Abstract
The colonic crypt is the functional unit of the colon mucosa with a central role in ion and water reabsorption. Under steady-state conditions, the distal colonic crypt harbors a single stem cell at its base that gives rise to highly proliferative progenitor cells that differentiate into columnar, goblet, and endocrine cells. The role of c-Myb in crypt homeostasis has not been elucidated. Here we have studied three genetically distinct hypomorphic c-myb mutant mouse strains, all of which show reduced colonic crypt size. The mutations target the key domains of the transcription factor: the DNA binding, transactivation, and negative regulatory domains. In vivo proliferation and cell cycle marker studies suggest that these mice have a progenitor cell proliferation defect mediated in part by reduced Cyclin E1 expression. To independently assess the extent to which c-myb is required for colonic crypt homeostasis we also generated a novel tissue-specific mouse model to allow the deletion of c-myb in adult colon, and using these mice we show that c-Myb is required for crypt integrity, normal differentiation, and steady-state proliferation.
Keywords: colon, hypomorphs, A33, stem cells, p27
Colonic crypts are exquisite structures that generate a vast and continuous epithelial surface allowing efficient water and ion reabsorption. Crypt cells are rapidly renewed, consisting of comparable numbers of columnar cells and mucin-producing goblet cells and fewer enteroendocrine cells. Mouse colonic crypts consist of ≈500 cells, of which stem cells located at the base of the crypt give rise to a dividing population that differentiate into the three lineages (1). This process takes 4–7 days, during which progenitor cells proliferate and progressively differentiate while migrating toward the top of the crypts where many of the cells undergo apoptosis (2). During development, differentiation of the murine intestine occurs in a proximal to distal wave between embryonic day 15 (E15) and E19. Crypts continue to lengthen and divide by fission until 28 days postnatal, after which time homeostasis is attained (3).
The rapid expansion of multiple lineages building up to homeostasis in the adult parallels processes in the hematopoietic system where stem cells give rise to progenitor cells and finally terminally differentiated cells. Colonic crypts offer an attractive model in which to study stem and progenitor cells because these cells occupy discrete positions within crypts. Specifically, distal colonic crypt stem cells reside at the crypt base from where progenitor cells arise. In the proximal colon, stem cells are found several cells above the crypt base, in parallel to that observed in small intestinal crypt-villi structures. Thus, understanding these apparently simple structural features affords the opportunity to identify defects in progenitor cell replacement and turnover and has been used in numerous examples of mutant and gene-knockout mouse lines.
c-myb is expressed early in the development of colonic mucosa and persists in the adult colonic epithelia in both mouse and human (4). We used c-myb−/− embryos (5) to indirectly investigate the role of c-myb in colon development (6). The effect of the c-myb deletion on intestinal development could not be assessed in situ because mice with targeted disruptions of both c-myb alleles die at E15 because of a severe defect in fetal liver hematopoiesis (5). However, dissecting colon and small intestine from c-myb−/− embryos at E14 and transplanting the tissue under the kidney capsule of recipient adult mice allowed the development of these tissues to proceed. We found that colon tissue from the c-myb−/− embryos developed with profound epithelial disorganization such that normal crypts failed to form (6).
In contrast to c-myb−/− embryos, c-myb+/− mice develop normally, but under cytotoxic stress produced by treatment with sublethal to lethal doses of cytotoxic drugs or radiation, profound differences from the wild-type response are evident (7). As such, this model has been a powerful means of revealing a biological role for c-Myb in the colon. Thus, we have found that both alleles of c-myb are required for crypt survival and recovery. In this study we have investigated a unique series of mouse mutants that affect the three key domains of the c-Myb protein as well as a novel tissue-specific inducible c-myb knockout model to show that c-myb is essential to normal colonic crypt proliferation and architectural integrity in adult mice.
Results
Fully Functional c-Myb Is Required for Normal Colonic Crypt Length.
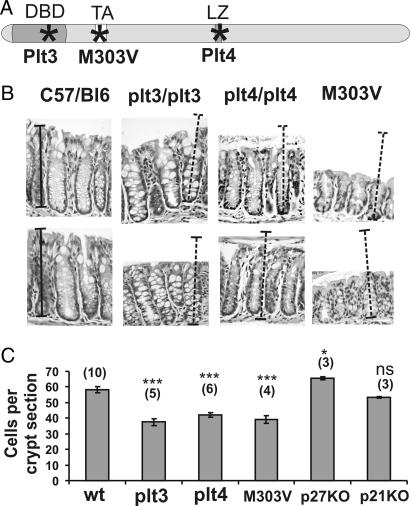
Three genetically distinct mouse lines with mutations in the c-myb locus have been generated in two separate studies after saturation mutagenesis with ENU (8, 9). These mice have mutations in the three well characterized functional domains of the c-Myb protein, the DNA binding domain (Plt3), transactivation domain (M303V), and negative regulatory domain/leucine-rich motif (Plt4) noted in Fig. 1A. The c-Myb proteins encoded by these mutant loci have suboptimal transactivation capacity (8, 9).
Fig. 1.
Hypomorphic c-myb mutant mice have shorter crypts than wild-type mice. (A) Murine c-Myb diagram marking (∗) the location of amino acid substitutions within the DNA binding domain (Plt3), the leucine zipper region (Plt4) (8), and the transactivation domain (M303V) (9). (B) H&E-stained sections of wild-type, plt3/plt3, plt4/plt4, and M303V/M303V distal colons show that the mutant crypts are shorter than wild type. Relative length size bars are shown layered over normal distal colonic crypts, and these have been transferred to panels representing the three hypomorphs. (C) Morphometric analysis of the sections indicates that the hypomorphic mutant crypts are significantly shorter than wild-type C57BL/6 crypts whereas p27−/− crypts are significantly longer. p21−/− crypts are indistinguishable from wild type. Bars represent mean ± SEM (number of mice analyzed). ∗∗∗, P > 0.0001 (ANOVA).
When longitudinal sections of colonic crypts were examined from each of the three hypomorphic mutants their reduced length was immediately obvious. Fig. 1B shows that, compared with wild-type distal colonic crypts, c-mybPlt3/plt3, c-mybPlt4/plt4, and c-mybM303/M303V crypts retain overall crypt architecture but are of reduced length and display an apparent excess of goblet cells compared with that observed in wild-type crypts as evidenced by the enlarged vacuoles after H&E staining.
Morphometric analysis showed that all three hypomorphic mutant mice had significantly reduced cell numbers per crypt compared with wild-type C57BL/6 mice (P < 0.0001, ANOVA; for each hypomorph) (Fig. 1C). When crypts were examined for spontaneous apoptosis there was no indication that increased cell death could account for the shortened crypt length as assessed by the presence of apoptotic bodies or activated caspase-3-positive cells (data not shown). Therefore, these observations suggested that there may be a role for c-myb in driving proliferation of colonic crypts.
To highlight the role of proliferation regulators in maintaining crypt length, we also examined the impact on crypt length when two negative regulators of growth were deleted. We first examined p27 because its expression appears to be a reciprocal to the high c-myb expression observed at the crypt base (4, 10) where p27 expression is low at the base and increases toward the colon lumen (11). Second, we examined p21 as there is an inverse relationship between c-myb expression and p21 expression during colon cell differentiation (12). To investigate whether the loss of expression of these genes had an effect on crypt length, cells per longitudinal section were quantified. Histological examination of p27−/− crypts showed they were significantly longer than wild type when sections were observed and this was confirmed by counting crypt cells in longitudinal sections (P = 0.05, ANOVA). In contrast, p21−/− crypts were indistinguishable from normal controls but markedly different to p27−/− crypts (P = 0.0004, ANOVA).
Disrupted Differentiation and Retarded Proliferation in Hypomorphic Crypts.
The observed defects in colonic crypt morphology raised the prospect that the crypts in hypomorphic c-myb mutant mice had a defect in cytodifferentiation and/or cell proliferation. To test this, sections were stained with periodic acid/Schiff reagent (PAS) that detects mucins thereby identifying goblet cells, one of the two predominant cell types within the colonic crypt. PAS-positive cells were readily observed in wild-type, plt3/plt3 and plt4/plt4 crypts indicating that the goblet cell lineage was generated in the presence of c-myb hypomorphic mutations. However, there was a consistent trend toward over-representation of this cell type in crypts whereby PAS-positive cells were predominant in mutant versus wild-type colonic crypts. In contrast, p27−/− crypts appeared to have reduced PAS staining [supporting information (SI) Fig. 6A]. Reduced cell density and spatial disorder of cells at the crypt base was also apparent. Determining the actual frequency of goblet cells for each mutant was problematic because the boundaries of the goblet cell cytoplasm and released mucin are ill-defined. Nevertheless, when mucin content was indirectly assessed by using MetaMorph software that allows relative levels of PAS staining compared with hematoxylin staining of nuclei, it was determined that mucin content was significantly increased in two of the hypomorphic crypts, and thus goblet cells were implicitly over-represented (SI Fig. 6B). The third lineage generated within the colonic crypt is the enteroendocrine cell type that is readily detected by immunohistochemistry after staining sections with anti-ChromograninA antibody. This cell type, like columnar cells, was in deficit in the hypomorphic mutant colonic crypts (SI Fig. 7).
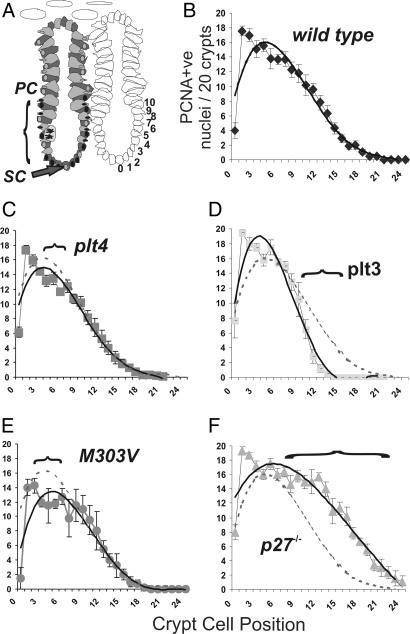
When sections were stained for proliferating cell nuclear antigen (PCNA) to assess cell proliferation it was evident that the three c-myb hypomorphs had fewer PCNA-positive cells when quantified by crypt position. Because the presumed stem cell is located at the very crypt apex of the distal colon this cell position was often PCNA-negative. For counting purposes such cells were allocated the zero cell position within the crypt (Fig. 2A). At least 20 crypts were assessed for PCNA stained nuclei enumerating positive cells on both sides of each crypt. When this was done plt4/plt4 and plt3/plt3 crypts showed significantly fewer PCNA-positive cells at positions 4–7 and 12–18, respectively (Figs. 2C and 3D), compared with wild type (Fig. 2B). In contrast M303V/M303V crypts showed significantly less PCNA closer to the base of the crypt (positions 2–7) (Fig. 2E). In accord with the significantly longer crypts in p27−/− mice the extent of PCNA staining was greater at positions 9–21 suggesting that in the absence of p27 proliferation is sustained for longer, as cells migrate to the luminal surface of the colon (Fig. 2F). Collectively these data imply that each hypomorphic c-myb mutant has a proliferative defect within the progenitor cell zone and perhaps these defects are in different positions within this zone.
Fig. 2.
Hypomorphic mutant crypts show reduced proliferation. (A) Model of a distal colonic crypt showing the presumed location of the stem cell (SC) and region of proliferation ascribed to the progenitor cell population (PC). The crypt cell position referenced to position zero represents the location of the presumptive stem cells. (B–F) When crypts were stained for proliferation marker PCNA, the extent and distribution of antigen-positive cells was significantly different in the plt3/plt3, plt4/plt4, M03V/M303V, and p27 knockout mice. Horizontal brackets demark where there are significant differences from wild-type crypts. c-myb hypomorphs show significantly less proliferation in these regions. The data also show that proliferation in the p27−/− crypts is more extensive than wild-type crypts, consistent with the longer crypts evident in these mice. Quadratic trend lines are shown to allow visual comparisons between the mutant and wild-type (hashed lined) crypts.
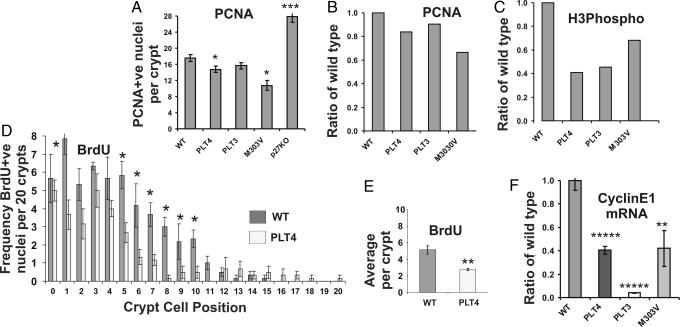
Fig. 3.
Proliferation markers indicate that hypomorphic c-myb mice have a defect in cell cycle progression. (A) Collectively, the total number of PCNA-positive cells per crypt for each strain of mice was significantly different for all mutants with the exception of PLT3 mutants. (B) To allow a further assessment of a potential defect in cell cycle progression each data set was analyzed as a ratio to wild type to show the reduced PCNA staining in mutants. (C) This type of analysis was extended to include phospho-histone-3 expression whereby it would appear that the ratios of this marker of G2/M phase cells are less than those observed for cycling PCNA-stained cells. (D and E) PLT4 mutant and wild-type mice were further assessed for the extent of BrdU staining on the basis of crypt position (D) and total per crypt (E). Both analyses suggested significantly less S-phase progression in PLT4 mutant mice compared with wild-type littermates. Bars represent mean ± SEM. ∗∗, P < 0.01; ∗, P < 0.05 (ANOVA). (F) Real-time RT-PCR studies on colonic crypt RNA isolated from wild-type (n = 6), PLT3 (n = 3), PLT4 (n = 3), and M303V (n = 3) mice show that Cyclin E1 expression is lower in these hypomorphic mutant mice, perhaps explaining the defect in cell cycle progression particularly at the crypt base.
c-myb Hypomorphic Crypts Show Different Defects in Proliferation.
From the proliferation profiles shown in Fig. 2 it would appear that the plt4 and plt3mutations affect the progenitor cell region of crypts while the M303V mutation influences proliferation closer to the crypt base where stem cells are located. Both plt4 and M303V mutants showed significantly fewer, and p27−/− mice had significantly more, PCNA-positive cells within the crypts when the total frequency was assessed (Fig. 3A). The relative PCNA ratio to wild type is also shown for each mutant (Fig. 3B) and when compared with Phospho-histone-3 staining, an antigen that marks cells within the M phase of the cell cycle, it was apparent that the c-myb hypomorphs were perturbed in their progression through the cell cycle (Fig. 3C).
Cell Cycle Progression Is Retarded in c-myb Hypomorphic Crypts.
To test the hypothesis that hypomorphic crypts had a defect in cell cycle progression, plt4/plt4 mice were injected with BrdU and killed 2 h later, and colon sections were prepared and then examined for the presence of cells in S-phase by immunohistochemistry with anti-BrdU antibodies. Fig. 3 D and E shows that there are significantly fewer cells in S-phase in plt4/plt4 distal colonic crypts when assessed by crypt position (positions 5–10) (P < 0.05, ANOVA) or by total positive cells per crypt (P = 0.01, ANOVA). Both distal and proximal (data not shown) crypts showed less BrdU incorporation. Collectively these results suggest that the colonic crypts in plt4/plt4 mice have a defect in cell cycle progression associated with shortened crypt length.
Cyclin E1 Expression Is c-Myb-Dependent.
To provide an explanatory mechanism for the defective cell cycle progression in hypomorphic mutant mice we examined the expression of various G1/S regulators in mouse embryonic fibroblasts (MEF) that serve as a useful cell type to study cell cycle progression because they are easily arrested by serum starvation and show synchronous cell cycle re-entry after serum (FCS) restoration. Under these conditions c-myb mRNA levels increase from a low base level within 30 min of FCS addition, while Cyclin E1 mRNA increased at 4 h. We next examined the effect on this process in c-myb−/− MEFs at 6 and 24 h after FCS restoration the most notable difference compared with wild-type MEFs was the reduced Cyclin E1 expression (data not shown). To link these in vitro data with the cell cycle defect observed in the plt3/plt3, plt4/plt4 and M303V/M303V mice, colonic crypts were isolated from three mutant mice per genotype and three matched littermate controls. RNA was extracted and subjected to real-time RT-PCR whereby a significant difference in Cyclin E1 was observed (Fig. 3F) (P < 0.007, ANOVA). These data suggest that one reason why c-myb hypomorphic mice have a defect in cell cycle progression is due in part to reduced Cyclin E1 expression.
Tissue-Specific, RU486-Inducible Cre-Dependent c-myb Deletion in Colon.
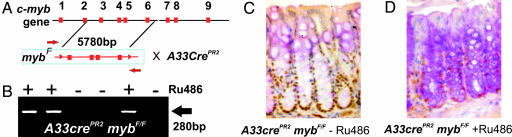
Several strategies are available for the tissue-specific deletion of genes flanked by loxP sites in mouse intestines mostly preferentially targeting the small intestine and were not considered ideal for asking the question about the role of c-myb in colonic crypt. Using a novel approach described here it has become possible to selectively target Cre expression to the distal colon and rectum to allow the study of c-myb deletion in adult mouse colon for the first time. This strategy employs an ORF encoding a Cre recombinase-progesterone receptor ligand domain fusion knocked into the intestine-specific A33 locus. The encoded protein is activated by the progesterone antagonist RU486. Most importantly this transgene shows faithful A33 promoter regulation of the Cre-recombinase (M.E., R.G.R., and J.K.H., unpublished data). The A33-Cre promoter configuration has been reported elsewhere (13) while the c-myb exons flanked by loxP sites maps the expected deletion of exons 3–6 described previously Fig. 4A (14). Mid colon and small intestine tissue sections were extracted for DNA and PCR analysis and results confirmed that c-myb locus-specific recombination had occurred. c-myb deletion in the colon tissues are shown in Fig. 4B using primer pairs diagrammed in Fig. 4A.
Fig. 4.
Tissue-specific conditional deletion of c-myb in distal colon leads to a reduction of proliferation in colonic crypts. (A) Schematic of the strategy for the conditional deletion of the c-myb locus (14) whereby exons 3–6 are excised after the activation of A33CrePR2 activity by RU486. (B) An image of an agarose gel with ethidium bromide-stained PCR products generated by using primers (marked by red arrows in A) indicated successful deletion of exons 3–6 in the presence of RU486 (+). Costaining of colonic crypts for c-Myb (C) and PAS (D) shows that the intensity of PAS staining (as an indicator of goblet cell differentiation) increases with the loss of c-Myb.
To document the recombination activity of the Cre-transgene, mice were crossed onto a Rosa26Cre-reporter mouse background. Mice were fed a paste of mouse chow and the anti-progesterone RU486 for up to 4 weeks. This route of administration has proven to be superior to daily intramuscular injection of the hormone in terms of recombination events in the colon (M.E., unpublished data, and data not shown). Sections were also stained for c-Myb protein expression to show antigen loss (Fig. 4 C and D) after prolonged ingestion of RU486. The most profound differences were the absence of c-Myb-positive cells at the crypt base.
The ablation of c-Myb expression was not absolute; nevertheless, regions of total c-Myb loss were extensive particularly in the distal colon (SI Fig. 8). Taken together with the observations that the loss of c-Myb was not absolute, that A33 expression is intestinal-restricted and that A33-driven recombination of the Ros-LacZ13 locus was absent in bone marrow and spleen it is unlikely that the colon defects reported here arose indirectly in response to c-Myb ablation in the hematopoietic system.
c-myb Deletion Leads to Reduced Proliferation and Disrupted Differentiation.
It was suggested above that crypts in the hypomorphic mutant mice had an over-representation of goblet cells. This distortion of differentiation was far more evident in the conditional deletion mutants. Furthermore, the expanded vacuoles associated with goblet cells appear to be a feature of the knockout crypts and as judged by PAS content there was significantly more mucin present in the c-Myb-deleted crypts (SI Fig. 8C). Typical examples of abnormal crypts present after recombination and disruption of the c-myb locus are shown Fig. 4 C and D.
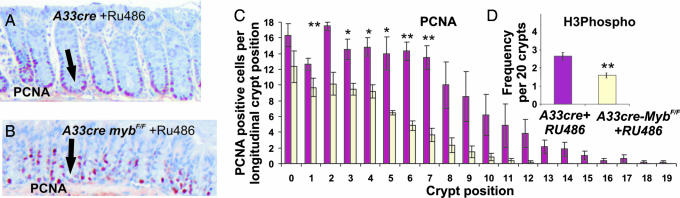
In view of the proliferation defects observed with the hypomorphic mutant mice colon sections A33cre × mybF/F mice plus RU486 were also stained for PCNA. Micrographs of colon sections stained for PCNA highlight the reduced proliferation after Cre-mediated recombination when compared with A33 transgenic mice alone (Fig. 5 A and B). Quantitation of PCNA staining in discernable crypts in mutant and control mice is shown for each crypt cell position, indicating that after Cre induction with RU486, the proportion of cells per crypt is significantly reduced (Fig. 5C). Crypt staining for phospho-histone-3 expression to assess cell cycle progression indicated that the crypts with deleted c-myb had fewer cells in G2/M phase of the cell cycle compared with control crypts (Fig. 5D).
Fig. 5.
c-myb is required for proliferation in colonic crypts. (A and B) Colon sections were stained for PCNA in nonrecombined colonic crypts (A) and in recombined crypts where reduced proliferation is evident (B). (C) Quantitation of PCNA staining in discernable crypts in nonrecombined and RU486-treated mice is shown for each crypt cell position, indicating that after Cre activation with RU486 the proportion of cells per crypt is significantly reduced. (D) Crypts stained for phospho-histone-3 to examine cell cycle progression into the G2/M phases of cell cycle indicate that the crypts with deleted c-myb had fewer cells in the G2/M phase of the cell cycle compared with unrecombined crypts. Bars represent mean ± SEM (n = 3 per treatment). ∗∗, P < 0.01; ∗, P < 0.05 (ANOVA).
Discussion
We have taken advantage of the unprecedented opportunity to employ mutant mice with specific c-myb mutations in the three key functionally defined domains to examine the effect of c-Myb functional loss in adult colonic crypts. The complete ablation of c-Myb function by germ-line deletion has until now precluded analysis of the role of c-myb in tissues other than the hematopoietic system (5). Indeed, the recent use of tissue-specific knockout and hypomorphic mutants has expanded the repertoire of approaches available for the analysis of c-Myb in adult mouse hematopoiesis (8, 9, 14–16). We further report here the tissue-specific ablation of c-myb in the colon showing a more severe phenotype than observed with the c-myb hypomorphs.
It would appear that c-myb mutations that affect the hematopoietic system in terms of progenitor cell production and lineage commitment also affect colonic crypt morphogenesis and homeostasis. The three hypomorphic mutants studied allow sufficient c-Myb functional activity to permit development beyond critical checkpoints like the ability to make sufficient definitive red blood cells and subsequent survival to adulthood. These biological defects do not appear to relate to mutations in any one functional domain. Perhaps the unifying basis for these mutations generating similar biological outcomes is due to disruptions of interactions that c-Myb has with other regulators of crypt morphology.
We found that in all three cases crypt lengths in distal colonic mucosa of c-myb hypomorphic mice were significantly shorter than wild type. Small intestine crypt and villus formation appear normal consistent with our previous studies (6) (data not shown). Moreover the proliferative activity of the colonic crypts was significantly reduced as measured by PCNA and phospho-histone-3 staining on the basis of overall proliferation and crypt cell position. Essentially plt3/plt3 and plt4/plt4 mutant mice showed premature termination of proliferation within the crypt by position 10 while the M303V/M303V mutant appears to have a defect in proliferation closer to the crypt base. These are indicative of either accelerated differentiation and/or retarded cell cycling. Defining the precise demarcation between stem and transit amplifying cells within the distal colonic crypt underpins an ongoing debate, however from the data presented here it is suggested that defective c-Myb function directly affects transit amplifying cell proliferation. Previous studies using irradiated and 5-Fluorouracil exposed c-myb heterozygous mice indicate that normal c-Myb function is also required for the replacement of transit amplifying cells and this need has to be met by stem cells (7). In contrast to shorter crypts in c-myb hypomorphs the deletion of the cell cycle regulator p27, but not p21, led to crypt lengthening. This 26% increase in length over normal controls may simply reflect the overall larger size (25%) manifested by the p27−/− mice reported elsewhere (17). Nevertheless, these data suggest that p27 and c-Myb are part of the network of cell cycle regulators that govern crypt homeostasis.
The role of c-myb in controlling differentiation in colon has to be considered based on the observations reported here and previous studies in mouse and human colon cancer cell lines where exogenous c-myb expression blocks cytodifferentiation (18). Consistent with this role is the observation that c-myb expression is found in immature columnar cells (4, 19) and reduced c-Myb function might be expected to allow an over-representation of goblet cells as well as mucin production. We also found that endocrine cell numbers as assessed by positive staining for ChromograninA were in deficit in the hypomorphic colonic crypts. The goblet defect in particular was most evident in the tissue-specific knockout crypts. As all three cell lineages arise from a single stem cell these differentiation data suggest that c-Myb may regulate the balanced differentiation in the colonic crypt.
Separate expression profiling studies in MEFs was consistent with Cyclin E1 being a c-Myb target and analysis of the mouse and human Cyclin E1 promoters identifies a number of high affinity c-Myb binding sites (data not shown). From the analysis of isolated colonic crypts from the hypomorphic mutant mouse crypts we propose that in colonic crypts Cyclin E1 is a direct c-Myb target gene required for cell cycle entry of stem and/or progenitor cells. Because the principle defect in the viable Cyclin E1 knockout is a failure to enter the cell cycle from quiescence (20), we suggest that a similar defect in cell cycle entry would account for some of the defects observed in the c-myb hypomorphic mutants described here.
Recently, another intestine-specific cre-mediated recombination mouse model has been used to delete c-myc expression, most particularly in the small intestine (21). This study unexpectedly showed that c-myc is required for small intestine crypt development but not homeostasis. In contrast, we show here that c-myb is required for homeostasis and, as previously suggested, c-myb is required for colonic crypt development but not small intestinal morphogenesis during embryogenesis (6). These contrasting observations are puzzling because c-myc is subject to c-myb regulation in reporter studies (22), but clearly the in vivo studies suggest that the regulation and role of both of these protooncogenes is distinct. It was also interesting to find that the deletion of c-myb expression as well as reactivation of the Rosa locus by A33-Cre recombination (data not shown) was incomplete within the colon. Rosa reactivation was not observed in bone marrow, spleen, lung, or liver (M.E., R.G.R., and J.K.H., unpublished data). Thus, the progressive and apparently stochastic c-Myb deletion in the crypts leads to reduced proliferation and distorted differentiation that is intrinsic to the colonic crypts.
Finally, the observations reported here might also be considered in the context that c-Myb is overexpressed in colon cancers (23), which may be due in rare cases to gene amplification (24) or mutations within the transcriptional attenuator region (25). Thus, the antithesis to the low c-myb expression and reduced proliferation in the mutant mice might be unrestricted proliferation in colon cancers in part driven by c-Myb.
Materials and Methods
Mice.
p27−/− (26) mice were maintained on a C57BL/6J background and housed in a specific pathogen-free Thoren racking system (Thoren Caging Systems, Hazelton, PA). p21−/− mice (27) were maintained on a mixed 129/C57BL/6J background and were a gift from David Vaux (The Walter and Eliza Hall Institute). C57BL/6J c-mybplt3/plt3 and c-mybplt4/plt4 mice (8) were also maintained at The Walter and Eliza Hall Institute. c-mybM303V/M303V mice (9) were housed at the Novartis Genomics Institute (San Diego, CA).
Conditional Deletion Construct and RU486.
To generate the intestine-specific c-myb mutants, we used mice in which c-myb exons 3–6 were flanked with loxP sites (14). Mice harboring two c-mybF alleles were crossed with transgenic mice possessing the Cre recombinase gene under the control of the endogenous A33 promoter (M.E., R.G.R., and J.K.H., unpublished data). The cre-cDNA was fused to a mutant progesterone ligand-binding domain to allow activation by RU486 (Mifepristone; Sigma. St. Louis, MO). RU486 was prepared in mouse chow (1.6 g/kg). Confirmation that the Cre was activated by RU486 was established by using Rosa reporter mouse and by deletion-specific c-myb PCR (14).
BrdU Labeling and Morphometric Analysis.
BrdU (Sigma) was delivered by i.p. injection at 100 mg/kg to mice, which were killed 2 h later. Colons were fixed in 4% buffered formalin, processed for sectioning, and stained with mouse monoclonal anti-BrdU (2 μg/ml; Roche) detected by biotinylated goat anti-mouse IgG (1:200; Vector Laboratories), and staining was visualized with diaminobenzidine and H2O2. BrdU-positive cells were counted under high power on a Nikon E800 microscope with Magnifire digital camera, and the image was displayed on a computer monitor. Results were expressed as the average number of BrdU-positive crypt cells (20) per animal and by crypt position.
For c-Myb and H&E staining colon sections were fixed in methacarn for 2 h and transferred to 70% ethanol, embedded, sectioned, and stained with H&E. Full crypts exposing a lumen and identifiable base were scored. Crypt cells per 40–50 longitudinal sections were scored as previously described (7). Sample groups were subjected to one-way ANOVA using Origin software. The anti-PCNA antibody PC-10 (Santa Cruz Biotechnology, Santa Cruz, CA) was used at a 1:100 dilution to identify PCNA followed by goat anti-mouse-HRP at 1:250 (Bio-Rad) and developed with Pierce metal enhanced detection reagent. c-Myb was visualized by using Mab1.1 and processed as described previously (4) using antigen retrieval by boiling slides in 1 mM EDTA in a pressure cooker for 3 min. Rabbit anti-ChromograninA was used at 1:100 (SC-13090; Santa Cruz Biotechnology) after citrate buffer antigen retrieval. Donkey anti-goat HRP at 1:250 (Santa Cruz Biotechnology) was used as a secondary antibody. Phospho-histone-3 (Upstate Biotechnology) staining was performed by immunofluorescence at a final titer of 1:200.
Mucin content was estimated by using integrated morphometric analysis based on pixel density of differential color staining of pink-PAS-positive cytoplasm corrected for blue-nuclear staining over five microscopic fields at ×40 magnification on at least three mice per genotype. This was done by using the MetaMorph computer program (Universal Imaging, Downingtown, PA) in a comparable manner to a previous report (28).
Real-Time RT-PCR.
Colonic crypts were isolated from mouse colons by using the method described elsewhere (29). cDNA was prepared in 20-μl volumes by the addition of 1 μl of random hexamers (1 μg/μl) and 6 μl of diethyl pyrocarbonate-treated H2O to 4 μl of RNA. cDNA was prepared by using SuperScript III (Promega) according to the manufacturer's instructions. Eight microliters of cDNA (1:10 dilution) was combined with 10 μl of SyBr Green PCR Master Mix (Applied Biosystems) and 200 nM each sense and antisense oligonucleotides (Geneworks, Adelaide, Australia) and amplified by using temperatures of 50°C for 2 min and 95°C for 10 min. These initial steps were followed by 45 cycles of 95°C for 15 sec and 60°C for 1 min. Expression of all genes was compared with β2-microglobulin to determine relative levels of mRNA transcripts. Cyclin E1 primers: sense, 5′TTT CTG CAG CGC CAT CCT; antisense, 5′- GCA CAC CTC CAT CAG CCA A-3′.
Supplementary Material
Acknowledgments
We thank Ms. Sarah Ellis for advice on the application of and analysis with MetaMorph software. We also thank Dr. Maree Overall for her critical reading of the manuscript. Dr. Melanie Trivett provided invaluable assistance in performing immunohistochemistry, and Ms. Sally Lightowler performed all of the genotyping and tissue processing. We are grateful to the animal facility staff for expert animal husbandry of the mice used in this study. The National Health and Medical Research Council supported this work, and R.G.R., M.E., W.A., and D.H. are recipients of National Health and Medical Research Council Research Fellowships.
Abbreviations
- PCNA
proliferating cell nuclear antigen
- PAS
periodic acid/Schiff reagent
- MEF
mouse embryonic fibroblast
- En
embryonic day n.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610055104/DC1.
References
- 1.Potten CS. Philos Trans R Soc London B. 1998;353:821–830. doi: 10.1098/rstb.1998.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavrieli Y, Sherman Y, Ben-Sasson SA. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon JI, Hermiston ML. Curr Opin Cell Biol. 1994;6:795–803. doi: 10.1016/0955-0674(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 4.Rosenthal MA, Thompson MA, Ellis S, Whitehead RH, Ramsay RG. Cell Growth Differ. 1996;7:961–967. [PubMed] [Google Scholar]
- 5.Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Jr, Potter SS. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 6.Zorbas M, Sicurella C, Bertoncello I, Venter D, Ellis S, Mucenski ML, Ramsay RG. Oncogene. 1999;18:5821–5830. doi: 10.1038/sj.onc.1202971. [DOI] [PubMed] [Google Scholar]
- 7.Ramsay RG, Micallef S, Lightowler S, Mucenski ML, Mantamadiotis T, Bertoncello I. Mol Cancer Res. 2004;2:354–361. [PubMed] [Google Scholar]
- 8.Carpinelli MR, Hilton DJ, Metcalf D, Antonchuk JL, Hyland CD, Mifsud SL, Di Rago L, Hilton AA, Willson TA, Roberts AW, et al. Proc Natl Acad Sci USA. 2004;101:6553–6558. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP. Dev Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Thompson MA, Rosenthal MA, Ellis SL, Friend AJ, Zorbas MI, Whitehead RH, Ramsay RG. Cancer Res. 1998;58:5168–5175. [PubMed] [Google Scholar]
- 11.Walsh S, Murphy M, Silverman M, Odze R, Antonioli D, Goldman H, Loda M. Am J Pathol. 1999;155:1511–1518. doi: 10.1016/S0002-9440(10)65466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gartel AL, Serfas MS, Gartel M, Goufman E, Wu GS, el-Deiry WS, Tyner AL. Exp Cell Res. 1996;227:171–181. doi: 10.1006/excr.1996.0264. [DOI] [PubMed] [Google Scholar]
- 13.Johnstone CN, White SJ, Tebbutt NC, Clay FJ, Ernst M, Biggs WH, Viars CS, Czekay S, Arden KC, Heath JK. J Biol Chem. 2002;277:34531–34539. doi: 10.1074/jbc.M204865200. [DOI] [PubMed] [Google Scholar]
- 14.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. EMBO J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen RD, III, Bender TP, Siu G. Genes Dev. 1999;13:1073–1078. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramsay RG. Growth Factors. 2005;23:253–261. doi: 10.1080/08977190500233730. [DOI] [PubMed] [Google Scholar]
- 17.Philipp J, Vo K, Gurley KE, Seidel K, Kemp CJ. Oncogene. 1999;18:4689–4698. doi: 10.1038/sj.onc.1202840. [DOI] [PubMed] [Google Scholar]
- 18.Ramsay RG, Ciznadija D, Sicurella C, Reyes N, Mitchelhill K, Darcy PK, D'Abaco G, Mantamadiotis T. DNA Cell Biol. 2005;24:21–29. doi: 10.1089/dna.2005.24.21. [DOI] [PubMed] [Google Scholar]
- 19.Thompson CB, Challoner PB, Neiman PE, Groudine M. Nature. 1986;319:374–380. doi: 10.1038/319374a0. [DOI] [PubMed] [Google Scholar]
- 20.Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 21.Bettess MD, Dubois N, Murphy MJ, Dubey C, Roger C, Robine S, Trumpp A. Mol Cell Biol. 2005;25:7868–7878. doi: 10.1128/MCB.25.17.7868-7878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagoshi H, Kanei-Ishii C, Sawazaki T, Mizuguchi G, Ishii S. Oncogene. 1992;7:1233–1240. [PubMed] [Google Scholar]
- 23.Ramsay RG, Barton AL, Gonda TJ. Expert Opin Ther Targets. 2003;7:235–248. doi: 10.1517/14728222.7.2.235. [DOI] [PubMed] [Google Scholar]
- 24.Alitalo K, Winqvist R, Lin CC, de la Chapelle A, Schwab M, Bishop JM. Proc Natl Acad Sci USA. 1984;81:4534–4538. doi: 10.1073/pnas.81.14.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugo H, Cures A, Suraweera N, Drabsch Y, Purcell D, Mantamadiotis T, Phillips W, Dobrovic A, Zupi G, Gonda TJ, et al. Genes Chromosomes Cancer. 2006;45:1143–1154. doi: 10.1002/gcc.20378. [DOI] [PubMed] [Google Scholar]
- 26.Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM, et al. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 27.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 28.Ruddell A, Mezquita P, Brandvold KA, Farr A, Iritani BM. Am J Pathol. 2003;163:2233–2245. doi: 10.1016/S0002-9440(10)63581-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitehead RH, Brown A, Bhathal PS. In Vitro Cell Dev Biol. 1987;23:436–442. doi: 10.1007/BF02623860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.