Abstract
Abnormal proliferation of vascular smooth muscle cells (VSMCs) constitutes a key event in atherosclerosis, neointimal hyperplasia, and the response to vascular injury. Estrogen receptor α (ERα) mediates the protective effects of estrogen in injured blood vessels and regulates ligand-dependent gene expression in vascular cells. However, the molecular mechanisms mediating ERα-dependent VSMC gene expression and VSMC proliferation after vascular injury are not well defined. Here, we report that the ER coactivator steroid receptor coactivator 3 (SRC3) is also a coactivator for the major VSMC transcription factor myocardin, which is required for VSMC differentiation to the nonproliferative, contractile state. The N terminus of SRC3, which contains a basic helix-loop-helix/Per-ARNT-Sim protein–protein interaction domain, binds the C-terminal activation domain of myocardin and enhances myocardin-mediated transcriptional activation of VSMC-specific, CArG-containing promoters, including the VSMC-specific genes SM22 and myosin heavy chain. Suppression of endogenous SRC3 expression by specific small interfering RNA attenuates myocardin transcriptional activation in cultured cells. The SRC3–myocardin interaction identifies a site of convergence for nuclear hormone receptor-mediated and VSMC-specific gene regulation and suggests a possible mechanism for the vascular protective effects of estrogen on vascular injury.
Keywords: steroid hormone receptors, transcriptional coregulators, vascular biology
In addition to an important role in growth and development of reproductive tissues, estrogen and its receptors regulate a variety of cardiovascular functions, including vascular tone and the response of blood vessels to injury (1, 2). The pleiotropic effects of estrogen are mediated through binding to its two cognate receptors, estrogen receptors α (ERα) and β (ERβ), members of the nuclear hormone receptor (NHR) superfamily. NHRs are ligand-dependent transcription factors, regulating target gene expression by recruiting coactivator or corepressor complexes to their target promoters (3). Steroid receptor coactivator (SRC) 3 (SRC3) (RAC3/AIB1/TRAM1/pCIP) is a member of the p160 family of NHR coactivators including SRC1 and TIF2 (thyroid hormone receptor interacting factor 2 or SRC2) (4, 5). These molecules were initially identified as ligand-dependent nuclear receptor coactivators that interact with ligand-bound nuclear receptors to modulate target gene activation (3, 6–9). The SRC proteins recruit protein acetyltransferases such as cAMP response element-binding protein binding protein (CBP)/p300 and p/CAF (10–12) and protein methytransferases such as coactivator-associated arginine methyltransferase 1 (CARM-1) and protein arginine methyltransferase-1 (PRMT-1) to nuclear receptor target gene promoters to modify chromatin structure and/or assemble transcription initiation (13, 14). Biochemical studies support that SRC complexes are essential for nuclear receptor activities. The SRC family of coactivators share a signature motif of LXXLL sequences (15) that are responsible for mediating interactions between these coactivators and liganded nuclear receptors (16–18). The SRCs also contain an intrinsic transcriptional activation function mediated through interactions with CBP/p300 and the CBP/p300-associated factor P/CAF. All three SRC family proteins share a highly conserved N-terminal basic helix–loop–helix (bHLH) and Per-ARNT-Sim (PAS) domain motif. The bHLH domain is a DNA-binding and dimerization motif shared among a number of transcription factors, including MyoD family proteins; whereas the PAS motif has been shown to be a binding motif for transcription factors such as AHR, ARNT, and HIF1α. The function of the bHLH–PAS motif in SRC-dependent nuclear receptor signaling remains less well understood. CoCoA, a recently identified GRIP1 (glucocorticoid receptor interacting protein-1) N-terminal interacting protein (19), has been found to potentiate liganded nuclear receptor activation, suggesting that the bHLH–PAS motif participates in receptor action through recruiting additional transcriptional coactivators.
Recent knockout mouse studies support that the SRC proteins each have distinct functions in physiology. Unlike SRC1 and SRC2, SRC3 knockout mice exhibit growth retardation from embryonic day 13.5 through adulthood, and delayed puberty, reduced female reproductive function, and blunted mammary gland development (20, 21). In ovariectomized SRC3 knockout mice, the vascular smooth muscle cell (VSMC) proliferative response to vascular injury is abnormal and unlike the effect of estrogen in wild-type mice, which fully inhibits VSMC proliferation after injury, estrogen replacement is far less effective in inhibiting the vascular injury response in SRC-3 null mice (22–24). The molecular events mediated by SRC3 in VSMC proliferation remain unclear.
VSMCs modulate their phenotypes in response to a wide range of extracellular cues, but the transcriptional mechanisms that regulate this plasticity are not fully understood. In vascular injury, VSMC de-differentiate to a synthetic, proliferative phenotype that is required for the VSMC proliferative response and the formation of neointima in response to injury. Myocardin is a serum response factor (SRF) cofactor that coactivates SRF-dependent genes, is required for SMC differentiation to a nonproliferative, contractile phenotype during development, and functions as a “master regulator” of VSMC gene expression (25–27). SRC proteins have been shown to regulate signaling by classes of DNA binding transcription factors other than NHR, including AP-1, NF-κB, HNF-1α, MEF2C, E2F1, and TEF (28–33). Here, we report that SRC3 is a coactivator for myocardin, thus identifying a site of convergence for NHR mediated- and VSMC-specific gene regulation of potential importance to ER regulation of vascular responses to injury.
Results
SRC3 Is Expressed in Cardiovascular Cells.
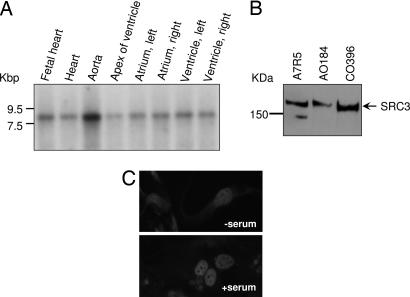
To investigate the role of SRC3 in human vascular cells, we first determined the expression pattern of SRC3 mRNA and protein in human cardiovascular cells and tissues. SRC3 transcripts were detected in human vascular tissues by Northern blot assays (Fig. 1A), with the highest expression levels detected in human aorta. SRC3 protein also was detected in cell lysates from a number of vascular cell lines, including human aortic and coronary VSMC cells (AO184 and CO393 cells, respectively) by immunoblotting (Fig. 1B). In the absence of serum, SRC3 is localized in both nuclear and cytoplasmic compartments (Fig. 1C). After serum exposure, SRC3 localized exclusively in the nuclei of VSMCs, consistent with the effect of serum seen on SRC3 localization in nonvascular cells, such as COS7 and MCF7 cells (34).
Fig. 1.
SRC3 transcript and protein are expressed in VSMCs. (A) Expression pattern of SRC3 in human cardiovascular tissues. A human cardiovascular tissue Northern blot (Clontech) was hybridized with a 32P-SRC3-specific DNA probe encoding a 292-aa fragment (amino acids 400–692). (B) Western blot analysis of SRC3 protein expression in multiple cell lines including human VSMCs. Equal protein loading was confirmed by normalization to GAPDH (data not shown). (C) Immunostaining of SRC3 in aortic VSMC in serum-free media (Upper) or normal DMEM containing 10% FBS (Lower).
SRC3 Interacts with Myocardin in Vitro.
SRC family proteins contain an N-terminal bHLH/PAS domain that mediates protein–protein interactions, such as those between SRC2 and the skeletal muscle-specific transcription factors MyoD and MEF2. SRF, a MADS box family protein, mediates expression of many CArG box-containing smooth muscle genes that characterize the differentiated state of VSMCs (25–27). We hypothesized that SRC3 in smooth muscle cells might influence VSMC differentiation and the VSMC responses seen in vascular injury by direct interactions between SRC3 and either SRF itself or myocardin, the smooth muscle-specific transcriptional coactivator of SRF (25, 26). GST pull-down assays with 35S-labeled myocardin demonstrated a direct interaction between the SRC3 N-terminal bHLH/PAS domain and myocardin, but not between SRC3 and SRF under conditions where the known myocardin–SRF interaction was readily detectable (Fig. 2). Three myocardin family members have been identified, including myocardin, and the myocardin-related transcription factors A and B (MRTF-A and MRTF-B). SRC3 also interacted with MRTF-A and MRTF-B (Fig. 2B). These proteins share high homology, although, unlike myocardin, expression of which is highly restricted to cardiac and smooth muscle cells, MRTF-A and MRTF-B are expressed in smooth and cardiac muscles and a broad range of adult tissues.
Fig. 2.
SRC3 interacts with myocardin family members in vitro. GST pull-down assays with SRF, the N-terminal domain of SRC3 that contains the bHLH and PAS domains [GST-SRC3(1–408)] or GST alone and in vitro-translated 35S- labeled proteins are shown. (A) (Left) GST-SRC3(1–408) interaction with 35S-myocardin. (Center) GST-SRF incubation with 35S-SRC3 (negative control). (Right) GST-SRF incubation with 35S-myocardin (positive control). (B) GST pull-down assays with GST-SRC3(1–408) and myocardin or the related myocardin family members MRTF-A and MRTF-B. The data are representative of at least three individual experiments.
The domains of myocardin mediating the in vitro myocardin–SRC3 interaction were also studied. Deletion of the C-terminal domain (amino acids 715–936) to create myocardin 128–715 leads to a dominant negative mutant shown previously to interfere with myocardial cell differentiation (27). This mutant also abolished interactions between myocardin and SRC3 (amino acids 1–408) (Fig. 3B Upper). This C-terminal domain of myocardin alone interacted directly with the amino-terminal bHLH/PAS domain of SRC3 (Fig. 3B Lower), but not with a control vector containing only the Gal-DNA binding domain (DBD) (data not shown). A construct containing this C-terminal domain [myocd(581–935)] also bound 35S-SRC3 (Fig. 3C). Myocardin also bound the other two SRC family members SRC1 and TIF2, although the interactions in vitro were much weaker than with SRC3 (data not shown). These studies support that the amino-terminal bHLH/PAS domain of SRC3 interacts in vitro with myocardin and related family members, and that the SRC3 interaction with myocardin is mediated through binding to its known C-terminal transactivation domain (TAD).
Fig. 3.
Myocardin C-terminal TAD interacts with SRC3. (A) Domain structure of myocardin, including the TAD (amino acids 715–935). RPEL, protein motif containing RPXXXEL sequence involved in actin binding; Q, poly(Q)-rich region; SAP, 35-aa motif conserved among nuclear scaffold attachment proteins SAF-A/B, Acinus, PIAS; LZ, leucine zipper domain. (B and C) 35S-labeled in vitro-translated myocardin domains including the dominant negative mutant [myocd(128–715)] (B Upper), TAD [myocd(715–935)] (B Lower), or SRC3 (C) were incubated with GST-SRC3(1–408) (B) or GST-myocd(581–935) (C) in pull-down assays. Input represents 12.5% of each in vitro-translated protein used in the pull-down assay.
We examined the functional domain of SRC3 involved in SRC3–myocardin interaction in more detail by using a series of GST-fusion SRC3 fragments representing SRC3 functional domains, including GST-SRC3 (amino acids 1–408); GST-SRC3 (amino acids 400–612); GST-SRC3 (613–752), a nuclear receptor interaction domain; and GST-SRC3 (amino acids 1017–1207), a domain of SRC3 known to interact with CBP/p300 (35). As shown in Fig. 4B, in addition to the N-terminal bHLH/PAS domain (SRC3–1-408), the SRC3 activation domain (amino acids 1017–1207) was found to interact with myocardin in this assay, suggesting the possibility that multiple domains of SRC3 may interact with myocardin.
Fig. 4.
Interaction of SRC3 domains with myocardin. (A) Schematic presentation of the SRC3 domain structure. The N-terminal bHLH/PAS domain, receptor interaction domain (RID), and activation domain (AD) are indicated. (B) GST pull-down assay of a series of GST-SRC3 fusion proteins interacting with 35S-myocardin is shown.
In Vivo Association of SRC3 with Myocardin.
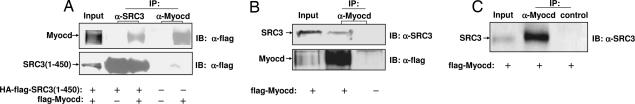
To explore the interaction of myocardin with SRC3 further, coimmunoprecipitation assays were performed. SRC3 (amino acids 1–450) protein was coprecipitated from HEK293 cells with full-length epitope-tagged myocardin (Fig. 5A). Coimmunoprecipitation of myocardin with endogenous SRC3 from MCF7 cells, which express high levels of SRC3 (7), was also observed from cells transiently transfected with myocardin (Fig. 5B). The SRC3–myocardin interaction was also detected in PAC1 cells, a well characterized pulmonary VSMC line (36) (Fig. 5C). These data support that myocardin interacts with SRC3 in intact cells.
Fig. 5.
Coimmunoprecipitiation of myocardin with SRC3 from cells. (A) Myocardin interacts with SRC3 from HEK293 cells transfected with mammalian flag-tagged Myocardin (flag-myocd) and HA-flag-tagged SRC3(1–450) [HA-flag-SRC3(1–450)]. Vector alone (−) was used as negative control. (B) Endogenous SRC3 interacts with myocardin in MCF7 cells. MCF7 cells were transfected with either flag-tagged myocardin (flag-myocd) or vector alone (−). (C) Coimmunoprecipitation of SRC3 with myocardin from vascular smooth muscle (PAC1) cells transfected with mammalian flag-tagged myocardin (flag-myocd) is shown.
SRC3 Is a Coactivator for Transcriptional Activation by Myocardin.
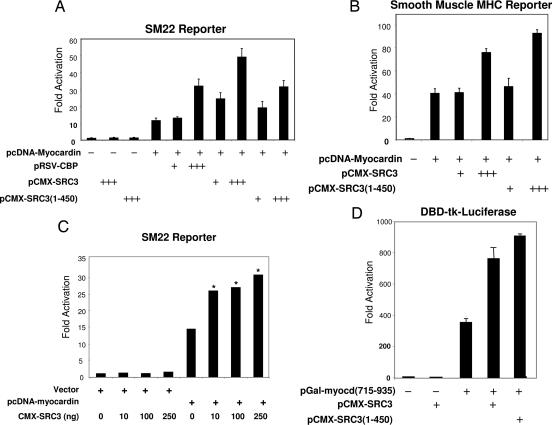
To assess the functional significance of the SRC3–myocardin interaction, we tested whether SRC3 affects myocardin-mediated transactivation of two SRF-responsive, CArG-containing smooth muscle-specific promoters, the SM22 and the smooth muscle MHC genes (37). Myocardin alone resulted in transactivation of the SM22 reporter in HEK293 cells (Fig. 6A). Cotransfection of plasmids encoding full-length SRC3 or the SRC3 bHLH/PAS domain [SRC(1–450)] further increased myocardin-mediated transcriptional activation in a dose-dependent fashion (Fig. 6A). SRC3 enhanced myocardin activity by 3.8-fold (P < 0.003, n = 3), and SRC3(1–450) alone enhanced myocardin activity 2.7-fold (P < 0.0001, n = 3). SRC3 or SRC3(1–450) alone did not transactivate the SM22–445-luc reporter (Fig. 6A). SRC3 also augmented activation of a second CArG-containing, SRF-dependent VSMC gene by myocardin, the smMHC-luc reporter [increase caused by full-length SRC3 coactivation of myocardin, P < 0.03; increase caused by SRC3(1–450) coactivation of myocardin, P < 0.05; Fig. 6B]. SRC3 also modestly coactivated myocardin transactivation of a VSMC-specific reporter in PAC1 smooth muscle cells (Fig. 6C). Myocardin interacts with SRF, an important VSMC transcription factor that controls the expression of a number of VSMC-specific genes. To exclude the possibility that the SRC3 effect on myocardin-mediated transactivation was indirect, through SRF, we used a Gal fusion reporter transcriptional assay to examine directly the effect of SRC3 on myocardin transactivation (Fig. 6D). Gal-myocd(715–935) activated a Gal4 DNA binding reporter DBD-tk-luciferase 382-fold, and cotransfection of SRC3 or SRC3(1–450) further significantly increased Gal-myocd(715–935) activity 2.35- or 2.1-fold on this reporter (P < 0.01 and P < 0.04, respectively). Together, these data support that SRC3 coactivates VSMC-specific gene expression through a direct interaction with the myocardin C-terminal TAD. However, despite numerous studies in which ERα was cotransfected with myocardin in the absence or presence of estradiol, no effect of estrogen on SRC3-mediated coactivation of myocardin on VSMC-specific reporters (see Discussion).
Fig. 6.
SRC3 augments myocardin transactivation of vascular smooth muscle-specific promoters. Dose-dependent coactivation of myocardin transcriptional activation by SRC3 is shown. (A and B) Increasing amounts of mammalian expression plasmids pRSV-CBP, pCMX-SRC3(1–450), or pCMX-SRC3 (−, 0 μg; +, 100 ng; +++, 300 ng) were transfected into HEK293 cells with 1 ng of pcDNA-myocardin and either the SM22–445lucifease reporter (A) or smMHC-luciferase reporter construct (B). (C) SRC3 enhances myocardin transactivation of the SM22 reporter in vascular smooth muscle PAC1 cells. Relative fold activation in A–C was determined by comparison with the basal SM22–445 luciferase activity or smMHC-luciferase when transfected with control CMX vector. Data are expressed as the mean ± SD of triplicates from a representative experiment of three independent experiments. Asterisks represent P for comparison of data between SRC3 coactivation and vector only. *, P < 0.02. (D) SRC3 enhances Gal-myocardin TAD activation in Gal4 DBD-luciferase reporter system. Myocardin C-TAD from amino acids 715–935 was expressed as Gal4-DBD fusion protein in the pCMX-Gal vector.
SRC3 siRNA Attenuates Myocardin Transactivation.
To examine the effect of SRC3 on myocardin transactivation in situ in HEK293 and PAC1 cells, we used small RNA interference to diminish endogenous SRC3 expression and performed reporter assays. A 21-bp double-stranded SRC3 siRNA oligo from SRC3 nucleotides 415–434 or nonspecific control (scrambled) RNA were transiently transfected into HEK293 or PAC1 cells, followed by cotransfection of pcDNA–myocardin or vector and the SM22 reporter. The inhibition of SRC3 expression resulted in a 55% reduction in myocardin transactivation of the SM22 reporter (Fig. 7A). Similar results were obtained in PAC1 cells (Fig. 7B), supporting further that SRC3 is required for the complete transactivation of the smooth muscle-specific, CArG-containing SM22 promoter by myocardin.
Fig. 7.
Knockdown of SRC3 with siRNA reduces myocardin transactivation of VSMC-specific reporters. HEK293 (A) or PAC1 (B) cells were transfected with 75 nM of SRC3 siRNA (siSRC3) or control scrambled oligo duplex and then cotransfected with 10 ng (A) or 50 ng (B) of pcDNA-myocardin or pcDNA vector and SM22–445luciferase reporter together with 50 ng CMX-β-gal as an internal control. Results shown are mean values of three independent experiments in triplicate. In A, *, P < 0.013; in B, *, P < 0.043. SRC3 protein was assayed by immunoblotting, and expression levels of GAPDH (A) or lamin A (B) were used as protein loading controls.
Discussion
Myocardin is a SAP (SAF-A/B, acinus, PIAS) domain-containing protein expressed exclusively in cardiac and smooth muscle cell lineages (27). Myocardin activates many VSMC differentiation marker genes in a strictly SRF-CArG box-dependent way. Recent reports from myocardin gain- and loss-of–function studies identify myocardin to be necessary to initiate smooth muscle differentiation (26). The interplay between differentiation and dedifferentiation of VSMC by growth factor stimulation relies in part on the displacement of myocardin from SRF (38). These findings provide a molecular basis for smooth muscle plasticity, which is a fundamental element of both vascular differentiation and the vascular response to injury. However, the mechanisms that regulate SMC differentiation and dedifferentiation are complex, involve the cooperation of multiple transcription factors and signaling pathways, and are poorly understood in the process of vascular injury in adult vessels. Previous structural and functional analyses revealed that the myocardin N terminus contains a conserved basic, polyglutamine-rich domain that interacts with SRF and a strong TAD located at its C terminus. Direct association of the myocardin TAD and p300 leads to acetylation of nucleosomal histones surrounding CArG box binding sites (39). However, the molecular mechanisms mediating transactivation of VSMC genes by myocardin are not fully understood. In this study, we find that the nuclear receptor coactivator SRC3 is a transcriptional coactivator for the key regulator of smooth muscle transcription and differentiation, myocardin. SRC3 protein possesses intrinsic histone acetyltransferase activities and also recruits other chromatin-modifying enzymes to the transcriptional complex assembled by ERs, such as protein acetyltransferase CBP/p300 and p/CAF, and protein methyltransferases CARM1 and PRMT1 (for review, see ref. 40). We hypothesize that SRC3 assembles chromatin remodeling complexes to a subset of smooth muscle-specific promoters through a direct interaction with myocardin. The cellular compartmentalization of SRC3 has been suggested to be regulated by serum in a number of cells (34). In mouse embryonic fibroblasts, serum deprivation results in the redistribution of p/CIP to the cytoplasmic compartment and stimulation with growth factors or tumor-promoting phorbol esters promotes p/CIP shuttling into the nucleus (41). However, in smooth muscle cells, we observed the nuclear localization of SRC3 in both the presence and absence of serum, with a more diffuse expression in cytoplasm as well in the absence of serum, suggesting that SRC3 function might also be regulated by its localization in VSMCs.
A number of diseases, including atherosclerosis, asthma, neointima formation, and hypertension, have been linked to abnormal proliferation of VSMCs in the adult. SRC3 coactivates a number of transcription factors, such as NF-κB, AP-1, STATs, ETS, p53, and E2F1, in addition to NHRs (for review, see ref. 4). SRC3 posttranslational modification also has been implicated in pleiotropic actions of the coactivator in response to various cellular cues. Recent studies from O'Malley's group (42) have characterized specific SRC3 phosphorylation sites required for its interactions with transcription factors, which forms the basis for the hypothesis that distinct cellular signals may differentially phosphorylate SRC3 to direct its coactivator activity to distinct cellular pathways. It will be important to determine whether posttranslational modification also regulates the interaction between SRC3 and myocardin. Smooth muscle cells are highly plastic in modulating their phenotype between contractile (differentiated) and synthetic (proliferative) states in response to different environmental cues (43).
Further experiments to explore the role of SRC3 in myocardin-dependent transactivation are warranted. It will be important to demonstrate that both myocardin and SRC3 occupy the same endogenous promoter in vivo and that silencing of SRC3 reduces the activity of an endogenous promoter as it does in the reporter assays shown. In the mouse carotid injury model, the vascular protective effects of estrogen are lost in mice harboring full-body disruption of ERα, but not in those lacking ERβ (ref. 25 and references therein). Estrogen inhibits the proliferation of VSMC both in vivo and in cell culture (24, 44) and inhibits PDGF-stimulated VSMC cell cycle progression (45). The lack of any effect of estrogen on the SRC3-dependent coactivation of VSMC-specific promoters suggests that any involvement of the SRC3–myocardin interaction in limiting estrogen's protective effects in SRC3 knockout mice likely involves the coactivation of VSMC genes that require SRC3 and myocardin, but not ERα, and/or a model in which forms of SRC3 are limiting in the regulation of a subset of VSMC genes. These models will need to be tested in mice harboring VSMC-specific deletions of ERα and SRC3.
In summary, these studies show that SRC3 is a coactivator for the VSMC master regulatory transcription factor myocardin and identify a site of convergence for NHR-mediated and VSMC-specific gene regulation that may be important in the vascular response to injury in a wide variety of human diseases.
Materials and Methods
Plasmids.
Mammalian expression plasmids pcDNA-myocardin and its dominant negative mutant pcDNA-myocardinDN have been described and express an N-terminal flag epitope (27), which was used to identify myocardin expression in all myocardin expression experiments shown. A myocardin C-terminal TAD fragment was PCR-amplified and subcloned into either pCMX-Gal vector for mammalian expression or pGEX-2TA vector for bacterial expression. The resulting fusion constructs have been verified by sequencing. All SRC3 constructs used have been described (6, 46).
Northern Blot Assay.
A random-primed 32P-labeled DNA probe specific for SRC3 was hybridized to a human cardiovasculature Northern blot according to the manufacturer's instructions (BD Biosciences/Clontech, Palo Alto, CA). A 876-bp SRC3 EcoR1/EcoR1 cDNA fragment from construct pCDG-SRC3(400–1204) was labeled with 32P-dCTP by using a random priming kit (Amersham, Piscataway, NJ) and used as a probe in the Northern blot assay. Each lane in the blot contains ≈1 μg of poly(A)+ RNA. Filters were exposed to x-ray film at −70°C.
Indirect Immunofluorescence Microscopy.
Human aortic smooth muscle cells AO184 were grown on coverglasses in 12-well plates. For serum-free conditions, 24 h after plating, the cells were changed to serum-free media for an additional 48 h. Immunostaining was performed as described (47). Briefly, cells were washed twice with PBS and fixed in methanol/acetone (1:1) for 1 min on dry ice. After rehydration, the cells were overlaid with mαSRC3 monoclonal antibody (BD Transduction Laboratory, San Jose, CA) and detected by rhodamine-conjugated goat anti-mouse secondary antibody. The cell nuclei were counterstained with DAPI (Sigma, St. Louis, MO). The coverglasses were mounted on microscopy slides and imaged on an epi-fluorescence microscope with an Axiocam CCD camera and Axiovision software (Carl Zeiss, Thornwood, NY).
GST Pull-Down Assay.
Plasmids that encode the fusion protein of SRC3 N terminus from amino acids 1–408 and myocardin C terminus (amino acids 581–935) with GST were transformed into BL21 DE3+ bacterial. The fusion proteins were induced by isopropyl β-d-thiogalactoside (10 μM) and purified with glutathione agarose beads (Sigma). Approximately 0.5 μg of purified GST fusion protein was incubated with 5 μl of in vitro-translated 35S-labeled receptors with moderate shaking at 4°C overnight in binding buffer (20 mM Hepes, pH 7.7/75 mM KCl/0.1 mM EDTA/2.5 mM MgCl2/0.5% Nonidet P-40/1 mM DTT/1 μg/ml BSA). The bound proteins were washed five times with binding buffer, and the beads were collected by microcentrifugation. The bound protein was eluted into SDS sample buffer, subjected to SDS/PAGE, and detected by autoradiography.
Coimmunoprecipitation and Western Blotting.
Coimmunoprecipitation was conducted according to the standard protocol using protein A/G sepherose beads (Amersham). HEK293, MCF7, or PAC1 cells were transfected by electroporation with mammalian expression plasmids as indicated in Fig. 5. The cells were changed to fresh media 6 h after transfection and maintained for an additional 30 h. The cells were then harvested in lysis buffer (20 mM Tris·Cl, pH 7.5/137 mM NaCl/2 mM EDTA/1% Triton/10% glycerol/25 mM β-glycerol phosphate/1 mM PMSF and protease inhibitor mixture). The immunoprecipitation was performed by the addition of 2 μg of antibodies to cell lysates: mαflag (Sigma) for IP myocardin protein, rabbit anti-HA antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for IP SRC3(1–450) protein, and mαSRC3 for endogenous SRC3 protein. The samples were incubated overnight at 4°C. Protein A/G beads (Amersham Bioscience) were added for an additional 2 h. The immunoprecipitants were washed five times with wash buffer (50 mM Tris, pH 7.5/7 mM MgCl2/2 mM EDTA/1 mM PMSF). Associated proteins were resolved by SDS/PAGE and immunoblotted with antibodies as indicated.
Cell Culture and Transient Transfection.
All cell lines used in this study including HEK293T cells, MCF7 cells, PAC1 cells, and AO187 cells were maintained in DMEM with 10% FBS and penicillin-streptomycin. The luciferase reporter constructs SM22–455-luciferase, SM-MHC-luciferase and DBD-tk-luciferase have been described (6, 27, 48). One day before transfection, cells were seeded in 12-well plates at 20,000 cells per well. A DNA mixture containing 1 ng of mammalian expression vector pcDNA-myocardin, 0.1 μg of CMX-SRC3 plasmid, 50 ng of internal control pCMX-β-Gal, 150 ng of luciferase reporter construct, and 0.2 μg of carrier DNA (pGEM), was prepared in a final volume of 50 μl. The DNA solution was mixed with 5 μl of polyfect reagent (Qiagen, Valencia, CA) before addition to each well. The cells were harvested in cell lysate buffer (Promega, Madison, WI) 48 h after transfection and subjected for luciferase and β-gal assays. All transfection assays were performed in triplicate and repeated two or more times.
siRNAs for SRC3 (5′-AGACTCCTTAGGACCGCTT-3′) and control scamble oligo were produced by Dharmacon (Lafayette, CO). siRNAs (75 nM) were transfected into HEK293 or PAC1 cells plated in 12-well plates with ≈40-∼50% cell density by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Twenty-four hours after transfection, the cells were retranfected with expression plasmids for myocardin or vector alone, SM22-luciferase reporter, and β-gal (internal control) for an additional 36 h before harvesting for luciferase and β-gal assays.
Luciferase and β-Gal Assay.
Transfected cells in each well were lysed in 300 μl of 1× cell lysis solution (Promega) and processed for luciferase and β-gal assay. Fifteen microliters of lysed cells was transferred into 96-well microtiter plates, and luciferase and β-gal reporter activities were assayed with a Luciferase Assay System (Promega) and Galacto-Light system (Applied Biosystems, Foster City, CA). The activities were measured in Luminoskan (Labsystems, Chicago). The luciferase activities were normalized to the β-gal activity expressed from the cotransfected pCMX-β-Gal plasmid.
Abbreviations
- SRC
steroid receptor coactivator
- ER
estrogen receptor
- VSMC
vascular smooth muscle cell
- NHR
nuclear hormone receptor
- CBP
cAMP response element-binding protein binding protein
- TAD
transactivation domain
- DBD
DNA binding domain
- bHLH
basic helix–loop–helix
- PAS
Per-ARNT-Sim
- SRF
serum response factor
- MRTF
myocardin-related transcription factor.
Footnotes
The authors declare no conflict of interest.
References
- 1.Mendelsohn ME, Karas RH. N Engl J Med. 1999;340:1801–1811. doi: 10.1056/NEJM199906103402306. [DOI] [PubMed] [Google Scholar]
- 2.Mendelsohn ME, Karas RH. Science. 2005;308:1583–1587. doi: 10.1126/science.1112062. [DOI] [PubMed] [Google Scholar]
- 3.Smith CL, O'Malley BW. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 4.Xu J, Li Q. Mol Endocrinol. 2003;17:1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- 5.Leo C, Chen JD. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Gomes PJ, Chen JD. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 9.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 10.Bannister AJ, Kouzarides T. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 11.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 12.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 13.Xu W, Cho H, Kadam S, Banayo EM, Anderson S, Yates JR, 3rd, Emerson BM, Evans RM. Genes Dev. 2004;18:144–156. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stallcup MR, Chen D, Koh SS, Ma H, Lee YH, Li H, Schurter BT, Aswad DW. Biochem Soc Trans. 2000;28:415–418. [PubMed] [Google Scholar]
- 15.Savkur RS, Burris TP. J Pept Res. 2004;63:207–212. doi: 10.1111/j.1399-3011.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- 16.Heery DM, Kalkhoven E, Hoare S, Parker MG. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 17.Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. Nature. 1997;387:677–684. [Google Scholar]
- 18.Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ, Stallcup MR. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 19.Kim JH, Li H, Stallcup MR. Mol Cell. 2003;12:1537–1549. doi: 10.1016/s1097-2765(03)00450-7. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Rose DW, Hermanson O, Liu F, Herman T, Wu W, Szeto D, Gleiberman A, Krones A, Pratt K, et al. Proc Natl Acad Sci USA. 2000;97:13549–13554. doi: 10.1073/pnas.260463097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O'Malley BW. Proc Natl Acad Sci USA. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan Y, Liao L, Tulis DA, Xu J. Circulation. 2002;105:2653–2659. doi: 10.1161/01.cir.0000018947.95555.65. [DOI] [PubMed] [Google Scholar]
- 23.Sullivan TR, Jr, Karas RH, Aronovitz M, Faller GT, Ziar JP, Smith JJ, O'Donnell TF, Jr, Mendelsohn ME. J Clin Invest. 1995;96:2482–2488. doi: 10.1172/JCI118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pare G, Krust A, Karas RH, Dupont S, Aronovitz M, Chambon P, Mendelsohn ME. Circ Res. 2002;90:1087–1092. doi: 10.1161/01.res.0000021114.92282.fa. [DOI] [PubMed] [Google Scholar]
- 25.Pipes GC, Creemers EE, Olson EN. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Wang DZ, Pipes GC, Olson EN. Proc Natl Acad Sci USA. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 28.Belandia B, Parker MG. J Biol Chem. 2000;275:30801–30805. doi: 10.1074/jbc.C000484200. [DOI] [PubMed] [Google Scholar]
- 29.Lee SK, Kim HJ, Na SY, Kim TS, Choi HS, Im SY, Lee JW. J Biol Chem. 1998;273:16651–16654. doi: 10.1074/jbc.273.27.16651. [DOI] [PubMed] [Google Scholar]
- 30.Chen SL, Wang SC, Hosking B, Muscat GE. Mol Endocrinol. 2001;15:783–796. doi: 10.1210/mend.15.5.0637. [DOI] [PubMed] [Google Scholar]
- 31.Werbajh S, Nojek I, Lanz R, Costas MA. FEBS Lett. 2000;485:195–199. doi: 10.1016/s0014-5793(00)02223-7. [DOI] [PubMed] [Google Scholar]
- 32.Ruas JL, Poellinger L, Pereira T. J Biol Chem. 2002;277:38723–38730. doi: 10.1074/jbc.M205051200. [DOI] [PubMed] [Google Scholar]
- 33.Louie MC, Zou JX, Rabinovich A, Chen HW. Mol Cell Biol. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu RC, Qin J, Hashimoto Y, Wong J, Xu J, Tsai SY, Tsai MJ, O'Malley BW. Mol Cell Biol. 2002;22:3549–3561. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 36.Rothman A, Kulik TJ, Taubman MB, Berk BC, Smith CW, Nadal-Ginard B. Circulation. 1992;86:1977–1986. doi: 10.1161/01.cir.86.6.1977. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Miano JM, Cserjesi P, Olson EN. Circ Res. 1996;78:188–195. doi: 10.1161/01.res.78.2.188. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 39.Cao D, Wang Z, Zhang CL, Oh J, Xing W, Li S, Richardson JA, Wang DZ, Olson EN. Mol Cell Biol. 2005;25:364–376. doi: 10.1128/MCB.25.1.364-376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenna NJ, O'Malley BW. Endocrinology. 2002;143:2461–2465. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- 41.Qutob MS, Bhattacharjee RN, Pollari E, Yee SP, Torchia J. Mol Cell Biol. 2002;22:6611–6626. doi: 10.1128/MCB.22.18.6611-6626.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O'Malley BW. Mol Cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida T, Owens GK. Circ Res. 2005;96:280–291. doi: 10.1161/01.RES.0000155951.62152.2e. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura Y, Suzuki T, Miki Y, Tazawa C, Senzaki K, Moriya T, Saito H, Ishibashi T, Takahashi S, Yamada S, Sasano H. Mol Cell Endocrinol. 2004;219:17–26. doi: 10.1016/j.mce.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Hisamoto K, Bender JR. Steroids. 2005;70:382–387. doi: 10.1016/j.steroids.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 46.Leo C, Li H, Chen JD. J Biol Chem. 2000;275:5976–5982. doi: 10.1074/jbc.275.8.5976. [DOI] [PubMed] [Google Scholar]
- 47.Li H, Leo C, Zhu J, Wu X, O'Neil J, Park EJ, Chen JD. Mol Cell Biol. 2000;20:1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L, Miano JM, Mercer B, Olson EN. J Cell Biol. 1996;132:849–859. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]