Abstract
Previously, we characterized a parity-induced mammary epithelial cell population that possessed the properties of pluripotency and self-renewal upon transplantation. These cells were lineally marked by the expression of β-galactosidase (LacZ) as a result of mammary-specific activation of a reporter gene through Cre-lox recombination during pregnancy. We used this experimental model to determine whether testicular cells would alter their cell fate upon interaction with the mammary gland microenvironment during pregnancy, lactation, and involution. Adult testicular cells, isolated from seminiferous tubules, were mixed with limiting dilutions of dispersed mammary epithelial cells and injected into epithelium-divested mammary fat pads. The host mice were bred 6–8 weeks later and examined 20–30 days postinvolution. This approach allowed for the growth of mammary tissue from the injected cells and transient activation of the whey acidic protein promoter-Cre gene during pregnancy and lactation, leading to Cre-lox recombination and constitutive expression of LacZ from its promoter. Here we show that cells from adult seminiferous tubules interact with mammary epithelial cells during regeneration of the gland. They adopt mammary epithelial progenitor cell properties, including self-renewal and the production of cell progeny, which differentiate into functional mammary epithelial cells. Our results provide evidence for the ascendancy of the tissue microenvironment over the intrinsic nature of cells from an alternative adult tissue.
Keywords: transdifferentiation, transplantation, niche, stem cell
Somatic stem cells are maintained and regulated by their surrounding microenvironment (niche). A tissue-specific niche is a restricted locale that supports self-renewing division of stem cells and prevents them from differentiating. A simple stem cell niche has three components: localized signaling cells and extracellular matrix-controlling stem cell behavior, a specified range of signaling, and stem cell(s) (1). Any portion of the mouse mammary gland can regenerate an entire functional gland upon transplantation into a cleared mammary fat pad (2–4). This capacity remains undiminished regardless of the donor's age or reproductive history (5). Therefore, mammary stem cells are stably maintained within specific microenvironments throughout the gland for life. Mammary regeneration also occurs when dispersed epithelial cells from the glands are transplanted, suggesting that complete mammary epithelial stem cell niches may be reconstituted de novo (6–8). Stepwise (limiting) dilution of dispersed mammary cells results in a reduction of the percentage of inoculated fat pads positive for mammary epithelial growth, implying reduction in the number of mammary epithelial stem cells (7, 9). We hypothesized that the remaining cells comprised the epithelial signaling components and might support glandular regeneration if supplied with an extraneous source of stem/progenitor cells when injected into the mammary stroma. To test this hypothesis, we isolated cells from an alternative adult tissue (testis) known to possess active stem/progenitor cells throughout the lifetime and introduced them, along with the diluted mammary epithelial cells, into epithelium-divested mammary fat pads.
Employing tissue-targeted, Cre-lox-mediated, conditional activation of a reporter gene, new evidence was recently developed indicating the formation of a previously unrecognized mammary epithelial cell population that may originate from differentiating cells during pregnancy. This population does not undergo cell death during involution following lactation and persists throughout the lifetime of the female. In transplantation studies, these cells show the capacity for self-renewal and contribute significantly to the reconstitution of the resulting mammary outgrowth. In limiting dilution assays, it was found that they could produce both luminal and myoepithelial cells, comprise both lobule- and duct-limited epithelial outgrowths, and differentiate into all of the cellular subtypes recognized within the murine mammary epithelium. These cells, named parity-induced mammary epithelial cells (PI-MEC), have the property of self-renewal, are multipotent, and contribute progeny directly to the formation of secretory acini upon subsequent pregnancies. Therefore, PI-MEC represent LacZ+ lobule-limited, pluripotent epithelial progenitors, one of three distinct multipotent cell types previously identified in the mouse mammary gland (7, 9). To determine whether LacZ+ PI-MEC could develop from uncommitted cells from another adult tissue upon interaction with a mammary microenvironment comprised of signaling mammary epithelial cells and the mammary fat pad stroma, we inoculated testicular cells isolated from the seminiferous tubules of adult whey acidic protein promoter (WAP)-Cre/Rosa26 R mice mixed with limiting dilutions of mammary epithelial cells into epithelium-cleared mammary fat pads and allowed pregnancy, lactation, and involution to proceed in the transplanted hosts. Our results indicate that testicular cells can indeed interact with the mammary epithelial cells within the mammary stroma, proliferate, and contribute epithelial cell progeny to the resulting mammary outgrowths with all of the characteristics previously described for LacZ+ PI-MEC and their progeny.
Results and Discussion
Formation of Chimeric Mammary Outgrowths upon Transplantation.
Seminiferous cords were isolated from the decapsulated adult male testes of bitransgenic WAP-Cre/Rosa26 R mice (10, 11) and dissociated in collagenase, trypsin, and DNase as described by Bellve et al. (12). Interstitial tissue was removed before the dissociation of the spermatic cords. Following dissociation, the preparation was filtered through 40-μm mesh to remove aggregates. Microscopy confirmed that this testicular cell mixture contained all of the cell types present in the adult male seminiferous cords as previously reported, i.e., ≈10% spermatogonia Types A and B, ≈28% Sertoli cells, and the remainder differentiating spermatocytes (13). Both freshly isolated mammary cells and those cultured on plastic for 3–5 days were used in attempts to develop chimeric mammary outgrowths comprised of both male and female cells.
Mammary and testicular cells were combined and immediately inoculated (10 μl) directly into epithelium-divested mammary fat pads of 3-week-old Nu/Nu females. Mammary ductal growth proceeded for 6–8 weeks following injection, whereupon some hosts were allowed to complete a full-term pregnancy (required for activation of the Rosa26 LacZ reporter gene via WAP-Cre expression) (10, 11). Pups were removed after 1 week. Complete glandular involution was allowed. Subsequently, the mammary outgrowths were removed and stained as whole mounts for LacZ activity by X-Gal. Only male cells contained both the WAP-Cre and Rosa26 reporter transgenes. Therefore, only male-derived cells, which survived tissue remodelling following lactation and possessed both the WAP-Cre and Rosa26 reporter transgenes during pregnancy, will be LacZ+. Following Cre-induced recombination, LacZ expression is constitutive and subsequently acts as a lineal marker, which can be used to trace the subsequent fate of the activated, surviving LacZ+ cells and their progeny.
The presence of LacZ+ (blue) cells signals the occurrence of testicular cell progeny within the mammary outgrowth. An important feature of the experimental design is the restriction of WAP-Cre expression to mammary epithelium during secretory differentiation (14). This conclusion is borne out by the absence of LacZ+ cells in the mammary outgrowths removed from unbred hosts (Table 1). Mammary epithelial cells from primary culture (5.0 × 104 and 1.0 × 105 cells) produced a 1/6 and 4/6 frequency of positive takes, respectively (Table 1). In the experimental groups, 21/38 fat pads implanted with either 5 × 104 mammary/5 × 104 or 1 × 105 testicular cells contained positive epithelial growth, and LacZ+ cells were found in a large percentage of these outgrowths (18 of 21).
Table 1.
Transplantation results from implantation of dispersed testicular cells, mammary epithelial cells, and mixtures of each into mammary fat pads
| No. of testicular cells | No. of mammary cells | Parity | No. of positive takes/no. of implants | No. of LacZ+ takes/no. of positive takes |
|---|---|---|---|---|
| 50,000 (fresh) | 50,000 (culture) | No | 5/8 | 0/5 |
| 50,000 (fresh) | 50,000 (culture) | Yes | 15/30 | 12/15 |
| 100,000 (fresh) | 50,000 (culture) | No | 7/8 | 0/7 |
| 100,000 (fresh) | 50,000 (culture) | Yes | 6/8 | 6/6 |
| 0 | 50,000 (culture) | Yes | 1/6 | 0/1 |
| 0 | 100,000 (culture) | Yes | 4/6 | 0/4 |
| 100,000* (fresh) | 0 | Yes | 0/6 | 0/0 |
| 0 | 100,000 (fresh) | Yes | 10/10 | 0/10 |
| 0 | 50,000 (fresh) | Yes | 9/10 | 0/9 |
| 50,000 (fresh) | 50,000 (fresh) | Yes | 29/32 | 0/29 |
| Second transplants/LacZ+ outgrowths† | No | 12/18 | 12/12 |
Seminiferous tubule cells and mammary epithelial cells were introduced into the epithelium-free mammary fat pads of 3-week-old female mice and allowed to grow for 6–8 weeks. Hosts were maintained as virgins or had one cycle of pregnancy, lactation, and involution. Whole mounts were stained by X-Gal to reveal β-galactosidase expression. Only outgrowths from impregnated hosts contained WAP-Cre-activated LacZ+ cells. Freshly isolated mammary cells gave more positive outgrowths either alone or mixed with testicular cells, but never contained LacZ+ epithelial cells.
*Implants of 1 × 106 testicular cells also failed to produce outgrowths (data not shown).
†Successful secondary transplants of LacZ+ outgrowths uniformly contained LacZ+ progeny throughout the ducts.
Freshly isolated mammary cells inoculated at 1.0 × 105 and 5.0 × 104 per fat pad gave 100 and 96.6% positive mammary outgrowths (Table 1), but no LacZ+ cells when combined with 5.0 × 104 testicular cells. This result is consistent with the conclusion that the testicular cells effectively competed for reforming mammary stem cell niches only when mammary stem cell numbers (and the frequency of positive takes) were reduced. Alternatively, it is very difficult to completely disperse mammary cells from fresh tissue as compared with cultures, and the increased regenerative capacity and absence of LacZ+ cells (96 and 100%) may result from the presence of cell aggregates. Our interpretation was confirmed when freshly isolated mammary cells were reduced to <10,000 before injection with testicular cells. At this concentration, freshly isolated mammary cells gave results similar to those obtained with the cultured mammary cells (data not shown). No LacZ+ cells appeared in outgrowths produced by injecting mammary epithelium alone (Table 1), and no outgrowths resulted from inoculating testicular cells alone.
LacZ+ Cells in the Chimeric Outgrowths Possess Mammary Characteristics.
LacZ+ cells were uniformly positioned along the mammary ducts and side branches (Fig. 1 a–c), suggesting that male cells had proliferated in concert with female cells during duct morphogenesis before activation of the LacZ reporter during pregnancy. In lactating hosts that had undergone a previous gestation, LacZ+ cells were found both among the luminal epithelium (secretory) and in basal locations, apparently adopting a myoepithelial fate (Fig. 2). The secretory lineage was established by colocalization of casein and β-galactosidase (Fig. 3a). The myoepithelial lineage of the LacZ+ cells was confirmed by colocalization of β-galactosidase and cytokeratins (K5, K14) and smooth muscle actin in Fig. 3 b and c.
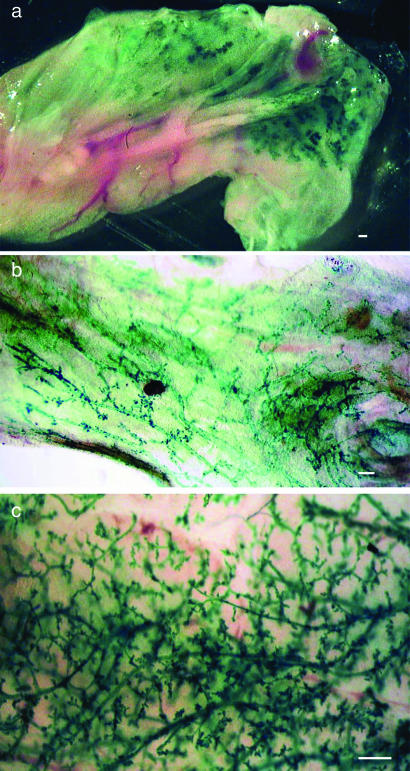
Fig. 1.
Detection of male-derived cells in chimeric outgrowths. (a) X-Gal-stained whole mammary fat pad from postpregnant host mouse reveals numerous LacZ+ cells within the gland. (b) After removal of fat, LacZ− cells were found uniformly distributed along the mammary ducts in this whole mount. (c) LacZ+ cells in a whole mount from a second-generation transplant showing the self-renewal of male progeny. (Scale bars: 1.0 mm.)
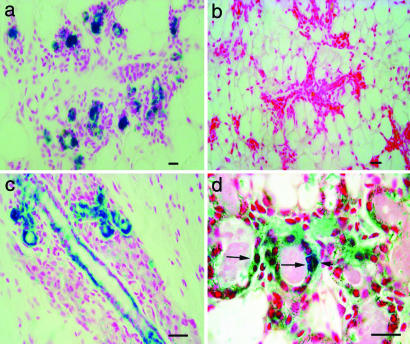
Fig. 2.
Tissue section analysis of X-Gal-stained outgrowths. (a) Cross-section through mammary ducts showing the distribution of LacZ+ epithelial cells among negative stroma. (b) Section from the host mammary gland from same mouse. (c) Longitudinal section of duct in chimeric outgrowth with LacZ+ epithelium. (d) Higher magnification of LacZ+ epithelial cells in secretory acinus of pregnant host showing blue luminal cells (long arrows) adjacent to blue myoepithelial cell (short arrow). (Scale bars: 20 μm.)
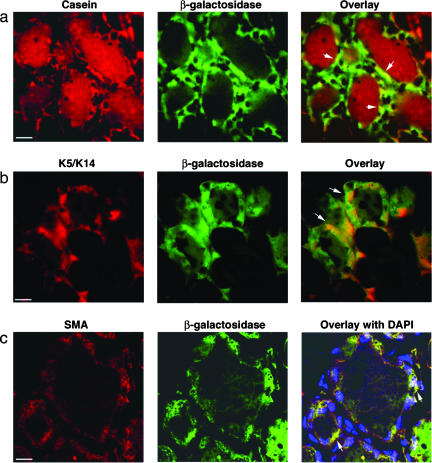
Fig. 3.
Male cell progeny may express luminal or myoepithelial cell markers. (a) Colocalization of casein and β-galactosidase in chimeric outgrowths of lactating murine mammary glands. Arrows indicate luminal cells that express both casein and β-galactosidase. (b and c) Male cells expressing β-galactosidase and keratin 5 (K5)/keratin 14 (K14) (b) or smooth muscle actin (SMA) (c), indicating a myoepithelial fate. Arrows in b indicate cells expressing both K5/K14 and β-galactosidase; arrows in c indicate cells expressing both β-galactosidase and smooth muscle actin. (Scale bars: a and b, 20 μm; c, 10 μm.)
Demonstration of Transgene and Y Chromosome-Specific Sequences in Chimeric Glands.
PCR analysis of the DNA isolated from the chimeric outgrowths demonstrated the presence of both transgenes and sequences specific to the Y chromosome, verifying the presence of testicular cell DNA. In addition, titration of male donor DNA with host female DNA indicated that at least 15% of the epithelial cells in the chimeric outgrowth possessed all three genes [supporting information (SI) Fig. 5]. This estimate was determined from previous studies (9, 15), which determined the relative epithelial cell DNA (≈14%) content represented in a fully filled mammary fat pad and the dilution factor at which the PCR signal for the transgenic and Y chromosome sequences were no longer detectable.
Appearance of LacZ+ Cells in Second-Generation Transplants of Chimeric Mammary Tissue.
Our previous studies of WAP-Cre-activated, LacZ+ PI-MEC in intact primiparous, involuted mammary glands showed that they were capable of self-renewal upon transplantation, in addition to giving rise to epithelial cell progeny of both luminal and myoepithelial cell lineages. Second-generation transplantation of fragments from the male/female chimeric outgrowths (Table 1 and Fig. 1c) confirmed that the LacZ+, male-derived epithelial cells were also capable of self-renewal and proliferation, activities analogous to those displayed by the PI-MEC in intact WAP-Cre/Rosa reporter mouse mammary glands.
The results clearly demonstrate that male cells from the seminiferous tubules of adult mice interact with female mammary epithelial cells upon inoculation into the epithelium-divested mammary fat pad and proliferate to contribute cells analogous to PI-MEC in the resulting epithelial outgrowths (SI Fig. 6 shows a summary of our results).
Fluorescent in Situ Hybridization (FISH) and Cell Cycle Analysis of the Chimeric Outgrowths.
Interaction of testicular and mammary epithelial cells in the outgrowths did not involve testicular-mammary cell fusion. To confirm this, FISH with X chromosome telomere-specific probe and Y chromosome locus-specific probe was carried out on the interphase nuclei of epithelium in chimeric outgrowths. Hybridization of sections containing LacZ+ and LacZ− postlactation epithelium in chimeric outgrowths with X and Y chromosome-specific probes indicated the absence of polyploidy and clearly demonstrated the presence of interspersed cells bearing either two X chromosomes or one X and one Y chromosome in the same acinus (Fig. 4 and SI Figs. 7 and 8). This result was obtained in both multiple primary outgrowths and second-generation transplants. To further support these observations, epithelium from chimeric outgrowths was dispersed and compared with dispersed seminiferous tubule cells, mammary cells from primary cultures used in the mixing experiments, and dispersed epithelium isolated from the intact glands of the transplanted host by flow cytometry after staining with propidium iodide (SI Fig. 9). This analysis confirmed that the chimeric tissue did not contain a greater percentage of 4N (or greater) nuclei than any of the nonchimeric populations.
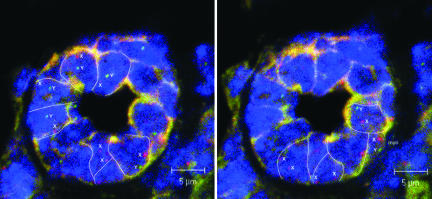
Fig. 4.
X and Y chromosome FISH analysis was performed on paraffin-embedded, 6-μm sections of mammary glands containing chimeric outgrowths. A mammary secretory structure is shown with the X chromosome labeled in red (x), the Y chromosome labeled in green (y), and the nuclei stained with DAPI. Serial 1.0-μm slices photographed under three-color confocal microscopy reveal the presence of male cells juxtaposed with female cells in the same acinus. (Scale bars: 5.0 μm.)
Alpha-6 Integrin and Testicular Cell Interaction with Mammary Cells.
Testicular cells were sorted for the presence or absence of alpha-6 integrin, a surface marker more prominently displayed on germinal stem cells (GSC) (16). Preliminary results show that testicular cell fractions depleted of alpha-6 integrin++ cells failed to produce positive mammary outgrowths (0/6) when mixed with mammary cells, whereas those sorted for the presence of alpha-6 integrin produced 3/6 positive outgrowths. This result suggests that GSC may play an important role in chimera formation. To determine the presence of undifferentiated GSCs, chimeric outgrowths containing LacZ+ cells were stained for germ cell nuclear antigen 1 (GCNA1), a nuclear antigen expressed exclusively within the germinal cells of the mouse seminiferous tubules (17, 18). LacZ− cells located basal to the differentiated LacZ+ epithelium lining the secretory mammary ducts stained positive for GCNA1, suggesting the persistence of GCNA expression in undifferentiated testicular cells. Conversely, LacZ+ male cells within the secreting acini of fully lactating chimeric outgrowths were negative for GCNA1 expression (SI Fig. 10). This result suggests extinction of GCNA1 expression in the testicular progeny during mammary secretory differentiation. Similarly, GCNA1 expression is lost during spermatogenic differentiation in the intact seminiferous tubules. No GCNA1 staining was present in wild-type mammary tissue.
The interaction between the testicular and mammary epithelial cells to form a regenerated mammary gland capable of fully functional differentiation is unprecedented. The present study demonstrates that testicular cells can enter reforming mammary niches that support the function of lobule-limited progenitors where they are maintained in subsequent mammary tissue expansion. Further study is required to determine the specific cellular cues that permit this interaction and its continued support in subsequent transplants. The result anticipates that stem/progenitor cells from other organ systems may also be able to substitute in a similar way in chimeric mammary outgrowths and perform mammary-specific stem cell functions. These experiments announce an apparent dominance of the tissue-specific stem cell niche over progenitor cell function and cell fate.
Materials and Methods
Mice.
The transgenic WAP-Cre/Rosa26 R mice were engineered and typed according to Wagner et al. (11). Female Nu/Nu/NCR mice were used as hosts for the transplantation studies. All mice were housed in Association for Assessment and Accreditation of Laboratory Animal Care-accredited facilities in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The National Cancer Institute Animal Care and Use Committee approved all experimental procedures.
Sperm/Germ Cell Dissociation Procedure.
Testes were excised from WAP-Cre/Rosa26 R males following the protocol in Bellve et al. (12) with a few modifications. They were then decapsulated to remove the tunica albuginea, placed in DMEM (Invitrogen, Carlsbad, CA) containing 0.5 mg/ml collagenase (Sigma–Aldrich, St. Louis, MO), and incubated at 33°C in a shaking water bath for 15 min at 120 rpm. The suspension was spun for 5 min at 1,200 rpm to pellet the seminiferous chords. The fatty top layer and the pellet were washed two times in straight DMEM. This procedure allowed for removal of the interstitial tissue. The dispersed seminiferous chords were then placed in 20 ml of DMEM containing 0.5 mg/ml trypsin and 1 μg/ml DNase (Invitrogen) and incubated as above. Any remaining cell aggregates were then sheared by pipetting 10–12 times. The cells were then spun and washed as above. They were resuspended in 10 ml of DMEM containing 0.5% BSA (Sigma–Aldrich) and filtered through a 40-micron filter to remove any remaining clumps. Viability was determined by trypan blue exclusion, and cell counts were done in a hemacytometer.
Mammary Epithelial Cell Preparation.
Mammary epithelial cells were collected from primary mammary cultures after 4–7 days on plastic culture flasks in Dulbecco's MEM supplemented with 10% FCS, 1.0 mg/ml Insulin, and 10 μg/ml epidermal growth factor. Fibroblasts were reduced before collection of the epithelial cells by differential trypsinization (7).
Cell and Tissue Transplantation.
The surgical techniques used to clear the mammary epithelium from the no. 4 fat pad of 3-week-old host mice, and the subsequent transplantation of tissue fragments or cell suspensions, have been described in detail (7, 8, 19). Briefly, the mice were anesthetized, and the clearing procedure was performed immediately before insertion of the transplanted fragment or cell suspension. Cell suspensions were implanted in 10-μl volumes with a Hamilton (Reno, NV) syringe equipped with a 30-gauge needle. The implanted females were placed with males 4–6 weeks following implantation to initiate pregnancy and secretory development.
X-Gal and Immunostaining of Mammary Gland Whole Mounts.
Whole mounts of the entire inguinal gland were fixed and stained as described earlier. Briefly, the gland was spread on a glass slide, fixed in paraformaldehyde (4.0%) for 1–2 h, permeabilized in 0.01% Nonidet P-40 in PBS overnight at 4°C, and subsequently processed for X-Gal as described. Stained glands were repeatedly rinsed in PBS, postfixed in Carnoy's fixative, cleared in 100% ethanol, and placed in xylenes before whole-mount analysis. For histological examination, X-Gal-stained whole mounts were embedded in paraffin, sectioned at 6.0 μm, and counterstained with nuclear fast red. Immunocytochemistry was carried out on deparaffinized sections. The antibodies used were against total purified mouse caseins (19) (rabbit polyclonal 1:500), cytokeratin 5 (Covance, 1:500), cytokeratin 14 (Covance; 1:500), smooth muscle actin (Zymed Laboratories, South San Francisco, CA; 1:100), and β-galactosidase (Invitrogen; 1:500). Primary antibodies were directly fluorescently labeled with Zenon IgG labeling kits (Invitrogen) according to the manufacturer's protocol. Antigen retrieval in boiling 0.01 M sodium citrate (pH 6.0) for 10 min was performed when investigating cytokeratins. For GCNA1, detection sections were exposed to the antibody overnight at 4°C, followed by secondary antibody conjugated with Alexa 647. Known positive and negative tissue controls fixed similarly were used to confirm the specificity of staining. Fluorescent images were collected with a Zeiss (Thornwood, NY) NLO confocal microscope.
DNA Extraction and PCR.
Genomic DNA was extracted from mammary tissue according to QIAGEN DNeasy kit (catalog no. 69506; Valencia, CA) protocol. PCR analysis was performed with WAP and Cre primers from Wagner et al. (10, 11), Y6 primers from Peters et al. (20), and standard mouse GAPDH primers (21).
FISH Analysis on Paraffin Sections.
Slides containing multiple 5–6-μm paraffin sections were deparaffinized 3 × 10 min in xylenes. The tissue was then rehydrated in an ethanol series (100%, 90%, 70%) 3 × 3 min each, followed by 2 × 3 min in 2× SSC with shaking. Slides were pepsin treated (Sigma–Aldrich, catalog no. P6887) in 0.01 M HCl at 37°C for 30 min and then washed in PBS 3 × 3 min. Fixation and dehydration were done simultaneously by reversing the ethanol series (70%, 90%, 100%) for 10 min each. Using an X chromosome telomere probe and a Y chromosome locus-specific BAC probe (bJKB4) (22) that were labeled by nick translation by using Spectrum Orange-dUTP (Vysis, Des Plaines, IL) and Oregon Green-dUTP (Invitrogen), respectively, analysis was performed according to a previously published protocol (23).
Determination of Ploidy by Propidium Iodide Staining.
Dispersed cell preparations were washed twice in DMEM without serum, resuspended at 1–2 × 107 cells/ml, and placed on ice for 15 min. Cells were fixed in 70% ethanol, treated with 100 units of RNase (Sigma–Aldrich) at 37°C for 20 min, and stained for 30 min at 4°C in a 50 μg/ml solution of propidium iodide (Molecular Probes, Eugene, OR) in PBS. Cell clumps were removed by filtering through 40-μm nylon mesh before analysis on a Calibur flow cytometer running CellQuest software (BD Biosciences, Franklin Lakes, NJ). Subsequent cell cycle and DNA content analyses were performed by using FlowJo software (Tree Star, Inc., San Carlos, CA).
Magnetic Enrichment for Alpha-6 Integrin Testicular Cells.
After cells were purified from the seminiferous tubules, they were counted and resuspended in 100 μl of primary antibody, alpha-6 integrin (Santa Cruz Biotechnologies, Santa Cruz, CA; 1:50 dilution). After incubating at 4° for 30 min, the cells were washed with 10× volume of buffer (autoMACS running buffer; Miltenyi Biotec, Auburn, CA) and spun at 300 times g for 10 min. The cell pellet was resuspended in 100 μl of secondary antibody, anti-goat IgG-B (1:100 dilution); incubated at 4° for 15 min; and washed with 10× volume buffer and spun as above. The pellet was then resuspended in 80 μl of buffer and 20 μl of antibiotin magnetic beads (Miltenyi Biotec Inc., Auburn, CA) per 10.0 × 106 cells. Cells were mixed with beads, incubated at 4° for 15 min, and then washed and spun as above. The cell/bead mixture was resuspended in 500 μl of buffer and run through large selection columns (Miltenyi Biotec Inc.) according to the manufacturer's instructions.
Supplementary Material
Abbreviations
- FISH
fluorescent in situ hybridization
- GCNA1
germ cell nuclear antigen 1
- GSC
germinal stem cells
- PI-MEC
parity-induced mammary epithelial cells
- WAP
whey acidic protein promoter.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611637104/DC1.
References
- 1.Li L, Xie T. Annu Rev Cell Dev Biol. 2005;21:605–631. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- 2.Deome KB, Faulkin LJ, Jr, Bern HA, Blair PB. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- 3.Daniel C, DeOme K, Young L, Blair P, Faulkin L. Proc Natl Acad Sci USA. 1968;61:53–60. doi: 10.1073/pnas.61.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniel CW, Young LJ. Exp Cell Res. 1971;65:27–32. doi: 10.1016/s0014-4827(71)80046-0. [DOI] [PubMed] [Google Scholar]
- 5.Young LJ, Medina D, DeOme KB, Daniel CW. Exp Gerontol. 1971;6:49–56. doi: 10.1016/0531-5565(71)90048-9. [DOI] [PubMed] [Google Scholar]
- 6.Daniel CW, DeOme KB. Science. 1965;149:634–636. doi: 10.1126/science.149.3684.634. [DOI] [PubMed] [Google Scholar]
- 7.Smith GH. Breast Cancer Res Treat. 1996;39:21–31. doi: 10.1007/BF01806075. [DOI] [PubMed] [Google Scholar]
- 8.Smith GH, Gallahan D, Zwiebel JA, Freeman SM, Bassin RH, Callahan R. J Virol. 1991;65:6365–6370. doi: 10.1128/jvi.65.11.6365-6370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kordon EC, Smith GH. Development (Cambridge, UK) 1998;125:1921–1930. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- 10.Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner KU, Boulanger CA, Henry MD, Sagagias M, Hennighausen L, Smith GH. Development (Cambridge, UK) 2002;129:1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- 12.Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romrell LJ, Bellve AR, Fawcett DW. Dev Biol. 1976;49:119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- 14.Boulanger CA, Wagner KU, Smith GH. Oncogene. 2005;24:552–560. doi: 10.1038/sj.onc.1208185. [DOI] [PubMed] [Google Scholar]
- 15.Nicoll CS, Tucker HA. Life Sci. 1965;4:993–1001. doi: 10.1016/0024-3205(65)90203-1. [DOI] [PubMed] [Google Scholar]
- 16.Ebata KT, Zhang X, Nagano MC. Mol Reprod Dev. 2005;72:171–181. doi: 10.1002/mrd.20324. [DOI] [PubMed] [Google Scholar]
- 17.Enders GC, May JJ., II Dev Biol. 1994;163:331–340. doi: 10.1006/dbio.1994.1152. [DOI] [PubMed] [Google Scholar]
- 18.Wang D, Enders GC. Cancer Lett. 1996;100:31–36. doi: 10.1016/0304-3835(95)04068-4. [DOI] [PubMed] [Google Scholar]
- 19.Smith GH, Vonderhaar BK, Graham DE, Medina D. Cancer Res. 1984;44:3426–3437. [PubMed] [Google Scholar]
- 20.Peters SO, Bauermeister K, Simon JP, Branke B, Wagner T. J Immunol Methods. 2002;260:109–116. doi: 10.1016/s0022-1759(01)00525-7. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen SB, Kordon E, Callahan R, Smith GH. Oncogene. 2001;20:5291–5301. doi: 10.1038/sj.onc.1204624. [DOI] [PubMed] [Google Scholar]
- 22.Simpson EM, Johnson KA, Shirley BJ, Fang GY, Bayleran JK, Lerner CP. Genesis. 2002;33:62–66. doi: 10.1002/gene.10093. [DOI] [PubMed] [Google Scholar]
- 23.Van Prooijen-Knegt AC, Van der Ploeg M. Cell Biol Int Rep. 1982;6:653. doi: 10.1016/0309-1651(82)90128-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.