Abstract
Chk1 is a checkpoint kinase and an important regulator of mammalian cell division. Because null mutation of Chk1 in mice is embryonic lethal, we used the Cre-loxP system and the Lck promoter to generate conditional mutant mice in which Chk1 was deleted only in the T lineage. In the absence of Chk1, the transition of CD4−CD8− double-negative (DN) thymocytes to CD4+CD8+ double-positive (DP) cells was blocked due to an increase in apoptosis at the DN2 and DN3 stages. Strikingly, loss of Chk1 activated the checkpoint kinase Chk2 as well as the tumor suppressor p53 in these thymocytes. However, the developmental defects caused by Chk1 deletion were not rescued by p53 inactivation. Significantly, even though Chk1 deletion is highly lethal in proliferating tissues, we succeeded in using in vivo methods to generate Chk1/Chk2 double-knockout T cells. Analysis of these T cells revealed an interesting interaction between Chk1 and Chk2 functions that partially rescued the apoptosis of the double-mutant cells. Thus, Chk1 is both critical for the survival of proliferating cells and engages in cross-talk with the Chk2 checkpoint kinase pathway. These factors have implications for the targeting of Chk1 as an anticancer therapy.
Keywords: apoptosis, cancer, T cells, p53, bcl2
Cell survival depends on continuous surveillance that ensures the maintenance of genomic integrity. Threats to genomic integrity take the form of mutations sustained either during normal cell proliferation or after exposure to environmental agents. Failure to detect and resolve these threats can have dire consequences, as evidenced by the predisposition to cancer observed in individuals bearing mutations of genes involved in DNA damage response pathways (1–3). The normal functions of these genes protect an organism from malignancy by halting the division of a cell with genomic damage and preventing the defect from being passed on to daughter cells.
The checkpoint pathways are conserved signaling cascades activated in response to genotoxic stress (4). The activated checkpoints delay cell cycle progression to facilitate DNA repair, or they induce cell death to eliminate damaged cells, thus protecting the organism against cancer. Depending on the type of genotoxic stress, either ataxia telangiectasia mutated (ATM) or ataxia telangiectasia and Rad3-related (ATR) becomes activated and phosphorylates a downstream checkpoint kinase, either Chk1 or Chk2. Although it was originally believed that Chk1 phosphorylation depended solely on ATR and that of Chk2 on ATM, recent reports have indicated that there is substantial cross-talk between these checkpoint pathways. For example, Chk1 can be phosphorylated in response to ionizing radiation (IR) in an ATM-dependent manner (5, 6), and Chk2 can be activated independently of ATM (7).
The ATR–Chk1 pathway is usually activated by DNA lesions that impair replication fork progression. Indeed, this pathway employs components of the replication fork as a DNA damage sensor (8). Recent studies have shown that optimal activation of Chk1 also requires the cooperative action of several other factors, including the tumor suppressor Brca1 (9), the claspin adaptor molecule (10), and the proliferating cell nuclear antigen-like DNA sliding clamp Rad9/Rad1/Hus1 (11). Once activated, Chk1 phosphorylates a cascade of downstream targets, including the cell cycle phosphatases Cdc25A and Cdc25C (12). The end result of this Chk1-mediated cascade is a delay of cell cycle progression through the S and G2 phases (8, 13, 14). The central role played by Chk1 in regulating cell cycling in response to DNA damage has made Chk1 an attractive target for anticancer therapy.
In contrast to Chk1, Chk2 is activated primarily by ATM in response to double-stranded breaks in DNA caused by IR. Activated Chk2 contributes to the stabilization of p53, which promotes either cell cycle arrest or apoptosis. Similar to Chk1, activated Chk2 phosphorylates multiple Cdc25 molecules that subsequently inhibit the activation of cyclin-dependent kinases (15).
Much of our understanding of the checkpoint kinase network and its role in mitigating DNA damage has been gained from studies of human cancer cell lines. To understand the functions of the checkpoint kinases in a normal developmental (noncancerous) context, knockout mice deficient for either Chk1 or Chk2 have been generated (7, 13, 16). Analyses of these mutant animals have shown that, despite their overlapping functions in checkpoint signaling, the biological outcomes of Chk1 and Chk2 depletion are very different. Chk2-deficient mice are viable, do not show any developmental defects, and do not spontaneously develop tumors. However, Chk2-deficient cells show significant defects in IR-induced apoptosis and G1/S arrest (7). In dramatic contrast, Chk1-deficient mice are embryonic lethal. Furthermore, Chk1-deficient mammary gland cells that are induced to proliferate soon undergo apoptosis (13, 16, 17). Although it has been proposed that Chk1 acts as a tumor suppressor, the extreme sensitivity of cells to complete loss of Chk1 has hindered confirmation of this hypothesis. Lam et al. (17) were able to demonstrate haploinsufficiency for a potential tumor suppressor function for Chk1 in that cells with inactivation of one Chk1 allele showed inappropriate entry into S phase, accumulation of DNA damage during replication, and a failure to restrain mitotic entry in the presence of a dysregulated S phase. These observations suggest that inactivation of one Chk1 allele may result in sufficient genomic instability to initiate tumorigenesis. Indeed, tumor incidence is increased in Chk1+/−;WNT-1 transgenic mice, in which the WNT-1 transgene is driven by the mouse mammary tumor virus promoter (13). Nevertheless, despite these findings, our understanding of the relevance of complete loss of Chk1 in normal cell proliferation and genotoxic stress responses remains limited.
T cell development occurs in the thymus through a complex process involving distinct stages of proliferation and cell death. Thus, tissue-specific deletion of a gene in the T lineage constitutes an easily monitored developmental system in which to examine the role of that gene in normal cell physiology over time and during proliferation and differentiation. Furthermore, gene targeting in T cells allows the examination of the role of the gene in tumorigenesis because mice are more highly prone to developing T cell lymphomas than any other form of cancer. To investigate the function of Chk1 in normal cell development and during tumorigenesis, we generated mice with a T cell-specific deletion of Chk1 (18) and analyzed cell populations from the thymus of these animals. By using the advantages of phospho-specific flow cytometry (19–21), we analyzed the function of Chk1 in lymphocyte subsets of various genetic backgrounds.
Our data indicate that Chk1 is crucial for T cell development and that the cytoprotective function of Chk1 depends on more than just the activation of p53 and Chk2. In addition, we show that complete loss of Chk1 in T cells is sufficient to reduce the life span of otherwise normal mice.
Results
Expression Pattern of Chk1 in Wild-Type (WT) T Cells.
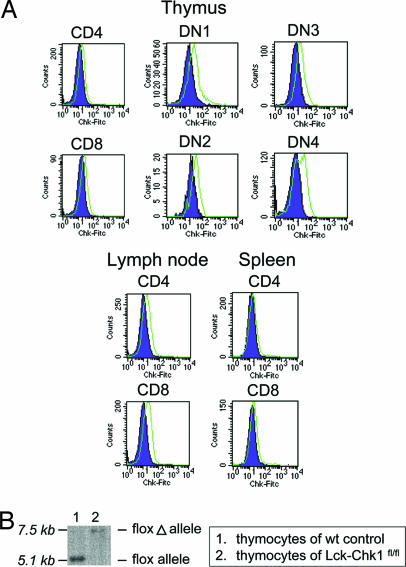
Because the expression pattern of Chk1 during T cell development was not known, we first used flow cytometry to examine Chk1 expression in various subpopulations of WT thymocytes and peripheral T cells (Fig. 1A). Increased levels of Chk1 were found in both double-negative (DN) and double-positive (DP) WT thymocytes as well as in mature CD4+ and CD8+ T cells in the lymph node and spleen. We then analyzed Chk1 expression during the differentiation of the four DN thymocyte stages (DN1–DN4) as defined by expression of the early developmental markers CD25 and CD44 (22). We found that all populations of DN cells had some level of Chk1 expression, suggesting that Chk1 functions throughout T cell development in the thymus.
Fig. 1.
Chk1 protein expression in various subsets of WT T cells and tissue-specific disruption of the Chk1 locus. (A) Chk1 protein expression in various T lineage cell populations. Surface staining of WT thymocytes with either anti-CD4 or anti-CD8 was followed by intracellular staining with anti-Chk1 Ab (green) or isotype control (purple) in samples of the thymus, spleen, or lymph node. To determine the Chk1 protein level in DN cells, thymocytes were stained with the anti-CD25, anti-CD44, and anti-Lin Abs. Lin+ cells were then gated out. (B) Southern blot analysis of DN thymocytes of Lck-Chk1fl/fl and WT mice revealed proper integration of the floxed Chk1 allele and its complete excision after Cre expression. Thymocytes were depleted of CD4+ and CD8+ cells, and the Southern blot was performed as described previously (13).
Deletion of Chk1 in T Lineage Cells.
To investigate the precise function of Chk1 in T cell development, we used a conditional gene-targeting approach in which loxP sites were engineered to flank exon 2 of the Chk1 gene. This exon contains the translational initiation sequence and encodes the ATP-binding site of the kinase (13). To delete Chk1 specifically in T cells, homozygous Chk1-floxed mice (Chk1fl/fl) were bred with Lck-Cre transgenic (Lck) mice (23) to generate Chk1flox/flox;Lck-Cre mice (Lck-Chk1fl/fl). To determine whether Cre-mediated recombination of the Chk1 locus had indeed occurred in the mutants, we performed Southern blot analysis of genomic DNA isolated from thymocytes of 8-week-old Lck-Chk1fl/fl mice and age-matched WT controls. A single band of 7.5 kb appeared in the blot of Lck-Chk1fl/fl thymocyte DNA but not in that of WT thymocyte DNA (Fig. 1B), confirming that complete Cre-mediated excision of the floxed Chk1 allele had occurred in Lck-Chk1fl/fl thymocytes.
Chk1 Is Essential for T Lineage Development.
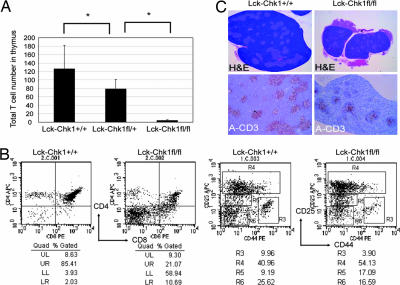
We next compared numbers of thymocytes and peripheral T cells in sex- and age-matched Lck-Chk1fl/fl, Lck-Chk1fl/+, and Lck-Chk1+/+ littermates. It was reported previously (23) that Lck-Cre is expressed during the DN stage of thymocyte development. We therefore examined total cell numbers in the thymi of Lck-Chk1fl/fl, Lck-Chk1+/fl, and Lck-Chk1+/+ littermates at 6–8 weeks of age. Compared with Lck-Chk1+/+ mice, thymocyte numbers were decreased by 97.5% in Lck-Chk1fl/fl mice (Fig. 2A), consistent with an essential role for Chk1 in thymocyte development. Furthermore, we noted a 64% decrease in total thymocyte numbers in Lck-Chk1fl/+ mice compared with Lck-Chk1+/+ mice, confirming that Chk1 is haploinsufficient for proper T cell development.
Fig. 2.
Flow cytometric and histological analyses of the effect of Chk1 depletion on murine thymus. (A) Defective thymocyte production in Chk1 heterozygotes. Total thymocytes from Lck-Chk1+/+, Lck-Chk1fl/+, and Lck-Chk1fl/fl mice (eight per group) were counted with a flow cytometer. Results shown are the mean number ± SD. P = 0.04 and P < 0.001, respectively. (B) Impaired development of Lck-Chk1fl/fl thymocytes. Thymocytes from Lck-Chk1+/+ and Lck-Chk1fl/fl mice were stained with anti-CD4 and anti-CD8 Abs (Left) or with anti-Lin, anti-CD25, and anti-CD44 Abs (Right). The Lin− population (only) was then examined by flow cytometry. (C) Reduced size and cellularity of Lck-Chk1fl/fl thymus. Hematoxylin/eosin and anti-CD3 staining of transverse sections of the thymus from a Lck-Chk1+/+ (Left) and a Lck-Chk1fl/fl (Right) mouse are shown. Data are representative of three thymi examined per group.
To determine at what stage thymocyte development was blocked in Lck-Chk1fl/fl mice, we carried out flow cytometric analysis of total thymocytes stained with anti-CD4 and anti-CD8 Abs. The proportion of thymocytes in the DN phase was increased in Lck-Chk1fl/fl thymus compared with Lck-Chk1+/+ thymus (59% versus 3.9%, respectively; n = 10), suggesting that the loss of Chk1 affected either the initial production of DN cells or the DN to DP transition (Fig. 2B Left). To distinguish between these possibilities, we used magnetic beads to deplete CD4+, CD8+, and B220+ cells from the total thymocytes of Lck-Chk1fl/fl and Lck-Chk1+/+ littermates mechanically, and we stained the remaining DN cells to detect CD25 and CD44 expression (Fig. 2B Right). This analysis revealed an increase in DN3 cells (R4 and R5) but a decrease in DN4 (R6) cells in the thymi of Lck-Chk1fl/fl mice. Histological findings confirmed the block in the DN3 to DN4 transition in the absence of Chk1. The thymus of Lck-Chk1+/+ mice had a typical structure, featuring a cortex containing mature T cells (Fig. 2C Upper Left; dark purple area) and a medulla containing smaller DN cells (Fig. 2C, light purple area). In contrast, the thymus of Lck-Chk1fl/fl mice was much smaller and lacked compartmentalization into a cortex and a medulla (Fig. 2C Upper Right). The vast majority of thymic cells in the mutants were large in size and stained light purple, which is characteristic of immature thymocytes. Moreover, histological staining with anti-CD3 revealed a massive reduction in the number of mature T cells in Lck-Chk1fl/fl thymus (Fig. 2C Lower Left and Right). Thus, in the absence of Chk1, DN thymocytes fail to expand and progress to the DP stage, abrogating the generation of mature T cells.
Chk1 Deficiency Leads to Increased Apoptosis During T Cell Development.
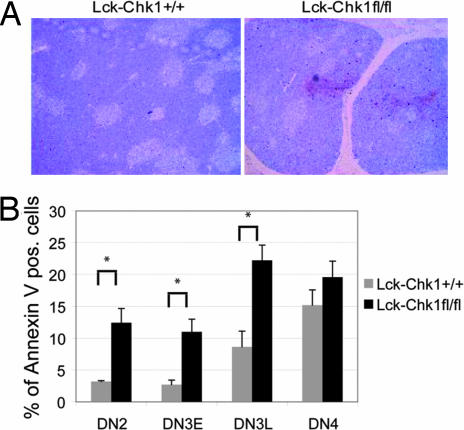
To define the nature of the developmental block induced by loss of Chk1, we used TUNEL and Annexin V staining to investigate whether the mutant thymocytes were undergoing apoptosis. Not surprisingly, we found that the thymi of Lck-Chk1fl/fl mice showed significantly greater numbers of TUNEL+ cells than the thymi of Lck-Chk1+/+ mice (Fig. 3A; P < 0.01). Quantitation of Annexin V staining levels indicated that a particularly high level of apoptosis was occurring within the DN2 and the early and late DN3 subpopulations (Fig. 3B; P < 0.01 for DN2, DN3E, and DN3L). In contrast, there was no difference in the number of apoptotic DN4 thymocytes in Lck-Chk1fl/fl and Lck-Chk1+/+ thymi (Fig. 3B; P > 0.5). These results suggest that either the phenotype imposed by Chk1 deficiency does not persist past the DN3 stage or that the population that progresses beyond DN3 has escaped Cre-induced deletion of Chk1.
Fig. 3.
T cells lacking Chk1 show increased cell death. (A) TUNEL staining. An increased number of TUNEL+ cells is present in the thymus of a Lck-Chk1fl/fl mouse compared with a Lck-Chk1+/+ mouse. Data are representative of three thymi examined per group. (B) DN thymocytes were prepared ex vivo; depleted of CD4+, CD8+, and B220+ cells with Dynabeads; and stained with anti-CD25, anti-CD44, and anti-Lin Ab followed by Annexin V. The number of Annexin V+ Lin− cells was assessed by flow cytometry. Results shown are the mean ± SD of eight mice per group and are representative of at least three experiments. P < 0.01 for DN2, DN3E, and DN3L; P > 0.5 for DN4.
Chk1 May Act as a Weak Tumor Suppressor.
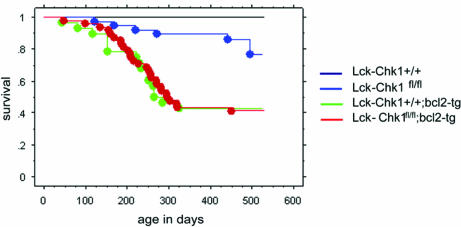
Using a mouse with a conditional deletion of Chk1 in the mammary glands, Lam et al. (17) demonstrated that Chk1 is haploinsufficient for the control of cell proliferation, genomic stability, and cell cycle coordination. Although these data suggest that Chk1 can act as a tumor suppressor, vigilant monitoring of Chk1+/− mice up to the age of 18 months failed to detect a predisposition to early tumorigenesis (13). To determine whether loss of Chk1 in T cells alone could decrease mouse life span, we monitored the survival of untreated Lck-Chk1fl/fl, Lck-Chk1+/fl, and Lck-Chk1+/+ littermate mice for 18 months. The Lck-Chk1fl/fl group showed a significantly lower survival rate compared with the Lck-Chk1+/+ group (Fig. 4; P = 0.001). However, when we compared the survival of Lck-Chk1fl/fl and Lck-Chk1+/+ littermates that had been irradiated with 5 Gy, no significant differences in survival were observed (data not shown; P = 0.1). To determine whether countering apoptosis could rescue the survival of Lck-Chk1fl/fl mice, we crossed these mutants into a bcl2-transgenic (tg) background. However, both WT;bcl2-tg and Lck-Chk1fl/fl;bcl2-tg mice showed equally curtailed life spans (Fig. 4). These results suggest that if Chk1 is a tumor suppressor, its effects are quite weak and are not enhanced by either IR or the bcl2-tg background.
Fig. 4.
Decreased survival of T cell-specific Chk1-deficient mice. Kaplan–Meier plots of survival of Lck-Chk1+/+ and Lck-Chk1fl/fl mice in a WT or bcl2-tg background are shown. Lck-Chk1+/+ (WT) mice (61), Lck-Chk1fl/fl mice (40), bcl2-tg mice (48), and Lck-Chk1fl/fl;bcl2-tg mice (28) were left untreated and monitored for survival. Survival is plotted against animal age in days.
Effect of Different Genetic Backgrounds on Lck-Chk1fl/fl T Cells.
Liu et al. (13) showed that null mutation of p53 neither delayed nor rescued the early embryonic lethality of Chk1−/− blastocysts. Furthermore, Chk1−/−;p53−/− blastocysts underwent apoptosis to the same degree as Chk1−/− blastocysts, suggesting that early apoptosis during embryogenesis is independent of p53. To test whether apoptosis during T cell development is also independent of p53, we crossed our Lck-Chk1fl/fl mice into the p21-null and p53-null backgrounds. The early defect in thymocyte development associated with conditional Chk1 deletion was still observed in the absence of either p21 or p53 (Fig. 5A). These results are consistent with the embryonic lethality of Chk1−/−;p53−/− blastocysts.
Fig. 5.
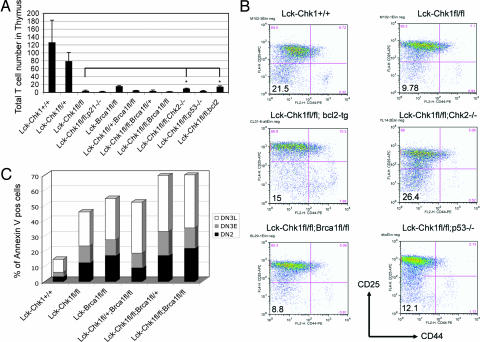
Effect of different genetic backgrounds on the Chk1 T cell phenotype. (A) Partial rescue of T cell numbers in Lck-Chk1fl/fl mice induced by loss of Chk2 or expression of bcl2. Lck-Chk1fl/fl mice were backcrossed to mice of different genetic backgrounds, including Chk2-, p53-, Brca1-, p21-null, and bcl2-tg mice. The total number of T cells in the thymi of the indicated double-mutant offspring and controls was counted by flow cytometry. ∗, P < 0.05. (B) Flow cytometric analysis of DN thymocytes of different genetic backgrounds. After depletion of CD4+, CD8+, and B220+ cells with Dynabeads, staining was performed with anti-CD25, anti-CD44, and anti-Lin Abs. Lin− cells of the indicated double-mutant mice and controls were analyzed for CD25 and CD44 expression. (C) Apoptosis in DN thymocyte subpopulations in Lck-Chk1fl/fl;Brcafl/fl double-mutant mice. Thymocytes from the indicated mutant mice and controls (at least three per group) were analyzed for DN stage and stained with Annexin V. Results shown are flow cytometric determinations of the percentages of Annexin V+ cells in the DN2, DN3E, and DN3L subpopulations.
Brca1 acts upstream of the checkpoint kinases and is involved in transducing DNA damage signals (24–26). Brca1 deficiency results in Chk2 phosphorylation and the Chk2-dependent accumulation and activation of p53 (27). We investigated the effect of Brca1 deletion on the development of Chk1-deficient thymocytes by crossing Brcafl/fl mice to Lck-Chk1fl/fl mice. However, T cell-specific deletion of Brca1 did not rescue the development of Chk1-deficient T cells (Fig. 5A). Interestingly, the Lck-Chk1+/fl;Brca1fl/fl thymocytes were sensitized to cell death as evidenced by a significant decrease in total thymocyte number as well as a reduction in the DN4 population compared with Lck-Chk1fl/+ or Lck-Brca1fl/fl mice (Fig. 5A and data not shown). The additional staining of the DN2 and DN3 subpopulations with Annexin V confirmed that thymocytes from Lck-Chk1fl/fl and Lck-Chk1fl/fl;Brca1fl/fl mice had succumbed to spontaneous cell death (Fig. 5C).
We next examined the effect of Chk2 deficiency on the development of Chk1-deficient T cells by crossing Chk2−/− mice with Lck-Chk1fl/fl mice. In our analysis of purified DN thymocytes, we found a small but statistically significant recovery of total T cell numbers in the thymus as well as a slight increase in the DN4 thymocyte population (Fig. 5 A and B; P < 0.05). These data indicate that Chk2 deficiency can mitigate the effects of Chk1 deletion on T cell development.
Chk1 Deficiency Is Associated with Increased Levels of p-Chk2 and p-p53 in T Cells.
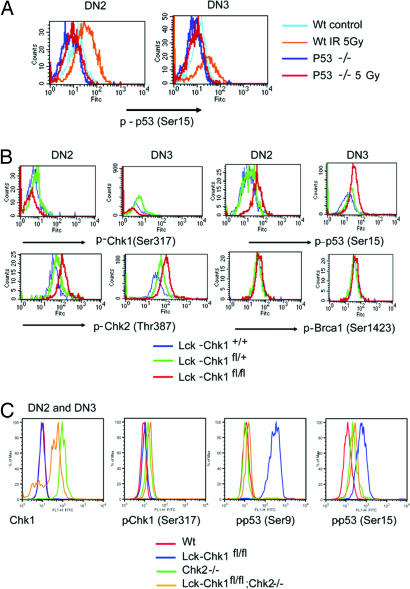
To better understand the mechanism underlying the defect in Chk1-deficient thymocyte development, we analyzed levels of known downstream targets of Chk1 in DN2 and DN3 thymocytes (the DN stages during which Cre is activated). We used the highly sensitive method of phospho-specific flow cytometry (19–21) to determine levels of individual phosphoproteins in T cells of various genetic backgrounds and at various developmental stages. Specifically, we examined levels of p53 phosphorylated at Ser-9 and Ser-15; Chk1 phosphorylated at Ser-317 and Ser-345; Chk2 phosphorylated at Ser-68, Ser-383, and Thr-387; and Brca1 phosphorylated at Ser-1423. Specificity of the phospho-flow antibodies was confirmed by using T cells from mice that were deficient for one of the proposed targets (Chk1, Chk2, p53, or Brca1) and subjected to 5-Gy irradiation (Fig. 6A and data not shown). As shown in Fig. 6B, Chk1 deficiency led to the presence of enhanced levels of phospho (p)-Chk2 and p-p53 in T cells, consistent with the increased apoptosis of these cells. Furthermore, p-Brca1 was increased in Chk1-deficient T cells, suggesting that Brca1 is a bona fide phosphorylation target downstream from Chk1. Interestingly, levels of p-Chk1 were increased in Chk2-deficient thymocytes (Fig. 6C). This finding supports earlier reports of cross-talk between these checkpoint pathways. Furthermore, when we analyzed Chk1/Chk2 double-knockout thymocytes, we found that the enhanced phosphorylation of p-p53 at Ser-9 that occurred in the absence of Chk1 was significantly less prominent (Fig. 6C). Thus, in the absence of Chk1, Chk2 is apparently required for this p53 phosphorylation event. A similar but less dramatic decrease in the phosphorylation of p53 at Ser-15 was also observed in Chk1/Chk2 double-knockout T cells. These findings suggest that absence of Chk1 disrupts checkpoint signaling such that p53 is activated by Chk2 and apoptosis of the mutant cell is induced.
Fig. 6.
Phospho-specific flow cytometric analysis of phosphoprotein expression in Chk1-deficient T cells. (A) Specificity of the anti-p-p53 Ab. Thymocytes from WT or p53-null mice that had been treated with 5 Gy or sham irradiation were stained with anti-CD25, anti-CD44, anti-Lin, and anti-p-p53 (Ser-15) Ab. Levels of phosphorylated p53 were determined by phospho-specific flow cytometry. (B) Phospho-specific flow cytometric analysis of DN2 and DN3 subpopulations in Lck-Chk1fl/fl mice. Intracellular staining for p-p53 (Ser-9 and Ser-15), p-Chk1 (Ser-317), p-Chk2 (Thr-387), and p-Brca1 (Ser-1423) was performed for mice of the indicated genetic backgrounds. (C) Phospho-specific flow analysis of DN2 and DN3 thymocytes from Lck-Chk1fl/fl mice in a Chk2-null background. Intracellular staining for p-Chk1 (Ser-317) and p-p53 (Ser-9 and 15) was performed for mice of the indicated genetic backgrounds.
Discussion
In this work, we have examined the effects of conditional deletion of Chk1 on T cell development. Consistent with previous reports on Chk1-deficient cells, we found that the vast majority of T lineage cells lacking Chk1 were nonviable and died of apoptosis, most likely because of excessive uncorrected DNA damage. Through an analysis of Chk1 depletion during early thymocyte development, we have uncovered evidence pointing to an intricate cross-talk between Chk1 and Chk2. Our findings also indicate that these checkpoint kinases can phosphorylate different in vivo targets.
Chk1 Deletion Results in Cell Death Caused by Apoptosis.
Previous in vivo studies have shown that Chk1 deficiency leads to the death of embryonic stem cells, to periimplantation embryonic lethality in mice, and to increased apoptosis in proliferating mammary cells (13, 16, 17). Using a T cell-specific knockout model, we have confirmed that the Chk1 gene is crucial for the survival of T lineage cells. Thymocytes developing in the absence of Chk1 readily undergo apoptosis at the DN2 stage, precluding the generation of significant numbers of mature T cells. Because the half-life of the Chk1 protein is very short (3.4 h) (28), it is not surprising that depletion of Chk1 in vivo at the DN2 stage leads to a rapid wave of thymocyte apoptosis. Thus, our results are consistent with the sudden apoptotic death of Chk1-depleted cells previously shown by others (13, 17). The main trigger for this apoptosis may be increased genomic instability because cells heterozygous for Chk1 show an uncoordinated cell cycle and an abnormally high level of Cdc25A (17). As well, cell proliferation appears to be required because Chk1-depleted lobuloalveolar mammary epithelial cells (which are nonproliferative) do not undergo apoptosis (17). This link to proliferation suggests that death resulting from Chk1 depletion may involve mitotic catastrophe (29). Our preliminary studies of Chk1 depletion in postmitotic glial cells support this hypothesis.
Genetic Interactions Between Chk1 and p53.
Deletion of p53 did not rescue the block in thymocyte development caused by Chk1 deficiency, suggesting that Chk1 can act on important alternative targets to ensure the survival of normal cells. Thus far, only the p53-deficient avian tumor cell line DT-40 has been shown to be viable in the absence of Chk1 (30). Thus, it is possible that a number of other unknown survival or antiapoptotic genetic aberrations could allow cells to survive in the absence of Chk1.
We found that in the absence of Chk1, p53 phosphorylation was strongly up-regulated in thymocytes such that these cells died, consistent with the finding published by Syljuasen et al. (31). However, in the absence of both Chk1 and Chk2, p53 phosphorylation was only slightly up-regulated, and more of the thymocytes survived. These findings suggest that an upstream kinase, most likely ATM or ATR, is modulating DNA damage signals and controlling the activation of perhaps a panel of checkpoint kinases that can act on p53 or other substrates to stimulate cell death. Although p53 signaling is not absolutely required for such death, any enhancement in its function would be expected to intensify the apoptotic result.
Chk1 May Act as a Weak Tumor Suppressor.
The genomic instability observed in Chk1+/− cells as well the increased tumor incidence in Chk+/−; MMTV-WNT-1 transgenic mice suggest that Chk1 may act as a tumor suppressor (13, 17). However, in humans, mutations of Chk1 have been found in only in a few sporadic endometrial and gastrointestinal cancers (32–34). In our longitudinal analysis of the survival of Lck-Chk1+/+ and Lck-Chk1fl/fl mice, we observed a significant decrease in the life span of the latter. Comparing these mice with untreated WT mice of a tumor-prone background, we have seen no differences in spontaneous tumorigenesis or leukemogenesis. Because T cell lymphomas are the most frequently encountered spontaneous tumor seen in experimental mice, our observation argues that although Chk1 heterozygosity causes defects in tumor-suppressive activities, Chk1 itself has only a weak tumor-suppressive function.
Functional Cross-Talk Between Chk1 and Chk2.
Chk1 and Chk2 are key mediators that link the sensors monitoring DNA integrity to components of the cell cycle machinery. After their phosphorylation, Chk1 and Chk2 activate unique downstream targets as well as targets that overlap, such as Cdc25A and p53 (12). Despite their parallel functions in checkpoint signaling, Chk1 and Chk2 are unrelated protein kinases with different biological roles. Most strikingly, Chk1 is essential for embryonic development and cell viability, whereas Chk2 is dispensable. In this work, we have used a phospho-specific flow cytometric technique and thymocytes of various genetic backgrounds to analyze known phosphoproteins within the checkpoint network. By sorting developmental subpopulations of T lineage cells, we have been able to examine cells lacking either Chk1 or Chk2 or both checkpoint kinases. Our data show that in Chk1-deficient thymocytes, the Chk2 pathway is activated and drives hyperphosphorylation of p53. Chk1/Chk2 double-knockout thymocytes contain lower levels of phospho-p53 than do Chk1-deficient thymocytes, correlating with a slight increase in total thymocyte numbers in Chk1/Chk2 double-mutant mice. One possible explanation of these findings is that apoptotic signaling in Chk1-depleted cells utilizes but does not require Chk2-mediated activation of p53. Although such cross-talk has been hypothesized previously (7), our work provides valuable direct evidence to support this theory.
Targeting Chk1 as a Cancer Therapy.
Because of the key role of Chk1 in controlling the cell cycle, it has been proposed that anticancer therapeutics be developed based on the inhibition of this checkpoint kinase. The fact that Chk1 depletion, in both malignant and nonmalignant cells, triggers their death sounds a note of caution. An effective anticancer agent should have a good therapeutic ratio; that is, the agent should be highly efficacious in killing cancer cells but of low toxicity to normal cells. Our results suggest that inhibition of Chk1 would have detrimental effects on both normal and tumor cells, making the cancer cell eradication index of an anti-Chk1 agent unacceptable.
Conclusions
In this work, we have shed some light on the complex functions of the important checkpoint kinase Chk1. We have provided genetic evidence that its depletion leads to cell death in early thymic development. Interestingly, this apoptosis cannot be rescued by deficiency of p53, p21, or Brca1, suggesting that an alternative route of cell death commitment is triggered by Chk1 loss. We have also provided evidence indicating that cross-talk between Chk1 and Chk2 is important for regulating normal T cell development and that Chk2 can at least partially compensate for an absence of Chk1 in some circumstances. These insights should prove useful when considering the design of anticancer agents that focus on checkpoint kinases as targets.
Materials and Methods
Mice.
Floxed Chk1 mice (M. Lam, Baylor College of Medicine, Houston, TX); floxed Brca1 mice (R. Hakem, Ontario Cancer Institute); Lck-Cre transgenic mice (Jamey Marth, Howard Hughes Medical Institute, La Jolla, CA); p21−/−, p53−/−, and bcl2-tg mice (The Jackson Laboratory, Bar Harbor, ME); and Chk2−/− mice were used in this work. Double-mutant mice were obtained by crossing these strains. Strains were typed by PCR using previously published primer sets. Mice were analyzed at 4–8 weeks of age. All experiments were performed in compliance with the guidelines of the Ontario Cancer Institute animal care committee.
Histological Analysis.
Thymi were fixed overnight in freshly prepared 4% paraformaldehyde/PBS and processed for histology. Serial sections were stained with hematoxylin/eosin, anti-CD3 Ab (AO452; Dako, Carpinteria, CA), or TUNEL by using standard protocols.
Flow Cytometry.
Thymocytes, lymph node cells, or peripheral blood cells were stained with mAbs (all from BD Biosciences, San Jose, CA) recognizing the following markers: CD4, CD8, CD25, CD44, B220, TCRαβ, CD11c, NK1.1, TER-119, Mac-1, and G1-1. The mAbs were directly coupled to FITC, PE, APC, Cy-5, or biotin. Surface marker expression by thymocytes and peripheral T cells was analyzed by using a flow cytometer (FACSCalibur; BD Biosciences), CellQuest, and FlowJo software according to standard protocols.
To analyze double-negative T cells, cells expressing CD4, CD8, or B220 were depleted by using a mixture of CD4/CD8/B220 magnetic beads (Invitrogen, Carlsbad, CA). After two rounds of depletion, the remaining DN T lineage cells were preincubated with Fc-Block (BD Biosciences); stained with anti-CD25, anti-CD44, and specific intracellular mAbs; and subjected to flow cytometry as described above. Intracellular staining was performed by using a cell fixation/permeabilization kit (BD Biosciences) and Abs specific for the following proteins: Chk1, phospho-Chk1, Chk2, phospho-Chk2, phospho-p53 (Ser-9), phospho-p53 (Ser-15) (all from Cell Signaling Technology, Inc., Beverly, MA); and phospho-Brca1 (Ser-1423) (Novus Biologicals, Cedarlane, Hornby, ON, Canada). At least three mice per group were used for all experiments.
Apoptosis Assay.
In vitro thymocyte apoptosis was evaluated by using an apoptosis detection kit (R & D Systems, Minneapolis, MN) and Annexin V according to standard protocols. At least three mice per group were used for all experiments.
Statistics.
Data are given as mean ± SD. Kaplan–Meier event-free survival curves were compared by log rank test (Mantel–Cox). Paired t tests or unpaired t tests, if appropriate, were used for comparisons. P values were Bonferroni-corrected for multiple comparisons. P < 0.05 was considered significant. Analyses were performed by using StatView version 5 (SAS Institute, Chicago, IL).
Acknowledgments
We thank Jamey Marth for the Lck-Cre transgenic mice; Gordan Duncan, Eliana Lucchinetti, and Simone Ringold for technical assistance; Jim Woodgett, Sam Benchimol, Pamela Ohashi, and members of the T.W.M. laboratory for helpful comments; and Mary Saunders for scientific editing. This work was supported by grants from the Canadian Institutes of Health Research (to T.W.M.) and from the National Institutes of Health and Center for Countermeasures against Radiation (to S.J.E.). S.J.E. is a Howard Hughes Medical Institute Investigator.
Abbreviations
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related
- DN
double-negative
- DP
double-positive
- IR
ionizing radiation
- tg
transgenic;
Footnotes
The authors declare no conflict of interest.
References
- 1.Harris SL, Levine AJ. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 2.Nevanlinna H, Bartek J. Oncogene. 2006;25:5912–5919. doi: 10.1038/sj.onc.1209877. [DOI] [PubMed] [Google Scholar]
- 3.Shiloh Y. Natl Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 4.Zhou BB, Elledge SJ. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 5.Gatei M, Sloper K, Sorensen C, Syljuasen R, Falck J, Hobson K, Savage K, Lukas J, Zhou BB, Bartek J, Khanna KK. J Biol Chem. 2003;278:14806–14811. doi: 10.1074/jbc.M210862200. [DOI] [PubMed] [Google Scholar]
- 6.Sorensen CS, Syljuasen RG, Falck J, Schroeder T, Ronnstrand L, Khanna KK, Zhou BB, Bartek J, Lukas J. Cancer Cell. 2003;3:247–258. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 7.Hirao A, Cheung A, Duncan G, Girard PM, Elia AJ, Wakeham A, Okada H, Sarkissian T, Wong JA, Sakai T, et al. Mol Cell Biol. 2002;22:6521–6532. doi: 10.1128/MCB.22.18.6521-6532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorensen CS, Syljuasen RG, Lukas J, Bartek J. Cell Cycle. 2004;3:941–945. [PubMed] [Google Scholar]
- 9.Yarden RI, Pardo-Reoyo S, Sgagias M, Cowan KH, Brody LC. Nat Genet. 2002;30:285–289. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- 10.Kumagai A, Dunphy WG. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- 11.Dang T, Bao S, Wang XF. Genes Cells. 2005;10:287–295. doi: 10.1111/j.1365-2443.2005.00840.x. [DOI] [PubMed] [Google Scholar]
- 12.Bartek J, Lukas J. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao H, Watkins JL, Piwnica-Worms H. Proc Natl Acad Sci USA. 2002;99:14795–14800. doi: 10.1073/pnas.182557299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuoka S, Huang M, Elledge SJ. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 16.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M, Nakayama K. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 17.Lam MH, Liu Q, Elledge SJ, Rosen JM. Cancer Cell. 2004;6:45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 18.Hennet T, Hagen FK, Tabak LA, Marth JD. Proc Natl Acad Sci USA. 1995;92:12070–12074. doi: 10.1073/pnas.92.26.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krutzik PO, Irish JM, Nolan GP, Perez OD. Clin Immunol. 2004;110:206–221. doi: 10.1016/j.clim.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Krutzik PO, Nolan GP. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 21.Perez OD, Nolan GP. Immunol Rev. 2006;210:208–228. doi: 10.1111/j.0105-2896.2006.00364.x. [DOI] [PubMed] [Google Scholar]
- 22.Okada H, Bakal C, Shahinian A, Elia A, Wakeham A, Suh WK, Duncan GS, Ciofani M, Rottapel R, Zuniga-Pflucker JC, Mak TW. J Exp Med. 2004;199:399–410. doi: 10.1084/jem.20032092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orban PC, Chui D, Marth JD. Proc Natl Acad Sci USA. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatei M, Zhou BB, Hobson K, Scott S, Young D, Khanna KK. J Biol Chem. 2001;276:17276–17280. doi: 10.1074/jbc.M011681200. [DOI] [PubMed] [Google Scholar]
- 26.Gudmundsdottir K, Ashworth A. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 27.McPherson JP, Lemmers B, Hirao A, Hakem A, Abraham J, Migon E, Matysiak-Zablocki E, Tamblyn L, Sanchez-Sweatman O, Khokha R, et al. Genes Dev. 2004;18:1144–1153. doi: 10.1101/gad.1192704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, Abraham RT. Mol Cell. 2005;19:607–618. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 29.Okada H, Mak TW. Natl Rev Cancer. 2004;4:592–603. doi: 10.1038/nrc1412. [DOI] [PubMed] [Google Scholar]
- 30.Zachos G, Rainey MD, Gillespie DA. EMBO J. 2003;22:713–723. doi: 10.1093/emboj/cdg060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertoni F, Codegoni AM, Furlan D, Tibiletti MG, Capella C, Broggini M. Genes Chromosomes Cancer. 1999;26:176–180. [PubMed] [Google Scholar]
- 33.Menoyo A, Alazzouzi H, Espin E, Armengol M, Yamamoto H, Schwartz S., Jr Cancer Res. 2001;61:7727–7730. [PubMed] [Google Scholar]
- 34.Vassileva V, Millar A, Briollais L, Chapman W, Bapat B. Cancer Res. 2002;62:4095–4099. [PubMed] [Google Scholar]