Abstract
The Escherichia coli envelope-stress response is a sensor system that increases transcription of stress genes in the cytoplasm when misfolded porins are detected in the periplasm. This response is initiated by DegS cleavage of the periplasmic domain of RseA, a transmembrane protein. Additional proteolysis of transmembrane and cytoplasmic portions of RseA then frees the σE transcription factor, which directs the transcriptional response. We show that RseB protein, a known negative regulator, inhibits proteolysis by DegS in vitro by binding tightly to the periplasmic domain of RseA. Inhibition of DegS cleavage requires RseB binding to a conserved region near the C terminus of the poorly structured RseA domain, but the RseA sequences that mediate DegS recognition and RseB binding do not overlap directly. Although DegS cleavage of RseA is normally activated by binding of the C termini of porins to the PDZ domain of DegS, RseB inhibition is independent of this activation mechanism.
Keywords: extra cytoplasmic stress, MucB, regulated intramembrane proteolysis
A variety of physiological sensor systems use proteolytic cleavage of a membrane-spanning regulatory protein as a key early step in initiating rapid changes in gene expression (1). This method of signal transduction has been named “regulated intramembrane proteolysis” (2). Although these systems permit information to be transmitted across membranes in diverse pathways and organisms, the biochemical mechanisms by which regulated intramembrane proteolysis is modulated remain largely undetermined.
The envelope-stress response pathway of Escherichia coli is a regulated intramembrane proteolysis system that includes the σE transcription factor, the RseA and RseB regulators, and the DegS and RseP (YaeL) proteases (3–7). σE controls expression of gene products that facilitate the refolding or degradation of misfolded periplasmic proteins (8–10). The association of σE with RNA polymerase is normally inhibited by formation of a tight complex between σE and the cytoplasmic domain of RseA, a transmembrane protein (11). At high temperature or under other conditions that result in protein misfolding, a series of proteolytic cleavages destroy RseA and liberate σE to activate gene expression (12). The periplasmic domain of RseA is initially cleaved by DegS, a protease anchored to the periplasmic face of the inner membrane (13, 14). This periplasmic cleavage event activates RseP cleavage within the transmembrane region of RseA (15), releasing the complex of σE and the cytoplasmic domain of RseA from the membrane. The final step in σE activation involves degradation of the RseA cytoplasmic domain by ClpXP or other intracellular proteases (16, 17).
The signaling cascade that activates σE can be initiated by misfolded outer membrane porins (OMPs) that have a C-terminal YxF tripeptide that is buried in native membrane-embedded OMPs but is likely to be accessible to other proteins in unassembled or denatured OMPs. Peptides containing a C-terminal YxF (OMP peptides) activate DegS cleavage of the periplasmic domain of RseA in vitro, and secretion of proteins bearing these C-terminal OMP sequences activates σE-mediated gene expression in vivo (18). DegS is a trimer, with each subunit consisting of a membrane anchor, a serine-protease domain, and a PDZ domain (18, 19). OMP peptides bind to the DegS PDZ domain (18), and crystallographic studies suggest that the bound peptide activates the protease (19). Alternatively, OMP-peptide binding to the DegS PDZ domain may relieve an inhibitory interaction with the DegS protease domain, because DegS that lacks the PDZ domain has OMP-independent activity in vivo (18). Regardless of uncertainty about the detailed mechanism, it is clear that misfolded OMPs or OMP peptides are required to activate cleavage of RseA by full-length DegS.
RseB is a periplasmic protein that negatively regulates the envelope-stress response. Mutational inactivation of RseB results in faster cleavage of RseA and increased activity of σE in the absence of stress (14, 20–23). RseB also appears to inhibit RseP proteolysis of full-length RseA (23). RseA and RseB, which interact with each other and are encoded in the same operon with σE, have orthologs in numerous bacterial species. In Pseudomonas aeruginosa, for example, MucA and MucB (the RseA and RseB orthologs) regulate both heat-stress response and alginate production by modulating the activity of the AlgU transcription factor (24–26). Interestingly, inactivation of either MucA or MucB can result in comparably large increases in AlgU activity (24, 26), supporting major regulatory roles for both proteins.
To understand the regulatory role of RseB in greater detail, we have carried out biochemical studies with purified components. Here, we show that RseB binds to the periplasmic region of RseA strongly and with 1:1 stoichiometry. RseB binding to RseA directly inhibits RseA cleavage by DegS, independently of OMP peptides and the PDZ domain of DegS. We find that RseB recognizes a small C-terminal region of RseA, which is released from the membrane after DegS cleavage. Our results suggest that RseB needs to be inactivated by a cellular signal that is distinct from C-terminal OMP peptides to allow RseA release and subsequent cleavage by the DegS and RseP proteases during the envelope-stress response.
Results
RseB Inhibition of RseA Cleavage by DegS.
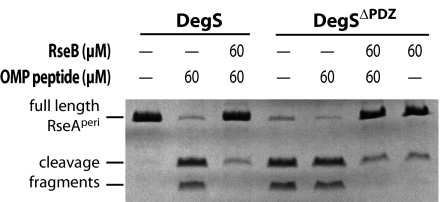
His6-RseB lacking its N-terminal signal sequence was cloned, overexpressed, and purified (see Materials and Methods). This variant contains residues 24–318 of the RseB precursor protein and should be similar to mature periplasmic RseB after signal-sequence cleavage. Soluble His6-tagged variants of DegS and the periplasmic region of RseA (RseAperi) were also purified (18). We tested the effect of RseB on DegS proteolysis of RseAperi monitored by SDS/PAGE and Coomassie staining (Fig. 1). As expected, DegS cleaved RseAperi in the presence of OMP peptide when RseB was absent. When RseB and OMP peptide were both present, however, cleavage of RseAperi by DegS was strongly inhibited (Fig. 1).
Fig. 1.
RseB inhibition of DegS cleavage of the RseA periplasmic domain. Results are shown for SDS/PAGE of samples after incubation of 20 μM full-length RseAperi and 32 μM DegS with or without OMP peptide and RseA at the concentrations indicated. The DegS and RseB bands have similar electrophoretic mobilities and are not shown.
DegSΔPDZ, which is a DegS variant lacking the PDZ domain, was used to test whether RseB inhibition of RseA cleavage was mediated by interactions with the OMP peptide or the PDZ domain. DegSΔPDZ cleaved RseAperi at comparable rates in both the presence and absence of OMP peptide (Fig. 1). Moreover, cleavage of RseAperi by DegSΔPDZ under both conditions was at least as fast as cleavage observed by using full-length DegS and OMP peptide. This result demonstrates that the serine-protease domain of DegS is sufficient for recognition and cleavage of RseA; neither the DegS PDZ domain nor OMP peptide is required for this reaction. RseB inhibited RseAperi cleavage by DegSΔPDZ in both the presence and absence of OMP peptide. We conclude that RseB inhibits cleavage of RseA by using a mechanism independent of the regulation of DegS activity by the PDZ domain and OMP peptide.
It has been suggested that unfolded or misfolded periplasmic proteins bind RseB, causing it to release RseA and providing a second physiological signal for initiation of the envelope-stress response (20, 21, 27). To test this model, we added excess quantities of four different largely unstructured proteins (α-casein, β-casein, an unfolded variant of the titin I27 domain, and an unfolded variant of RNase H) to reaction mixtures containing DegS, OMP peptide, RseAperi, and RseB. None of these nonnative proteins prevented DegS cleavage of RseAperi in the absence of RseB, and none of them allowed cleavage in the presence of RseB (data not shown).
RseB·RseA Binding.
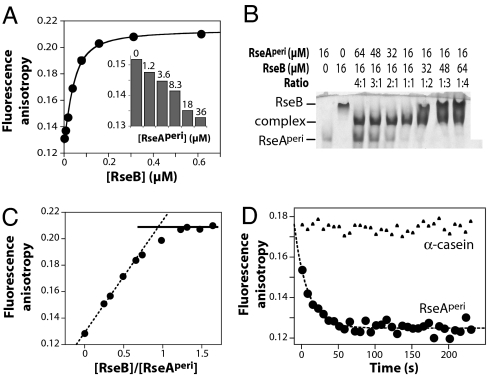
We covalently modified RseAperi by attaching a fluorescein (fl) dye (fl-RseAperi) and assayed RseB binding using changes in fluorescence anisotropy (Fig. 2A). Fitting the binding data gave an equilibrium dissociation constant (Kd) of 20 nM at 25°C (200 mM KCl, pH 7.4). Unlabeled RseAperi competed efficiently for fl-RseAperi binding to RseB (Fig. 2A Inset). RseB·RseA binding was ≈3-fold stronger at 5°C and 15°C than at 25°C and was ≈3- to 4-fold weaker at 35°C and 45°C (data not shown). Although RseB·RseA binding was weaker at higher temperatures, at 50°C, RseB had a native structure (Fig. 3B) and inhibited DegS cleavage of RseAperi (data not shown).
Fig. 2.
RseB interactions with the RseA periplasmic domain. (A) Increasing RseB was added to 43 nM fl-RseAperi, and binding at 25°C was assayed. The solid line is a nonlinear least-squares fit of the data with a Kd of 20 nM. (Inset) Unlabeled RseAperi competed efficiently for binding of fl-RseAperi to RseB. (B) RseB binding to RseA assayed by native-gel electrophoresis saturated at a stoichiometry of ≈1:1 (subunit equivalents). (C) Stoichiometry of RseB·RseA binding assayed by using changes in fluorescence anisotropy. Increasing RseB was added to a mixture of 43 nM fl-RseAperi and 1.54 μM unlabeled RseAperi. The dotted line is a fit to the first four data points. The solid line is the maximal binding anisotropy. (D) Dissociation kinetics. We preincubated 60 nM RseB and 43 nM fl-RseAperi at 25°C, and 5.3 μM excess RseAperi or 32 μM α-casein was added ≈10 s before monitoring dissociation.
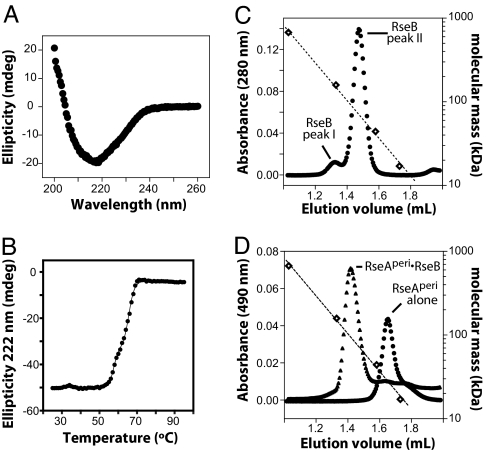
Fig. 3.
Characterization of RseB and its complex with RseA. (A) CD spectrum of 1 μM RseB at 25°C in 0.007× cleavage buffer. (B) Thermal unfolding of 3 μM RseB in 0.02× cleavage buffer was assayed by using changes in CD ellipticity. The solid line is a fit for a three-state denaturation model. (C) Gel filtration of RseB on Superdex 200 (Amersham Biosciences) at 4°C in cleavage buffer. Open diamonds mark the elution positions of molecular-weight standards. The dotted line is an exponential fit of molecular mass versus elution volume. The relative proportions of peak-I and peak-II RseB varied in different preparations. In the chromatogram shown, RseB was urea-denatured and refolded immediately before chromatography. (D) Gel filtration (same column and conditions as C) of fl-RseAperi (circles) or fl-RseAperi with excess RseB (triangles).
To determine the binding stoichiometry, we performed nondenaturing gel electrophoresis of mixtures of RseAperi and RseB at concentrations well above Kd. Formation of the RseB·RseAperi complex was maximal when the concentrations of RseAperi and RseB were equal (Fig. 2B), supporting a 1:1 binding stoichiometry. The same result was obtained when increasing RseB was titrated against fixed RseAperi, and binding was assayed by changes in anisotropy of trace amounts of fl-RseAperi (Fig. 2C).
The kinetics of dissociation of the fl-RseAperi·RseB complex at 25°C (200 mM KCl, pH 7.4) were determined after addition of excess unmodified RseA (Fig. 2D). Fitting of these kinetic data gave a dissociation rate constant (kdiss) of 0.055 s−1, corresponding to a half-life of ≈13 s. The association rate constant (kassn) calculated as kdiss/Kd was 2.8·106 M−1·s−1. Addition of excess α-casein did not cause dissociation of the fl-RseAperi·RseB complex (Fig. 2D). This result and those discussed above indicate that RseB is still able to bind RseA and to inhibit its cleavage by DegS in the presence of significant quantities of nonnative proteins.
RseB Secondary and Quaternary Structure.
The CD spectrum of RseB was consistent with a predominately β-sheet structure (Fig. 3A). In thermal denaturation monitored by CD, RseB melted cooperatively, but the unfolding transition was biphasic (Fig. 3B), suggesting that RseB contains multiple domains. Supporting this idea, PSI-BLAST searches revealed statistically significant sequence homology (E < 10−20) between the N-terminal 180 residues of mature RseB and LolA, a periplasmic protein of known structure that transports lipoproteins to the outer membrane.
Two forms of RseB were observed in gel-filtration experiments. One form eluted at a position expected for globular dimer (Fig. 3C, peak II) and inhibited DegS cleavage of RseA (data not shown). Another form eluted at a position corresponding to a species with a molecular weight 2- to 3-fold larger (Fig. 3C, peak I) and did not inhibit DegS (data not shown). Both species had identical CD spectra (data not shown). Peak I typically represented 40–50% of the freshly purified RseB, but this species increased and peak II decreased during storage. These observations suggest that peak I is formed from peak II by oligomerization. Consistent with this model, urea denaturation followed by renaturation of inactive peak-I material produced protein that largely eluted as peak II and was active in inhibition of DegS cleavage of RseA. Based on these observations, we believe that peak-II RseB represents the protein species that is biologically active in RseA binding and inhibition of DegS cleavage. RseAperi is monomeric in solution (18). When a mixture of RseB and fl-RseAperi was analyzed by gel-filtration, fl-RseAperi eluted at a position expected for a globular complex containing two RseB molecules and two RseAperi molecules (Fig. 3D).
RseA Residues Required for Interaction with RseB.
In sequence comparisons with orthologs, residues 165–189 of E. coli RseA displayed higher conservation than most parts of the periplasmic region (Fig. 4A). To determine whether this RseA region binds RseB, we synthesized a peptide containing RseA residues 160–189. A fl-labeled variant of this peptide bound RseB with a Kd of ≈6 μM (Fig. 4B), and the unlabeled peptide competed for RseB binding to fl-RseAperi (Fig. 4C). Thus, RseA residues 160–189 compose a major site of interaction with RseB. We also created C-terminally truncated variants of RseAperi (residues 121–161 and 121–175) in which most or part of the sequence from residues 160–189 was removed. Neither truncated variant competed substantially with fl-RseAperi for RseB binding (Fig. 4B). Taken together, these results demonstrate that RseA residues 160–189 are sufficient for the RseB·RseA interaction and suggest that residues between 176 and 191 play a key role in binding. Because the RseB affinity of the 160–189 peptide was lower than that of full-length RseAperi, regions of RseA outside of the 160–189 sequence must also contribute to binding.
Fig. 4.
Fragments of the periplasmic domain of RseA bind RseB. (A) Sequence alignment of the periplasmic domains of RseA from E. coli (Ec; GI 1173288), Salmonella typhimurium (St; GI 16765959), Yersinia pestis (Yp; GI 16122916), Haemophilus influenzae (Hi; GI 1173289), Pasteurella multocida (Pm; GI 15603653), and Vibrio cholerae (Vc; GI 15642462). The positions of fragments of the E. coli protein are shown below the alignment. (B) RseB binding to fl-labeled RseA 160–189 (50 nm) assayed by using fluorescence anisotropy. The solid line is a fit with a Kd of 6.5 μM. (C) Competition experiments. First, 160 nM RseB and 43 nM fl-RseAperi were mixed, and then fluorescence anisotropy was assayed 5 min after the addition of competitor proteins.
Effect of RseB Binding on RseA Cleavage by DegS.
Several experiments were performed to determine whether RseB binding to RseA is the mechanism of inhibition of DegS cleavage. First, we assayed RseB inhibition of DegS cleavage of the C-terminally truncated RseAperi variants. DegS cleaved both truncated variants, albeit somewhat less efficiently than it cleaved full-length RseAperi, but RseB did not inhibit DegS cleavage of either substrate (Fig. 5A). Thus, RseA mutations that severely impair RseB binding prevent RseB inhibition of DegS cleavage. Second, we tested whether the 160–189 peptide could relieve RseB-mediated inhibition of DegS cleavage of RseAperi. This peptide did not affect DegS cleavage of RseAperi in the absence of RseB but largely reversed the inhibitory effect of RseB (Fig. 5B). These experiments show that RseB does not inhibit DegS cleavage of RseA when binding is impaired by mutations in RseA or by peptide competition. We conclude that RseB exerts its inhibitory effect by binding to RseA and making RseA a poor substrate for DegS.
Fig. 5.
Effects of mutations in RseA or the presence of competitor on RseB inhibition of DegS cleavage of RseA. (A) DegS cleavage of 20 μM RseAperi or truncated variants was assayed by SDS/PAGE in the presence or absence of OMP peptide and RseB. (B) Relief of RseB-mediated inhibition of RseAperi cleavage by the RseA 160–189 peptide. (C) RseAperi variants with insertions between the DegS-cleavage site and RseB-binding site were incubated with DegS in the presence or absence of OMP peptide and RseB. The DegS concentration in all lanes was 32 μM.
DegS cleaves RseA between Val148 and Ser149 (18), whereas our results indicate that residues farther toward the C terminus of RseA participate in RseB binding. To test the importance of the spacing between the scissile peptide bond and the RseB binding determinants, we created RseA variants in which 8, 16, or 24 residues were inserted between the DegS cleavage site and the known site of contact with RseB. Each insertion mutant was cleaved by DegS (Fig. 5C). Moreover, RseB inhibited DegS cleavage of each of these insertion variants. Thus, moving the cleavage site and the primary site of RseB contact farther apart in RseA does not prevent RseB binding from inhibiting DegS cleavage of RseA.
Discussion
Our results demonstrate that RseB binds directly to the periplasmic region of RseA and that this binding prevents or severely slows RseA cleavage by DegS. The precise mechanism by which RseB binding prevents cleavage of RseA by DegS remains to be determined, but the results presented here and previously constrain potential models. Free RseAperi is molten-globule-like, with little stable tertiary structure (18). Thus, the Val148–Ser149 cleavage site and any other parts of the periplasmic domain of RseA that are required for DegS recognition should be freely available to the enzyme in the absence of RseB. The primary RseB-binding site and the DegS-cleavage/recognition sites in RseA do not overlap to any substantial degree. For example, RseB binds the RseA160–189 peptide, whereas DegS still cleaves the RseA121–161 fragment. Moreover, inserting up to 24 residues between the Val148–Ser149 cleavage site and the primary RseB-binding site in RseA did not prevent bound RseB from inhibiting DegS cleavage. This result and the low degree of homology between periplasmic domain sequences of RseA orthologs from closely related organisms make it unlikely that RseB binding causes the entire periplasmic region of RseA to fold into a compact structure that is resistant to DegS cleavage.
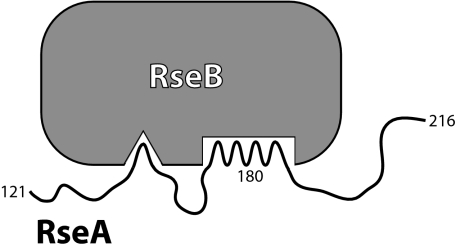
Nevertheless, RseB binding must in some fashion shield RseA sequences that are required for proteolysis by DegS. This could occur by the model depicted in Fig. 6, in which RseB bound to the primary RseA site also interacts with and blocks DegS access to more distant, secondary RseA sequences. This model does not require the intervening RseA sequences to be folded. In fact, these intervening sequences would need to be sufficiently flexible to allow formation of both sets of RseB·RseA interactions without significant strain. A model of this type would also explain why full-length RseAperi binds RseB more tightly than the RseA160–189 peptide. The secondary contacts could contribute to overall binding affinity, because of effective concentration considerations but not be sufficiently strong to allow RseB binding in the absence of the primary RseA binding site. We note that DegS cleavage of RseA would release a C-terminal fragment that retains affinity for RseB. If active RseB were limiting in the cell, then increasing quantities of the C-terminal RseA fragment could compete with intact RseA for RseB binding, potentially acting to enhance the rate of DegS cleavage.
Fig. 6.
Model for RseB binding to the RseA periplasmic domain. A primary site in RseA between residues 160 and 189 mediates binding to RseB. A secondary RseA site (overlapping determinants required for DegS recognition) strengthens binding to RseB.
Independently of its effects on DegS cleavage of RseA, RseB also appears to inhibit RseA cleavage by the second-site protease RseP in vivo (23). RseA variants that lack the primary RseB-binding site or have mutations in or near this site are cleaved by RseP in the absence of DegS (28). Moreover, RseP possesses two predicted PDZ domains (29), one of which has been shown to regulate RseP activity. RseP variants with mutations in the C-terminal PDZ domain cleave full-length RseA in the absence of DegS (23, 28, 30), but RseB does not inhibit this cleavage efficiently (23). Intriguingly, the region of RseA to which RseB has a strong affinity is between two glutamine-rich regions that play some role in conferring resistance to RseP proteolysis (28). A model consistent with these results is one in which RseB binding to RseA allows binding of this complex to the PDZ domain(s) of RseP, which in turn negatively regulates RseA cleavage by RseP. Alternatively, the added bulk of bound RseB might block RseA recognition by RseP, possibly as a result of steric clashes between RseB and the PDZ domains of RseP. In a different intramembrane proteolysis system, addition of bulky structured domains to the eukaryotic endoplasmic reticulum stress sensor ATF6 inhibited proteolysis by a second-siteprotease (31).
In our studies in vitro, little DegS cleavage of RseA occurred when sufficient RseB was present, even when DegS activity was fully induced by OMP peptide. Moreover, the affinity of RseB for RseA should ensure efficient complex formation in the cell. For example, there are ≈5,000 molecules of σE in an E. coli cell (32), and RseA is likely to be present in comparable quantities, which would correspond to a concentration of ≈80 μM in the periplasm. Because the Kd for the RseB–RseA interaction is <1 μM, most molecules of RseB in the periplasm should be bound to RseA in the absence of competing interactions. However, E. coli mutants lacking RseB show only modest increases in σE-mediated gene expression compared with mutants lacking RseA (22, 23). These results suggest that the negative regulatory role of RseB in vivo is smaller than might be expected from our biochemical results. Several factors could explain these differences. (i) The intracellular RseB level is not known, and there may be less RseB than RseA. (ii) Other cellular signals may reduce RseB binding to RseA, diminishing its inhibitory ability. (iii) Another regulator of envelope stress in E. coli may slow DegS cleavage of RseA in the absence of RseB. (iv) Levels of denatured or unassembled OMPs may be too low under nonstress conditions to activate DegS fully in the absence of RseB.
In P. aeruginosa, elimination of the RseB or RseA ortholog causes a comparable increase in AlgU-mediated gene expression under some conditions (24, 26). This result shows that RseB orthologs can have major regulatory roles and suggests that cellular mechanisms must be present to allow relief of RseB inhibition and efficient induction of the envelope-stress response. Our experiments show that temperature increases within the physiological range for E. coli growth (up to 50°C) do not prevent RseB inhibition of DegS cleavage of RseA. Thus, temperature per se is unlikely to represent an inducing signal for relief of RseB inhibition. Nevertheless, high temperatures and other environmental conditions that cause envelope stress might result in denatured macromolecules, fragments, or the accumulation of other molecular species that prevent RseB binding to RseA. In principle, stress-induced molecular signals could drive formation of the inactive peak-I RseB species observed in our studies or could compete reversibly for RseA binding to RseB. Because dissociation of RseB from RseA is relatively fast (half-life ≈ 13 s), a competition mechanism could rapidly inactivate RseB on a time scale consistent with the transcriptional response to envelope stress, which is detected within minutes of the initial stimulus (22).
Competition between RseA and other nonnative proteins for RseB binding has been proposed because RseB is recovered with the unstable MalE31 mutant protein in inclusion bodies (27). However, Grigorova et al. (23) found that MalE31 overproduction relieved RseB inhibition only modestly. Moreover, denatured OMPs are unlikely to be the stress signal that affects RseB activity. We observed RseB inhibition of DegS cleavage of RseA in the presence of high concentrations of C-terminal OMP peptides in vitro, and overproduction of OmpC did not affect RseB inhibition of RseP cleavage of RseA in vivo (23). In addition, our studies show that RseB binds and inhibits DegS cleavage of RseA, even in the presence of significant concentrations of several different nonnative and unfolded proteins. Thus, bulk unfolded protein in the periplasm is unlikely to prevent RseB binding to RseA. RseB shares homology with LolA, which transports lipoproteins to the outer membrane (33, 34), and antibiotic-induced changes in the structure of the LPS in the outer membrane of E. coli are sufficient to induce the σE-stress response (35). Hence, unassembled lipoproteins, periplasmic lipids, or modified LPS molecules or fragments might provide inputs into the envelope-stress response by regulating RseB activity. The biochemical assays described here should provide a useful tool for future studies of the control of RseB activity.
Materials and Methods
Proteins and Peptides.
DNA encoding RseB lacking its periplasmic localization sequence (residues 2–23) was cloned between the NdeI and XhoI sites of pET21b, appending an LEHHHHHH tag to the C terminus. Transformants of E. coli strain X90(DE3) were grown at 37°C in LB medium with 100 μg/ml ampicillin to an OD600 of ≈0.6, and protein expression was induced by addition of 100 μg/ml isopropyl β-d-thiogalactoside. Cells were harvested after 2 h, resuspended in a 1/50 volume of lysis buffer [50 mM sodium phosphate (pH 8)/500 mM KCl/20 mM imidazole], and lysed by using sonication. The cell lysate was spun for 30 min at 23,000 × g, and the supernatant was applied to a Ni-NTA column equilibrated in lysis buffer. The column was washed with 60 volumes of lysis buffer before addition of elution buffer [50 mM sodium phosphate (pH 8)/500 mM KCl/500 mM imidazole]. Fractions containing the most concentrated RseB as determined by using the Bradford stain assay were combined and dialyzed overnight against 1,000 volumes of buffer A [50 mM sodium phosphate (pH 6)/100 mM NaCl]. The dialysate was loaded onto a MonoS column equilibrated in buffer A, and RseB was eluted by using a linear gradient from buffer A to buffer B [50 mM sodium phosphate (pH 6)/1 M NaCl]. Fractions contained purified RseB were visualized by using SDS/PAGE, pooled, dialyzed against 1,000 volumes of cleavage buffer [50 mM sodium phosphate (pH 7.4)/200 mM KCl/10% glycerol], and stored frozen at −80°C.
DegSΔPDZ, which contains an N-terminal His6 tag but lacks the DegS membrane anchor (residues 2–26) and PDZ domain (residues 257–355), was cloned and expressed in strain X90(DE3). Purification and storage were similar to that for RseB, except that after Ni-NTA chromatography, appropriate fractions were pooled and dialyzed overnight into cleavage buffer containing 5 mM EDTA, and the ion-exchange step was not performed. RseAperi, DegS, and OMP peptide (NH2-DNRDGNVYYF-COOH) were purified as described (18). For labeling, Ser154 in RseAperi was mutated to cysteine; the mutation did not affect purification. Purified RseAperi-C154 was reduced with Tris(2-carboxyethyl)phosphine hydrochloride, mixed with a 10-fold molar excess of fl-5-maleimide, and incubated overnight at 4°C. Products were separated by using reverse-phase HPLC. The fl-modified protein, which ran at the same position as RseAperi in SDS/PAGE, was lyophilized and resuspended in cleavage buffer. Solid-phase peptide synthesis was performed at the biopolymers laboratory at Massachusetts Institute of Technology. Peptides were purified by using reverse-phase HPLC with a Vydac C18 column. α- and β-casein were purchased from Sigma (St. Louis, MO). Unfolded carboxymethylated titin and unfolded L78D/L112D RNase H* were prepared as described (36, 37).
To create RseAperi insertion mutants, the Gly151 codon was changed from GGG to GGA to introduce a unique KpnI site without altering the protein sequence. Oligonucleotides (5′-CTTCTGAAGCGACCGCAAAGGTAC-3′ and 5′-CTTTGCGGTCGCTTCAGAAGGTAC-3′) were phosphorylated, annealed, and ligated into vector cut with KpnI. Transformants were screened by PCR for insertions of one to three cassettes.
Cleavage Assays.
Proteolysis reactions were performed in cleavage buffer at 37°C for 16 h. Reactions were quenched by addition of SDS/PAGE loading buffer, boiled, electrophoresed on 12% or 15% Tris-tricine gels, and stained with Coomassie brilliant blue.
RseB Characterization.
CD spectra were taken with an Aviv 60DS instrument (Aviv Biomedical, Lakewood, NJ). RseB protein (145 μM in cleavage buffer) was diluted with water to the desired concentration and placed in a 1-cm path-length cuvette. Spectra were taken at 25°C with 1-s integration time. Temperature melts were performed in increments of 1°C with 30 s of equilibration and 10 s of integration.
Gel filtration was performed on a SMART system (Amersham Biosciences, Piscataway, NJ) at 4°C with a Superdex 200 column. Elution of RseB was monitored by absorbance at 280 nm; elution of fl-labeled RseAperi and fl-labeled RseAperi·RseB was monitored by absorbance at 490 nm. For some experiments, RseB or RseAperi·RseB mixtures were added to an equal volume of 9 M urea, diluted with an equal volume of cleavage buffer, filtered, and then loaded onto the column. Molecular-weight standards (catalog no. 151-1901) were from Bio-Rad (Hercules, CA).
Binding and Kinetic Assays.
Binding of fl-RseAperi to RseB was monitored by changes in fluorescence anisotropy at 25°C with a PTI QM-2000–4SE spectrofluorometer (Photon Technology International, Birmingham, NJ). The fl-RseAperi protein was diluted in cleavage buffer to a final concentration of 43 nM in a 60 μl volume cuvette. RseB protein was serially diluted into cleavage buffer. Increasingly concentrated RseB dilutions were titrated into the cuvette in 1-μl increments. The sample was excited at 467 nm, and emission was monitored at 520 nm. Anisotropy was calculated based on the scattering correction of a sample containing an identical amount of RseB but no fl-RseAperi.
To determine binding stoichiometry, different amounts of RseB and RseAperi were mixed in 4.65 M urea and electrophoresed on a native 10% Tris·glycine polyacrylamide gel. This procedure eliminated most inactive peak-I RseB material and resulted in reproducible binding. The native gel was stained with Coomassie brilliant blue. In a second experiment, a mixture of 43 nM fl-RseAperi and 1.54 μM unlabeled RseAperi was added to a cuvette, and fluorescence anisotropy was measured (excitation, 467 nm; emission, 520 nm). Another mixture with 43 nM fl-RseAperi, 1.54 μM RseAperi, and 10 μM RseB was prepared. The RseB protein used for this experiment was urea-denatured, diluted, and buffer-exchanged, resulting in >95% of active peak-II RseB as judged by native gel and gel filtration. A fraction of the cuvette mixture was withdrawn and replaced with an equal volume of the mixture containing RseB, and fluorescence anisotropy was measured again. This procedure was repeated to obtain data for a range of RseA/RseB ratios.
For measurement of dissociation kinetics, 50 nM fl-RseAperi and 164 nM RseB were mixed and fluorescence anisotropy was measured. Unlabeled RseAperi or α-casein were then added and mixed thoroughly, and anisotropy was measured as a function of time. For equilibrium competition assays, RseAperi, RseA 121–161, RseA 121–175, or the RseA 160–189 peptide were added in a volume of 1 μl to 61 μl of fl-RseAperi·RseB sample, and fluorescence anisotropy was measured after 5 min.
Acknowledgments
We thank Jon Kenniston (Massachusetts Institute of Technology) for protein and Sarah Ades and members of the R.T.S. laboratory for advice and discussions. This work was supported by National Institutes of Health Grant AI-16892.
Abbreviations
- OMPs
outer membrane porins
- fl
fluorescein.
Footnotes
The authors declare no conflict of interest.
References
- 1.Urban S, Freeman M. Curr Opin Genet Dev. 2002;12:512–518. doi: 10.1016/s0959-437x(02)00334-9. [DOI] [PubMed] [Google Scholar]
- 2.Brown MS, Ye J, Rawson RB, Goldstein JL. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 3.Ades SE. Curr Opin Microbiol. 2004;7:157–162. doi: 10.1016/j.mib.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Alba BM, Gross CA. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 5.Ehrmann M, Clausen T. Annu Rev Genet. 2004;38:709–724. doi: 10.1146/annurev.genet.38.072902.093416. [DOI] [PubMed] [Google Scholar]
- 6.Duguay AR, Silhavy TJ. Biochim Biophys Acta. 2004;1694:121–134. doi: 10.1016/j.bbamcr.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz N, Silhavy TJ. Curr Opin Microbiol. 2005;8:122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Kabir MS, Yamashita D, Koyama S, Oshima T, Kurokawa K, Maeda M, Tsunedomi R, Murata M, Wada C, Mori H, Yamada M. Microbiology. 2005;151:2721–2735. doi: 10.1099/mic.0.28004-0. [DOI] [PubMed] [Google Scholar]
- 9.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dartigalongue C, Missiakas D, Raina S. J Biol Chem. 2001;276:20866–20875. doi: 10.1074/jbc.M100464200. [DOI] [PubMed] [Google Scholar]
- 11.Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Mol Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 12.Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. Genes Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alba BM, Zhong HJ, Pelayo JC, Gross CA. Mol Microbiol. 2001;40:1323–1333. doi: 10.1046/j.1365-2958.2001.02475.x. [DOI] [PubMed] [Google Scholar]
- 14.Ades SE, Connolly LE, Alba BM, Gross CA. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akiyama Y, Kanehara K, Ito K. EMBO J. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn JM, Levchenko I, Sauer RT, Baker TA. Genes Dev. 2004;18:2292–2301. doi: 10.1101/gad.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaba R, Grigorova IL, Flynn JM, Baker TA, Gross CA. Genes Dev. 2007;21:124–136. doi: 10.1101/gad.1496707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 19.Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T. Cell. 2004;117:483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- 20.Missiakas D, Mayer MP, Lemaire M, Georgopoulos C, Raina S. Mol Microbiol. 1997;24:355–371. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- 21.De Las Penas A, Connolly L, Gross CA. Mol Microbiol. 1997;24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- 22.Ades SE, Grigorova IL, Gross CA. J Bacteriol. 2003;185:2512–2519. doi: 10.1128/JB.185.8.2512-2519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grigorova IL, Chaba R, Zhong HJ, Alba BM, Rhodius V, Herman C, Gross CA. Genes Dev. 2004;18:2686–2697. doi: 10.1101/gad.1238604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schurr MJ, Yu H, Martinez-Salazar JM, Boucher JC, Deretic V. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schurr MJ, Deretic V. Mol Microbiol. 1997;24:411–420. doi: 10.1046/j.1365-2958.1997.3411711.x. [DOI] [PubMed] [Google Scholar]
- 26.Rowen DW, Deretic V. Mol Microbiol. 2000;36:314–327. doi: 10.1046/j.1365-2958.2000.01830.x. [DOI] [PubMed] [Google Scholar]
- 27.Collinet B, Yuzawa H, Chen T, Herrera C, Missiakas D. J Biol Chem. 2000;275:33898–33904. doi: 10.1074/jbc.m006214200. [DOI] [PubMed] [Google Scholar]
- 28.Kanehara K, Ito K, Akiyama Y. EMBO J. 2003;22:6389–6398. doi: 10.1093/emboj/cdg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinch LN, Ginalski K, Grishin NV. Protein Sci. 2006;15:84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohn C, Collier J, Bouloc P. Mol Microbiol. 2004;52:427–435. doi: 10.1111/j.1365-2958.2004.03985.x. [DOI] [PubMed] [Google Scholar]
- 31.Shen J, Prywes R. J Biol Chem. 2004;279:43046–43051. doi: 10.1074/jbc.M408466200. [DOI] [PubMed] [Google Scholar]
- 32.Grigorova IL, Phleger NJ, Mutalik VK, Gross CA. Proc Natl Acad Sci USA. 2006;103:5332–5337. doi: 10.1073/pnas.0600828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuyama S, Tajima T, Tokuda H. EMBO J. 1995;14:3365–3372. doi: 10.1002/j.1460-2075.1995.tb07342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narita S, Matsuyama S, Tokuda H. Arch Microbiol. 2004;182:1–6. doi: 10.1007/s00203-004-0682-4. [DOI] [PubMed] [Google Scholar]
- 35.Tam C, Missiakas D. Mol Microbiol. 2005;55:1403–1412. doi: 10.1111/j.1365-2958.2005.04497.x. [DOI] [PubMed] [Google Scholar]
- 36.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 37.Kenniston JA, Burton RE, Siddiqui SM, Baker TA, Sauer RT. J Struct Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]