phosphomimetic property of the S136D mutation, we tested whether this amino acid exchange also altered the catalytic activation of PARP-1 observed for WT NMNAT-1. As shown in Fig. 5C, this mutant did not increase PARP-1 activity. However, although inactive, the catalytically dead W169A mutant fully retained the capacity to activate PARP-1 (Fig. 5C). The notion that a structure-based interaction with NMNAT-1 is required for PARP-1 activation was further corroborated when human NMNAT-3, a mitochondrial isoform (2), was tested in these experiments. In fact, PARP-1 activity was approximately the same in the presence of NMNAT-3 and the S136D mutant of NMNAT-1 (Fig. 5C). These observations indicate that NMNAT-1 is a specific activator of PARP-1 and that phosphorylation at serine 136 serves to down-regulate this interaction.
Discussion
Here we reveal a functional and physical interaction between the major NAD-consuming pathway, poly(ADP-ribosyl)ation by PARP-1, and NMNAT-1, a key enzyme of NAD biosynthesis. NMNAT-1 stimulates poly(ADP-ribosyl)ation by associating with automodified PARP-1. Strikingly, these interactions can be abrogated by a single amino acid substitution in NMNAT-1, S136D, that resembles PKC-mediated phosphorylation of serine 136.
Poly(ADP-ribosyl)ation is known to be an immediate early event that follows genotoxic assault. Among other functions, this modification serves as an assembly signal for DNA repair factors such as XRCC1 (27, 45, 46). In view of the limited availability of NAD within the nucleus (47), the vicinity of PARP-1 and NMNAT-1 may result in locally confined substrate supply and thereby selectively feed poly(ADP-ribosyl)ation. Such a mechanism could be of particular importance for the signaling of alterations that require immediate responses such as DNA damage. In this context, the interaction between NMNAT-1 and PARP-1 would require the initial automodification of PARP-1 that is triggered by DNA single-strand breaks, and the subsequent binding of NMNAT-1 might serve to amplify the signal. The observation that the overexpression of NMNAT-1 enhanced the relocation of AIF from the mitochondria in response to oxidant treatment further corroborates this notion. It has been demonstrated that AIF translocation is directly triggered by PAR in a dose-dependent manner (39, 40). Importantly, NMNAT-1 overexpression does not alter the steady-state level of NAD+ (48, 49). Consequently, the stimulation of PARP-1 and the ensuing AIF translocation are unlikely to be brought about by a general increase of the substrate for PARP-1. Rather, it would seem that PARP-1 activity is enhanced by another mechanism, which is in line with our in vitro observations of a physical interaction with NMNAT-1.
Apparently, under some conditions the activation of PARP-1 by NMNAT-1 is undesirable and can be abrogated by phosphorylation. Our results support the conclusion that serine 136 in NMNAT-1 is a site of PKC-specific phosphorylation and that this modification eliminates the activation of PARP-1. The phosphorylation of serine 136 in NMNAT-1 diminishes the affinity of NMNAT-1 for poly(ADP-ribosyl)ated PARP-1. It may be that the effect of the S136D mutation is attributable to the homohexameric structure of NMNAT-1, resulting in a 6-fold pseudophosphorylation of the assembled protein. Consequently, our results indicate that PKC-mediated phosphorylation of NMNAT-1 could gradually decrease the extent of poly(ADP-ribosyl)ation by PARP-1.
Although the protein kinase catalyzing the phosphorylation of serine 136 in NMNAT-1 has not been identified, several lines of evidence support the conclusion that it belongs to the PKC family. First, the modification of NMNAT-1 was prevented in the presence of 1 μM BIM and stimulated by phorbol myristate acetate (PMA) when using nuclear extracts to phosphorylate the protein. Second, 1 μM BIM was also effective in inhibiting the phosphorylation of NMNAT-1 in living cells. Third, commercial PKC (α and β isoforms) phosphorylated NMNAT-1 in the same peptide as the kinase activity in nuclear extracts. We also noted that serine 136 in NMNAT-1 represents a highly probable consensus site for PKC-dependent phosphorylation.
The observed stimulation of PARP-1 seems to be mediated by noncovalent association of PARP-1-bound PAR and NMNAT-1. Two different protein motifs have been identified that specifically bind PAR. First, a sequence consisting of ≈22 amino acids with basic residues in the N-terminal part followed by a stretch containing nonpolar and basic residues was identified in myristoylated alanine-rich C kinase substrate (MARCKS) proteins. A similar motif mediating PAR binding was also found in several other proteins (50, 51). Second, macrodomains specifically bind ADP-ribose derivatives including PAR (52). Although it seems that there is no macrodomain in NMNAT-1, we found a stretch of 20 amino acids (residues 56–75) that, according to previously established criteria (50, 51), could resemble a PAR-binding motif. According to the crystal structure of NMNAT-1, this part of the sequence is located at the outer surface [see supporting information (SI) Fig. 6]. Moreover, the identified phosphorylation site S136 is part of an unresolved external loop (amino acids 108–147) that has its exit and entry points into the core structure in the vicinity of amino acids 56–75. Therefore, the possibility exists that the phosphorylation of S136 might modulate an interaction of PAR with the potential PAR-binding motif. However, this remains to be investigated.
Collectively, our results provide evidence for a mechanism regulating PARP-1 activity. It involves the interaction of NMNAT-1 with activated PARP-1, leading to increased poly(ADP-ribosyl)ation and possibly including locally confined substrate supply. PKC-dependent phosphorylation of NMNAT-1 prevents this interaction and thereby the stimulation of PARP-1. Therefore, NMNAT-1 is not only essential for poly(ADP-ribosyl)ation, because it generates the substrate, NAD+, but it might also serve as a transmitter of PKC-mediated signaling processes to regulate PARP-1-dependent functions.
Materials and Methods
Cloning, Mutagenesis, Overexpression, and Purification of Recombinant Proteins.
Human recombinant NMNAT-1 (2) and PARP-1 (53) were overexpressed and purified as described. NMNAT-1 cDNA was mutated by using the QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Cells were cultured in DMEM (Sigma, St. Louis, MO) and analyzed 48 h after transfection.
Measurement of Enzymatic Activities and Synthesis of PAR.
The activity of NMNAT-1 was measured by a photometric assay (3). To generate poly(ADP-ribosyl)ated PARP-1, human recombinant PARP-1 was incubated with 1 μCi of 32P-NAD+ (1 Ci = 37 GBq) and 10 μM NAD+ in reaction buffer [20 μg/ml sonicated herring sperm DNA, 10 mM DTT, 6 mM MgCl2, and 50 mM Tris·HCl (pH 8.0)]. For oligo(ADP-ribosyl)ation of PARP-1, the reaction mixture contained no unlabeled NAD+. After 20 min at 37°C, samples were analyzed by SDS/PAGE followed by autoradiography. PAR was synthesized as described previously (54).
PARP-1 activity was determined by the amount of nicotinamide released during PARP-1 automodification. PARP-1 was incubated with 1–100 μM NAD+ containing 0.05 μCi of nicotinamide/[14C]NAD+ in reaction buffer. After 10 min at room temperature, nucleotides and nicotinamide were separated from the proteins by centrifugation through a microfiltration device (Millipore, Billerica, MA) with a size exclusion of 5 kDa. The filtrate was analyzed by reverse-phase HPLC as described (2). Nicotinamide/[14C]NAD+ and the liberated [14C]nicotinamide were detected by using a radioflow detector (LB 509; Berthold Technologies, Bad Wilbad, Germany) and quantified by peak integration.
Blot-Overlay Assays.
Proteins (0.1–2.7 μg) were spotted onto a nitrocellulose membrane. After blocking for 30 min with 0.5% BSA in Tris-buffered saline containing 0.5% Tween 80 (TBST) and washing with TBST PAR (5 μM), oligo(ADP-ribosyl)ated (0.25 μg/ml) or poly(ADP-ribosyl)ated (0.25 μg/ml) PARP-1 were incubated with the membrane for 30 min. The membrane was washed with TBST containing 300 mM NaCl, dried, and then analyzed by autoradiography.
In Vitro Phosphorylation of NMNAT-1.
NMNAT-1 (500 ng) was incubated with 2–5 μg of nuclear extracts or 10–20 units of PKC or casein kinase II (CKII) (Calbiochem, Darmstadt, Germany), 100 μM ATP, 1 μCi of [γ-32P]ATP, 10% glycerol, 10 mM MgCl2, 2 mM CaCl2, 50 mM KCl, 100 mM NaCl, 0.2 mM EDTA, 1 mM DTT, and 10 mM Hepes/KOH (pH 7.9) for 10 min at 37°C. The proteins were then immediately processed for SDS/PAGE, and the gels were subjected to autoradiography.
Phosphorylation of Endogenous NMNAT-1.
Human fibroblasts (WI38) were grown in a six-well plate to ≈80% confluence. After three washings with phosphate-free DMEM, the cells were incubated with 0.2 mCi of [32P]orthophosphate per milliliter for 8 h. BIM was added 1 h before cell lysis. Cells were lysed in 10 mM MgCl2, 2 mM CaCl2, 50 mM KCl, 100 mM NaCl, 0.2 mM EDTA, 1 mM DTT, and 10 mM Hepes/KOH (pH 7.9) and passed 30 times through a 14-gauge needle. The suspension was centrifuged for 10 min at 14,000 × g at 4°C. The supernatant was used for immunoprecipitation.
Identification of the Phosphorylation Site.
Recombinant NMNAT-1 was 32P-phosphorylated and then precipitated by diluting the sample 10-fold with water. Under these conditions, >90% of NMNAT-1 precipitated, whereas PKC or proteins of the nuclear extract, as well as unreacted [32P]ATP, remained in the supernatant. The sample was centrifuged for 15 min at 14,000 × g, and the pellet was washed three times with water. The pellet was resuspended in 100 mM NH4HCO3 and 10 mM 2-mercaptoethanol, and the protein was digested with trypsin (0.2 μg/μg of NMNAT-1). After 1 h at 37°C, the peptides were diluted 1:10 with 0.1% trifluoroacetic acid and 3% acetonitrile and separated by reverse-phase HPLC. Fractions of 300 μl were collected and analyzed for radioactivity by Cherenkov counting. Radioactive fractions were analyzed by MALDI-TOF mass spectrometry by using a Bruker (Billerica, MA) Reflex MALDI mass spectrometer and α-cyano-4-hydroxy cinnamic acid as matrix.
Induction and Detection of AIF Translocation.
HeLa S3 cells were transfected to express FLAG-tagged NMNAT-1 (2) or EGFP localized to the nucleus. After 48 h, the cells were stressed for 10 min with 100 μM H2O2 in PBS containing 0.5 mM MgCl2, 1 mM CaCl2, and 1 mg/ml glucose. Thereafter, the cells were kept in growth medium for 110 min. The cells were then fixed with 4% formaldehyde, permeabilized with 0.5% Triton X-100, and immunostained with a polyclonal antibody against AIF (Santa Cruz Biotechnology, Santa Cruz, CA) and overexpressed NMNAT-1 by using a monoclonal antibody against the FLAG epitope (Sigma).
Supplementary Material
Acknowledgments
We thank Dr. P. Franke for the mass spectrometric analyses, Dr. P. Punterroll for help with structural analyses, and Drs. G. de Murcia and J.-C. Amé for helpful discussions. This work was supported by funds from the Norwegian Research Council and by Deutsche Forschungsgemeinschaft ZI 541/4-2 and the Norwegian Cancer Society Kreftforeningen A05128/005.
Abbreviations
- NMNAT
NMN adenylyl transferase
- PAR
poly(ADP-ribose)
- PARP-1
PAR polymerase 1
- AIF
apoptosis-inducing factor
- BIM
bisindolylmaleimide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609211104/DC1.
References
- 1.Magni G, Amici A, Emanuelli M, Raffaelli N, Ruggieri S. Adv Enzymol Relat Areas Mol Biol. 1999;73:135–182. doi: 10.1002/9780470123195.ch5. [DOI] [PubMed] [Google Scholar]
- 2.Berger F, Lau C, Dahlmann M, Ziegler M. J Biol Chem. 2005;280:36334–36341. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- 3.Schweiger M, Hennig K, Lerner F, Niere M, Hirsch-Kauffmann M, Specht T, Weise C, Oei SL, Ziegler M. FEBS Lett. 2001;492:95–100. doi: 10.1016/s0014-5793(01)02180-9. [DOI] [PubMed] [Google Scholar]
- 4.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. J Biol Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 5.Berger F, Ramirez-Hernandez MH, Ziegler M. Trends Biochem Sci. 2004;29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Corda D, Di Girolamo M. EMBO J. 2003;22:1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virag L, Szabo C. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 8.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 9.Blander G, Guarente L. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 10.Guse AH. Curr Med Chem. 2004;11:847–855. doi: 10.2174/0929867043455602. [DOI] [PubMed] [Google Scholar]
- 11.Lee HC. Curr Mol Med. 2004;4:227–237. doi: 10.2174/1566524043360753. [DOI] [PubMed] [Google Scholar]
- 12.Galione A, Churchill GC. Sci STKE. 2000;2000:1–6. doi: 10.1126/stke.2000.41.pe1. [DOI] [PubMed] [Google Scholar]
- 13.Clapper DL, Walseth TF, Dargie PJ, Lee HC. J Biol Chem. 1987;262:9561–9568. [PubMed] [Google Scholar]
- 14.Denu JM. Trends Biochem Sci. 2003;28:41–48. doi: 10.1016/s0968-0004(02)00005-1. [DOI] [PubMed] [Google Scholar]
- 15.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 17.Tissenbaum HA, Guarente L. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 18.Lin SJ, Defossez PA, Guarente L. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 19.Araki T, Sasaki Y, Milbrandt J. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 20.Ruggieri S, Gregori L, Natalini P, Vita A, Emanuelli M, Raffaelli N, Magni G. Biochemistry. 1990;29:2501–2506. doi: 10.1021/bi00462a010. [DOI] [PubMed] [Google Scholar]
- 21.Uhr ML, Smulson M. Eur J Biochem. 1982;128:435–443. doi: 10.1111/j.1432-1033.1982.tb06983.x. [DOI] [PubMed] [Google Scholar]
- 22.Bouchard VJ, Rouleau M, Poirier GG. Exp Hematol. 2003;31:446–454. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 23.Bürkle A. BioEssays. 2001;23:795–806. doi: 10.1002/bies.1115. [DOI] [PubMed] [Google Scholar]
- 24.De Murcia G, Menissier de Murcia J. Trends Biochem Sci. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 25.Wielckens K, George E, Pless T, Hilz H. J Biol Chem. 1983;258:4098–4104. [PubMed] [Google Scholar]
- 26.Dantzer F, de La Rubia G, Menissier-De Murcia J, Hostomsky Z, de Murcia G, Schreiber V. Biochemistry. 2000;39:7559–7569. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
- 27.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Mol Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. Proc Natl Acad Sci USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassa PO, Hottiger MO. Cell Mol Life Sci. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valenzuela MT, Guerrero R, Nunez MI, Ruiz De Almodovar JM, Sarker M, de Murcia G, Oliver FJ. Oncogene. 2002;21:1108–1116. doi: 10.1038/sj.onc.1205169. [DOI] [PubMed] [Google Scholar]
- 31.Conde C, Mark M, Oliver FJ, Huber A, de Murcia G, Menissier-de Murcia J. EMBO J. 2001;20:3535–3543. doi: 10.1093/emboj/20.13.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver FJ, Menissier de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC, de Murcia G. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitacre CM, Hashimoto H, Tsai ML, Chatterjee S, Berger SJ, Berger NA. Cancer Res. 1995;55:3697–3701. [PubMed] [Google Scholar]
- 34.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, Jacobson EL. J Biol Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 36.Goodwin PM, Lewis PJ, Davies MI, Skidmore CJ, Shall S. Biochim Biophys Acta. 1978;543:576–582. doi: 10.1016/0304-4165(78)90312-4. [DOI] [PubMed] [Google Scholar]
- 37.Skidmore CJ, Davies MI, Goodwin PM, Halldorsson H, Lewis PJ, Shall S, Zia'ee AA. Eur J Biochem. 1979;101:135–142. doi: 10.1111/j.1432-1033.1979.tb04225.x. [DOI] [PubMed] [Google Scholar]
- 38.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 39.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Proc Natl Acad Sci USA. 2006;103:18314–18329. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Nemoto M, Xu Z, Yu SW, Shimoji M, Andrabi SA, Haince JF, Poirier GG, Dawson TM, Dawson VL, Koehler RC. Neuroscience. 2007;144:56–65. doi: 10.1016/j.neuroscience.2006.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garavaglia S, D'Angelo I, Emanuelli M, Carnevali F, Pierella F, Magni G, Rizzi M. J Biol Chem. 2002;277:8524–8530. doi: 10.1074/jbc.M111589200. [DOI] [PubMed] [Google Scholar]
- 42.Panzeter PL, Realini CA, Althaus FR. Biochemistry. 1992;31:1379–1385. doi: 10.1021/bi00120a014. [DOI] [PubMed] [Google Scholar]
- 43.Griesenbeck J, Oei SL, Mayer-Kuckuk P, Ziegler M, Buchlow G, Schweiger M. Biochemistry. 1997;36:7297–7304. doi: 10.1021/bi962710g. [DOI] [PubMed] [Google Scholar]
- 44.Malanga M, Pleschke JM, Kleczkowska HE, Althaus FR. J Biol Chem. 1998;273:11839–11843. doi: 10.1074/jbc.273.19.11839. [DOI] [PubMed] [Google Scholar]
- 45.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia S, de Murcia G. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Q, Piston DW, Goodman RH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- 48.Mack TGA, Reiner M, Beirowski B, Mi W, Emanuelli M, Wagner D, Thomson D, Gillingwater T, Court F, Conforti L, et al. Nat Neurosci. 2001;4:1199–1206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- 49.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. J Biol Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 50.Schmitz AA, Pleschke JM, Kleczkowska HE, Althaus FR, Vergères G. Biochemistry. 1998;37:9520–9527. doi: 10.1021/bi973063b. [DOI] [PubMed] [Google Scholar]
- 51.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 52.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AD. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oei SL, Griesenbeck J, Buchlow G, Jorcke D, Mayer-Kuckuk P, Wons T, Ziegler M. FEBS Lett. 1996;397:17–21. doi: 10.1016/s0014-5793(96)01137-4. [DOI] [PubMed] [Google Scholar]
- 54.Griesenbeck J, Oei SL, Mayer-Kuckuk P, Ziegler M, Buchlow G, Schweiger M. Biochemistry. 1997;36:7297–7304. doi: 10.1021/bi962710g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.