Abstract
A procedure is described to discover genes that are specifically expressed in human prostate. The procedure involves searching the expressed sequence tag (EST) database for genes that have many related EST sequences from human prostate cDNA libraries but none or few from nonprostate human libraries. The selected candidate EST clones were tested by RNA dot blots to examine tissue specificity and by Northern blots to examine the transcript size of the corresponding mRNA. The computer analysis identified 15 promising genes that were previously unidentified. When seven of these were examined in an RNA hybridization experiment, three were found to be prostate specific. The genes identified could be useful in the targeted therapy of prostate cancer. The procedure can easily be applied to discover genes specifically expressed in other organs or tumors.
Expressed sequence tags (ESTs) (1) are sequences of cDNA fragments prepared from different tissue sources. There are now well over one million of these sequences in the publicly available database, and these sequences are believed to represent more than half of all human genes (2). Although still incomplete, this large database now can be used to obtain valuable genetic information. The recently announced Cancer Genome Anatomy Project includes, among other features, an analysis of the EST database (refs. 3 and 4, for further information, see http://www.ncbi.nlm.nih.gov/dbEST/; and Cancer Genome Anatomy Project at http://www.ncbi.nlm.nih.gov/ncicgap/). We present herein one example of the way this store of information can be used to identify genes specifically expressed in a particular tissue.
The ESTs belong to different cDNA libraries, each of which was prepared from one particular cell type, organ, or tumor. Therefore, the presence or absence of ESTs in different libraries provides information about the organ, cell type, or tumor specificity of expressed genes. Also, a gene is often represented by many ESTs; generally, the more a gene is expressed in a given tissue, the more ESTs for that gene will be found in the library. Thus, the number of ESTs that represent the same gene in a given library is a rough indication of the expression level of the gene in the tissue from which the library was derived. We use these characteristics of the EST database to identify genes that are specifically expressed in one particular tissue or organ; in this report we use the human prostate as an example. Such genes could be useful in the diagnosis or therapy of cancer.
Data Preparation.
There are two sources from which the EST information can be obtained (ftp://ncbi.nlm.nih.gov/repository/dbEST), the report file generated from the dbEST database and the EST-FASTA file made from GenBank (http://www.ncbi.nlm.nih.gov/Web/GenBank/index.html). We used the dbEST report file because the EST-FASTA file contained many entries with no library name information. A human EST file was generated by collecting ESTs from all libraries that contained the words “Homo sapiens” in the organism field of the library.‡ A separate human prostate EST file also was generated by collecting ESTs from all human libraries that contained the word “prost” in the library name, organism, tissue type, organ, or cell line field of the library.‡
Identification of Prostate-Specific ESTs.
After these files were prepared, the sequence homology searching program blastn (ref. 5, for further information, see http://www.ncbi.nlm.nih.gov/BLAST/) was run sequentially for each human prostate EST sequence against all of the human ESTs. The homology stringency was set high [S = 300; V = 300; B = 300; n = −20, see the blast manual available through e-mail (toolbox@ncbi.nlm.nih.gov)] so that the procedure would select identical rather than homologous sequences, but not so high as to disallow mismatches caused by possible sequencing errors. The ESTs that produce more than 300 selections were discarded because these contained repetitive elements.§
For each query EST, the search produced a list of EST entries (hits) that had one or more stretches of high sequence identity. Each hit list was separated into two groups, one for hits among the prostate ESTs and another for those among the nonprostate ESTs. The prostate hit list was used to group the ESTs (see below). The nonprostate hit list was used to determine the specificity. We define the specificity index of a prostate EST as the number of different tissues represented in its nonprostate hit list. The lower the specificity index (fewer organs hit), the higher is the specificity of the EST for prostate.
Collecting Prostate ESTs That Belong to the Same cDNA Clone.
The prostate ESTs were grouped into clusters so that two or more of ESTs that shared one or more stretches of high sequence identity belonged to one cluster. This was done by an iterative algorithm in which a cluster was formed by including one EST and all of its neighbors (those in its prostate hit list) and then all the neighbors of the neighbors, and so on. The iteration stopped when no new members were found for any cluster.
Most ESTs come in pairs that have the same name, except for the endings, which are either r1 or s1. These pairs, which we call partners, come from opposite ends of the same insert in one clone and may or may not overlap. To include as many ESTs from one transcript as possible in one cluster, we combined two clusters into one if they shared more than one partner pair between them. We used more than one partner pair as the criterion, because the opposite ends of one insert may, sometimes, come from different cDNAs caused by a ligation error or a computer control tracking error. If two clusters shared only one partner pair, we combined them only if the specificities of the two partners and those of the two clusters (see below) were similar.¶
Sorting for the Frequent and Differentially Expressed cDNA Candidates.
Once the prostate ESTs were clustered in the manner described, a specificity index was assigned to each cluster. The cluster specificity index was defined as the number of different tissues represented in the nonprostate hit list of all the ESTs in the cluster. We then selected only those clusters that had specificity index of 0, if detected in no other tissue; 1, if detected in one other tissue; 2, if detected in two other tissues; or 3, if detected in three other tissues. There are several reasons that clusters with less than complete specificity for prostate (those with a specificity index of 1, 2, or 3) were considered. One reason is that the gene may be expressed in nonprostate tissues only at a low expression level, in which case it may still be considered relatively prostate-specific. Another reason is that a cluster may represent more than one gene transcript, as will be described later, in which case additional examination of the constituent ESTs may reveal a more specific gene. Also, an EST from one prostate gene transcript can have a hit to an EST from a different gene transcript, in which case the false hit should be disregarded. The fourth reason is that the gene may be expressed in a cancer but not in the normal nonprostate tissue in which the cancer developed, because genes are often activated in cancer. The clusters that met the specificity requirements were sorted in decreasing order of their size, i.e., number of individual ESTs. Therefore, the most expressed cDNA candidates will be on the top of the list. A table was then produced from the sorted list in which we kept only those clusters with at least five or more ESTs.
Computer Analysis Results.
The results presented below were obtained by using the dbEST file provided by National Center for Biotechnology Information as of July 26, 1997. The database contained 1,137,304 EST entries in 907 cDNA libraries. There were 539 human libraries, of which 16 were from the prostate. Clustering of the human prostate ESTs resulted in 7,200 clusters made of 10,865 sequences. Another 6,703 prostate ESTs were rejected because they had more than 300 ESTs each.
The first three clusters have more than 100 inserts each. As expected they contain ESTs with putative identifications [putative identifiers (ID) in the dBest file] from known genes that are expressed in the prostate (Table 1). The largest cluster contains ESTs from the prostate specific antigen (PSA) and from the glandular kallikrein. The reason that two different proteins appear in the same cluster is that their DNA sequences share stretches that are highly homologous. This is one mechanism by which more than one gene becomes grouped into one cluster. Although PSA is considered to be prostate-specific (ref. 4, for further information, see Cancer Genome Anatomy Project at http://www.ncbi.nlm.nih.gov/ncicgap/), it had hits in two tumor libraries, breast and lung. Two nearly identical ESTs from the glandular kallikrein have hits to an EST from the pancreas, but these are probably false hits as the overall homology between the prostate and pancreatic sequences is low. The second largest cluster in Table 1 contains ESTs from the prostate-secreted seminal plasma protein (PSSPP). This cluster is also listed as being prostate-specific in the Cancer Genome Anatomy Project web page, but we found by the computer analysis that it was also expressed in lung cancer libraries. The third largest cluster contains ESTs from the prostatic acid phosphatase with matches in lung tumor and fetal heart libraries.
Table 1.
Clusters with five or more ESTs and cluster specificity index of 0, 1, 2, or 3
| Selected cluster name | No. ESTs | No. of ESTs with zero hits in other organs | Prostate source | Specificity index (hits in other organs or tumors) | Putative ID or EST IDs |
|---|---|---|---|---|---|
| 274 | 115 | n/t | 3 (lung, breast, pancreas) | PSA Glandular kallikrein | |
| 234 | 5 | n/t | 1 (lung) | PSSPP* | |
| 133 | 35 | n/t | 2 (heart, lung) | Prostatic acid phosphatase | |
| 16 | 0 | t | 2 (brain, placenta) | T-cell receptor γ chain C region | |
| 14 | 0 | n/t | 3 (senescent fibroblast, testis, lung) | nc35a03, nj93c05, nj65e04, ng89g12, nh69c06, nc35h08, ng75f05, ng95e05, nj09a12, nj15a05, ng77b09, nh52g10 | |
| 13 | 12 | n | 1 (kidney) | Semenogelin 1 and semenogelin II | |
| C1 | 12 | 12 | n/t | 0 | nc19c11, nc13d11, nc46c10, nc45d06, nc47f03, nc16a06 |
| 11 | 8 | n | 1 (colon) | Adenylate kinase isoenzyme 1 | |
| C2 | 8 | 8 | n | 0 | nc14b02, nc04c08, nc71b05, nc06e12, nc71b06 |
| C3 | 7 | 7 | t | 0 | nc39f01, nc39e12, nc39f02, nc39f08, nc39f10 |
| 7 | 7 | t | 0 | nc45b05, nc47g07, nc50c01, nc50e02, nc46b12, rep‡/na† | |
| 7 | 1 | n/t | 2 (muscle, fet. liver–spleen) | EST82997, EST82999, nc14b12, nc35g10, nc13f10 | |
| 6 | 6 | n | 0 | nc21c02, nc21g08, nc25h05, nc27d09, rep‡ | |
| C4 | 6 | 6 | n | 0 | nc09h02, nc11c01, nc27e02, nc11g03 |
| C5 | 6 | 6 | n | 0 | nc09b07, nc26f02, nc19c02 |
| 6 | 6 | t | 0 | nc45b06, nc47e09, nc47c02 na† | |
| C6* | 6 | 6 | t | 0 | nc49e12, nc50a11, nc50a10 |
| 6 | 0 | n | 2 (fetal heart, melanocyte) | SNAP-23 protein | |
| 6 | 0 | n/t | 3 (colon, colon mucosa, pancreatic islet) | nc08h07, nj62g06, nj52h03, nj97a07, nc35e08 | |
| 6 | 0 | n/t | 2 (placenta, fet. liver spleen) | nc27g01, nc79f08, nc33g02, nh32c06 | |
| 6 | 0 | t | 1 (lung) | nc44a10, nc44f02, nc50d07 | |
| C7 | 5 | 5 | t | 0 | ng90g10, nc44h02, nc51d11, nc45e06 |
| C8 | 5 | 5 | t | 0 | nc47a06, nc51c05, nc47d03 |
| 5 | 5 | n | 0 | ERBB-3 receptor protein-tyrosine kinase | |
| 5 | 5 | n | 0 | nc09b11, nc26c10, nc17g01 | |
| 5 | 5 | n/t | 0 | nc35f10, nc78a06, nj45e02 | |
| 5 | 5 | n/t | 0 | nc74f10, nc75f10, nj94a02 | |
| 5 | 5 | t | 0 | SYT | |
| 5 | 5 | t | 0 | Androgen receptor | |
| 5 | 5 | t | 0 | ni72d09, ni72e05, ni67h04, ni62e03, ni75f09 na† | |
| 5 | 2 | n | 3 (brain, placenta, melanocyte) | Contains MER2.b2, MER2 repetitive element | |
| 5 | 0 | t | 2 (breast, fetal liver-spleen) | nc76b03, nc76b02, nc78c03, nc78a03 | |
| 5 | 0 | n/t | 2 (Ewing’s sarcoma, colon | Homeobox protein HOX-D13 |
Tumor libraries are underlined. n, These clusters contain ESTs from normal prostate libraries; t, these clusters contain ESTs from tumor prostate libraries. A more extended table can be found at http://www.nci.nih.gov/RESEARCH/basic/lmb/mms.htm. The EST clones that were selected and experimentally tested are in boldface type. The dBEST clone numbers are listed as IDs in this table because a search in the dBEST library under these names will list the r1 as well as the s1 sequences. (The Genbank accession numbers will list only the r1 or the s1 sequence, and additional searches need to be done to find the other partner.)
The C6 cluster was not analyzed because the selected clone (nc50a10) contained the wrong insert. The actual insert was found to belong to the PSSPP cluster.
EST clones from these clusters were not available.
These clusters contain ESTs with a warning for the presence of some kind of repetitive element.
EST Clusters Specific for Prostate.
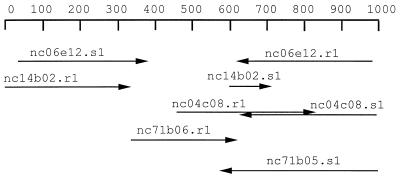
There are 18 clusters in Table 1 that have a specificity index of zero, i.e., no hits in any other tissue, indicating they were not found in nonprostate libraries. All but three of these have no putative IDs assigned to any of their ESTs. The 15 clusters with complete specificity and no putative ID represent candidates for genes specifically expressed in prostate that have not yet been characterized. We selected eight of these, mostly from the top of the list, for the experimental tests. The clusters chosen are designated C1–C8 in Table 1. The C1 cluster is represented in both normal prostate and prostate cancer libraries; the C2, C4, and C5 clusters are represented only in normal prostate libraries; and C3, C6, C7, and C8 are found only in prostate cancer libraries. We assembled a combined maximal sequence for each of these clusters. For example, about 1 kb of sequence could be assembled for the C2 cluster (Fig. 1).
Figure 1.
Assembly of the maximal sequence for candidate cluster C2. Each arrow represents an EST sequence in the cluster. The assembly shows, surprisingly, that both partners of the nc14b02 insert run in the same direction.
Analysis of Selected Clones by RNA Hybridization.
An EST was selected from each of the selected clusters and the corresponding clone (Table 1, indicated in boldface type) was obtained and verified by DNA sequencing. The inserts were radiolabeled and used for RNA hybridization. The hybridization results are summarized in Table 2.
Table 2.
Hybridization results for the selected prostate specific clusters
| Cluster | Clone name | Library | Insert size, bp | Dot blot | Northern blot |
|---|---|---|---|---|---|
| C1 | nc46c10 | Pr1, Pr3 | 550 | Prostate specific | ∼600 bases (dominant) 1,600, 2,400 bases (weak) |
| C2 | nc06e12 | Pr1 | 980 | Prostate specific | 10,000 bases |
| C3 | nc39f10 | Pr2 | 500 | No hybridization | No hybridization |
| C4 | nc09h02 | Pr1 | 550 | No hybridization | No hybridization |
| C5 | nc26f02 | Pr1 | 650 | Prostate specific | ∼600 bases |
| C7 | nc44h02 | Pr3, Pr6 | 550 | No hybridization | No hybridization |
| C8 | nc47d03 | Pr3 | 250 | No hybridization | No hybridization |
| PSSPP | (nc50a10) | All except Pr20 | 400 | Prostate, trachea, (lung, stomach, salivary gland) | ∼600 bases (dominant) 9,500 bases (weak) |
EST clone with the largest insert was chosen as a probe from each cluster. Prostate dbEST libraries as of July 26, 1997 are NCI_CGAP_Pr1, 2, 3, 5, 6, 8, 9, 10, 11, 12, 20, 21, 22; Tigr_human prostate gland, prostate gland I, prostate gland V. Tumor libraries are underlined.
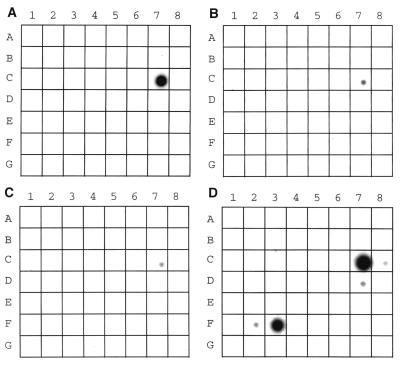
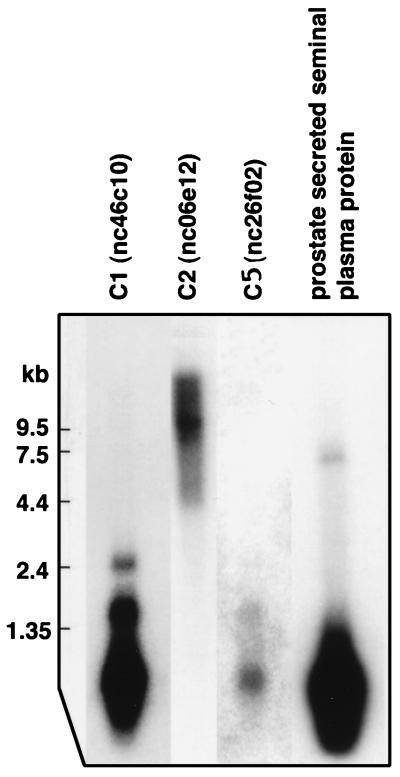
The prepared EST clone inserts were evaluated for specificity by hybridizing them with filters containing normalized amounts of mRNA from 50 different human tissues. As shown in Figs. 2 A–C, inserts from the C1 (nc46c10), C2 (nc06e12), and C5 (nc26f02) clusters are all prostate-specific, as assessed by the RNA dot blot. For nc46c10 from C1, a Northern blot shows a major band at approximately 600 bases and two minor bands at 1.6 and 2.4 kb (Fig. 3). The lower bands are probably splice variants or degradation products. The insert in nc06e12 from C2 is 980 bp long and hybridized with a 10-kb full-length message (Fig. 3). The insert in nc26f02 from C5 shows one band at approximately 600 bp on the Northern blot and it is likely that the EST clone contains the full-length transcript (Fig. 3).
Figure 2.
cDNA inserts representing candidate clusters C1 (A), C2 (B), and C5 (C) and from the PSSPP (D) were hybridized to RNA dot blots (Human RNA Master blot, CLONTECH) containing mRNAs from 50 normal human cell types or tissues. The mRNA dots are from whole brain, amygdala, caudate nucleus, cerebellum, cerebral cortex, frontal lobe, hippocampus, medulla oblongata, occipital lobe, putamen, substantial nigra, temporal lobe, thalamus, subthalamic nucleus, spinal cord, heart, aorta, skeletal muscle, colon, bladder, uterus, prostate (position C7 in A–D), stomach (position C8), testis, ovary, pancreas, pituitary gland, adrenal gland, thyroid gland, salivary gland (D7), mammary gland, kidney, liver, small intestine, spleen, thymus, peripheral leukocyte, lymph node, bone marrow, appendix, lung (F2), trachea (F3), placenta, fetal brain, fetal heart, fetal kidney, fetal liver, fetal spleen, fetal thymus, and fetal lung. Each EST clone (Genome Systems, St. Louis) was first confirmed by sequencing, and the clone inserts were isolated as EcoRI–NotI fragments and labeled with 32P (Lofstrand Laboratories, Gaithersburg, MD).
Figure 3.
Northern blots (CLONTECH) of mRNA from normal prostate probed with cDNA inserts represent candidates C1, C2, and C5 and PSSPP. Indicated on the left are the positions of the mRNA size markers.
Four inserts from the C3 (nc39f10), C4 (nc09h02), C7 (nc44h02), and C8 (nc47d03) clusters showed no hybridization in either the RNA dot blots or the Northern blots in repeated experiments (Table 2). Additional investigation is needed to determine why these clones show no hybridization.
There is a mismatch between the name and the actual insert in some of the EST clones for the C3 cluster. When we obtained and sequenced the nc39f01, nc39f02, nc39f08, and nc39f10 clones that belong to this cluster (Table 1), we found that the s1 sequences matched the vector inserts from the named clones but none of the r1 sequences did. Thus, the r1 and s1 partners do not belong to the same insert in these cases and the number of inserts in the cluster is reduced to two to four, depending on how the mismatch was produced. Although this does not explain why the nc39f10 clone did not hybridize with any RNA, it shows that errors in the database can force unrelated clusters into a larger cluster.
The insert in the nc50a10 clone from the C6 cluster (Table 1) did not have the sequence given in the dbEST. After sequencing the clone, we did a blast search and found it to match the PSSPP sequence. Hybridization experiments with the nc50a10 insert showed that it hybridized strongly with mRNAs from prostate and trachea (Fig. 2D). In addition it hybridized weakly with lung, stomach, and salivary gland mRNAs. The Northern blot shows one major band at approximately 600 bases and a possible minor band at 9.5 kb (Fig. 3). The fact that the PSSPP gene is highly expressed in trachea has not been previously observed and is an unexpected finding.
DISCUSSION
These experimental results indicate that an analysis of the publicly available EST database can identify potential candidates for genes specifically expressed in human prostate. The procedure involves identifying ESTs from the prostate tissues through the use of annotations that come with each cDNA library and grouping them into clusters of related ESTs. Normally, each cluster contains only the ESTs from one gene transcript and the size of the cluster serves as a rough measure of the expression level of the gene. The specificity information for a cluster is obtained from the hit list for each ESTs in the cluster, which is a list of all nonprostate human ESTs that are related (share one or more highly homologous stretches) to the prostate EST. To obtain relatively specifically expressed clones, clusters that have hits to four or more different nonprostate organs are discarded. To select for frequently expressed cDNAs, the remainder are sorted according to the cluster size.
A look at the top of this list (Table 1) shows that the procedure produced the intended result; the well-known PSA tops the list and the first three large size clusters all correspond to genes known to be highly expressed in the prostate. We included ESTs with a specificity index of up to three to include PSA that is highly expressed in prostate but also expressed in nonprostate tumors (6). A more definite proof is provided by the experimental tests: When seven EST clones were selected from different clusters that have no hits in other organs and that have not been previously characterized, three turned out to be prostate-specific.
At the same time, our study uncovered various problems, some algorithmic (e.g., separation of highly homologous cDNAs that are from different genes) but most others related to the database. The most obvious problem is the incompleteness of the EST database, which makes our clusters appear more specific than they really are. An example is the EST clone nc50a10, which was selected from C6 but turned out to be from the gene for the PSSPP (the PSSPP cluster in Table 1). The cDNA hybridized with RNA from trachea and weakly with lung, stomach, and salivary gland. The PSSPP cluster shows no hit to trachea, probably because there is only a very small library from tracheal tumors in the database. Such a problem has, of course, been anticipated. Indeed, we are encouraged by the fact that at least three of the seven with apparent high specificity did turn out to be specific.
Any prostate ESTs that have zero hits in the nonprostate hit list are potentially from genes that are specifically expressed in prostate. However, because EST sequences are rather short DNA fragments, the probe and the target sequences often do not match even when both are from the same gene. Therefore, one obtains a false impression of high specificity if single ESTs are used. We pooled as many ESTs as possible that appeared to be from the same gene and used a specificity measure that applied to the whole group. The group specificity measure should be more reliable than that of individual ESTs. Another advantage of such clustering is that the number of ESTs in a cluster gives a rough measure of the relative expression level of the gene represented by the cluster. This information is useful because the specificity information becomes unreliable when the gene expression is low. Generally, there is no doubt that clustering will produce more reliable information on the specificities than if no attempt is made to cluster. However, clustering is subject to numerous problems, as described below.
Ideally, we would have liked to produce one cluster for each gene transcript. However, ESTs were prepared from a mixed pool of cDNAs from many different gene transcripts, and it was not possible to sort them perfectly into separate genes. The procedure we used was to cluster ESTs from all prostate libraries that shared one or more stretches of high homology and to include the partners (those that have the same clone name) in the same cluster if certain criteria were met. Despite this effort to include as many related ESTs as possible into one cluster, the ESTs from one gene may still be split among two or more clusters if there is no EST that connects them. Such a distribution will produce several small size clusters and tend to make one ignore the corresponding gene; at the same time, the apparent specificity of the smaller clusters may increase, giving rise to candidates for false positive clones.
On the other hand, depending on the degree of homology used for the clustering, this procedure can put highly similar but different genes in the same cluster, as happened for the largest cluster in Table 1. We also have seen cases wherein the EST sequences in one cluster could not be assembled into a single sequence. This can happen because of a ligation error, which puts the r1 and s1 partners from two different genes into one insert or, in rare cases, makes one EST sequence by joining cDNAs from two different genes. We have presented an example of another case, the C3 cluster, wherein two unrelated sequences were included in the same cluster because both were assigned the same clone name, probably by a computer control tracking error (i.e., the actual DNA sequence has been assigned to a wrong EST clone). When a cluster contains ESTs from more than one gene, the number of inserts in the cluster increases, giving a false impression of high expression for the underlying gene, while its apparent specificity can be lower than that of the individual genes, causing one to miss some specific genes. However, unlike the case when the clustering is incomplete, an overclustering will not produce false positives.
A similar problem exists when assessing the specificity. When an EST from the prostate has a hit to an EST from a nonprostate library, the underlying genes can still be different if the two genes are related but not identical or if the hit is produced accidentally because of an error in the database.
The incompleteness of the EST database, and the various problems listed above, indicate that the specificity and the cluster size information given in Table 1 should be used with caution; they only give a semiquantitative measure of the specificity and expression level. Nevertheless, our experimental tests show that a database analysis with the methods described here gives a useful guide for selecting promising clones among more than 17,000 ESTs from the prostate library. The procedure has been completely automated and can easily be extended to examine those specific for other organs or tumors.‖
Acknowledgments
M.E. is the recipient of a fellowship from the Swedish Cancer Society.
ABBREVIATIONS
- EST
expressed sequence tag
- ID
identifier
- PSA
prostate specific antigen
- PSSPP
prostate-secreted seminal plasma protein
Note Added in Proof
We found many ESTs from the untranslated and constant region of the T-cell receptor γ chain in prostate libraries (Table 1), indicating that this gene is highly expressed in prostate. Interestingly, ESTs representing TCR α, β, or δ chains were not found in any prostate library. Hybridization analyses with a radioactive probe from the TCR γ cluster (ng79d11) confirmed that TCR γ mRNA is present in RNA preparations from normal prostate and prostate cancer tissue. However, in mRNA preparations for LNCaP, PC-3, or DU145, cell lines (epithelial origin), TCR γ was not detectable. Immunohistochemistry with a mAb specific to the human TCR γ chain constant region (CγM1, ENDOGEN) provided an explanation for this descrepancy: TCR γ is found in cells in the interstitium of prostate, but not in the epithelial cells from which cancers and cancer cell lines are derived.
Footnotes
For information on the computer programs used in this study, contact G.V. or B.L.
When one of the fields was missing in the dbEST report file, the information was obtained from another source (http://www-bio.llnl.gov/bbrp/image/humlib_info.html).
ESTs from extraordinarily abundant transcripts that do not have repetitive elements also will be lost by this screening, but we have not encountered such ESTs in prostate so far.
A similarity score between specificities of two ESTs or clusters was calculated by adding two points for each organ they shared and subtracting one point for each unmatched organ. The specificities of two ESTs or clusters were judged similar if the calculated similarity score was zero or more.
References
- 1.Adams M D, Kelley J M, Gocayne J D, Dubnick M, Polymeropoulos M H, et al. Science. 1991;252:1651–1651. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 2.Hillier L D, Lennon G, Becker M, Bonaldo M F, Chiapelli B, et al. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- 3.Boguski M S, Lowe T M, Tolstoshev C M. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 4.Pennisi E. Science. 1997;276:1023–1024. doi: 10.1126/science.276.5315.1023. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Zarghami N, Levesque M, D’Costa M, Angelopoulou K, Diamandis E P. Am J Clin Pathol. 1997;108:184–190. doi: 10.1093/ajcp/108.2.184. [DOI] [PubMed] [Google Scholar]