Abstract
We present evidence that heterotrimeric G protein signaling is involved in cell death associated with the unfolded protein response (UPR) in Arabidopsis. Seedlings of homozygous agb1-2 (Gβ-null mutation) mutant plants are markedly more resistant to growth inhibition by the protein glycosylation inhibitor tunicamycin (Tm) than either wild-type plants or gpa1-4 (Gα-null mutation) mutants. Leaves of older Gβ mutant plants show much less cell death when infiltrated with Tm than leaves of wild-type plants. The transcriptional response of Gβ mutant plants to Tm is less pronounced than that of wild-type plants, as is the accumulation of BiP chaperone proteins. A majority of the Arabidopsis Gβ protein is associated with the endoplasmic reticulum (ER) and cofractionates with membrane-associated ER luminal BiP. Consistent with its ER localization, Gβ protein is degraded during the UPR, whereas Gα protein is not. Taken together, these observations imply that the Gβ protein, which forms a stable heterodimer with the Gγ subunit, is involved in the signaling events that trigger UPR-associated cell death. The different Tm sensitivities of Gα and Gβ mutants, the ER localization of Gβ, and the differential stabilities of Gα and Gβ proteins during the UPR suggest that the Gβγ complex serves a signaling function in the ER independent of its function in the Gαβγ heterotrimer.
Keywords: endoplasmic reticulum, tunicamycin
Endoplasmic reticulum (ER) stress occurs when protein folding and modification are disrupted, triggering a complex protective response termed the unfolded protein response (UPR) (1). In yeast and mammalian cells, the transcriptional and translational changes that comprise the UPR are set in motion by the release of three ER transmembrane proteins, the inositol-requiring transmembrane kinase and endonuclease 1 (Ire1), the basic leucine zipper transcription factor ATF6, and the PKR-like ER kinase (PERK), from inactive complexes with the ER-resident chaperone immunoglobulin heavy chain-binding protein (BiP, also known as glucose-regulated protein GRP78) (1). Release from BiP activates PERK, which phosphorylates the translation elongation factor eIF2α, inhibiting translation of most mRNAs, but activating translation of bZIP transcription factor 4 (ATF4) mRNA. ATF6 is translocated to the Golgi, where it is activated by protease cleavage; ATF6 then moves to the nucleus, up-regulating expression of genes that code for BiP and other ER chaperones (1). Release from BiP activates the ribonuclease domain of Ire1 to cleave the mRNA precursor encoding the transcription factor X-box binding protein 1 (XBP1) mRNA to its mature form, the XBP1 mRNA. XBP1, in turn, activates expression of additional genes coding for ER proteins, including p58IPK, an inhibitor of PERK that down-regulates the UPR (1). The component pathways of the UPR, which also includes ER-associated protein degradation (ERAD), enhance the capacity of the cell to promote protein folding and to dispose of misfolded proteins, but when such measures fail, the UPR activates apoptotic cell death (2).
Although the UPR has not been characterized as extensively in plants as it has been in yeast and mammalian cells, plants also respond to ER stress by up-regulating expression of genes encoding protein-folding enzymes and chaperones, such as protein disulfide isomerase (PDI) and BiP, as well as vesicle transport proteins and proteins involved in ERAD (3–8). Activation of the UPR in cultured plant cells by the antibiotic tunicamycin (Tm), an inhibitor of N-linked protein glycosylation, triggers programmed cell death (9, 10). Only a few homologs of proteins centrally involved in the UPR in other systems have been identified in plants: Ire1 (11–13), eIF2α (14, 15), and p58IPK (16). Although the Arabidopsis genome has no immediate homologs of the ER stress-induced XBP1 or ATF6 genes, the Tm-inducible bZIP60 transcription factor contains a transmembrane domain and is activated by cleavage; hence, it may function in a manner analogous to that of ATF6 (17).
Many of the characteristic proteins of the UPR are up-regulated in animal cells specialized for secretion, including plasma cells, pancreatic β cells, hepatocytes, and osteoblasts (18). Not surprisingly, dysregulation of the ER stress pathways contributes to many human diseases, including diabetes, Alzheimer's disease, atherosclerosis, and cancer (19) In plants, pathogens and abiotic stress stimulate the production of pathogenesis-related (PR) proteins, which are generally secreted or vacuole-targeted proteins, including chitinases, β-glucanases, proteinases, and proteinase inhibitor (20). The redox-regulated NPR1 protein that is required for PR gene expression also mediates the activation of a number of Arabidopsis genes encoding proteins involved in protein folding and secretion, such as BiP chaperones and PDIs (21). Moreover, mutations that affected ER chaperone and secretory protein genes compromise the ability of plants to withstand pathogen attack, indicating that the ability to up-regulate the secretory machinery is a critical part of the plant's defense response (21).
PR gene expression is up-regulated in response to both pathogen attack and abiotic stresses (22, 23), and previous studies have identified a central role for the Arabidopsis heterotrimeric G protein as an early mediator of plant stress signaling (24–26). The Arabidopsis genome encodes a single canonical heterotrimeric G protein α and β subunit and two γ subunits (27). We recently reported that the Gα and Gβ subunits serve both separable and synergistic functions in signaling by reactive oxygen species in the oxidative stress response (26). Here we provide evidence that signaling through the Gβ subunit of heterotrimeric G protein triggers the cell death response in the UPR. We show that plants homozygous for a null mutation of the Gβ subunit of heterotrimeric G protein are substantially more resistant to Tm-induced cell damage and death than either wild-type plants or plants homozygous for a null mutation of the Gα subunit. We also show that the transcriptional response of the agb1-2 mutant plants (Gβ-null mutation) is reduced and delayed compared with that of wild-type plants and that the large protein aggregates characteristic of the ER stress response are not observed in mutant plants. Although a fraction of the cellular Gβ protein is detected with the Gα protein in plasma membranes, a majority of the Gβ protein is in the ER and cofractionates with ER lumenal BiP chaperone proteins. Moreover, consistent with its ER localization, the Gβ protein but not the Gα protein is degraded during the UPR. These observations strongly suggest an independent role for the Gβγ complex of the heterotrimeric G protein in cell death signaling in the UPR.
Results
Plants Lacking the Heterotrimeric Gβ Subunit Are Tm Resistant.
To compare the Tm sensitivity of G protein mutants and wild-type Arabidopsis, seeds of plants homozygous for null mutations in the genes encoding the Gα and Gβ subunits, as well as doubly mutant plants, were plated on Tm-containing medium for 6 days and then transferred to Tm-free agar medium for 10 days. As shown in Fig. 1, homozygous gpa1-4 (Gα-null mutation) seedlings are as sensitive as wild-type seedlings to growth inhibition by Tm, whereas homozygous agb1-2 (Gβ-null mutation) seedlings are markedly less sensitive than either wild-type or gpa1-4 seedlings. Moreover, doubly mutant gpa1-4/agb1-2 seedlings show a resistance phenotype similar to that of the agb1-2 seedlings.
Fig. 1.
Plants homozygous for null mutations in genes coding for the α and β subunits of the heterotrimeric G protein exhibit different sensitivities to Tm-induced growth inhibition. Homozygous gpa1-4 (Gα-null mutation), agb1-2 (Gβ-null mutation), and gpa1-4/agb1-2 double mutants were germinated on agar medium containing 0.3 μg/ml Tm for 6 days and then transferred to medium without Tm, allowed to recover for 10 days, and photographed.
Leaves of older wild-type Col-0 plants infiltrated subepidermally with Tm show large areas of leaf senescence and cell death 6–8 days after injection (Fig. 2). In contrast, leaves of agb1-2 plants show small areas of damage (Fig. 2A). Close-up views of injected wild-type leaves show large areas of tissue collapse and cell death by 8 days after injection (Fig. 2B), whereas agb1-2 mutant leaves show small areas of damage confined to the immediate vicinity of the injection site. Trypan blue staining to detect cell death also revealed much larger areas of dead cells in wild-type than in agb1-2 mutant leaves (Fig. 2B). Similar results were obtained with leaves of plants homozygous for a second mutant allele, agb1-1, of the gene coding for the Gβ subunit of the heterotrimeric G protein (data not shown) (28).
Fig. 2.
Leaf senescence and cell death in wild-type and agb1-2 mutant plants. A 15 μg/ml Tm solution in 1.6% DMSO was infiltrated into leaves as described in Materials and Methods. Plants were photographed at 6 days after infiltration (A), and leaves were photographed at 8 days (B Left) or cleared and stained with trypan blue (B Right); leaves were harvested for determination of chlorophyll content at 8 days (C) or daily (D) to determine electrolyte release as described in Materials and Methods.
Tm-treated wild-type leaves exhibit much more marked loss of chlorophyll, a leaf senescence indicator, than do leaves of agb1-2 mutant plants (Fig. 2C). Cell damage was also quantified by measuring the release of intracellular electrolytes to the extracellular space. Wild-type plants show an increase in electrolyte release a day after Tm treatment, recover transiently, and then show a progressive increase in the release of cellular electrolyte content as cells die (Fig. 2D). Homozygous agb1-2 mutant plants exhibit little evidence of enhanced ion leakage after Tm administration. Thus, mutant plants lacking the Gβ subunit of heterotrimeric G protein are more resistant to Tm-induced leaf cell death than wild-type plants, suggesting that the Gβ subunit plays a role in the cell death response induced by ER stress.
Transcriptional and Translational Responses to Tm Are Muted in Gβ Mutants.
The abundance of several characteristic ER stress markers was followed in wild-type and agb1-2 mutant plants after Tm infiltration. As shown in Fig. 3A, the abundance of BiP transcripts, particularly that for BiP3, increases rapidly, peaking at ≈3 h after Tm infiltration. PDI mRNA also increases in abundance, but peaks somewhat later at 6 h after Tm infiltration. Both BiP and PDI mRNA levels decline somewhat at 9 h and increase again at 12 h. Although both BiP and PDI transcript levels increase in agb1-2 mutant plants after Tm infiltration, they increase more slowly than in wild-type plants and do not reach the levels observed in wild-type plants. Moreover, BiP proteins do not accumulate to the same levels in the agb1-2 mutants as in wild-type plants (Fig. 3B). Large aggregates migrating more slowly than the BiP monomers can be detected in extracts from wild-type plants but are much less prominent in agb1-2 mutant plants. Thus, the transcriptional and translational responses to Tm-induced ER stress is much less marked in agb1-2 mutant plants than in wild-type plants.
Fig. 3.
The Tm-induced UPR is attenuated in agb1-2 mutant plants. (A) Northern blot analyses of Col-0 and agb1-2 mutant plants treated with 15 μg/ml Tm. Total RNA was extracted at the indicated times, and 10 μg of total RNA was loaded into each lane. UPR marker probes are indicated on the right. 25S rRNA was used as an internal loading control. (B) Western blot analysis of Col-0 and agb1-2 mutant plants treated as in A. Total protein was extracted at the indicated times, and 30 μg of protein was loaded into each lane. Antibodies against AtBiP1 and 2 were used to probe the membrane.
Gβ Protein Is in the ER and Cofractionates with BiP.
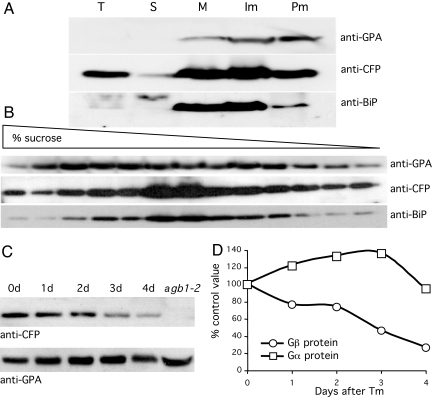
Heterotrimeric G proteins are generally assumed to carry out their signaling function in the plasma membrane by virtue of their association with G protein-coupled receptors (29). In animal cells, Gβγ heterodimers are assembled and modified in the ER and then move to the plasma membrane by virtue of their association with the Gα subunit (30, 31). In rice, Gβγ complexes have been reported to occur both as free 60-kDa heterodimers, as well in association with Gα proteins in ≈400-kDa complexes (32). In the present experiments, the location of the Arabidopsis Gβ protein was determined by subcellular fractionation of leaf tissue from plants stably transformed with a cyan fluorescent protein (CFP)-Gβ fusion construct expressed from the Gβ promoter. The microsomal fraction of cell homogenates was collected by ultracentrifugation and further separated by aqueous two-phase partitioning into plasma membrane and intracellular membrane fractions (32–34). Both Gα and Gβ proteins are enriched in the microsomal fraction (Fig. 4A). The Gα protein partitions relatively equally between the plasma membrane and intracellular membrane fractions, whereas a majority of the Gβ protein is associated with intracellular membranes, primarily ER, identified by using anti-BiP antibodies (Fig. 4A). The intracellular membrane fraction was subjected to sucrose density gradient centrifugation, aliquots of which were then analyzed by PAGE and Western blotting. The Gβ protein cosediments predominantly with the ER marker BiP, although a significant fraction of it cosediments with the Gα protein as well (Fig. 4B). Gα protein is detected throughout the gradient but shows peaks that sediment both more rapidly and more slowly than the bulk of the Gβ protein. Thus, a significant fraction of the intracellular Gβ protein is ER-associated in Arabidopsis, suggesting that it may serve a separate signaling function in the ER.
Fig. 4.
Gβ protein colocalizes with BiP in the ER and is degraded during the UPR. (A) Aliquots of total cell extracts (T) and soluble (S) supernatant and pelleted microsomal (M) fractions as well the upper plasma membrane (Pm) fraction and lower intracellular membrane (Im) fractions after aqueous two-phase partitioning were analyzed together by PAGE and Western blotting using anti-GPA antibodies to detect Gα protein, anti-CFP antibodies to detect the CFP-Gβ fusion protein in extracts of transgenic plants expressing a Gβ-CFP fusion protein from the native Gβ promoter, and anti-BiP antibodies for the ER markers BiP1 and 2. (B) The Im fraction was sedimented through a 10–50% sucrose density gradient, and aliquots of each fraction were analyzed by PAGE and Western blotting as in A. (C) Gβ and Gα proteins were detected as in A in total extracts from Gβ-CFP transgenic plants injected with Tm at 30 μg/ml and sampled after 1–4 days. A comparable protein extract from agb1-2 mutant plants was used in the last lane and shows that the Gα protein is expressed in the absence of the Gβ protein. (D) Band intensities from C are expressed as a percentage of control values.
Gβ Protein Is Degraded During the UPR.
The amount of Gβ protein declines to roughly 25% of its initial value by 4 days after infiltration of leaves with Tm (Fig. 4 C and D). In contrast, the amount of Gα protein remains relatively unchanged, even transiently increasing somewhat. Thus, consistent with its ER localization, Gβ protein appears to be subject to ERAD during the Tm-induced UPR. It should be noted that the Gα protein is present in extracts of agb1-2 mutant plants, consistent with previous reports of separable functions for the Gα and Gβ subunits of the Arabidopsis heterotrimeric G protein (26).
Discussion
We have presented evidence that implicates the Gβ subunit of the single Arabidopsis heterotrimeric G protein in triggering UPR-associated cell death in response to treatment with the protein glycosylation inhibitor Tm. Homozygous Gβ-null mutants are markedly more resistant to growth inhibition by Tm than either wild-type or Gα-null mutants when germinated on Tm-containing medium. Leaves of older Gβ mutant plants show less cell death when infiltrated with Tm than wild-type plants. The transcriptional response of Gβ mutant plants to Tm is less pronounced than that of wild-type plants, as is the accumulation of the BiP chaperone proteins. A majority of the Arabidopsis Gβ protein is associated with ER membranes and cofractionates with membrane-associated ER lumenal BiP markers. Consistent with its ER localization, Gβ protein is degraded during the UPR, whereas Gα protein is not. Taken together, these observations imply that the Gβ protein, which forms a stable heterodimer with the Gγ subunit in plants as it does in animals (32), is involved in the signaling events that trigger UPR-associated cell death.
Previous studies have shown that the Arabidopsis heterotrimeric G protein is involved in development as well as in oxidative stress and hormone-mediated stress responses (26, 27, 35). The results of the present study establish a connection between heterotrimeric G protein signaling and the UPR. Several studies have reported that ER stress-related proteins, including chaperones, BiP, and p58IPK are important in plant responses to viral and bacterial pathogens (16, 21, 36), but although a phospholipase D and the small GTPase Rac1 have been identified as downstream effectors of the Gα subunit (25, 37), no downstream targets of Gβγ signaling have yet been identified in plants.
In animal systems, inactive Gα-GDP/Gβγ heterotrimers associate with G protein-coupled receptors in the plasma membrane. Upon agonist binding, the G protein-coupled receptor promotes the dissociation of the heterotrimer and exchange of GDP for GTP, releasing active Gα-GTP and Gβγ to initiate signaling through a variety of effectors (29). Among its several signaling functions, Gβγ is a key component of phosphoinositide signaling in mammalian cells by virtue of its ability to interact with and activate phospholipase C (PLC) β2 and PLCβ3 to generate the signaling molecules diacylglycerol and inositol (1,4,5)-triphosphate (IP3) (38, 39). IP3 interacts with receptors in the ER to activate Ca2+ release (40). Gβγ-stimulated, IP3-mediated Ca2+ release from the ER contributes to normal physiological processes and stress responses, as well as apoptotic and necrotic cell death triggered by a variety of agents (41). PLC activation, IP3 production, and Ca2+ spikes are also among the earliest events in plant responses to pathogens, abiotic stress, and the stress hormone abscisic acid (42). A recent study provides evidence that a brief, transient burst of IP3 generated by PLC is one of the earliest signals arising from the recognition by a resistant host of a plant pathogen's avirulence protein (43). Heat shock of Arabidopsis tissue culture cells likewise elicits a brief transient IP3 burst that peaks within 3 min and can be suppressed by the PLC inhibitor U73122 (44). Thus, it is a reasonable conjecture that PLC or ER IP3 receptors are Gβγ complex effectors in plants.
Despite the presence of the Gβγ complex in the ER (31), the possibility that signals originate in the ER has not been explored. The simplicity of the G protein family in plants and the relatively mild phenotypic effect of Gα- and Gβ-null mutations make Arabidopsis an excellent system in which to examine the question of Gβγ signaling in the ER. The observation that Gβ-null mutant plants are Tm resistant but Gα-null mutant plants are not implies that the contribution of the Gβγ complex to activating the cell death program in response to Tm does not depend on either the formation or the function of the Gαβγ heterotrimer. The finding that a majority of the Gβ protein is in the ER even in wild-type Arabidopsis plants provides some evidence, albeit circumstantial, that it may function in the ER. Several observations lend credence to the concept that the Gβγ complex functions in the ER in mammalian cells, as well. Gβγ and the Gβ-like protein RACK1 bind directly to IP3 receptors to modulate their activity (45, 46). Because IP3 receptors are primarily in ER and other membranes, such as the Golgi, surrounding Ca2+ stores (40), these are the most likely sites of physical interactions between Gβγ and IP3 receptors. In addition, the conventional view that G protein signaling is confined to the plasma membrane has begun to expand with evidence that Ras signaling occurs both in the plasma membrane and in the ER and Golgi (47). The concept that signaling interactions occur in intracellular compartments is particularly compatible with recent reports identifying intracellular plant hormone receptors, including the ER-localized ethylene receptor (34).
Materials and Methods
Plant Materials, Growth Conditions, and Tm Treatment.
We used Arabidopsis thaliana Col-0 plants, homozygous agb1-1, agb1-2, gpa1-4, and agb1-2/gpa1-4 double mutants, all characterized as transcript-null mutants in the Col-0 background (28, 48). Plants were grown in MetroMix 200 (Scotts–Sierra Horticultural Products Company, Marysville, OH) in 5-cm pots at 65% humidity under fluorescent light at 30 W/m2 per s with a 14-h light/10-h dark photoperiod for 5 weeks. Five rosette leaves of similar size were infiltrated subepidermally on the lower surface with 15 or 30 μg/ml Tm (Sigma–Aldrich, St. Louis, MO) in 1.6% DMSO using a needleless syringe. Controls were infiltrated with the same DMSO solution lacking Tm.
Quantification of Leaf Senescence and Cell Death.
Chlorophyll degradation, an indicator of leaf senescence, was measured 8 days after Tm injection as described (49). The chlorophyll measurement was normalized to the wet leaf weight. Release of electrolytes was used to quantify cell damage and death (50). To measure electrolyte release, five injected leaves from a single plant were collected each day after inoculation and shaken in 15 ml of distilled water at 100 rpm for 4 h at room temperature. The conductivity of the wash solution (millisiemens per centimeter) was determined by using a portable conductivity meter (Control Company, Friendswood, TX). The total electrolyte content was obtained by measuring the same leaf-containing solution after autoclaving. Six replicates were averaged to calculate the fraction of electrolytes released as a fraction of the total electrolyte content. Relative electrolyte leakage is the ratio of the fraction obtained with leaves of treated plants to that obtained with leaves from untreated control plants. Cell death was detected histochemically by trypan blue staining as described (51).
RNA Isolation and Northern Blot Hybridization.
Leaves were flash-frozen in liquid N2. Total RNA was isolated by using TRIzol reagent (Invitrogen, Carlsbad, CA). Ten micrograms of total RNA from control and Tm-injected leaf tissue was fractionated on a 1.2% agarose/0.4 M formaldehyde gel and transferred to Hybond N+ nylon membrane (Amersham Pharmacia, Piscataway, NJ). Membranes were stained with methylene blue to visualize rRNA as loading controls. Probes were cloned from A. thaliana Col-0 cDNA, which was made as previously described (52), with the following gene-specific primers: BiP3, 5-ATGATTTTTATCAAGGAA AACACAGCG-3 and 5-TTACCAAGGGCTTTGTGATCC; BiP1 and 2, 5-GGCTCGCTCGTTTGGAGCAAA-3 and 5-CCGTTATCAATGGTCAAGACACT-3; and PDI, 5-ATG GCGAAATCTCAGATCTGGTT-3 and 5-GTCCCTGCTGG TCCCAGATTT-3. The probes were labeled by using the ReadyPrime DNA labeling kit (Amersham Pharmacia) with [α-32P]dCTP (MP Biomedicals, Irvine, CA). Blots were hybridized and washed according to the manufacturer's instructions (Sigma–Aldrich).
Cell Fractionation.
Five-week-old transgenic plants containing a CFP-Gβ fusion protein expressed from the AGB1 promoter were homogenized by using buffer containing 50 mM Tris-Mes, (pH 7.5), 1 mM DTT, 5 mM EDTA, 0.3 M sucrose, and protease inhibitors. The homogenate was filtered through a double layer of Miracloth (EMD Biosciences, La Jolla, CA) and centrifuged at 3,200 × g for 10 min (JA 20; Beckman Coulter, Fullerton, CA). The supernatant was further pelleted by centrifugation at 100,000 × g (TY70 Ti; Beckman Coulter) to obtain microsomal membranes, which were then subjected to aqueous two-phase partitioning (33, 34). The microsomal pellet was suspended in a solution containing 0.3 M sucrose, 3 mM KCl, and 5 mM KH2PO4 and adjusted to 6.4% (wt/wt) each of Dextran T500 and PEG3350. The phases were separated by centrifugation at 750 × g (IEC 243; Thermo Scientific, Waltham, MA), and the phase partitioning was repeated three times. The upper and lower phases were diluted separately with buffer containing 0.3 M sucrose, 3 mM KCl, and 1 mM EDTA and centrifuged at 100,000 × g (TY70 Ti; Beckman Coulter) to obtain plasma membrane and intracellular membrane fractions. The plasma membrane fraction was dissolved in SDS/PAGE sample buffer for analysis, and the intracellular membrane fraction was resuspended in buffer containing 25 mM Tris (pH 7.5), 10% sucrose, 1 mM DTT, 2 mM EDTA, and protease inhibitors and sedimented through a 10–50% sucrose density gradient at 100,000 × g for 16 h (SW28 3707; Beckman Coulter). Fractions (0.8 ml) were collected, concentrated using StrataClean Resin (Stratagene, Cedar Creek, TX), and dissolved in SDS/PAGE sample buffer for analysis. Isolated membrane fractions were loaded on a 10% polyacrylamide discontinuous gel (Mini Electrophoresis System; Bio-Rad, Hercules, CA). After electrophoresis, proteins were transferred to Hybond-P PVDF membrane (Amersham Biosciences, Piscataway, NJ) using a Mini TransBlot electrophoretic transfer cell (Bio-Rad). Immunoblotting was performed as described below.
Protein Analysis.
Total proteins were extracted using extraction buffer [50 mM Tris·HCl, (pH 7.6), 1 mM EDTA, 1% Triton ×-100, 2% SDS, 5 mM DTT, and 1 mM PMSF] and quantified using a DC protein assay kit (Bio-Rad), which is compatible with Triton and SDS. Equal amounts of total membrane protein were loaded on a 12% polyacrylamide discontinuous gel (Mini Electrophoresis System; Bio-Rad). After electrophoresis, proteins were transferred to Hybond-P PVDF membrane (Amersham Biosciences) using a Mini TransBlot electrophoretic transfer cell (Bio-Rad). Immunoblotting was performed with goat polyclonal anti-BiP antibodies (1:250; Santa Cruz Biotechnology, Santa Cruz, CA) and horseradish peroxidase-conjugated anti-goat IgG antibodies (Santa Cruz Biotechnology) for BiP protein analysis. The immunoblot was incubated with mouse monoclonal anti-green fluorescent protein antibodies (1:2,000; BD Biosciences, Mountain View, CA) and horseradish peroxidase-conjugated anti-mouse IgG antibodies for Gβ protein level measurement. The above blot was striped and probed with rabbit polyclonal anti-GPA antibodies (26) and horseradish peroxidase-conjugated anti-rabbit IgG antibodies (Invitrogen). Proteins were detected with ECL Plus protein gel blotting detection reagent (Amersham Biosciences) according to the manufacturer's instructions.
Acknowledgments
This work was supported by National Science Foundation Grant MCB-0447506 and endowment funds supporting the Willaman Professorship (to N.F.).
Abbreviations
- CFP
cyan fluorescent protein
- ER
endoplasmic reticulum
- ERAD
ER-associated protein degradation
- IP3
inositol (1,4,5)-triphosphate
- PR
pathogenesis-related
- PDI
protein disulfide isomerase
- PERK
PKR-like ER kinase
- PLC
phospholipase C
- Tm
tunicamycin
- UPR
unfolded protein response
- XBP1
X-box binding protein 1.
Footnotes
The authors declare no conflict of interest.
References
- 1.Schroder M, Kaufman RJ. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 2.Kim R, Emi M, Tanabe K, Murakami S. Apoptosis. 2006;11:5–13. doi: 10.1007/s10495-005-3088-0. [DOI] [PubMed] [Google Scholar]
- 3.D'Amico L, Valsasina B, Daminati MG, Fabbrini MS, Nitti G, Bollini R, Ceriotti A, Vitale A. Plant J. 1992;2:443–455. doi: 10.1111/j.1365-313x.1992.00443.x. [DOI] [PubMed] [Google Scholar]
- 4.Ceriotti A, Duranti M, Bollini R. J Exp Bot. 1998;49:1091–1103. [Google Scholar]
- 5.Sparvoli F, Faoro F, Daminati MG, Ceriotti A, Bollini R. Plant J. 2000;24:825–836. doi: 10.1046/j.1365-313x.2000.00933.x. [DOI] [PubMed] [Google Scholar]
- 6.Kamauchi S, Nakatani H, Nakano C, Urade R. FEBS J. 2005;272:3461–3476. doi: 10.1111/j.1742-4658.2005.04770.x. [DOI] [PubMed] [Google Scholar]
- 7.Martinez IM, Chrispeels MJ. Plant Cell. 2003;15:561–576. doi: 10.1105/tpc.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirst ME, Meyer DJ, Gibbon BC, Jung R, Boston RS. Plant Physiol. 2005;138:218–231. doi: 10.1104/pp.105.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosti P, Malerba M, Bianchetti R. Protoplasma. 2001;216:31–38. doi: 10.1007/BF02680128. [DOI] [PubMed] [Google Scholar]
- 10.Iwata Y, Koizumi N. Planta. 2005;220:804–807. doi: 10.1007/s00425-004-1479-z. [DOI] [PubMed] [Google Scholar]
- 11.Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ. Plant Physiol. 2001;127:949–962. [PMC free article] [PubMed] [Google Scholar]
- 12.Noh SJ, Kwon CS, Chung WI. Biochim Biophys Acta. 2002;1575:130–134. doi: 10.1016/s0167-4781(02)00237-3. [DOI] [PubMed] [Google Scholar]
- 13.Okushima Y, Koizumi N, Yamaguchi Y, Kimata Y, Kohno K, Sano H. Plant Cell Physiol. 2002;43:532–539. doi: 10.1093/pcp/pcf063. [DOI] [PubMed] [Google Scholar]
- 14.Chang LY, Yang WY, Browning K, Roth D. Plant Mol Biol. 1999;41:363–370. doi: 10.1023/a:1006379623534. [DOI] [PubMed] [Google Scholar]
- 15.Chang LY, Yang WY, Roth D. Biochem Biophys Res Commun. 2000;279:468–474. doi: 10.1006/bbrc.2000.3964. [DOI] [PubMed] [Google Scholar]
- 16.Bilgin DD, Liu Y, Schiff M, Dinesh-Kumar SP. Dev Cell. 2003;4:651–661. doi: 10.1016/s1534-5807(03)00125-4. [DOI] [PubMed] [Google Scholar]
- 17.Iwata Y, Koizumi N. Proc Natl Acad Sci USA. 2005;102:5280–5285. doi: 10.1073/pnas.0408941102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Kaufman RJ. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 19.Zhao L, Ackerman SL. Curr Opin Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Van Loon LC, Van Strien EA. Physiol Mol Plant Pathol. 1999;55:85–97. [Google Scholar]
- 21.Wang D, Weaver ND, Kesarwani M, Dong X. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- 22.Grimmig B, Gonzalez-Perez MN, Leubner-Metzger G, Vogeli-Lange R, Meins F, Jr, Hain R, Penuelas J, Heidenreich B, Langebartels C, Ernst D, et al. Plant Mol Biol. 2003;51:599–607. doi: 10.1023/a:1022385104386. [DOI] [PubMed] [Google Scholar]
- 23.Stintzi A, Heitz T, Prasad V, Wiedemann-Merdinoglu S, Kauffmann S, Geoffroy P, Legrand M, Fritig B. Biochimie. 1993;75:687–706. doi: 10.1016/0300-9084(93)90100-7. [DOI] [PubMed] [Google Scholar]
- 24.Wang XQ, Ullah H, Jones AM, Assmann SM. Science. 2001;292:2070–2072. doi: 10.1126/science.1059046. [DOI] [PubMed] [Google Scholar]
- 25.Suharsono U, Fujisawa Y, Kawasaki T, Iwasaki Y, Satoh H, Shimamoto K. Proc Natl Acad Sci USA. 2002;99:13307–13312. doi: 10.1073/pnas.192244099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joo JH, Wang S, Chen JG, Jones AM, Fedoroff NV. Plant Cell. 2005;17:957–970. doi: 10.1105/tpc.104.029603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perfus-Barbeoch L, Jones AM, Assmann SM. Curr Opin Plant Biol. 2004;7:719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Lease KA, Wen J, Li J, Doke JT, Liscum E, Walker JC. Plant Cell. 2001;13:2631–2641. doi: 10.1105/tpc.010315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS. Cell Mol Life Sci. 2005;62:551–577. doi: 10.1007/s00018-004-4462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michaelson D, Ahearn I, Bergo M, Young S, Philips M. Mol Biol Cell. 2002;13:3294–3302. doi: 10.1091/mbc.E02-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takida S, Wedegaertner PB. J Biol Chem. 2003;278:17284–17290. doi: 10.1074/jbc.M213239200. [DOI] [PubMed] [Google Scholar]
- 32.Kato C, Mizutani T, Tamaki H, Kumagai H, Kamiya T, Hirobe A, Fujisawa Y, Kato H, Iwasaki Y. Plant J. 2004;38:320–331. doi: 10.1111/j.1365-313X.2004.02046.x. [DOI] [PubMed] [Google Scholar]
- 33.Basboa O, Das J, Sharma CB. Indian J Biochem Biophys. 1987;24(Suppl):24–28. [PubMed] [Google Scholar]
- 34.Chen YF, Randlett MD, Findell JL, Schaller GE. J Biol Chem. 2002;277:19861–19866. doi: 10.1074/jbc.M201286200. [DOI] [PubMed] [Google Scholar]
- 35.Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, Botella JR. Plant Physiol. 2006;140:210–220. doi: 10.1104/pp.105.069625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jelitto-Van Dooren EP, Vidal S, Denecke J. Plant Cell. 1999;11:1935–1944. doi: 10.1105/tpc.11.10.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishra G, Zhang W, Deng F, Zhao J, Wang X. Science. 2006;312:264–266. doi: 10.1126/science.1123769. [DOI] [PubMed] [Google Scholar]
- 38.Bonacci TM, Ghosh M, Malik S, Smrcka AV. J Biol Chem. 2005;280:10174–10181. doi: 10.1074/jbc.M412514200. [DOI] [PubMed] [Google Scholar]
- 39.McCullar JS, Malencik DA, Vogel WK, Crofoot KM, Anderson SR, Filtz TM. Biochem Pharmacol. 2007;73:270–278. doi: 10.1016/j.bcp.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson RL, Boehning D, Snyder SH. Annu Rev Biochem. 2004;73:437–465. doi: 10.1146/annurev.biochem.73.071403.161303. [DOI] [PubMed] [Google Scholar]
- 41.Berridge MJ. Cell Calcium. 2002;32:235–249. doi: 10.1016/s0143416002001823. [DOI] [PubMed] [Google Scholar]
- 42.Lecourieux D, Ranjeva R, Pugin A. New Phytol. 2006;171:249–269. doi: 10.1111/j.1469-8137.2006.01777.x. [DOI] [PubMed] [Google Scholar]
- 43.Andersson MX, Kourtchenko O, Dangl JL, Mackey D, Ellerstrom M. Plant J. 2006;47:947–959. doi: 10.1111/j.1365-313X.2006.02844.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu HT, Gao F, Cui SJ, Han JL, Sun DY, Zhou RG. Cell Res. 2006;16:394–400. doi: 10.1038/sj.cr.7310051. [DOI] [PubMed] [Google Scholar]
- 45.Zeng W, Mak DO, Li Q, Shin DM, Foskett JK, Muallem S. Curr Biol. 2003;13:872–876. doi: 10.1016/s0960-9822(03)00330-0. [DOI] [PubMed] [Google Scholar]
- 46.Patterson RL, van Rossum DB, Barrow RK, Snyder SH. Proc Natl Acad Sci USA. 2004;101:2328–2332. doi: 10.1073/pnas.0308567100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Philips MR. Biochem Soc Trans. 2005;33:657–661. doi: 10.1042/BST0330657. [DOI] [PubMed] [Google Scholar]
- 48.Ullah H, Chen JG, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM. Plant Cell. 2003;15:393–409. doi: 10.1105/tpc.006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnon DI. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker CJ, O'Neill NR, Keppler LD, Orlandi EW. Phytopathology. 1991;81:1504–1507. [Google Scholar]
- 51.Pasqualini S, Piccioni C, Reale L, Ederli L, Della Torre G, Ferranti F. Plant Physiol. 2003;133:1122–1134. doi: 10.1104/pp.103.026591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han M-H, Goud S, Song L, Fedoroff N. Proc Natl Acad Sci USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]