Abstract
Production of transgenic animals has raised concern regarding their potential ecological impact should they escape or be released to the natural environment. This concern has arisen mainly from research on laboratory-reared animals and theoretical modeling exercises. In this study, we used biocontained naturalized stream environments and conventional hatchery environments to show that differences in phenotype between transgenic and wild genotypes depend on rearing conditions and, critically, that such genotype-by-environment interactions may influence subsequent ecological effects in nature. Genetically wild and growth hormone transgenic coho salmon (Oncorhynchus kisutch) were reared from the fry stage under either standard hatchery conditions or under naturalized stream conditions. When reared under standard hatchery conditions, the transgenic fish grew almost three times longer than wild conspecifics and had (under simulated natural conditions) stronger predation effects on prey than wild genotypes (even after compensation for size differences). In contrast, when fish were reared under naturalized stream conditions, transgenic fish were only 20% longer than the wild fish, and the magnitude of difference in relative predation effects was much reduced. These data show that genotype-by-environment interactions can influence the relative phenotype of transgenic and wild-type organisms and that extrapolations of ecological consequences from phenotypes developed in the unnatural laboratory environment may lead to an overestimation or underestimation of ecological risk. Thus, for transgenic organisms that may not be released to nature, the establishment of a range of highly naturalized environments will be critical for acquiring reliable experimental data to be used in risk assessments.
Keywords: coho salmon, phenotypic plasticity, risk-assessment
Transgenes with a variety of functions have now been inserted into a wide range of animals with foreseeable applications ranging from small-scale basic laboratory research and applied medicine to large-scale disease control and commercial meat production (1–6). The latter application, in particular, has raised concern regarding the potential negative impact that transgenic animals may have on the natural environment whether they escape from rearing facilities or are purposefully released into the wild (7).
This concern has led to numerous articles on the conceptual problem of ecological risk-assessment (8–10), and several laboratory and theoretical studies have addressed the fitness and/or ecological consequences of transgenic animals (11–17). However, except for a few field trials on nematodes and mites, few empirical studies to evaluate the direct ecological effects of transgenic animals in more natural environments have been undertaken (18, 19).
The phenotype of a transgenic animal is likely to depend not only on the desired biological effect of the transgene but also will be influenced by the interaction of the transgene with environmental conditions during development (20). Transgenic and wild conspecifics may show different phenotypic responses to altered environmental conditions (i.e., nonparallel reaction norms). As a consequence of such genotype-by-environment (G×E) interactions, phenotypes of laboratory-reared animals may not be predictive of phenotypes that would develop in nature. Further, developmental plasticity may influence subsequent fitness and ecological consequences of individuals when they enter a novel wild environment (21), thereby making risk assessments complicated (22, 23) or inaccurate (Fig. 1).
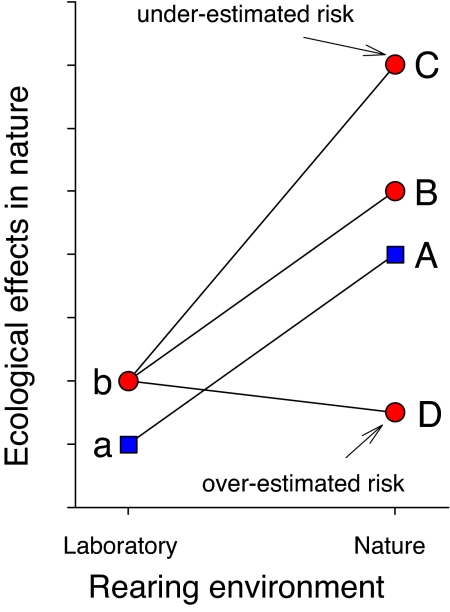
Fig. 1.
Conceptual model of G×E interactions and their potential implications for ecological risk assessment. Blue squares and line display the reaction norm of hypothetical ecological effects of wild genotypes in nature after being reared under laboratory conditions (a) and in nature (A). Red circles and line display the reaction norms of potential ecological effects of transgenic genotypes in nature after being reared under laboratory conditions (b) and in different natural environments (B–D). Only effects of a, b, and A can be observed with certainty because the transgenic genotype cannot be reared in nature. B displays the effects of transgenic genotypes reared under natural conditions when a G×E interaction is absent (i.e., the reaction norm of b–B is parallel to a–A). Under such a scenario (i.e., lacking G×E effects), the effect of a transgenic genotype reared in nature (B) can be predicted by looking at the magnitude of difference in effects between genotypes reared in the laboratory (b–a) and extrapolating from a wild genotype reared in nature [hence B = A + (b − a)]. C and D display the effects of the same transgenic genotypes (b) but with G×E interactions present. In C, the G×E interaction causes the transgenic genotype to have a stronger effect (at a magnitude of C–B) than would be predicted by the relationship between a, b, and A, and hence leads to an underestimation of risk. In D, the G×E interaction causes the transgenic genotype to have a reduced effect relative to that extrapolated (at a magnitude of B–D) and hence leads to an overestimation of risk.
Fast-growing growth hormone (GH) transgenic coho salmon (Oncorhynchus kisutch) are a relevant model for use in evaluating the effects of G×E on risk assessment. Several species of fish have been modified genetically and currently are being explored as a way to increase the production efficiency and yield in aquaculture (24–28). These transgenic fish overexpress GH and require enhanced feed consumption to achieve the increased growth rates (29). Observations under laboratory conditions have shown GH transgenic fish to be more competitive (30), less discriminate in choosing prey (31), less affected by predator presence when making foraging decisions (32), more likely to attack novel prey (31), and better at using lower-quality food (29) compared with genetically wild relatives. In the natural environment, such physiological and behavioral alterations have the potential to directly influence not only conspecifics through increased competition for food but also prey populations and previously unaffected species through increased predation.
Previous work on GH transgenic coho salmon conducted in simple laboratory environments has shown the effects of G×E on survival (33) and potentially on reproductive fitness (17). Currently, the release of genetically engineered fish to natural systems is associated with uncertain potential ecological consequences and, thus, it is not known whether G×E interactions would influence ecological consequences of transgenic organisms. To examine this scenario, we have developed biocontained simulated natural environments to rear and study transgenic and wild fish that can be compared with fish reared in conventional hatchery environments. The predation effects of these fish were then tested within simulated natural environments. Under hatchery conditions, wild fish were fed to satiation with commercial fish feed, and transgenic fish were reared under either satiation–ration conditions or restricted growth conditions (so that they were the same size as age-matched wild genotype fish). In experiment I, we assessed the predation effects of the three groups of hatchery-reared fish (wild, fast-growing transgenic, and restricted growth transgenic) under simulated natural conditions. In experiment II, we assessed the predation effects of the wild and transgenic fish reared in a simulated natural environment and also of wild fish captured from nature. In this report, we show that (i) phenotypic plasticity can lead to significant differences in phenotype between transgenic animals reared under more natural conditions relative to individuals reared under laboratory conditions and (ii) G×E interactions can preclude an accurate determination of ecological consequences from phenotypes developed in simple laboratory facilities to that of transgenic animals developing in nature.
Results
In experiment I, growth rates and predation effects of fish reared only in hatchery conditions were examined. Fish groups (all of the same age) included wild genotype salmon, transgenic salmon that were fed to satiation and experienced rapid growth, and transgenic salmon that were fed a restricted ration so that they were the same size and had experienced the same growth rate as the wild fish. Transgenic fish that were fed to satiation were able to sustain their increased potential for growth and, as a consequence, they were 2.7 times longer and 25 times heavier than the wild and restricted growth transgenic fish (Table 1 and Fig. 2c).
Table 1.
Initial and final length (L, millimeters), weight (W, grams), and condition factor (105 × W × L−3) of predator coho salmon in experiments I and II
| Exp. | Predator | Length |
Weight |
CF |
|||
|---|---|---|---|---|---|---|---|
| Initial | Final | Initial | Final | Initial | Final | ||
| I | Wsat | 106 ± 1.9a | 110 ± 1.9 | 14.0 ± 0.8a | 14.5 ± 0.8 | 1.2 ± 0.01a | 1.1 ± 0.01 |
| Tres | 107 ± 1.4a | 115 ± 1.3 | 12.4 ± 0.4a | 14.8 ± 0.2 | 1.0 ± 0.02b | 1.0 ± 0.03 | |
| Tsat | 289 ± 3.1b | 294 ± 3.7 | 322 ± 13b | 300 ± 13 | 1.3 ± 0.02c | 1.2 ± 0.02 | |
| II | Wnat | 105 ± 4.3a | 109 ± 4.1 | 11.8 ± 1.3a | 13.0 ± 1.4 | 1.0 ± 0.02a | 1.0 ± 0.00 |
| Wsim | 99 ± 4.5a | 106 ± 4.6 | 11.5 ± 1.3a | 12.8 ± 1.5 | 1.2 ± 0.02b | 1.1 ± 0.02 | |
| Tsim | 126 ± 5.3b | 130 ± 5.3 | 21.9 ± 2.4b | 22.8 ± 2.4 | 1.1 ± 0.02ab | 1.0 ± 0.02 | |
Experiments were conducted in naturalized conditions for 20 days and 14 days for experiments I and II, respectively. For experiment I, all predators were reared only in hatchery conditions. Wild genotype predators received satiation rations (Wsat), whereas transgenic genotypes received either satiation rations (Tsat) or growth-restricting rations (Tres), resulting in them being the same size at age as Wsat predators. For experiment II, wild-genotype predators either were reared in simulated natural conditions in the laboratory (Wsim) or were obtained from nature (Wnat), and transgenic predators were reared in the same simulated natural environment as Wsim salmon. Initial measurements within experiments with different superscript letters are significantly different (Scheffé's pairwise post hoc test with P < 0.05). Statistics for differences in specific growth in weight and length and change in condition are given in the text.
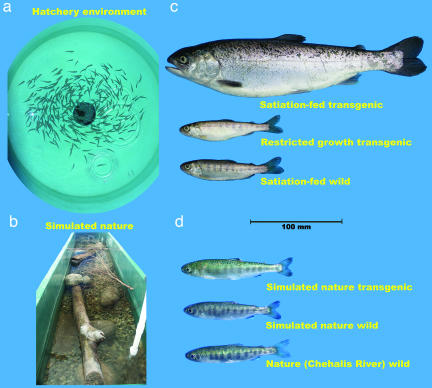
Fig. 2.
Rearing conditions and phenotypes of experimental fish in experiments I (a and c) and II (b and d). (a) Hatchery environment. (b) Simulated natural environment (these environments also were used during the predation experiments). (c) Phenotypes of fish reared in the hatchery environment. (d) Phenotypes of fish reared in the simulated natural environment or caught from nature (Chehalis River, British Columbia, Canada).
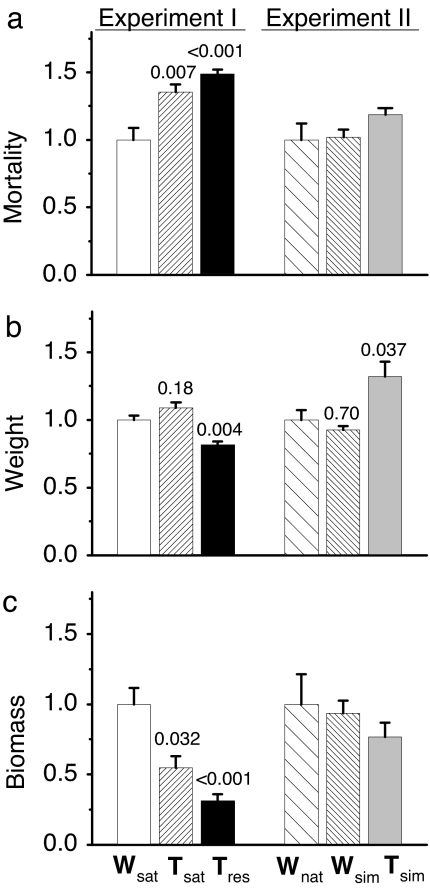
Although fish in experiment I were reared under hatchery conditions, actual experimentation was undertaken in naturalized stream environments. Differences in prey (rainbow trout fry) consumption rate were observed among the three hatchery-reared predator types (F2,9 = 15.3, P = 0.001; Fig. 3a) within their respective habitats. Wild fish consumed less prey (57.5 ± 5.1% SEM) than did large transgenic (77.8 ± 3.3%, P = 0.007) and restricted-growth transgenic predators (85.5 ± 1.9%, P < 0.001). The growth rate of the prey was influenced by predator type (F2,9 = 19.7, P < 0.001; Fig. 3b). Mean final weight of prey in the presence of wild predators (0.25 ± 0.004 g) was not different from the mean weight of prey in the presence of large transgenic predators (0.26 ± 0.006 g, P = 0.18) but was significantly greater than the mean weight of prey in the presence of restricted-growth transgenic predators (0.23 ± 0.003 g, P = 0.004). Differences in survival and growth of prey contributed to differences in final prey population biomass among predator types (F2,9 = 15.4, P = 0.001; Fig. 3c). In the presence of wild predators, final prey biomass (10.7 ± 1.2 g) was greater than the prey biomass for both the large transgenic (5.9 ± 0.9 g, P = 0.032) and the restricted growth transgenic predators (3.3 ± 0.5 g, P < 0.001).
Fig. 3.
Standardized predation effects (±SE) of transgenic and genetically wild coho salmon predators on fry prey under simulated natural conditions. (a) Predation rate. (b) Final weight of prey. (c) Final biomass of prey. In experiment I, predators were of wild (W) genotype or of transgenic (T) genotypes reared either under standard hatchery conditions (Wsat and Tsat) or growth-restricted conditions (Tres; restricted by feeding the same amount that satiated wild fish). In experiment II, predators were reared under simulated-natural (Wsim and Tsim) conditions or caught from the wild (Wnat). Data are standardized to 1 within each experiment relative to wild reference groups (Wsat in experiment I and Wnat in experiment II). Actual values are given in text. Number above a single bar indicates P value from a pairwise comparison between that group and the reference group (Dunnett's post hoc). P values are only provided when the one-way ANOVA was significant (i.e., P < 0.05).
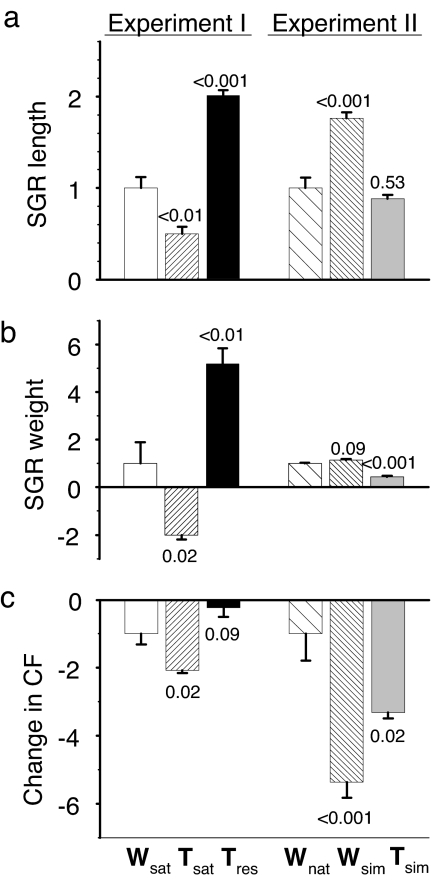
During experiment I, all three types of hatchery-reared predators increased in length, but the large transgenic fish lost weight whereas the other two predator types increased in weight (Table 1). Specific growth rate in length (SGRL) differed (F2,9 = 76.3, P < 0.001), with wild predators growing faster than the large transgenic predators (P = 0.006) but slower than the restricted growth transgenic predators (P < 0.001; Fig. 4a). Specific growth rate in weight (SGRW) differed among predator types in a similar way as SGRL (F2,9 = 31.0, P < 0.001), with large transgenic predators losing weight relative to the wild predators (P = 0.017) and restricted-growth transgenic predators growing faster than the wild predators (P = 0.003; Fig. 4b). Loss of condition varied among the predator types (F2,9 = 14.6, P < 0.001; Table 1), and the large transgenic fish lost more in condition than wild predators (P = 0.022), with no difference between wild and restricted-growth transgenic predators (Fig. 4c).
Fig. 4.
Standardized growth and change in condition of transgenic and genetically wild predators foraging on live prey under simulated natural conditions. (a) Specific growth in weight. (b) Specific growth in length. (c) Change in condition. See Fig. 3 legend for further explanation. Actual values are given in text.
In experiment II, we sought to mimic natural conditions during rearing as well as during experimentation. In simulated natural stream environments, transgenic fish did not realize their full growth potential (as observed under hatchery conditions) and were only ≈20% longer and twice as heavy compared with same-age wild fish at the onset of the predation experiment (Table 1 and Fig. 2d). Wild-genotype salmon reared in the artificial streams were the same size as wild fish collected from nature, indicating that the facilities created an environment that was capable of simulating growth rates that would be observed in nature. Therefore, transgenic fish reared in these simulated natural environments are anticipated to display predation effects similar to what would be observed if they were reared in nature. In this experiment, there were no differences in prey (coho salmon fry) consumption rates among the predator types (F2,9 = 1.55, P = 0.27; Fig. 3a) within their respective habitats, with wild predators caught from nature consuming 62.0 ± 7.4% of prey and wild and transgenic predators reared in the simulated natural environment consuming 63.2 ± 3.6% and 73.5 ± 3.1%, respectively. Predator type had an effect on final mean size of the prey (F2,9 = 6.9, P = 0.015; Fig. 3b): Prey in the presence of wild predators from nature (0.45 ± 0.01 g) were the same size as prey in the presence of the simulated naturally reared wild fish (0.44 ± 0.004 g) but were smaller than prey in the presence of transgenic predators (0.50 ± 0.02 g; P = 0.037). Final prey population biomass was not affected by predator type (F2,9 = 0.81; P = 0.48; Fig. 3c). Final biomass of prey was 17.1 ± 3.69 g in the presence of wild fish from nature, 16.2 ± 1.57 g in the presence of simulated-naturally reared wild predators, and 13.2 ± 1.73 g in the presence of transgenic predators.
During predation experiment II, all predators increased in length and weight (Table 1). Predators differed in SGRL (F2,9 = 34.7, P < 0.001) and SGRW (F2,9 = 69.1, P < 0.001). Wild predators from nature had lower SGRL than wild predators reared in the simulated natural environment (P < 0.001), with no difference relative to the transgenic predators (P = 0.53; Fig. 4a). SGRW of the wild-caught predators was similar to that of wild fish reared in the simulated natural environment (P = 0.086) and greater than that of transgenic fish (P < 0.001; Fig. 4b). The growth differences led to differences in loss of condition among predator types (F2,9 = 16.9, P < 0.001; Table 1). Wild-caught predators had a smaller decrease in condition than both the wild fish reared in the simulated natural environment (P < 0.001) and the transgenic predators (P = 0.024; Fig. 4c).
Discussion
These experiments show that GH transgenic coho salmon reared under simulated natural conditions have a reduced growth advantage compared with transgenic salmon reared under culture conditions. Further, the relative prey consumption rates of transgenic and wild predators in simulated natural habitats were affected by previous rearing conditions. Because transgenic organisms show evidence of phenotypic plasticity that in turn influences their predation ability, it is clear that forecasting ecological consequences in nature of transgenic organisms reared and assessed in simple laboratory facilities could be inaccurate.
Errors in extrapolation of ecological consequences could lead to both overestimation and underestimation of risk. In the present study, extrapolation of data from hatchery-reared fish would lead to an overestimation of predation effects posed by transgenic fish reared in nature (case D in Fig. 1). However, our results cannot be taken as evidence that growth-enhanced transgenic salmonids reared in nature will not influence prey population survival and growth differently than wild salmonids. Although development in a simulated natural environment reduced the difference in predation effects between transgenic and wild predators, the small number of replicates (n = 4 per predator type as the result of logistic reasons) reduced the likelihood of detecting small, but significant, differences (i.e., low power of test). In addition, the present experiments have only mimicked one type of “natural” environment during both rearing and experimental monitoring (e.g., for only the fresh-water phase of the salmon's life history). Because environment appears to have a strong influence on phenotype, other rearing conditions not tested in this experiment (e.g., prey density) may still affect phenotypic development of a transgenic animal and subsequent ecological consequences.
This study focused on one aspect of risk assessment of transgenic organisms: the direct ecological effect of altered predation levels on prey populations. There are several other factors that would have to be considered for a complete risk assessment. For example, even small differences in predation ability might have negative effects on an ecosystem over time whether transgenic animals have greater fitness relative to wild genotypes or whether the relative survival of the two genotypes varies at different life stages (11). Even if transgenic animals have lower levels of fitness than wild-type ones in nature, repeated and/or large-scale escapes of transgenic animals from culture conditions still may have negative ecological impacts on the ecosystem. Dispersal behavior also may affect the spatial extent of effects (34), which for salmonids may range anywhere from small streams to large areas of the Pacific Ocean. These and other evolutionary and ecological processes must be taken into consideration before a full assessment of risk of a transgenic organism can be made.
In experiment I, in which we compared hatchery-reared fish, large transgenic predators consumed more prey than wild predators. However, the difference in prey consumption rates cannot be explained by the size differences alone because restricted-growth transgenic predators (matched by size and growth with wild-type salmon) consumed even more prey than the large transgenic predators during the experiment. Further, when transgenic fish were reared under simulated natural stream conditions (which restricted their growth from rates they could achieve in the hatchery), their predation effects relative to wild fish were much reduced. Clearly, body size alone does not explain the relative differences in predation effects observed among transgenic and wild predators produced in different environments, suggesting that other factors (e.g., behavioral and physiological) must be responsible.
Fish learn some of their feeding behavior (35), and hatchery-reared fish often initiate feeding on natural prey soon after release into the wild (36). However, transgenic fish appear to adjust to new environments and learn to feed on novel prey faster than wild conspecifics (31). If differences in predation behavior between hatchery-reared wild and transgenic genotypes mainly are the result of altered learning ability, differences in ecological effects of these two predator types may diminish over the course of time. Alternatively, differences in rearing conditions may affect gene expression (including the transgene) and development (37–39), resulting in structural changes that could have long-lasting effects on traits, such as feeding behavior (40). For example, transgenic coho salmon have structural alterations of their pituitary gland (where GH is produced in wild fish) relative to genotypically wild fish (41) in addition to cranial abnormalities (42, 43), which might affect brain development with irreversible effect on behavior. Currently, it is not known whether expression of the GH transgene used in these studies is modified by environmental conditions. For transgenic mice strains, environmental enrichment can provide morphological compensation in the brain for the effects caused by a transgene (44–46). In general, environmental complexity reduces hormonal effects on behavior (47), and GH treatment of nontransgenic brown trout (Salmo trutta) had stronger phenotypic effects in the hatchery than under natural conditions (48).
Given the multitude of transgenes and organisms being modified, it is difficult to make general predictions on the implications of G×E interactions for ecological risk assessment. In some cases, the modified species may be inherently less plastic than salmon and thus might display limited G×E effects. Further, different transgenes will likely affect a variable number of traits, with those having the greatest pleiotropic effects anticipated to be the most sensitive to environmental influence. For risk assessments of transgenic animals that cannot be released to nature, it is important to examine the animals under a range of contained naturalized environments that yield the full breadth of phenotypes and ecological scenarios that transgenic animals would experience in the wild. Further, determining whether rearing conditions cause permanent or reversible phenotypic changes is critical, because this determination will influence the potential of animals from a rearing facility to become more “wild-like” which, in turn, will influence their long-term fitness and ecological impact in nature.
Methods
The experiments were conducted at the Department of Fisheries and Oceans/University of British Columbia Centre for Aquaculture and Environmental Research, West Vancouver, Canada, and approved by the Department of Fisheries and Oceans Pacific Region Animal Care Committee (no. 05–016). This noncommercial research facility is specially designed to prevent the escape of genetically modified fish to the natural environment.
Wild predators caught in the Chehalis River (British Columbia, Canada) were of unknown parentage, whereas all other predators originated from crosses involving at least five females and five males of either wild or transgenic genotype, where all males were crossed with all females to produce at least 25 half-sibling groups. Wild families were produced from wild parents caught in the Chehalis River. Transgenic fish were produced from wild-caught females, which were crossed with males homozygous for the transgene. Details of the production and subsequent performance of the transgenic line (M77) used in the present study can be found in Devlin et al. (27). Both predator genotypes were then reared under hatchery (Fig. 2a) or simulated natural (Fig. 2b) conditions for ≈1 year before being used in the experiments.
For experiment I, wild and transgenic fish were reared under standard hatchery conditions (10°C well water and artificial lighting after natural photoperiod) and fed to satiation three times a day with commercial salmon feed (Skretting Inc., Bayside, NB, Canada). A transgenic size-control group of predators was fed an amount of food that resulted in growth rates and feeding opportunity (in terms of amount of available food) being similar to that of the wild group.
For experiment II, 15 individuals each of the wild and transgenic genotype were reared together in three flow-through stream tanks (5 × 1 × 0.4 m) landscaped as similarly as possible with gravel, sand, numerous large rocks, a large log, and some debris. Temperature and light conditions varied with local weather conditions (natural creek water 10–17°C in July to October and 1–8°C in October to February). Fish were fed exclusively with natural food items (frozen mysis shrimp and blood worms, live tubifex, earthworms, crickets, and fruit flies, as well as various natural prey entering the system with the creek water) between one and three times per day, on average 3 days of 4. One week before the predation experiment, fish were moved from the rearing tanks into two 200-liter tanks, with genotypes kept separate. In addition, wild fish were trapped from the Chehalis River and kept in a separate 200-liter tank. In this way, all three predator types experienced the same environment directly before the onset of the experiment.
The two predation experiments were conducted sequentially in the same 12 flow-through stream tanks (5 × 1 × 0.4 m) landscaped as similarly as possible with gravel, sand, numerous large rocks, and a large log that provided refuge for both prey and predators. Water flow was generated by the water inflow (≈0.8 m3 h−1 of 15% well water and 85% creek water) and one submerged pump per tank (≈2.5 m3 h−1). Natural light through a semitransparent building cover provided natural photoperiods for the season (February to April).
On the first day of experiment I (using hatchery-reared predators), two individuals of the same predator type were introduced into each of the 12 habitats, resulting in 4 replicates for each predator type. To acclimate these predators to the simulated natural environment, we provided them with limited amounts of natural food four times during the initial 2-week period. Thereafter, 100 rainbow trout fry (average length 28.0 mm) were introduced into each tank. Twice a day, trout fry were fed newly hatched brine shrimp (which are too small for the predators to profitably feed on), whereas predators were not provided any additional food. After 20 days with both predators and prey present in the habitats, weight and length were measured on all survivors.
On the first day of experiment II (using predators reared in a simulated natural environment in addition to fish captured from the wild), 100 coho salmon fry (average 36.1 mm) were released into each of the 12 stream tanks. The next day, four predators were placed into each of the habitats, resulting in four replicates for each of the three predator types. Twice a day, fry were fed newly hatched brine shrimp, whereas predators where not provided any additional food. The predators and prey remained in the habitats together for 14 days, at which point all survivors were measured in weight and length.
In both experiments, we analyzed the effect of predator type on prey mortality, prey final weight (ln transformed to normalize data and make variances more homogenous), and prey biomass production (ln transformed) by using one-way ANOVA followed by Dunnett's post hoc test (pairwise comparisons between a reference category and all other categories). Genetically wild fish reared in the hatchery (in experiment I) and wild fish caught from the Chehalis River (in experiment II) were used as the reference categories.
Acknowledgments
We thank Larry Kahl for providing wild fish from the Chehalis River; Carlo Biagi for fertilizing the fish reared at the Centre for Aquaculture and Environmental Research (CAER); and Geoff Harrison, Nicole Hofs, Jörgen Johnsson, Rupert Marshall, Doug and Edna Vandersteen, and Morgan Williams for assisting with sampling. This work was supported by the Canadian Regulatory System for Biotechnology (RHD), by a postdoctoral grant from the Swedish Research Council FORMAS (to L.F.S.), and by a Marie Curie Outgoing International Fellowship under contract MOIF-CT-2005-8141 from the European Community's Sixth Framework Program.
Abbreviations
- G×E
genotype-by-environment
- GH
growth hormone
- SGRL
specific growth rate in length
- SGRW
specific growth rate in weight.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Tatar M. Trends Ecology Evol. 2000;15:207–211. doi: 10.1016/s0169-5347(00)01834-6. [DOI] [PubMed] [Google Scholar]
- 2.Nottle MB, Boquest AC, Harrison SJ, Grupen CG, Faast RA, Ashman RJ, McIlfatrick SM. Aust J Exp Agric. 2004;44:1113–1117. [Google Scholar]
- 3.Piedrahita JA, Mir B. Am J Transplant. 2004;4:43–50. doi: 10.1111/j.1600-6135.2004.0344.x. [DOI] [PubMed] [Google Scholar]
- 4.Niemann H, Kues W, Carnwath JW. Sci Tech Rev Int Office Epizootics. 2005;24:285–298. [PubMed] [Google Scholar]
- 5.Soler E, Le SA, Guinut F, Passet B, Cohen R, Merle C, Charpilienne A, Fourgeux C, Sorel V, Piriou A, et al. Transgenic Res. 2005;14:833–844. doi: 10.1007/s11248-005-1771-0. [DOI] [PubMed] [Google Scholar]
- 6.Alphey L, Beard CB, Billingsley P, Coetzee M, Crisanti A, Curtis C, Eggleston P, Godfray C, Hemingway J, Jacobs-Lorena M, et al. Science. 2002;298:119–121. doi: 10.1126/science.1078278. [DOI] [PubMed] [Google Scholar]
- 7.Check E. Nature. 2002;418:805. doi: 10.1038/418805a. [DOI] [PubMed] [Google Scholar]
- 8.Kapuscinski AR, Hallerman EM. Fisheries. 1990;15:2–11. [Google Scholar]
- 9.Tiedje JM, Colwell RK, Grossman YL, Hodson RE, Lenski RE, Mack RN, Regal PJ. Ecology. 1989;70:298–315. [Google Scholar]
- 10.Snow AA, Andow DA, Gepts P, Hallerman EM, Power A, Tiedje JM, Wolfenbargerh LL. Ecol Appl. 2005;15:377–404. [Google Scholar]
- 11.Muir WM, Howard RD. Proc Natl Acad Sci USA. 1999;96:13853–13856. doi: 10.1073/pnas.96.24.13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundström LF, Lõhmus M, Devlin RH. Evolution (Lawrence, Kans) 2005;59:1560–1569. [PubMed] [Google Scholar]
- 13.Sundström LF, Lõhmus M, Johnsson JI, Devlin RH. Proc Biol Sci. 2004;271:S350–S352. doi: 10.1098/rsbl.2004.0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muir WM, Howard RD. Am Naturalist. 2001;158:1–16. doi: 10.1086/320860. [DOI] [PubMed] [Google Scholar]
- 15.Dunham RA, Chitmanat C, Nichols A, Argue B, Powers DA, Chen TT. Mar Biotechnol. 1999;1:545–551. doi: 10.1007/pl00011809. [DOI] [PubMed] [Google Scholar]
- 16.Howard RD, DeWoody JA, Muir WM. Proc Natl Acad Sci USA. 2004;101:2934–2938. doi: 10.1073/pnas.0306285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bessey C, Devlin RH, Liley NR, Biagi CA. Trans Am Fisheries Soc. 2004;133:1205–1220. [Google Scholar]
- 18.Gaugler R, Wilson M, Shearer P. Biol Control. 1997;9:75–80. [Google Scholar]
- 19.Hoy MA. Exp Appl Acarol. 2000;24:463–495. doi: 10.1023/a:1006401225083. [DOI] [PubMed] [Google Scholar]
- 20.West-Eberhard MJ. Developmental Plasticity and Evolution. New York: Oxford University Press; 2003. [Google Scholar]
- 21.Yeh PJ, Price TD. Am Naturalist. 2004;164:531–542. doi: 10.1086/423825. [DOI] [PubMed] [Google Scholar]
- 22.Devlin RH, Sundström LF, Muir WM. Trends Biotechnol. 2006;24:89–97. doi: 10.1016/j.tibtech.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Kareiva P. Ecology. 1996;77:1651–1652. [Google Scholar]
- 24.Nam YK, Noh JK, Cho YS, Cho HJ, Cho KN, Kim CG, Kim DS. Transgenic Res. 2001;10:353–362. doi: 10.1023/a:1016696104185. [DOI] [PubMed] [Google Scholar]
- 25.Cook JT, McNiven MA, Richardson GF, Sutterlin AM. Aquaculture. 2000;188:15–32. [Google Scholar]
- 26.Fu C, Cui Y, Hung SSO, Zhu Z. J Fish Biol. 1998;53:115–129. [Google Scholar]
- 27.Devlin RH, Biagi CA, Yesaki TY. Aquaculture. 2004;236:607–632. [Google Scholar]
- 28.Venugopal T, Anathy V, Kirankumar S, Pandian TJ. J Experimental Zoolog A Comp Exp Biol. 2004;301A:477–490. doi: 10.1002/jez.a.78. [DOI] [PubMed] [Google Scholar]
- 29.Raven PA, Devlin RH, Higgs DA. Aquaculture. 2006;254:730–747. [Google Scholar]
- 30.Devlin RH, Johnsson JI, Smailus DE, Biagi CA, Jönsson E, Björnsson BT. Aquaculture Res. 1999;30:479–482. [Google Scholar]
- 31.Sundström LF, Lõhmus M, Devlin RH, Johnsson JI, Biagi CA, Bohlin T. Ethology. 2004;110:381–396. [Google Scholar]
- 32.Abrahams MV, Sutterlin A. Anim Behav. 1999;58:933–942. doi: 10.1006/anbe.1999.1229. [DOI] [PubMed] [Google Scholar]
- 33.Devlin RH, D'Andrade M, Uh M, Biagi CA. Proc Natl Acad Sci USA. 2004;101:9303–9308. doi: 10.1073/pnas.0400023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundström LF, Lõhmus M, Johnsson JI, Devlin RH. Ethology. 2007 in press. [Google Scholar]
- 35.Warburton K. Fish Fisheries. 2003;4:203–215. [Google Scholar]
- 36.Munakata A, Björnsson BT, Jönsson E, Amano M, Ikuta K, Kitamura S, Kurokawa T, Aida K. J Fish Biol. 2000;56:163–172. [Google Scholar]
- 37.Kotrschal K, Van Staaden MJ, Huber R. Rev Fish Biol Fisheries. 1998;8:373–408. [Google Scholar]
- 38.Würbel H. Trends Neurosci. 2001;24:207–211. doi: 10.1016/s0166-2236(00)01718-5. [DOI] [PubMed] [Google Scholar]
- 39.Rampon C, Jiang CH, Dong H, Tang Y-P, Lockhart DJ, Schultz PG, Tsien JZ, Hu Y. Proc Natl Acad Sci USA. 2000;97:12880–12884. doi: 10.1073/pnas.97.23.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchetti MP, Nevitt GA. Environ Biol Fishes. 2003;66:9–14. [Google Scholar]
- 41.Mori T, Devlin RH. Mol Cell Endocrinol. 1999;149:129–139. doi: 10.1016/s0303-7207(98)00248-2. [DOI] [PubMed] [Google Scholar]
- 42.Ostenfeld TH, Mclean E, Devlin RH. J Fish Biol. 1998;52:850–854. [Google Scholar]
- 43.Devlin RH, Yesaki TY, Donaldson EM, Hew CL. Aquaculture. 1995;137:161–169. [Google Scholar]
- 44.Hockly E, Cordery PM, Woodman B, Mahal A, Van Dellen A, Blakemore C, Lewis CM, Hannan AJ, Bates GP. Ann Neurol. 2002;51:235–242. doi: 10.1002/ana.10094. [DOI] [PubMed] [Google Scholar]
- 45.Lazic SE, Grote HE, Blakemore C, Hannan AJ, van Dellen A, Phillips W, Barker RA. Eur J Neurosci. 2006;23:1829–1838. doi: 10.1111/j.1460-9568.2006.04715.x. [DOI] [PubMed] [Google Scholar]
- 46.Lazarov O, Robinson J, Tang Y-P, Hairston IS, Korade-Mirnics Z, Lee VMY, Hersh LB, Sapolsky RM, Mirnics K, Sisodia SS. Cell. 2005;120:701–713. doi: 10.1016/j.cell.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 47.Sloman KA, Armstrong JD. J Fish Biol. 2002;61:1–23. [Google Scholar]
- 48.Johnsson JI, Jönsson E, Petersson E, Järvi T, Björnsson BT. J Fish Biol. 2000;57:326–336. [Google Scholar]