Abstract
A genomic region on distal mouse chromosome 1 and its syntenic human counterpart 1q23–42 show strong evidence of harboring lupus susceptibility genes. We found evidence of linkage at 1q32.2 in a targeted genome scan of 1q21–43 in 126 lupus multiplex families containing 151 affected sibpairs (nonparametric linkage score 2.52, P = 0.006). A positional candidate gene at 1q32.2, complement receptor 2 (CR2), is also a candidate in the murine Sle1c lupus susceptibility locus. To explore its role in human disease, we analyzed 1,416 individuals from 258 Caucasian and 142 Chinese lupus simplex families and demonstrated that a common three-single-nucleotide polymorphism CR2 haplotype (rs3813946, rs1048971, rs17615) was associated with lupus susceptibility (P = 0.00001) with a 1.54-fold increased risk for the development of disease. Single-nucleotide polymorphism 1 (rs3813946), located in the 5′ untranslated region of the CR2 gene, altered transcriptional activity, suggesting a potential mechanism by which CR2 could contribute to the development of lupus. Our findings reveal that CR2 is a likely susceptibility gene for human lupus at 1q32.2, extending previous studies suggesting that CR2 participates in the pathogenesis of systemic lupus erythematosus.
Keywords: linkage, disease susceptibility, antoimmunity, single-nucleotide polymorphisms, syntenic conservation
Systemic lupus erythematosus [SLE (OMIM 152700)] is a chronic autoimmune disease characterized by circulating autoantibodies to nuclear antigens. Epidemiology, linkage, and association studies have provided compelling evidence for a genetic contribution to SLE susceptibility (reviewed in ref. 1). Several loci linked to SLE reside on chromosome 1q, including 1q23 (2–4), 1q31 (2, 5), and 1q41–43 (6–8). Human chromosome 1q21–43 is syntenic to the distal end of mouse chromosome 1, where a recessive locus termed Sle1 has been strongly associated with lupus susceptibility (9). Syntenic conservation in susceptibility intervals between mice and humans suggests the possibility that the same genes may confer risk for both murine and human lupus.
Sle1 corresponds to at least three loci (Sle1a, Sle1b, and Sle1c) (9). The Sle1c interval contains the gene Cr2, which encodes complement receptors 1 and 2 (CR1 and CR2, CD35/CD21) by alternative splicing of a single mRNA transcript (10). Cr2 is a major positional candidate gene of the murine Sle1c lupus susceptibility interval (11). Its protein products are structurally and functionally altered because of a nonsynonymous amino acid change in the ligand-binding domain of CR2 that introduces a novel glycosylation site (11).
In humans, several positional candidate genes on chromosome 1q have been associated with SLE (reviewed in ref. 12). However, there are no published reports of linkage or association at 1q32.2, where CR2 is mapped. The human CR2 gene (OMIM 120650) encodes a membrane glycoprotein, consisting of 15 repeating structures termed short consensus repeats (SCRs), that is expressed on mature B cells and follicular dendritic cells, as well as an alternatively spliced 16 SCR variant that is expressed primarily on follicular dendritic cells (13). Its relative expression is primarily controlled at the level of transcription by the proximal promoter (14–17), and cell and lineage specificity of expression is regulated by an intronic silencer (18, 19). CR2 binds C3 degradation products covalently bound to antigen in the process of complement activation, as well as EBV (20), the immunomodulatory protein CD23 (21), and IFN-α (22). Cumulative studies suggest that CR2 plays a major role in immunity (reviewed in ref. 23). We report here linkage with lupus susceptibility in humans at 1q32.2 and provide compelling evidence that CR2 is an important gene in that region.
Results
Linkage to SLE at 1q32.2.
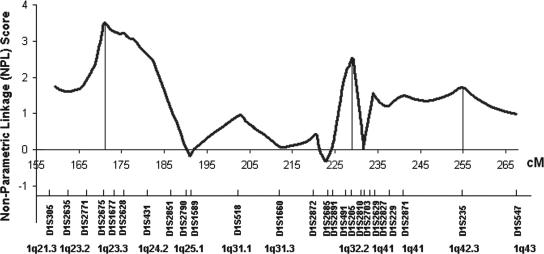
A targeted genome scan of chromosome 1q21–43 for lupus susceptibility loci, which spanned the 108-cM interval at an average distance of 4 cM, showed evidence for linkage at 1q23 [peak at D1S2675, nonparametric linkage (NPL) = 3.51, P = 0.0002], 1q32.2 (peak at D1S205, NPL = 2.52, P = 0.006), and 1q41–42 (peak at D1S235, NPL = 1.73, P = 0.04) in 126 families containing 151 SLE-affected sibpairs (Fig. 1). This sample was extended from our previous cohort of multiethnic affected sibpair families (3, 4, 6), in which we reported linkage to SLE at 1q23 and 1q41–42. In addition to providing support for the previously identified loci, this linkage analysis revealed a chromosomal region, 1q32.2, that may harbor SLE susceptibility genes.
Fig. 1.
Linkage to SLE within 1q21–43. The vertical axis represents the NPL score for linkage analysis, and the horizontal axis corresponds to genetic distance in centimorgans. Microsatellite markers tested are shown.
Characterization of CR2.
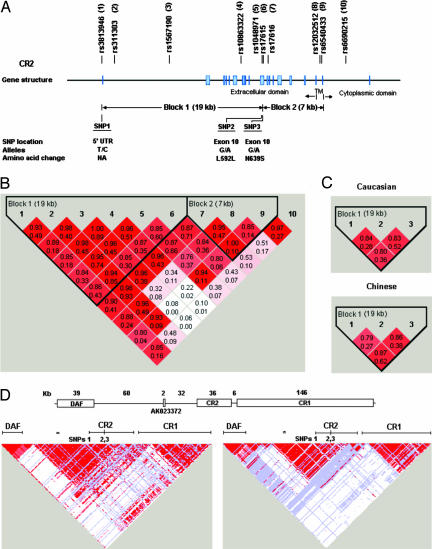
To determine whether CR2 might contribute to the linkage at 1q32.2, we evaluated 10 CR2 single-nucleotide polymorphisms (SNPs) in a pilot study of 105 Caucasian SLE simplex families [including 38 SLE probands affected with lupus nephritis (LN) and 67 without LN] to characterize CR2 haplotype blocks. We selected 10 informative SNPs (minor allele frequency between 0.2 and 0.5) that included 5 potentially functional SNPs, e.g., 1 regulatory SNP [rs3813946 in the 5′ untranslated region (UTR)] and 4 exonic SNPs (rs1048971 and rs17615 in exon 10, rs17616 in the alternatively spliced exon 10a, and rs6540433 in exon 17), and 5 haplotype-tagging intronic SNPs that tagged the two haplotype blocks (Fig. 2 A and B). Only a single SNP (rs17615 G allele, G/A, S639N), located in the 19-kb block 1, showed evidence for association with SLE when the transmission-disequilibrium test (TDT) was used (transmission:nontransmission = 44:26, P = 0.03). SNP2 showed a trend of association (transmission:nontransmission = 28:16, P = 0.07) in the 67 non-LN families. Because none of block 2 SNPs (SNP 7–10) exhibited evidence for association, these data supported further analysis of haplotype block 1 SNPs for association with SLE.
Fig. 2.
CR2 SNP locations and haplotype blocks. (A) The CR2 gene is composed of 19 exons (37). Ten SNPs in exon 1 (5′ UTR), intron 1, intron 6, exon 10 (SCR9 and SCR10), the alternatively spliced exon 10a (SCR11), intron 16, exon 17 (transmembrane domain), and intron 17 were genotyped. (B) Two haplotype blocks were constructed. D′ (the top number in each box) and r2 (the bottom number) of each SNP pair are depicted. Based on the strength of LD among SNP pairs, the first 6 SNPs formed the 19-kb block 1 and the next 3 SNPs formed the 7-kb block 2, which excluded the 10th SNP. (C) SNPs 1, 2, and 3 located in block 1 were assessed in extended Caucasian and Chinese SLE simplex families. (D) Pairwise LD of 374 SNPs from the 321-kb genomic region containing CR2 and its nearest known genes DAF and CR1. (Upper) Relative positions and sizes of CR2, the 5′ flanking hypothetical transcript AK023372 and DAF, and the 3′ flanking CR1. (Lower) Evidence of LD in D′ from 30 Caucasian trios and 45 Chinese individuals (HapMap Phase II, July 2006). ∗, Position of a single SNP (rs2135924) located within AK023372.
Characterization of CR2 SNPs in Caucasian and Chinese Cohorts.
We selected three SNPs in block 1 for further haplotype analyses in 258 Caucasian and 142 Chinese SLE simplex families. The SNP in exon 1 (rs3813946, SNP1; +21 T/C) lies within the 5′ UTR and could affect gene regulation, the synonymous SNP in exon 10 (rs1048971, SNP2; G/A, L592L) is a haplotype-tagging SNP in Caucasian and Chinese populations in the HapMap Phase I database (June 2005, www.hapmap.org), and the nonsynonymous SNP in exon 10 (rs17615, SNP3; G/A, S639N) was positively associated with SLE in our pilot study. The physical distance is 19 kb between SNP1 and SNP2 and 0.1 kb between SNP2 and SNP3 (Fig. 2A). The allele distribution of parental genotypes showed no deviation from Hardy–Weinberg equilibrium for each of these three SNPs. The minor allele frequency of each SNP in these families is shown in Table 1. Parental allele frequencies were not different from those of the same ethnic group in the current HapMap Phase II database, but were significantly different between these two ethnic family collections (P = 0.01, 0.001, and <0.0001 for SNP1, SNP2, and SNP3, respectively).
Table 1.
Preferential transmission of the major allele of CR2 SNPs in the Caucasian, Chinese, and combined samples
| Locus | Caucasian (n = 258) |
Chinese (n = 142) |
Total (n = 400) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| MAF | T:NT | P | MAF | T:NT | P | T:NT | P | ||
| SNP1 | 0.17 | 63:40 | 0.02 | 0.12 | 29:19 | 0.1 | 92:59 | 0.007 | |
| SNP2 | 0.32 | 102:89 | 0.3 | 0.24 | 57:33 | 0.01 | 159:22 | 0.03 | |
| SNP3 | 0.26 | 99:70 | 0.03 | 0.14 | 37:23 | 0.07 | 136:93 | 0.005 | |
MAF, minor allele frequency; T:NT, transmission:nontransmission (major allele of each SNP).
Association of CR2 SNPs and SNP Haplotypes with SLE Susceptibility.
Using the TDT family-based association method (24), we found that the major allele of each of the three CR2 SNPs was transmitted preferentially from heterozygous parents to their affected offspring in the Caucasian and Chinese families, and this overtransmission reached statistical significance in the combined sample (transmission:nontransmission = 92:59, 159:122, 136:93, P = 0.007, 0.03, and 0.005 for T at SNP1, G at SNP2, and G at SNP3, respectively, Table 1). Consistent with these results, the major alleles were not preferentially transmitted to unaffected offspring in these families (data not shown).
The two-locus CR2 SNP haplotypes span 18.6 kb (SNP1 and SNP2), 0.14 kb (SNP2 and SNP3), and 18.7 kb (SNP1 and SNP3). Pairwise linkage disequilibrium (LD) in both Caucasian (D′ = 0.80–0.84, r2 = 0.28–0.52) and Chinese (D′ = 0.79–0.87, r2 = 0.27–0.62) cohorts (Fig. 2C) showed that, in both populations, these three SNPs were independently informative and were contained within a single haplotype block, which was similar to the block structure in the current HapMap Phase II database (January 2006) and consistent with our pilot study. Table 2 gives the frequencies of major allele SNP haplotypes in both Caucasian and Chinese parents. Although parental haplotype distributions of these two cohorts were significantly different (P from <0.0001 to 0.0007 for two- or three-locus haplotypes), SNP haplotypes formed by the major allele of these three SNPs were preferentially transmitted to affected offspring in both cohorts, as well as in the combined 400 families (haplotype P = 0.00001–0.0008, global P = 0.0001–0.002, Table 2). As with the individual SNPs, the major allele SNP3 haplotype was not preferentially transferred to unaffected siblings in these families (data not shown). These data provided evidence that CR2 is a good candidate gene for SLE risk in 1q32.2.
Table 2.
Preferential transmission of CR2 haplotypes formed by the major allele of each SNP in the Caucasian, Chinese, and combined samples
| Loci | Caucasian (n = 258) |
Chinese (n = 142) |
Total (n = 400) |
|||||
|---|---|---|---|---|---|---|---|---|
| Haplotype frequency* | Haplotype P† | Global P‡ | Haplotype frequency* | Haplotype P† | Global P‡ | Haplotype P† | Global P‡ | |
| SNP1-2 | 0.64 | 0.04 | 0.09 | 0.74 | 0.002 | 0.008 | 0.0008 | 0.002 |
| SNP2-3 | 0.64 | 0.02 | 0.01 | 0.74 | 0.002 | 0.006 | 0.0002 | 0.0001 |
| SNP1-3 | 0.71 | 0.002 | 0.02 | 0.85 | 0.02 | 0.04 | 0.0001 | 0.0008 |
| SNP1-2-3 | 0.62 | 0.005 | 0.03 | 0.73 | 0.0003 | 0.009 | 0.00001 | 0.001 |
*Frequencies of two- or three-locus haplotypes formed by the major allele of CR2 SNPs in each ethnic group.
†Haplotype P represents the P value for the transmission of the specific haplotype.
‡Global P represents the overall significance using all possible haplotypes.
Differences in CR2 SNP Allele Frequencies in Family-Based Cases and Controls.
The affected family-based controls approach (25) can create controls by subtracting alleles in affected SLE children from parental alleles in complete trios. The case versus control allele frequencies are estimated from the parental transmitted versus nontransmitted alleles to the affected children. As expected from the TDT results, the major allele frequencies of the CR2 SNPs were significantly higher in SLE offspring (88%, 74%, and 82% for SNP1, SNP2, and SNP3, respectively) when compared with the controls (82%, 69%, and 75%) in the total cohort {odds ratio = 1.53 [95% confidence interval (C.I.) = 1.11–2.10], 1.32 [95% C.I. = 1.04–1.68], and 1.47 [95% C.I. = 1.13–1.92]}. Although no direct estimate of the frequency of the major allele CR2 haplotype can be made from the trio data, the frequency was estimated by using the expectation-maximization algorithm in the affected offspring, and the parental frequencies were inferred by subtraction. When this approach was used, the major allele CR2 haplotype frequency was higher in SLE offspring compared with controls (71% vs. 62%). Treating these inferred haplotypes as determined, their frequencies are significantly different [P = 0.0007, odds ratio = 1.54 (95% C.I. = 1.21–1.96)]. These results supported our hypothesis that the major alleles for the CR2 SNPs in haplotype block 1 conferred risk for the development of SLE.
Preferential Transmission of the Major CR2 SNP Alleles to SLE Patients Without Renal Involvement.
Involvement of the kidneys in SLE is one of the most serious complications of this disease. To assess whether the major allele CR2 SNP haplotype was associated with the development of LN, we stratified the SLE families by the presence or absence of LN (26) in the SLE proband within each family. Overtransmission of the haplotype formed by the major alleles of CR2 SNPs was observed mainly in SLE patients without renal involvement (haplotype P = 0.0001–0.002, global P = 0.002–0.02) (Table 3). Although various other clinical features of SLE might not be independently manifested, stratification on each SLE subphenotype revealed a strong positive association of the identified CR2 haplotype with malar rash, photosensitivity, oral ulcers, serositis, anti-cardiolipin antibodies, anti-Sjögren's Syndrome A antibodies, and anti-dsDNA antibodies (data not shown).
Table 3.
Skewed transmission of CR2 haplotypes formed by the major allele CR2 SNPs in non-LN families
| Loci | LN (n = 189) |
Non-LN (n = 211) |
||
|---|---|---|---|---|
| Haplotype P | Global P | Haplotype P | Global P | |
| SNP1-2 | 0.2 | 0.3 | 0.0009 | 0.004 |
| SNP2-3 | 0.1 | 0.04 | 0.0002 | 0.002 |
| SNP1-3 | 0.03 | 0.08 | 0.002 | 0.02 |
| SNP1-2-3 | 0.2 | 0.3 | 0.0001 | 0.01 |
SNP1 Regulates CR2 Transcription.
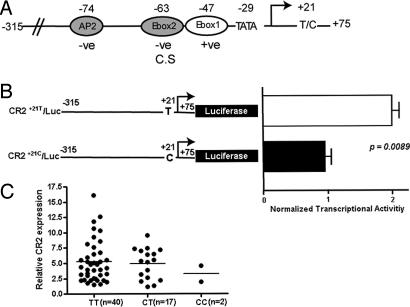
Several motifs in the human CR2 proximal promoter are involved in controlling promoter activity (Fig. 3A). These motifs include cell type-specific elements (16), as well as activator and repressor motifs that function in all tested cell lines (17). Although the −315/+75 region of the CR2 promoter is known to contain all of the factors necessary for basal transcription (17), sequences in the 5′ UTR of the gene have never been examined for functional relevance. To evaluate the effect of SNP1 on basal CR2 transcription, luciferase (Luc) constructs containing the CR2 promoter region spanning −315/+75 and expressing either the major T allele or the minor C allele at position +21 were generated and transiently transfected into the CR2-expressing Raji B lymphoblastoid cell line. Expression of the major +21 T (Fig. 3B, CR2+21T/Luc, n = 6) allele resulted in a 2-fold increase in transcriptional activity compared with expression of the minor C allele (Fig. 3B, CR2+21C/Luc, n = 10; P = 0.0089). In addition, although mean CR2 mRNA expression in peripheral blood mononuclear cells from healthy controls homozygous for the major T allele was not significantly higher than that of the other two groups (Fig. 3C; P = 0.59 for TT vs. CT + CC), all of the individuals expressing the highest levels of CR2 mRNA were homozygous for this allele. These data suggest that sequences within the CR2 5′ UTR, and specifically those surrounding SNP1 at position +21, regulate CR2 promoter activity, and this regulation may have implications in lupus pathogenesis.
Fig. 3.
Transcriptional effects of SNP1. (A) Transcription factor binding sites within the proximal CR2 promoter critical for regulation of basal transcription (17). Shown are nucleotide positions and functional role of localized elements (−ve, repressor motif; +ve, activator motif; C.S, cell type-specific), TATA box, transcriptional initiation site (arrow), and position of SNP1. (B) Transcriptional activity of CR2 constructs expressing the major (CR2+21T/Luc, n = 6) or minor (CR2+21C/Luc, n = 10) SNP1 allele. Results shown represent mean promoter activity ± SEM and are expressed as normalized transcriptional activity of the minor allele construct relative to the major allele construct. (C) Correlation of peripheral blood CR2 mRNA levels with SNP1 genotype. T, major allele of SNP1; C, minor allele of SNP1. The fold expression of CR2 mRNA normalized to CD19 mRNA for each subject relative to the lowest normalized CR2 mRNA level is shown. The bar marks mean fold expression for each genotype.
Discussion
We describe here evidence of linkage for human lupus susceptibility at chromosome 1q32.2, where CR2 is mapped, and the positive association between CR2 SNPs and a CR2 SNP haplotype with lupus susceptibility in two distinct ethnic groups. Furthermore, SNP1 in the associated haplotype, which is located in the 5′ UTR of the CR2 promoter, may be functionally relevant and could affect susceptibility to SLE. Together with previous data suggesting that Cr2 is a strong candidate gene for lupus susceptibility in a syntenic region of the mouse genome, and multiple lines of evidence suggesting that CR2 plays an important role in immunity and autoimmunity, our findings strongly support the hypothesis that CR2 variants predispose their carriers to SLE.
Family-based TDT analysis showed significant association of the major allele CR2 haplotype with SLE susceptibility [haplotype P = 0.00001, permuted P = 0.0047 (from 1,000,000 random iterations of the haplotype data by Haploview software)]. We estimated that the risk of developing SLE is 1.54-fold higher for individuals who carry the major allele CR2 haplotype. Although these studies also supported an association of the major allele CR2 haplotype with non-LN SLE, they lacked the statistical power to rule out an association of this haplotype with LN. Our initial analysis of the functional effects of SNPs in this haplotype revealed that the major T allele for SNP1 results in a 2-fold increase in promoter activity. This allele is also associated with increased transcription factor binding at the DNA, but not the RNA level (D.U., unpublished data). Individuals expressing the highest levels of CR2 were all homozygous for this allele, suggesting that this SNP alters transcriptional activity of the promoter in vivo.
Increased expression of CR2 regulated by SNP1 could promote the development of autoimmunity by several mechanisms. CR2 lowers the threshold for B cell activation, and autoreactive B cells in individuals genetically predisposed to express more CR2 may be easily activated when they encounter complement-coated immune complexes containing self-antigen. Furthermore, increased CR2 expression may alter B cell responses to IFN-α and EBV, two alternative ligands for human CR2 that have been implicated to play a role in SLE. Finally, alterations in CR2 expression have a variety of different effects on manifestations of disease in animal models of autoimmunity (27–30), suggesting that tight regulation of this receptor is critical in the induction and maintenance of tolerance. The specific effects of dysregulated expression likely depend on the cell type affected and the time in ontogeny when expression is altered. Although CR2 levels are decreased by ≈50% on B cells of patients with SLE (31, 32), this decrease may be the result of multiple factors, including alterations in B cell homeostasis, immune complex formation and deposition, and alterations in the cytokine milieu associated with active disease. The presence of low CR2 levels in individuals with established disease does not exclude the possibility that levels were increased before disease onset.
Other SNPs in the identified risk CR2 haplotype may also have functional effects that could predispose to SLE. For example, SNP3, which is located in exon 10 of the CR2 gene, was the only SNP that was independently associated with SLE in our pilot study. Exon 10 is positioned directly 5′ of the alternatively spliced exon 10a, which is found in a long CR2 isoform almost exclusively expressed on follicular dendritic cells (13). SNPs in coding domains can alter pre-mRNA splicing and message stability (33), and an SNP3 allelic variant may regulate the relative level of the long and short isoform of CR2. Furthermore, the major allele for SNP3 substitutes an asparagine for a serine, which is conserved in mice, rats, and sheep, suggesting that it may be important in receptor function.
These data provide initial evidence for a role for CR2 in human lupus susceptibility. We have identified a risk CR2 haplotype containing a major allele for a regulatory SNP that may influence disease development as a result of increased receptor expression, as well as two other major allele SNPs that may also be functional. Nevertheless, we cannot rule out the possibility that these SNPs are in LD with SNPs in this or other CR2 haplotype blocks, or in other genes surrounding CR2 at chromosome 1q32.2, including the 94-kb upstream decay accelerating factor (DAF) CD55 and the 6-kb downstream CR1 (Fig. 2D Upper). Data from the HapMap project (Phase II, July 2006; Fig. 2D Lower) show no/low LD of SNP1–3 with 39 variants of DAF (pairwise D′ = 0.07-1, r2 = 0–0.1 for Caucasian; D′ = 0.6–1, r2 = 0.001–0.2 for Chinese), strong LD with SNPs located within the ≈50-kb intergenic upstream region, a break in LD with SNPs located within the 3′ region of CR2, and some LD with 5′ CR1 SNPs in Caucasians and Chinese and 3′ CR1 SNPs in Caucasians only (r2 ≥ 0.3, D′ > 0.5). Therefore, although these data most strongly implicate the haplotype block containing SNP1–3 in the association with SLE that we demonstrate here, it is possible that CR1 SNPs or CR2 SNPs in other haplotype blocks are involved in this association. Only after a thorough and careful analysis of the haplotype block structure of CR2 and CR1, in combination with functional assessments of any associated SNPs, will we be able to assign causality to a specific CR2 SNP.
Methods
SLE Families.
SLE patients were enrolled after informed consent had been obtained. All SLE patients met the American College of Rheumatology criteria for the classification of SLE (26). Family collections in the linkage study consisted of 126 SLE multiplex families [60 Caucasian, 34 Asian (including 26 Chinese), 19 Hispanic, 10 African American, and 3 mixed ethnicity families] with 563 individuals, 273 of whom were SLE patients. There were 151 affected sibpairs (109 families had 2 affected siblings, 12 families had 3, and 1 family had 4) and 15 other affected family members. The association study consisted of 1,416 individuals from 258 Caucasian SLE simplex families, including 204 complete trios, and 142 ethnic Chinese SLE simplex families, including 131 complete trios. Self-reported ethnic origins of the four grandparents of the SLE proband were used to classify ethnic Chinese or European Caucasian cohorts. According to the American College of Rheumatology criteria for LN applied to the SLE proband in each family, 82 of 142 Chinese and 107 of 258 Caucasian SLE simplex families were designated as LN families. This study was reviewed and approved by the appropriate institution review board.
Microsatellite Marker Genotyping.
Genomic DNA was isolated from peripheral blood mononuclear cells by using a standard protocol. A total of 25 microsatellite markers (D1S305, D1S2635, D1S2771, D1S2675, D1S1677, D1S2628, D1S431, D1S2851, D1S2790, D1S1589, D1S518, D1S1660, D1S2872, D1S2685, D1S2891, D1S491, D1S205, D1S2810, D1S2703, D1S2629, D1S2827, D1S229, D1S2871, D1S235, and D1S547) spanning 108 cM on 1q21–43 were genotyped by using fluorescent-based PCR. The marker information and sequences of PCR primers for each marker are as shown on the public website (http://www.ncbi.nlm.nih.gov/genome/unists). Microsatellite PCR and genotyping analysis were conducted as described in ref. 3.
CR2 SNP Genotyping.
CR2 SNPs were genotyped by pyrosequencing (34). PCR primers were designed by using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) to generate amplicons of < 200 base pairs. Sequencing primers were designed by using SNP Primer Design Software Version 1.01 (http://primerdesign.pyrosequencing.com/jsp/TemplateInput.jsp). PCR and pyrosequencing of each SNP were performed as described in ref. 35. Forward primer 5′-CGTGTGCCGGACACTATTTA-3′, reverse primer 5′-CGACGAGAGCCAAGAAA-ACC-3′, and sequencing primer 5′-GCAGCTCTGGGAGG-3′ were used for SNP1; forward primer 5′-GAGAGAGCACCATCCGTTGT-3′, reverse primer 5′-GCAAGGAGGGAAAGTT-3′, and sequencing primer 5′-GCAGCTCTGGGAGG-3′ were used for SNP2; and forward primer 5′-TTGCTGTCCAGTGCTCACAT-3′, reverse primer 5′-TGGTATTTCAGGATCCCAGGT-3′, and sequencing primer 5′-TTTACTTTGAAGGGCA-3′ were used for SNP3. Seven non-Mendelian Interitance were detected in the Caucasian and Chinese cohorts, and those samples for that marker were excluded from further analysis.
Creation of CR2 Promoter/Luciferase Fusion Constructs.
An NheI/XhoI fragment of the CR2 promoter containing nucleotides −315/+75 with the major T allele at position +21 was previously cloned into the luciferase reporter pGL3-basic vector (Clontech, Mountain View, CA) (16). Site-directed mutagenesis was performed by using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA) to incorporate the minor C allele at position +21. Accuracy of mutagenesis was confirmed by sequence analysis.
Transfection and Quantification of Promoter Activity.
Raji cells were grown in logarithmic phase to 5 × 105 cells per ml and plated in 5 ml of medium in a six-well plate at a concentration of 5 × 105 to 1 × 106 cells per ml. Ten milligrams of CR2 promoter/luciferase fusion plasmid DNA and 300 ng of pRL-TK control vector were complexed together with Superfect transfection reagent (QIAGEN, Valencia, CA) for 10 min at room temperature and then added drop-wise to the cells. After 48 h at 37°C, cell lysates were prepared and assayed for firefly and control Renilla luciferase according to the supplier's instructions (Promega, Madison, WI). Data are representative of 3–10 independent transfections using at least two independent preparations of DNA. Promoter activity is expressed as firefly luciferase activity normalized to Renilla luciferase activity.
Sample Collection and RNA Processing.
Healthy controls were selected from a pool of volunteers at the University of California, Los Angeles, Medical Center. Peripheral blood was collected in BD Vacutainer tubes containing acid/citrate/dextrose (ACD) solution A (BD Biosciences, Mountain View, CA) or in PAXgene tubes (QIAGEN). For BD Vacutainer tubes, total RNA was extracted immediately by using TRIzol (Invitrogen, Carlsbad, CA). For PAXgene tubes, total RNA was extracted by using the PAXgene 96 Blood RNA Kit (QIAGEN). One to 2 μg of RNA was reverse transcribed into cDNA by using Omniscript reverse transcriptase (QIAGEN). RNA and cDNA samples were stored at −70°C before use.
Reverse Transcriptase PCR.
mRNA expression levels of CR2 and CD19 were assessed by using TaqMan gene expression Assays-on-Demand (assay ID for CR2 Hs00153398_m1, assay ID for CD19 Hs00174333_m1; Applied Biosystems, Foster City, CA). Data were displayed by using SDS Version 1.9 software (Applied Biosystems). CR2 mRNA expression level was normalized to that of CD19 for each sample. The lowest normalized CR2 mRNA expression level was used to determine the relative fold expression levels of the other samples.
Statistical Analysis.
Linkage.
Model-free multipoint linkage analysis was performed to assess the evidence of linkage with SLE by using GENEHUNTER software (36), in which identical-by-descent allele-sharing information among pairs of affected family members was evaluated. A multipoint NPL score was generated for each marker at 4-cM intervals.
Association.
The TDT (24) was used to investigate whether the alleles of each individual SNP were preferentially transmitted from heterozygous parents to affected offspring by using GENEHUNTER software (36). TRANSMIT software was used to assess preferential transmission of SNP haplotypes from parents to affected offspring, and unknown phase was determined with the expectation-maximization algorithm (http://www-gene.cimr.cam.ac.uk/clayton/software/transmit.txt). The strength of LD among the pairs of SNPs was assessed with Haploview 3.32 software (www.broad.mit.edu/mpg/haploview/index.php). Haplotype blocks were defined by using the approach of solid spine of LD by Haploview 3.32 software. Parental allele distributions of CR2 SNPs were tested for Hardy–Weinberg disequilibrium and for differences by an exact two-tailed t test. The affected family-based controls approach (25) was used to obtain allele frequencies of CR2 SNPs in family-based controls, and the odds ratio was estimated by using Fisher's exact test. Transcriptional activity of CR2 promoter alleles was compared by using an unpaired t test. CR2 mRNA levels among CR2 genotypes were compared by one-way ANOVA and an unpaired t test. Statistical analyses were performed with Prism4 (GraphPad, San Diego, CA). P values <0.05 were considered to indicate statistical significance.
Acknowledgments
We thank the families involved in this study and the physicians who referred them and V. Michael Holers for helpful discussions. This work was supported by the Meyer Young Investigator Award (to H.W.), Arthritis National Research Foundation (H.W.), Lupus Research Institute (S.A.B.), National Health and Medical Research Council (Australia) (D.U.), Arthritis Australia (D.U.), National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant R01AR43814 (to B.P.T.), Southern California Chapter of the Arthritis Foundation (B.P.T.), and the Paxson Family Foundation (B.P.T.).
Abbreviations
- CR1
complement receptor 1
- CR2
complement receptor 2
- LD
linkage disequilibrium
- LN
lupus nephritis
- NPL
nonparametric linkage
- SCR
short consensus repeat
- SLE
systemic lupus erythematosus
- TDT
transmission-disequilibrium test.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Tsao BP. Trends Immunol. 2003;24:595–602. doi: 10.1016/j.it.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Moser KL, Neas BR, Salmon JE, Yu H, Gray-McGuire C, Asundi N, Bruner GR, Fox J, Kelly J, Henshall S, Bacino D, et al. Proc Natl Acad Sci USA. 1998;95:14869–14874. doi: 10.1073/pnas.95.25.14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsao BP, Cantor RM, Grossman JM, Kim SK, Strong N, Lau CS, Chen C-J, Shen N, Ginzler EM, Goldstein R, et al. Arthritis Rheum. 2002;46:2928–2936. doi: 10.1002/art.10590. [DOI] [PubMed] [Google Scholar]
- 4.Cantor RM, Yuan J, Napier S, Kono N, Grossman JM, Hahn BH, Tsao BP. Arthritis Rheum. 2004;50:3203–3210. doi: 10.1002/art.20511. [DOI] [PubMed] [Google Scholar]
- 5.Johanneson B, Lima G, von Salomé J, Alarcón-Segovia D. Collaborative Group on the Genetics of SLE, BIOMED II Collaboration on the Genetics of SLE and Sjögrens Syndrome, Alarcón-Riquelme ME. Am J Hum Genet. 2002;71:1060–1071. doi: 10.1086/344289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsao BP, Cantor RM, Kalunian KC, Chen C-J, Badsha H, Singh R, Wallace DJ, Kitridou RC, Chen S-L, Shen N, et al. J Clin Invest. 1997;99:725–731. doi: 10.1172/JCI119217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moser KL, Gray-McGuire C, Kelly J, Asundi N, Yu H, Bruner GR, Mange M, Hogue R, Neas BR, Harley JB. Arthritis Rheum. 1999;42:1902–1907. doi: 10.1002/1529-0131(199909)42:9<1902::AID-ANR16>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 8.Graham RR, Langefeld CD, Gaffney PM, Ortmann WA, Selby SA, Baechler EC, Shark KB, Ockenden TC, Rohlf KE, Moser KL, et al. Arthritis Res. 2001;3:299–305. doi: 10.1186/ar319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morel L, Blenman KR, Croker BP, Wakeland EK. Proc Natl Acad Sci USA. 2001;98:1787–1792. doi: 10.1073/pnas.031336098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molina H, Kinoshita T, Inoue K, Carel J-C, Holers VM. J Immunol. 1990;145:2974–2983. [PubMed] [Google Scholar]
- 11.Boackle SA, Holers VM, Chen X, Szakonyi G, Karp DR, Wakeland EK, Morel L. Immunity. 2001;15:775–785. doi: 10.1016/s1074-7613(01)00228-x. [DOI] [PubMed] [Google Scholar]
- 12.Shen N, Tsao BP. Curr Rheumatol Rep. 2004;6:391–398. doi: 10.1007/s11926-004-0014-3. [DOI] [PubMed] [Google Scholar]
- 13.Liu YJ, Xu J, de Bouteiller O, Parham CL, Grouard G, Djossou O, de Saint-Vis B, Lebecque S, Banchereau J, Moore KW. J Exp Med. 1997;185:165–170. doi: 10.1084/jem.185.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vereshchagina LA, Tolnay M, Tsokos GC. J Immunol. 2001;166:6156–6163. doi: 10.4049/jimmunol.166.10.6156. [DOI] [PubMed] [Google Scholar]
- 15.Tolnay M, Vereshchagina LA, Tsokos GC. J Immunol. 2002;169:6236–6243. doi: 10.4049/jimmunol.169.11.6236. [DOI] [PubMed] [Google Scholar]
- 16.Ulgiati D, Holers VM. J Immunol. 2001;167:6912–6919. doi: 10.4049/jimmunol.167.12.6912. [DOI] [PubMed] [Google Scholar]
- 17.Ulgiati D, Pham C, Holers VM. J Immunol. 2002;168:6279–6285. doi: 10.4049/jimmunol.168.12.6279. [DOI] [PubMed] [Google Scholar]
- 18.Makar KW, Pham CTN, Dehoff MH, O'Connor SM, Jacobi SM, Holers VM. J Immunol. 1998;160:1268–1278. [PubMed] [Google Scholar]
- 19.Makar KW, Ulgiati D, Hagman J, Holers VM. Int Immunol. 2001;13:657–664. doi: 10.1093/intimm/13.5.657. [DOI] [PubMed] [Google Scholar]
- 20.Fingeroth JD, Weis JJ, Tedder TF, Strominger JL, Biro PA, Fearon DT. Proc Natl Acad Sci USA. 1984;81:4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aubry J-P, Pochon S, Graber P, Jansen KU, Bonnefoy J-Y. Nature. 1992;358:505–507. doi: 10.1038/358505a0. [DOI] [PubMed] [Google Scholar]
- 22.Delcayre AX, Salas F, Mathur S, Kovats K, Lotz M, Lernhardt W. EMBO J. 1991;10:919–926. doi: 10.1002/j.1460-2075.1991.tb08025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holers VM. Springer Semin Immunopathol. 2005;26:405–423. doi: 10.1007/s00281-004-0186-y. [DOI] [PubMed] [Google Scholar]
- 24.Spielman RS, Ewens WJ. Am J Hum Genet. 1996;59:983–989. [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson G. Am J Hum Genet. 1995;57:487–498. [PMC free article] [PubMed] [Google Scholar]
- 26.Hochberg MC. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 27.Prodeus AP, Georg S, Shen LM, Pozdnyakova OO, Chu L, Alicot EM, Goodnow CC, Carroll MC. Immunity. 1998;9:721–731. doi: 10.1016/s1074-7613(00)80669-x. [DOI] [PubMed] [Google Scholar]
- 28.Wu X, Jiang N, Deppong C, Singh J, Dolecki G, Mao D, Morel L, Molina HD. J Immunol. 2002;169:1587–1592. doi: 10.4049/jimmunol.169.3.1587. [DOI] [PubMed] [Google Scholar]
- 29.Kaya Z, Afanasyeva M, Wang Y, Dohmen KM, Schlichting J, Tretter T, Fairweather D, Holers VM, Rose NR. Nat Immunol. 2001;2:739–745. doi: 10.1038/90686. [DOI] [PubMed] [Google Scholar]
- 30.Del Nagro CJ, Kolla RV, Rickert RC. J Immunol. 2005;175:5379–5389. doi: 10.4049/jimmunol.175.8.5379. [DOI] [PubMed] [Google Scholar]
- 31.Wilson JG, Ratnoff WD, Schur PH, Fearon DT. Arthritis Rheum. 1986;29:739–747. doi: 10.1002/art.1780290606. [DOI] [PubMed] [Google Scholar]
- 32.Marquart HV, Svendsen A, Rasmussen JM, Nielsen CH, Junker P, Svehag S-E, Leslie RGQ. Clin Exp Immunol. 1995;101:60–65. doi: 10.1111/j.1365-2249.1995.tb02277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maquat LE. Nat Genet. 2001;27:5–6. doi: 10.1038/83759. [DOI] [PubMed] [Google Scholar]
- 34.Ronaghi M. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- 35.Wu H, Cantor RM, Graham DS, Lingren CM, Farwell L, Jager PL, Bottini N, Grossman JM, Wallace DJ, Hahn B, et al. Arthritis Rheum. 2005;52:2396–2402. doi: 10.1002/art.21223. [DOI] [PubMed] [Google Scholar]
- 36.Kruglyak L, Lander ES. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 37.Fujisaku A, Harley JB, Frank MB, Gruner BA, Frazier B, Holers VM. J Biol Chem. 1989;264:2118–2125. [PubMed] [Google Scholar]