Abstract
Mechanisms regulating CNS pattern formation and neural precursor formation are remarkably conserved between Drosophila and vertebrates. However, to date, few direct connections have been made between genes that pattern the early CNS and those that trigger neural precursor formation. Here, we use Drosophila to link directly the function of two evolutionarily conserved regulators of CNS pattern along the dorsoventral axis, the homeodomain protein Ind and the Sox-domain protein Dichaete, to the spatial regulation of the proneural gene achaete (ac) in the embryonic CNS. We identify a minimal achaete regulatory region that recapitulates half of the wild-type ac expression pattern in the CNS and find multiple putative Dichaete-, Ind-, and Vnd-binding sites within this region. Consensus Dichaete sites are often found adjacent to those for Vnd and Ind, suggesting that Dichaete associates with Ind or Vnd on target promoters. Consistent with this finding, we observe that Dichaete can physically interact with Ind and Vnd. Finally, we demonstrate the in vivo requirement of adjacent Dichaete and Ind sites in the repression of ac gene expression in the CNS. Our data identify a direct link between the molecules that pattern the CNS and those that specify distinct cell-types.
Keywords: proneural genes, Sox-domain proteins, transcriptional regulation, dorsoventral patterning
Genes of the proneural achaete–scute (ac/sc) class encode conserved transcription factors whose expression and function signal the transition from early patterning events that subdivide the neuroectoderm to the cell-fate specification steps that create discrete cell-types in the CNS (1–3). ac/sc genes encode basic helix–loop–helix transcription factors that promote the neural precursor fate in higher metazoans. Analysis of the expression, regulation, and function of ac/sc genes serves as a model for the genetic and molecular basis of cell-type-specific patterning (1–3). Many parallels exist between flies and vertebrates in the events that pattern the CNS and in the downstream functions of ac/sc genes. However, such studies have yet to link factors that pattern the CNS directly to the spatial regulation of ac/sc gene expression in the CNS.
In the early Drosophila embryonic CNS, the ac and sc proneural genes are coexpressed in four cell clusters per hemisegment [Fig. 2; (4)]. Within each cluster, genetic interactions between ac and sc and genes of the Notch pathway select one cell as the neuroblast (1), the precursor or stem cell of the Drosophila CNS. Thus, the pattern of proneural gene expression forecasts the later pattern of neuroblasts.
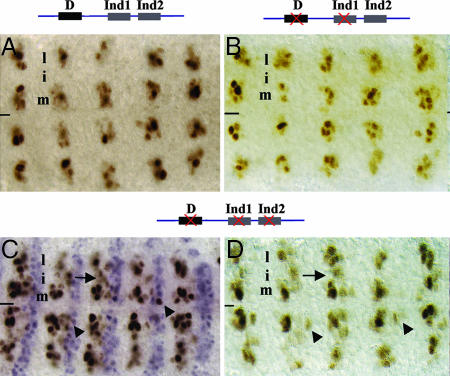
Fig. 2.
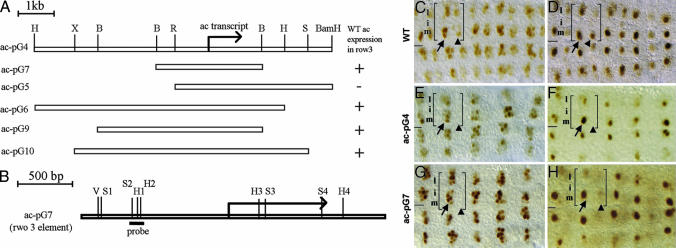
Identification of a CNS regulatory region of ac. (A) Genomic map of the 8.15-kb ac-pG4 minigene, including the restriction sites used to create each derivative construct. H, HindIII; B, BglII; X, XbaI; R, EcoRI. (B) Genomic map of ac-pG7 (the row 3 element) showing the locations of predicted Vnd- (V), Sox- (S), and homeodomain- (HD) binding sites. The DNA fragment used as the probe for gel-shift assays is underlined (see also Fig. 4C). (C–H) Expression of endogenous ac (C and D) and ac expression as driven by ac-pG4 (E and F) and ac-pG7 (G and H) during stages 8 (C, E, and G) and 9 (D, F, and H). Embryos in E–H are homo- or hemizygous for In (1)y[3pl]sc[8R]. (C) In wild-type embryos, ac is expressed in cell clusters in the medial and lateral columns of row 3 (arrow) and row 7 (arrowhead). (D) By stage 9, ac expression resolves to a single cell per cluster, and this cell will acquire the neuroblast fate. (E and G) ac minigenes pG4 and pG7 drive ac expression in cell clusters in the medial and lateral clusters of row 3 (arrow), but not row 7 (arrowhead), at stage 8, and restriction of ac expression to a single cell occurs by stage 9 (F and H). In C–H, a single hemisegment is bracketed.
Factors that initially pattern the embryonic CNS are prime candidates to regulate directly the spatial limits of ac/sc gene expression and with it the neuroblast pattern. For example, ventral nervous system defective (vnd) and intermediate neuroblasts defective (ind) encode conserved homeodomain proteins that pattern, respectively, the medial and intermediate columns of the developing CNS (5–9). The functions of vnd and ind define the initial dorsoventral limits of ac/sc gene expression in the CNS. vnd is expressed in the medial column before ac and sc expression (8, 9). vnd activates ac and sc expression in the medial column and in so doing promotes the formation of medial column neuroblasts. ind is expressed throughout the intermediate column before ac and sc expression in the CNS. ind represses ac and sc expression in the intermediate column, thus restricting ac/sc-expressing cell clusters and the neuroblasts that arise from them to the medial and lateral columns of the CNS (7).
Dichaete encodes a Sox B-type transcription factor thought to act with vnd and ind to pattern, and to regulate ac/sc expression, in the CNS (10–13). Sox-domain proteins regulate target gene transcription by associating with transcription factors from diverse gene families (13, 14), with the identity of target genes and the nature of regulation (activation vs. repression) depending on the constituents of the complexes. The proclivity of Sox proteins to pair with other transcription factors combined with the demonstration that the CNS phenotype of Dichaete is similar to those of vnd and ind and the identification of dosage sensitive interactions between Dichaete and vnd and ind led to the model that Dichaete associates with Vnd and Ind on target gene promoters to regulate directly gene expression in the CNS (10).
Here, we focus on the regulation of the ac gene and use genetic, molecular, and biochemical assays to test this model. Our results link directly the actions of Dichaete and Ind to the spatial regulation of ac in the CNS. In so doing, they identify a direct connection between transcription factors that pattern the CNS and those that directly control the neural precursor fate.
Results
Dichaete Can Physically Interact with Vnd and Ind.
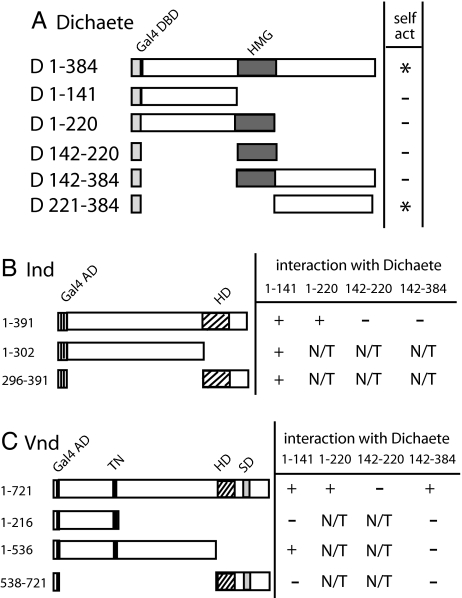
Sox-domain proteins physically associate with other transcription factors to regulate gene transcription. Thus, the identification that Dichaete genetically interacts with Vnd and Ind suggested that Dichaete associates with Vnd and Ind to regulate gene expression in the CNS (10). To test this model, we asked whether Dichaete can interact with Ind or Vnd in the yeast two-hybrid assay. Control experiments revealed that the full-length Dichaete protein as well as the region C-terminal to the high-mobility-group (HMG) DNA-binding domain (amino acids 221–384) activate transcription on their own when fused to the Gal4 DNA-binding domain (Fig. 1), suggesting that the C-terminal region contains transcriptional activation activity. As a result, we tested a number of distinct Dichaete fusion constructs for self-activation of transcription and identified four that were transcriptionally inert (Fig. 1). One of these contained the HMG domain and the C-terminal region, indicating that the presence of the HMG domain may mask the transactivation properties of the C-terminal region (Fig. 1). A prior study mapped a transactivation domain to the N-terminal region of Dichaete (15), yet we failed to identify transactivation properties of this domain. Consistent with a transactivation domain residing in the C-terminal region of Dichaete, all other identified transactivation domains in Sox-family proteins map C-terminal to the HMG domain (16, 17).
Fig. 1.
Dichaete physically interacts with Ind and Vnd. (A) Identification of Dichaete-Gal4 DNA-binding domain (DBD) fusion proteins appropriate for the yeast two-hybrid assay. The left column indicates regions of Dichaete contained in each construct, and the right column shows the ability of each construct to self-activate reporter gene expression. (B) The Dichaete N-terminal region interacts with two regions of Ind. The left column indicates region of Ind tested for binding to Dichaete, and the right column shows the ability of different regions of Ind to interact with indicated regions of Dichaete (D). (C) The Dichaete N- and C-terminal regions interact with Vnd. The left column indicates regions of Vnd tested for binding to Dichaete (D). The right column shows the ability of these regions to interact with the indicated regions of Dichaete. N/T, not tested; Gal4DBD, Gal4 DNA-binding domain; Gal4AD, Gal4 activation domain; HD, homeodomain; TN, tinman domain; SD, NK-2-specific domain.
By using the four Dichaete bait constructs, we found that the N-terminal region of Dichaete (amino acids 1–141) specifically interacted with full-length Ind protein. In a reciprocal manner, we tested the ability of the Dichaete N-terminal region to interact with two different regions of Ind: the region N-terminal to the homeodomain (amino acids 1–302) and the region including the homeodomain and all residues C-terminal to it (296–391; Fig. 1). Both regions of Ind interacted strongly with the Dichaete N-terminal region, suggesting that this region of Dichaete can interface with two distinct regions of Ind.
In a similar manner, we determined that two distinct regions of Dichaete, the regions N-terminal (amino acids 1–141) and C-terminal (amino acids 221–384) to the HMG domain, interact with the full-length Vnd protein (Fig. 1). We used three different Vnd prey constructs to localize the regions of Vnd that interact with Dichaete. We determined that the region of Vnd located between the TN domain (a domain common to Tinman/NK-2 proteins) and the homeodomain (amino acids 217–536) interacts with the Dichaete N-terminal domain. This result confirms and extends those of Yu et al. (18) who found that Vnd and Dichaete coprecipitate and that a Vnd deletion lacking the first 408 amino acids interacts with Dichaete. We were unable to define the region of Vnd that interacts with the Dichaete C-terminal region, perhaps because the constructs interrupt the domain to which the C-terminal region of Dichaete binds or disrupt the general topology of this domain. Nonetheless, our yeast two-hybrid results indicate that Dichaete can interact with Ind and Vnd consistent with the model that Dichaete complexes with Ind and Vnd on target gene promoters to regulate transcription in the CNS.
Identification and Delimitation of a CNS Regulatory Region of ac.
A molecular understanding of how Dichaete, Ind, and Vnd pattern the CNS requires the identification and characterization of the regulatory regions of candidate direct target genes. One such candidate is the ac gene. Prior studies on ac suggested that regulatory regions important for its spatial regulation exist both 5′ and 3′ to the ac gene (4, 19, 20). Thus, we generated an 8.15-kb minigene that contains the ac transcription unit as well as ≈4.8 kb of DNA 5′ to the transcription start and ≈2.4 kb of DNA 3′ to the polyadenylation site and tested its ability to drive ac expression in an In (1)y3PLsc8R mutant background. This genetic background carries a deletion of ac and also deletes the regulatory regions necessary to drive sc expression in row 3 (20). Thus, it allows us to visualize ac expression as driven by the minigene in the absence of endogenous ac/sc gene expression in row 3. The ac minigene drives ac expression in half of its wild-type CNS pattern because ac is expressed normally in the medial and lateral clusters of row 3 but is not expressed in row 7 (Fig. 2). The dynamics of ac expression as driven by the minigene in row 3 mirror those of endogenous ac expression because ac expression in each cluster quickly becomes restricted to a single cell, the presumptive neuroblast, which then delaminates into the interior of the embryo and extinguishes ac gene expression before its first division. Thus, the DNA contained within the minigene is sufficient to activate ac in its wild-type expression pattern in row 3 and to mediate the Notch-dependent restriction of ac to the presumptive neuroblast.
By creating a series of 5′ and 3′ deletions of the initial minigene, we delimited the regulatory regions sufficient to drive ac expression in row 3 to a 2.84-kb genomic fragment (pG7; Fig. 2), which we refer to as the row 3 element. This element contains the ac transcription unit, 1.34 kb of DNA 5′ to the start of transcription and 542 base pairs of DNA 3′ to the end of the transcription unit (Fig. 2). Below, we characterize ac minigenes for their ability to respond to the functions of Dichaete, ind, and vnd and for the presence and in vivo relevance of putative binding sites for these factors.
Identification of Putative Dichaete-, Vnd-, and Ind-Binding Sites in the Row 3 Element.
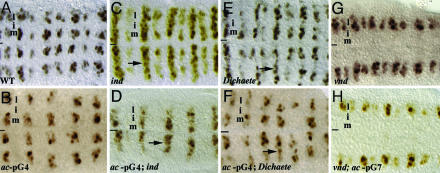
In support of Dichaete, Vnd, and Ind acting directly on the row 3 element to regulate ac expression, loss of Dichaete, vnd, or ind function affects ac expression as driven by ac-pG4 or ac-pG7 in the same way, and these defects are identical to those observed for endogenous ac expression in these mutant backgrounds. For example, loss of ind or Dichaete causes, respectively, strong or modest derepression of ac expression in the intermediate column, whereas loss of vnd results in the absence of ac expression in the medial column (Fig. 3).
Fig. 3.
ind, dichaete, and vnd regulate ac minigene expression. Endogenous ac expression (A, C, E, and G) or ac expression as driven by pG4 or pG7 (B, D, F, and H) in wild-type (A and B), ind (C and D), Dichaete (E and F), and vnd (G and H) embryos. Embryos in B, D, F, and H are also homo- or hemizygous for In (1)y3PLsc8R. (A) ac is normally expressed in the medial and lateral columns of rows 3 and 7. (B) The pG4 minigenes only drive ac expression in row 3. (C and D) Loss of ind function causes derepression of endogenous ac and ac-pG4 as well as ac-pG7 (data not shown) expression in the intermediate column (arrows); loss of Dichaete causes modest derepression of endogenous ac and ac-pG4 expression in this domain (E and F). (G and H) In vnd embryos, endogenous ac expression and ac-pG7 expression is lost from the medial column. Anterior, left; line, ventral midline. m, i, and l indicate positions of medial, intermediate, and lateral columns, respectively.
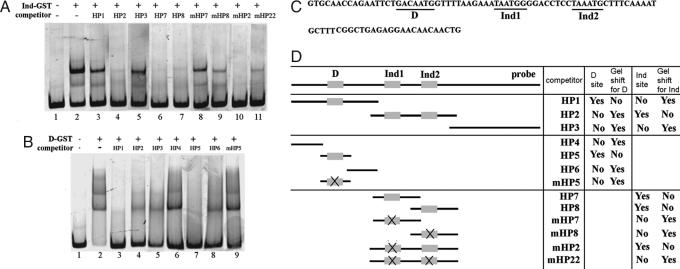
To see whether Dichaete, Ind, or Vnd act directly on the row 3 element to control ac expression, we searched this element for perfect matches to the consensus Vnd [CAAGTG; (21)], Sox-domain [(A/T)(A/T)CAA(A/T)G; (13)] and homeodomain [TAATGG; (22)] binding sites. We used the canonical Sox-domain and homeodomain binding site sequences because the consensus sites for Dichaete and Ind have not been determined. This search identified one match for Vnd (V) and three each for Dichaete (S1, S3, and S4) and Ind (H1, H3, and H4; Fig. 2). Notably, predicted Dichaete/Sox-binding sites tend to reside close to predicted Vnd or Ind sites (Figs. 2 and 4), consistent with Dichaete acting with Vnd and Ind to regulate ac expression. The sole exception is the Ind site (H1) located upstream of the transcriptional start site of ac. However, as detailed below, gel-shift assays identify a Dichaete-binding site 11 bp 5′ of this Ind site (S2; Figs. 2 and 4).
Fig. 4.
Ind and Dichaete bind to a trio of sites upstream of the achaete promoter. (A and B) Gel-shift assays showing binding of Ind (A) or Dichaete (B) to the HP probe (C) and the ability of different unlabeled probes to compete for binding (D). (C) Sequence of HP probe and the location of Dichaete and Ind sites. (D) Schematic of HP probe and unlabeled competitor DNA and a tabular representation of the results shown in A and B. (A) Ind binds two adjacent sites in this region. Unlabeled probes HP7 and HP8 compete for binding of Ind to the labeled HP probe. Mutation of the consensus or near-consensus homeodomain binding sites in HP7 or HP8 (mHP7 and mHP8) block the ability of these probes to compete for binding to Ind. (B) Dichaete binds a single site in this region. Probes HP1 and HP5 contain a sequence similar to the consensus Sox-binding site and compete for Dichaete binding to the HP probe. Mutation of this sequence (mHP5) renders the probe ineffective at competing for Dichaete binding.
Because the precise binding specificity of Ind is unknown, we first tested whether Ind can bind the predicted sites by using gel-shift assays. We focused on the predicted Ind site located upstream of the transcription start site because it is the only location where Dichaete and Ind sites are found adjacent to each other (see below). We found that Ind specifically binds this site in vitro (Fig. 4A). During these experiments, we also identified a second Ind-binding site (TAAATG; H2 in Fig. 2) 8 bp 3′ to this site, which differs slightly from the consensus homeodomain site (Fig. 4). Thus, Ind can bind to two sites located within 1 kb of the ac promoter, suggesting a possible molecular mechanism for Ind-dependent repression of ac.
Our initial search for Dichaete-binding sites required a perfect match to the consensus Sox-binding site. However, bona fide transcription factor-binding sites often differ from the experimentally defined consensus by a few base pairs (23), indicating that our search likely underpredicted possible Dichaete-binding sites. Because of this, we used gel-shift assays to search for Dichaete-binding sites throughout the entire row 3 element (pG7; Fig. 2). We identified three sites to which Dichaete bound specifically. Two of these correspond to sites identified in our consensus sequence search (sites S1 and S3 in Fig. 2); whereas the third resides 11 bp 5′ of the first of the two Ind sites near the transcriptional start of ac (S2 in Fig. 2); this site (GACAATG) differs from the consensus by one base pair. We failed to detect binding of Dichaete to one predicted Sox site (S4 in Fig. 2). Because Dichaete and ind are known to repress ac expression (7, 24), the three binding sites for Ind and Dichaete upstream of the ac promoter identify a likely site of action through which these factors repress ac.
Ind and Dichaete Directly Regulate ac Expression in the CNS.
The clustering of binding sites for Dichaete, Vnd, and Ind, together with the ability of Dichaete to interact with Vnd and Ind, supports the idea that Dichaete acts with these factors to regulate ac expression in the CNS. To test this model directly, we assayed the in vivo relevance of the adjacent Vnd and Dichaete sites as well as the adjacent Dichaete and Ind sites on ac expression. ac expression was unaltered when we mutated the Vnd-binding site, the adjacent Dichaete site, or both sites. Thus, vnd either does not regulate ac expression directly or other Vnd binding sites in the row 3 element compensate for the loss of this site (see Discussion).
We also tested the relevance of the three Dichaete- and Ind-binding sites located ≈850 bp upstream of the start of ac transcription. Mutating any single site or any combination of two sites had no effect on ac expression (Fig. 5). However, mutating all three sites derepressed ac expression in the intermediate column, a phenotype similar to that found in embryos mutant for ind or Dichaete (Figs. 3 and 5). This result provides direct link between genes that pattern the CNS and those that specify distinct cell types. Because the derepression of ac is less severe than that observed in ind mutant embryos (see Fig. 3), Ind and Dichaete likely act through additional sites in this element to repress ac expression fully in the intermediate column.
Fig. 5.
Dichaete and Ind directly repress ac expression in vivo. (A) The pG7 minigene drives ac expression in the medial and lateral, but not the intermediate, columns of row 3. (B) Mutating the Dichaete and first Ind sites does not affect ac expression. (C and D) Mutating all three sites causes moderate derepression of ac expression in the intermediate column (arrows) and immediately posterior to row 3 (arrowheads). Engrailed expression (purple) is shown in C to mark the posterior compartment of each segment. Anterior, left; line, ventral midline. m, i, and l indicate positions of medial, intermediate, and lateral columns, respectively.
Unexpectedly, we also observed derepression of ac expression posterior to row 3 upon mutation of the three sites. This posterior expansion of ac mimics the effect that removal of gooseberry function has on the expression of ac (24, 25), suggesting that Gooseberry, another homeodomain protein, may bind the same sites as Ind and act with Dichaete to repress ac expression in its expression domain.
Discussion
Prior genetic studies on Dichaete, ind, and vnd led to the model that Dichaete associates with Vnd and Ind on target promoters to regulate gene expression in the CNS directly. Our work supports this model by demonstrating that removal of three adjacent Ind- and Dichaete-binding sites upstream of ac derepresses ac expression in the intermediate column, thus identifying Dichaete and Ind as direct repressors of ac. However, these data are also consistent with a model in which Dichaete and Ind act redundantly to repress ac, because derepression was observed only when all three sites were removed. Moreover, it remains plausible that Ind can also act in conjunction with Dichaete bound at more distant sites; thus, adjacent positioning of Dichaete and Ind on target promoters may not be crucial for their ability to regulate target gene expression. Regardless of the precise mechanism through which Dichaete and Ind repress ac, our observation that Dichaete and Ind directly repress ac connects the activity of the transcription factors that pattern the CNS to those that promote the neural precursor fate.
The identification of a bona fide direct target of Dichate and Ind in the developing CNS represents an important first step in clarifying the molecular basis through which Dichaete and Ind pattern the CNS. However, in their CNS patterning roles, Dichaete and Ind must regulate directly the transcription of many genes in addition to ac. Genes expressed in region-specific patterns in the CNS, such as sc, lethal of sc, seven-up, and runt, are excellent candidates to be additional direct targets of Dichaete and Ind. Clearly, an in-depth understanding of the molecular basis of Dichaete and Ind function requires identifying most of their direct targets in the CNS and elucidating the molecular mechanism through which these factors direct the expression of these genes in the CNS. Recent advances in functional genomic techniques render it feasible to identify target genes of individual transcription factors on a genome-wide scale, whereas the integration of computational and molecular methods facilitates construction of genome-wide regulatory networks. In the future, it will be essential to use such methods to identify the batteries of genes regulated by Dichaete and Ind, as well as other transcriptional regulators that pattern the CNS, and to link together these genes in regulatory networks. Such approaches should help set the foundation for a system-wide view of the transcriptional networks that act progressively to construct a functional integrated nervous system.
Our results did not identify the motifs required to activate ac expression in the CNS. Although vnd is a clear candidate to activate ac expression in the medial column, removal of the single consensus Vnd site did not effect ac expression in row 3. Our failure to detect a direct role for Vnd may have arisen because of the presence of multiple Vnd-binding sites in the row 3 element, all of which would differ slightly from the consensus. Consistent with this scenario, three sites similar to the Nk-2/Vnd-consensus binding site exist in the row 3 element. Alternatively, vnd may permit ac expression in the medial column indirectly by repressing ind expression in this domain. In support of this hypothesis, ac is expressed in the medial column in embryos that lack vnd and ind (data not shown). Future experiments that identify in vivo targets of Vnd and dissect the row 3 element in greater detail will distinguish these models.
Do Vertebrate Homologs of Dichaete and Ind Directly Regulate Proneural Gene Expression?
Our work demonstrates a direct link between early patterning genes and genes that promote neural precursor formation. Although such a link has not yet been made in vertebrates, several lines of evidence support the direct regulation of proneural genes by SoxB-type and Gsh-1/2 proteins, the vertebrate orthologs of D/SoxNeuro and Ind, respectively. (i) SoxB proteins and Gsh-1/2 are expressed in the right time and place to regulate ac/sc gene expression in vertebrates. For example, SoxB-type genes are expressed widely in the early CNS (24, 26–29) and Gsh-1/2 are expressed in the intermediate domain of the neural tube (24, 30, 31); (ii) expression of these factors precedes ac/sc expression (24, 32); (iii) Gsh-1/2 repress the expression of the proneural bHLH genes Neurogenin-1 and Neurogenin-2 to regulate cell fate in the vertebrate spinal cord (24, 32); (iv) mutations in Sox1 and Gsh-1 function yield similar phenotypes in the ventral telencephalon, suggesting that Sox1, and other SoxB factors, may partner with Gsh-1/2 to regulate neuronal identity (24, 33); (v), SoxB factors regulate directly the expression of genes that act with ac/sc proneural genes to regulate neural precursor formation. For example, Sox1 can bind directly to the HES1 promoter and suppress its transcription (24, 32). Thus, SoxB proteins and Gsh-1/2 are strong candidates to regulate proneural gene expression directly and, thus, neuronal formation in vertebrates, perhaps by acting in association with each other.
Additional experiments are needed to see whether a direct link exists between SoxB and Gsh-1/2 proteins and proneural gene regulation in vertebrates, to identify the batteries of targets regulated by these factors in Drosophila and vertebrates and to reveal the precise roles these targets play in CNS development. Nonetheless, the identification of physical interactions between Dichaete and Ind and the demonstration that these proteins act together to regulate ac expression indicate that experiments along these lines will help clarify the molecular links between the events that pattern the early CNS and those that specify discrete cell types.
Methods
Drosophila Strains.
We used the following fly lines in the study: indRR108, D87, vndΔ38, In (1)y3PLsc8R, In (1)y3PLsc8R; indRR108, In (1)y3PLsc8R; D87. Oregon R is wild type.
Yeast Two-Hybrid Assays.
The yeast two-hybrid vectors pGBKT7 and pACT2 (Clontech, Mountain View, CA) were used to generate bait and prey constructs. Baits contained the Gal4 DNA-binding domain fused to the protein of interest, and the prey contained fusions to the Gal4 transactivation domain. A chromosomally integrated UAS-Ade reporter gene was used that contains Gal2 operator sites fused to the ADE2 gene. The bait and prey constructs were cotransformed into PJG69 4A host cells (MATa trp1 leu2 his3 ura3 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ). The transformed cells were plated on yeast–peptone–dextrose (YPD) (−Leu, −Trp) medium and incubated at 30°C for 3–4 days. Resulting transformants were plated on YPD (−Leu, −Trp, −Ade) medium and incubated at 30°C for 3–4 days to assay for the presence of colonies, an indicator of an interaction between bait and prey proteins.
We used standard molecular techniques to clone the appropriate bait genes into pGBKT7. We created five bait constructs for Dichaete, including one full-length construct and the following four truncated constructs: D1–141 contains the N-terminal 141 aa; D1–220 contains the 79-aa HMG domain as well as the N-terminal 141 aa; D142–220 contains only the HMG domain, D142–384 contains the HMG all amino acids C-terminal to it; and D221–384 contains the C-terminal 164 aa. John R. Nambu (Department of Biology, University of Massachusetts, Amherst, MA) kindly provided DNA corresponding to each of ORF except for the N-terminal region (15, 24). We used the same primers as reported in Ma et al. (15, 24) to make the N-terminal construct.
We used standard molecular methods to clone the full-length Ind prey construct, the N-terminal Ind prey construct containing amino acids 1–302 (Ind1–302), and the C-terminal Ind prey construct into pACT2. To make a full-length Vnd prey construct, we cloned the appropriate EcoRI/XhoI fragment from Vnd-pAC5.1 (kindly provided by Marshall Nirenberg, National Institutes of Health, Bethesda, MD) (24, 34) into pACT2, resulting in an out-of-frame fusion between Vnd and the GAL4 activation domain. To put Vnd in-frame, we digested the construct with EcoRI, treated with mung bean nuclease and then religated the construct. We verified construct integrity by sequencing and used standard methods to create three truncated versions of Vnd fused in-frame to the activation domain: the Vnd1–536 construct (amino acids 1–536) contains all amino acids N-terminal to the homeodomain; this region includes the Tinman domain (TN, 97–208); Vnd1–216 contains the first 216 amino acids; Vnd538–721 contains the homeodomain (HD, 544–603) and the NK-2-specific domain (SD, 628–650).
Generation of ac Minigenes.
Standard molecular techniques were used to generate the ac minigenes. The restriction sites used to create each construct are indicated in Fig. 2. The corresponding DNA fragments were cloned into pPCaSpeR2, and multiple independent transgenic lines were generated and assayed for each construct. ac expression was assayed by immunohistochemical staining with mouse anti-Ac monoclonal antibody (4).
Electrophoretic Mobility-Shift Assays.
To create the GST-Dichaete fusion protein, the DNA fragment encoding the full-length Dichaete protein was purified from an EcoRI restriction digest of pEG202-Fish (4, 15) and cloned in-frame into pGEX-4T (Amersham, Piscataway, NJ). To create a full-length GST-Ind fusion protein, we amplified and cloned the entire Ind coding region in-frame into pGEX-4T and verified construct integrity by sequencing. GST-Ind and GST-Dichaete proteins were purified from bacterial cell lysates by using glutathione-Sepharose beads following manufacturer's instruction (Amersham).
Gel-shift assays were carried out as described in ref. 35. Briefly, we used PCR to create DNA probes by using fluorescein HOX (IDT DNA) labeled primers and ac-pG7 as the template. The location of the probe is shown in Fig. 2. Labeled probe was incubated with either GST-Dichaete or GST-Ind at 25°C for 30 min in binding buffer. Immediately after incubation, complexes were resolved on 8% native polyacrylamide gels. Gels were immediately scanned by using a Typhoon 8600 Variable Mode Imager (Amersham). Sequences of the oligonucleotides used as unlabeled competitor DNA are provided in supporting information (SI) Data.
Site-Directed Mutagenesis.
We used the SOE method (4, 36) to perform site-directed mutagenesis of predicted Ind-, Dichaete-, and Vnd-binding sites in ac pG7. After SOE PCR, we cloned the generated product, which contained flanking restriction sites corresponding to endogenous restriction sites in pG7, into pGEMT and verified its sequence. We then replaced the wild-type fragment with the mutated one in ac-pG7. For the clustered Ind and Sox sites, we used the SalI and BamHI restriction sites that flank the ac transcriptional start, and for the Vnd site, we used the BglII and EcoRI sites that flank the predicted Vnd site. The sequence of the primers used to mutate Vnd, Ind, and Dichaete sites are provided in SI Data.
Supplementary Material
Acknowledgments
We thank Heather Broihier, Kristen Kroll, and Hemlata Mistry for comments on the manuscript; Drs. John Nambu and Marshall Nirenberg for supplying many reagents and helpful advice; Min-Ho Lee and Jiajian Liu for helpful discussions; the Bloomington Stock Center, Bloomington, IN, for providing fly lines; and the reviewers for their excellent suggestions. This work was supported by National Science Foundation Grant IBN0212282 (to J.B.S.) and National Institutes of Health Grants NS-3657 (to J.B.S.) and GM59871 (to G.B.-F.).
Abbreviations
- ac
achaete
- sc
scute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611700104/DC1.
References
- 1.Bertrand N, Castro DS, Guillemot F. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 2.Arendt D, Nubler-Jung K. Development (Cambridge, UK) 1999;126:2309–2325. doi: 10.1242/dev.126.11.2309. [DOI] [PubMed] [Google Scholar]
- 3.Skeath JB, Carroll SB. FASEB J. 1994;8:714–721. doi: 10.1096/fasebj.8.10.8050670. [DOI] [PubMed] [Google Scholar]
- 4.Skeath JB, Carroll SB. Development (Cambridge, UK) 1992;114:939–946. doi: 10.1242/dev.114.4.939. [DOI] [PubMed] [Google Scholar]
- 5.Chu H, Parras C, White K, Jimenez F. Genes Dev. 1998;12:3613–3624. doi: 10.1101/gad.12.22.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDonald JA, Holbrook S, Isshiki T, Weiss J, Doe CQ, Mellerick DM. Genes Dev. 1998;12:3603–3612. doi: 10.1101/gad.12.22.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss JB, Von Ohlen T, Mellerick DM, Dressler G, Doe CQ, Scott MP. Genes Dev. 1998;12:3591–3602. doi: 10.1101/gad.12.22.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jimenez F, Martin-Morris LE, Velasco L, Chu H, Sierra J, Rosen DR, White K. EMBO J. 1995;14:3487–3495. doi: 10.1002/j.1460-2075.1995.tb07355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mellerick DM, Nirenberg M. Dev Biol. 1995;171:306–316. doi: 10.1006/dbio.1995.1283. [DOI] [PubMed] [Google Scholar]
- 10.Zhao G, Skeath JB. Development (Cambridge, UK) 2002;129:1165–1174. doi: 10.1242/dev.129.5.1165. [DOI] [PubMed] [Google Scholar]
- 11.Overton PM, Meadows LA, Urban J, Russell S. Development (Cambridge, UK) 2002;129:4219–4228. doi: 10.1242/dev.129.18.4219. [DOI] [PubMed] [Google Scholar]
- 12.Buescher M, Hing FS, Chia W. Development (Cambridge, UK) 2002;129:4193–4203. doi: 10.1242/dev.129.18.4193. [DOI] [PubMed] [Google Scholar]
- 13.Wilson M, Koopman P. Curr Opin Genet Dev. 2002;12:441–446. doi: 10.1016/s0959-437x(02)00323-4. [DOI] [PubMed] [Google Scholar]
- 14.Kamachi Y, Uchikawa M, Kondoh H. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 15.Ma Y, Niemitz EL, Nambu PA, Shan X, Sackerson C, Fujioka M, Goto T, Nambu JR. Mech Dev. 1998;73:169–182. doi: 10.1016/s0925-4773(98)00050-1. [DOI] [PubMed] [Google Scholar]
- 16.Kamachi Y, Uchikawa M, Kondoh H. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 17.Bowles J, Schepers G, Koopman P. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 18.Yu Z, Syu LJ, Mellerick DM. Nucleic Acids Res. 2005;33:1–12. doi: 10.1093/nar/gki140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Doren M, Powell PA, Pasternak D, Singson A, Posakony JW. Genes Dev. 1992;6:2592–2605. doi: 10.1101/gad.6.12b.2592. [DOI] [PubMed] [Google Scholar]
- 20.Skeath JB, Panganiban G, Selegue J, Carroll SB. Genes Dev. 1992;6:2606–2619. doi: 10.1101/gad.6.12b.2606. [DOI] [PubMed] [Google Scholar]
- 21.Weiler S, Gruschus JM, Tsao DH, Yu L, Wang LH, Nirenberg M, Ferretti JA. J Biol Chem. 1998;273:10994–11000. doi: 10.1074/jbc.273.18.10994. [DOI] [PubMed] [Google Scholar]
- 22.Gehring WJ, Affolter M, Burglin T. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 23.Stormo GD. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 24.Zhao G, Skeath JB. Development (Cambridge, UK) 2002;129:1165–1174. doi: 10.1242/dev.129.5.1165. [DOI] [PubMed] [Google Scholar]
- 25.Skeath JB, Zhang Y, Holmgren R, Carroll SB, Doe CQ. Nature. 1995;376:427–430. doi: 10.1038/376427a0. [DOI] [PubMed] [Google Scholar]
- 26.Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, Stevanovic M, Goodfellow PN, Lovell-Badge R. Development (Cambridge, UK) 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- 27.Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Development (Cambridge, UK) 1998;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- 28.Rex M, Orme A, Uwanogho D, Tointon K, Wigmore PM, Sharpe PT, Scotting PJ. Dev Dyn. 1997;209:323–332. doi: 10.1002/(SICI)1097-0177(199707)209:3<323::AID-AJA7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 29.Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. Mech Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- 30.Valerius MT, Li H, Stock JL, Weinstein M, Kaur S, Singh G, Potter SS. Dev Dyn. 1995;203:337–351. doi: 10.1002/aja.1002030306. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh-Li HM, Witte DP, Szucsik JC, Weinstein M, Li H, Potter SS. Mech Dev. 1995;50:177–186. doi: 10.1016/0925-4773(94)00334-j. [DOI] [PubMed] [Google Scholar]
- 32.Kriks S, Lanuza GM, Mizuguchi R, Nakafuku M, Goulding M. Development (Cambridge, UK) 2005;132:2991–3002. doi: 10.1242/dev.01878. [DOI] [PubMed] [Google Scholar]
- 33.Ekonomou A, Kazanis I, Malas S, Wood H, Alifragis P, Denaxa M, Karagogeos D, Constanti A, Lovell-Badge R, Episkopou V. PLoS Biol. 2005;3:e186. doi: 10.1371/journal.pbio.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nirenberg M, Nakayama K, Nakayama N, Kim Y, Mellerick D, Wang LH, Webber KO, Lad R. Ann NY Acad Sci. 1995;758:224–242. doi: 10.1111/j.1749-6632.1995.tb24830.x. [DOI] [PubMed] [Google Scholar]
- 35.Ma Y, Certel K, Gao Y, Niemitz E, Mosher J, Mukherjee A, Mutsuddi M, Huseinovic N, Crews ST, Johnson WA, Nambu JR. J Neurosci. 2000;20:4596–4605. doi: 10.1523/JNEUROSCI.20-12-04596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horton RM. Mol Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.