Abstract
The Vpr accessory protein of HIV-1 induces a response similar to that of DNA damage. In cells expressing Vpr, the DNA damage sensing kinase, ATR, is activated, resulting in G2 arrest and apoptosis. In addition, Vpr causes rapid degradation of the uracil-DNA glycosylases UNG2 and SMUG1. Although several cellular proteins have been reported to bind to Vpr, the mechanism by which Vpr mediates its biological effects is unknown. Using tandem affinity purification and mass spectrometry, we identified a predominant cellular protein that binds to Vpr as the damage-specific DNA-binding protein 1 (DDB1). In addition to its role in the repair of damaged DNA, DDB1 is a component of an E3 ubiquitin ligase that degrades numerous cellular substrates. Interestingly, DDB1 is targeted by specific regulatory proteins of other viruses, including simian virus 5 and hepatitis B. We show that the interaction with DDB1 mediates Vpr-induced apoptosis and UNG2/SMUG1 degradation and impairs the repair of UV-damaged DNA, which could account for G2 arrest and apoptosis. The interaction with DDB1 may explain several of the diverse biological functions of Vpr and suggests potential roles for Vpr in HIV-1 replication.
Keywords: proteasomal degradation, UNG2, DNA repair, G2 arrest
Vpr is a small nuclear accessory protein of HIV-1 and other lentiviruses that, when expressed in cells, causes G2 cell-cycle arrest and apoptosis. Although Vpr is not required for virus replication in vitro, it is well conserved in primary virus isolates and contributes to pathogenesis (1). Whereas Δvpr SIV was found to be nearly as pathogenic as wild-type, a vpr/vpx double mutant was highly attenuated, suggesting an important and partially redundant role in pathogenesis (2). Vpr is the only HIV-1 accessory protein to be specifically packaged into virions and localizes to the nucleus of infected cells. Several roles for Vpr in virus replication and pathogenesis have been proposed (reviewed in refs. 3 and 4). It causes a modest increase in virion production (5) and may facilitate nuclear import of the preintegration complex (6). Vpr has been found both to cause packaging of UNG2 (7) and to cause its proteasomal degradation (8). Vpr activates the ataxia telangiectasia-mutated (ATM) and Rad3-related protein (ATR) (9–13) causing the phosphorylation of the ATR substrates Chk1 (10) and the histone 2A variant H2AX (11), resulting in G2 arrest. Vpr also induces BRCA-1 and GADD45a (14) and generates nuclear foci containing H2AX and BRCA-1 (11). These phenotypes resemble that of DNA damage although Vpr itself does not seem to damage DNA.
Activation of ATR is the furthest activity upstream in the cell-cycle regulatory pathway known to be affected by Vpr, but this activity is not thought to be caused by a direct interaction. Several cellular binding partners for Vpr have been reported including UNG2, HSP70, HHR23A, p300, and VprBP (reviewed in ref. 15), but these interactions have not been shown to induce G2 arrest or cause apoptosis. The identification of the immediate target of Vpr would provide valuable insight into its function. Using tandem affinity purification (TAP) coupled with mass spectrometry, we identified the damaged DNA-binding protein (DDB1) as a specific cellular binding partner for Vpr. The interaction with DDB1 mediates the proteasomal degradation of UNG2 and SMUG1, is required for Vpr-induced apoptosis, and prevents the cells from repairing UV damage. Interestingly, viruses such as simian virus 5 (SV5) and hepatitis B also target DDB1 (16–18).
Results
Vpr Interacts with DDB1.
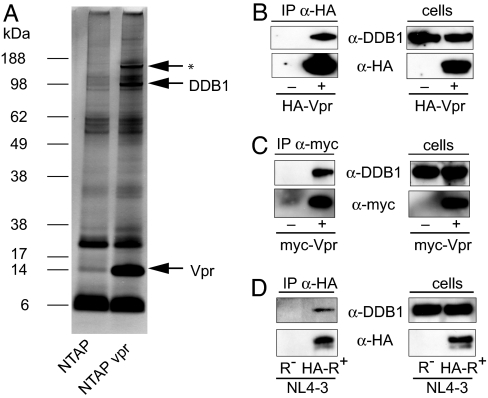
To identify interacting cellular proteins, a TAP-tagged Vpr expression vector was generated in which tandem streptavidin and calmodulin-binding sites were linked to the N terminus of Vpr. The TAP-tagged Vpr was expressed in transiently transfected 293T cells and shown to be functional for G2 arrest by DNA content profiling [supporting information (SI) Fig. 6]. Complexes of the tagged Vpr were purified from lysates of the transfected cells by TAP over streptavidin and calmodulin columns and the purified protein complexes were visualized by Coomassie staining after SDS/PAGE. Protein bands specific to the TAP-tagged Vpr (Fig. 1A) were excised and subjected to mass spectrometry. Two proteins were identified in the 100-kDa band: DDB1 and VprBP, a previously reported binding partner for Vpr that was affinity purified on a bacterially synthesized recombinant Vpr column (19). In repeated analyses, we could not identify the 150-kDa protein.
Fig. 1.
Identification of DDB1 as Vpr interacting protein. (A) Protein complexes containing TAP-tagged Vpr from lysates of transfected 293 cells were purified over streptavidin and calmodulin and a portion was visualized on a silver-stained SDS/PAGE. DDB1 and Vpr are indicated with arrows. The three bands were excised and subject to MALDI-TOF mass spectrometry. Two of the bands were identified as DDB1 and Vpr. This identification was reproduced in three independent analyses with separate preparations on different instruments. The third (indicated by the asterisk) was not identified. (B) HA-Vpr was expressed in transfected 293T cells and immunoprecipitated with anti-HA mAb. Coimmunoprecipitated DDB1 was detected on an immunoblot probed with anti-DDB1. (C) Myc-tagged SIVmac Vpr was expressed in transfected cells, immunoprecipitated with anti-Myc mAb and the coimmunoprecipitated DDB1 was detected on an immunoblot. (D) CEMss cells were infected with a replication-competent NL4-3 [NL4-3(HA-Vpr)] engineered to express HA-tagged Vpr or with control Δvpr NL4-3. After 4 days, HA-Vpr was immunoprecipitated with anti-HA mAb. Coimmunoprecipitated DDB1 was detected on an immunoblot probed with anti-DDB1. (B, C, and D Right) Immunoblot analysis of the cell lysates to confirm expression of the relevant proteins.
To confirm the association of Vpr with DDB1 in cells, the complex was coimmunoprecipitated with DDB1 in transfected cell lysates. An HA-tagged Vpr (HA-Vpr) was expressed in HeLa cells and immunoprecipitated with anti-HA mAb. The coimmunoprecipitated DDB1 was detected on an immunoblot. DDB1 was found to specifically coimmunoprecipitate with Vpr (Fig. 1B). DDB1 also coimmunoprecipitated with a myc-tagged SIV Vpr (Fig. 1C). In addition, the complex was coimmunoprecipitated from HIV-1-infected T cells. For this analysis, CEMss cells were infected with replication-competent NL4-3 HIV-1 that encodes a HA-tagged Vpr or with Δvpr NL4-3 (Fig. 1D).
Vpr Interacts with the DDB1-Cul4-Roc1 E3 Ubiquitin Ligase.
DDB1 is a 128-kDa UV-damaged DNA-binding protein involved in nucleotide excision repair (NER) that forms a heterodimer with the 48-kDa protein DDB2 (17, 18). Upon damage to DNA by UV light, the DDB1/DDB2 complex (UV-DDB) translocates to the nucleus where it binds with high affinity to UV-damaged DNA to mediate repair. In addition to its role in NER, DDB1 forms part of a ubiquitin ligase containing Cul4A and Roc1 that targets cell-cycle regulatory proteins such as Cdt1 and p27kip for proteasomal degradation (20, 21).
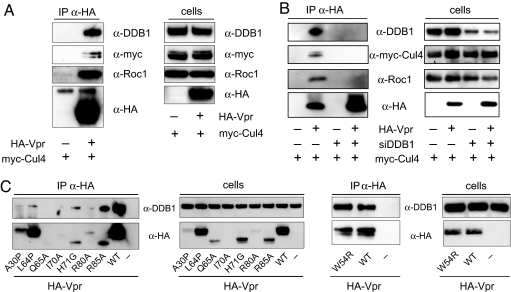
Vpr induces the degradation of UNG2 and SMUG1 in association with Cul4A (8). To determine whether DDB1 might play a role in this degradation, we tested for the presence of a complex of Vpr with DDB1, Cul4A, and Roc1. 293T cells were cotransfected with HA-Vpr and myc-Cul4A expression vector. HA-Vpr was immunoprecipitated, and the coimmunoprecipitated DDB1, Cul4A, and Roc1 were detected on an immunoblot (Fig. 2A). The analysis confirmed the presence of a complex containing Vpr, DDB1, Cul4A, and Roc1.
Fig. 2.
Vpr interacts with DDB1-Cul4A E3 ubiquitin ligase complexes. (A Left) HA-Vpr and Myc-Cul4 were expressed in cotransfected 293T cells, and HA-Vpr was immunoprecipitated with anti-HA mAb. Coimmunoprecipitated DDB1, myc-Cul4, and Roc1 were detected on an immunoblot. (Right) Expression of the proteins was confirmed by immunoblot analysis of the cell lysates. (B Left) DDB1 was knocked down by siRNA transfection. To detect the association of Vpr with DDB1 Cul4A and Roc1, Vpr was immunoprecipitated, and coimmunoprecipitated proteins were detected on an immunoblot. (Right) The efficiency of DDB1 knockdown was determined by immunoblot analysis of the cell lysates. (C) HA-Vpr point mutants were expressed in transfected 293T cells. HA-Vpr was immunoprecipitated, and the coimmunoprecipitated DDB1 was detected on an immunoblot (Left). Stability of the mutant Vpr was determined by immunoblot analysis of the cell lysates (second panel from the Left). The W54R Vpr mutant was tested in a separate experiment (Right two panels).
To determine whether the interaction with Cul4A and Roc1 was mediated by the interaction of Vpr with DDB1, siRNA transfection was used to knockdown DDB1. DDB1 was knocked down with specific or control siRNA, and the next day the cells were transfected with myc-Cul4A with or without HA-Vpr. Vpr complexes were immunoprecipitated, and the coimmunoprecipitated DDB1, Cul4A, and Roc1 were detected on an immunoblot. siRNA knocked down DDB1 by 80–90% (Fig. 2B). DDB1 knockdown prevented the association with Cul4A and Roc1. Complexes of Vpr with Cul4A and Roc1 were detected in control-transfected cells but not in those knocked down for DDB1. These findings demonstrated the dependence of DDB1 on formation of the complex with the Cul4A-Roc1 E3 ubiquitin ligase.
Identification of a Stable Vpr Point Mutant that Fails to Bind DDB1.
To probe the specificity of the interaction, a panel of mutated Vpr proteins was tested for DDB1 binding. HA-Vpr point mutants were expressed in transfected 293T cells and immunoprecipitated. The coimmunoprecipitated DDB1 was detected on an immunoblot. The majority of the mutants, including A30P, I70A, R80A, Q65A, H71G, and R85A were poorly expressed (Fig. 2C, second panel), a property that is typical of Vpr point mutants (our unpublished observations). Only L64P and W54R were stably expressed. Importantly, L64P was stably expressed but failed to interact efficiently with DDB1. The L64P mutant has been described by Jian and Zhao (22), who previously showed that this mutant was particularly proapoptotic. Two mutants, H71G and R85A, were expressed at low levels but maintained their interaction with DDB1. W54R, a mutant that has been shown previously not to bind UNG2 (23), was stably expressed but maintained its interaction with DDB1. Two mutants, H71G and R85A, maintained their interaction with DDB1 despite their reduced expression. No conclusion could be drawn for A30P, Q65A, I70A, and R80A, which were poorly expressed.
Vpr Induces the DDB1-Cul4A-Roc1 Ubiquitin Ligase to Target UNG2 for Degradation.
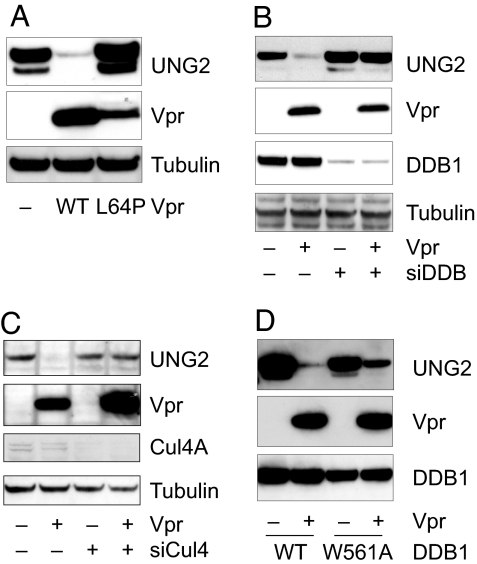
The association of Vpr with the DDB1-containing E3 ligase suggested that this complex might mediate the degradation of UNG2 and SMUG1. To determine whether Vpr binding to DDB1 was required for UNG2 degradation, we tested whether the DDB1-binding-defective Vpr mutant, L64P, could degrade UNG2. HA-UNG2 was expressed in cotransfected 293T cells with wild-type or L64P Vpr and detected on an immunoblot. The results showed that Vpr, but not L64P Vpr, caused a significant reduction in UNG2 expression (Fig. 3A). To test whether Vpr-induced degradation of UNG2 depends on DDB1, we knocked down DDB1 and tested for Vpr-induced degradation of UNG2. Knockdown of DDB1 was efficient and specific (Fig. 3B) and prevented the Vpr-induced degradation of UNG2. Similar results were obtained with SMUG1 (data not shown). These findings suggested that the interaction with DDB1 is required for Vpr-induced degradation of UNG2 and SMUG1.
Fig. 3.
Vpr recruits the DDB1-Cul4A E3 ubiquitin ligase to target UNG2 for proteasomal degradation. (A) 293T cells were cotransfected with HA-UNG2 expression vector and empty vector, Vpr, or L64P Vpr expression vector. UNG2, Vpr, and tubulin in cell lysates were detected on an immunoblot. (B) DDB1 was knocked down with siRNA. The cells were then transfected with Vpr and UNG2 expression vector. UNG2, Vpr, DDB1, and tubulin were detected on an immunoblot. (C) Cul4 was knocked down in 293T cells. The cells were then transfected with Vpr and UNG2 expression vector. UNG2, Vpr, Cul4A, and tubulin were detected on an immunoblot. (D) 293T cells were cotransfected with UNG2, Vpr, or empty expression vector and myc-DDB1 or W561A myc-DDB1 expression vector. UNG2, Vpr, and DDB1 in the cell lysates were detected on an immunoblot.
We further knocked down Cul4 to determine whether it is involved in Vpr-induced UNG2 degradation. Cul4 knockdown with specific siRNA, but not control siRNA prevented Vpr-induced UNG2 degradation (Fig. 3C). We further tested the DDB1 mutant W561A that does not bind Cul4A (24) and found that it was impaired for Vpr-induced UNG2 degradation (Fig. 3D). The effect was modest, probably because of the endogenous DDB1. Taken together, the results suggested that Vpr interacts with the DDB1-Cul4A-Roc1 E3 ubiquitin ligase to target UNG2 for proteasomal degradation.
Vpr Interferes with UV Damage-Induced Nuclear Localization of DDB1.
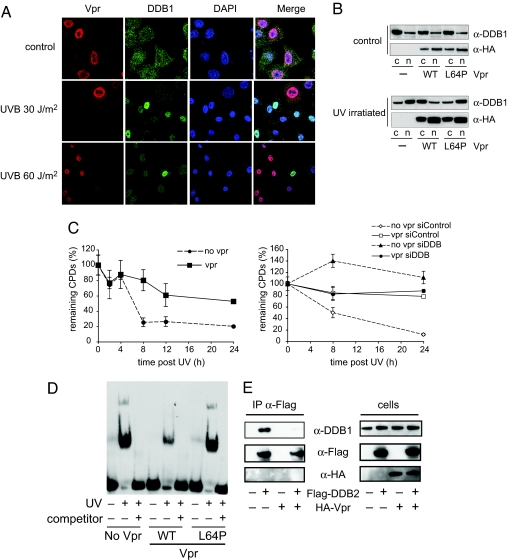
UV damage causes the DDB1/DDB2 complex to translocate to the nucleus (25, 26). To determine whether Vpr might affect the translocation of DDB1 to the nucleus, HA-Vpr was expressed in transfected HeLa cells. The cells were exposed to UV light and subsequently Vpr and DDB1 were visualized by immunofluorescence. In unirradiated cells, Vpr localized to the nucleus (Fig. 4A) whereas DDB1 was present throughout the cell. In low dose UV irradiated cells (UV-B 30 J/m2) without Vpr, DDB1 accumulated in the nucleus (cells lacking Vpr are shown in SI Fig. 7). Expression of Vpr prevented the nuclear accumulation of DDB1 in cells exposed to low and high doses of UV light. Biochemical analysis of DDB1 localization confirmed these results. Cells were transfected with Vpr expression vector and then fractionated into nucleus and cytoplasm. Effective fractionation was confirmed by immunoblot quantitation of the nuclear protein, heterogeneous nuclear ribonucleoprotein C, and cytoplasmic protein, GAPDH (SI Fig. 8). In unirradiated cells, Vpr was present in the nuclear fraction and DDB1 was in the cytoplasm (Fig. 4B). UV irradiation caused DDB1 to accumulate in the nucleus but in the presence of Vpr, DDB1 remained in the cytoplasm. L64P Vpr did not affect DDB1 translocation to the nucleus. These data suggest that Vpr blocks the UV damage induced nuclear accumulation of DDB1.
Fig. 4.
Interaction of Vpr with DDB1 disrupts the UV-DDB complex. (A) Cells were UV irradiated with the indicated dose, and 6 h later Vpr (red) and DDB1 (green) were visualized by immunofluorescence with specific antibody and labeled second antibody. The nuclei (blue) were stained with DAPI. (B) 293T cells were transfected with wild-type or L64P HA-Vpr expression vector or empty vector control. After 2 days, the cells were UV irradiated, and, after an additional 6 h, nuclear and cytoplasmic fractions were prepared. DDB1 and Vpr in the fractions were detected on an immunoblot. (C) HeLa cells transfected with Vpr expression vector or empty vector (Left). The cells were irradiated with 10 J/m2 UV-C and harvested at the indicated time points. The signal from the control cells was subtracted, and counts at t0 were set to 100%. At the indicated time points, genomic DNA was prepared, and the CPD content was measured by ELISA. (Right) The cells were transfected with control or DDB1-specific siRNA before irradiation. The results are representative of at least two repetitions. (D) HeLa cells were transfected with wild-type, L64P Vpr, or empty vector, and, after 2 days, nuclear extracts were prepared. The extracts were incubated with irradiated or unirradiated 5′-biotinylated double stranded oligonucleotide in the presence or absence of a 100-fold molar excess of unlabeled competitor oligonucleotide. Interaction of the oligonucleotide with the UV-DDB complex was detected by electrophoresis mobility shift assay (EMSA). (E) 293T cells were transfected with Flag-DDB2 and HA-Vpr or empty vector. Flag-DDB2 was immunoprecipitated, and coimmunoprecipitated DDB1 and Vpr were detected by immunoblot analysis (Left). An immunoblot on the cell lysates is shown (Right).
Vpr Impairs DNA Repair.
UV light causes the formation of cyclobutane pyrimidine dimers (CPD) and pyrimidine-pyrimidone (6-4) photoproducts (6-4PP) (17, 18). To test whether the binding of Vpr to DDB1 might interfere with DNA repair, HeLa cells were transfected with Vpr expression vector and two days later UV irradiated. Resulting CPDs in the genomic DNA was then quantitated by ELISA. In control UV irradiated cells, UV light induced CPDs which were rapidly repaired (Fig. 4C). In cells that expressed Vpr, CPDs remained relatively stable over the time course. To confirm that the repair was DDB1-mediated, the experiment was repeated in cells in which DDB1 was knocked down. In control cells, 80% of CPDs generated by UV irradiation were repaired within 24 h whereas, in cells expressing Vpr, repair was prevented. In cells in which DDB1 had been knocked down, the CPDs failed to be repaired (Fig. 4C Right). These results suggested that Vpr interferes with DDB1-mediated repaired of UV-damaged DNA.
Vpr Prevents the Binding of DDB to UV-Damaged DNA.
The impaired ability to repair UV-damaged DNA in the presence of Vpr could be caused by a failure of UV-DDB to bind to damaged DNA. To determine whether Vpr interfered with the ability of DDB1 to bind UV-damaged DNA, nuclear extracts were prepared from cells that expressed a transfected Vpr expression vector and incubated with a biotinylated UV irradiated DNA oligonucleotide. The DDB:DNA complexes were detected by an electrophoretic mobility shift assay (EMSA). A shifted band was observed with the UV irradiated but not the unirradiated control oligonucleotide probe (Fig. 4D). Nuclear extract from cells that expressed L64P Vpr or empty vector resulted in two bands on the EMSA. These bands were much weaker in cells that expressed Vpr. Addition of a 100 M excess of unlabeled CPD prevented the shift. We concluded that Vpr reduces the affinity of the UV-DDB complex for CPD containing DNA.
Vpr Disrupts the UV–DDB Complex.
The DNA repair function of DDB1 requires the interaction with DDB2. To determine whether Vpr might block this association, we transfected 293T cells with Flag-DDB2 expression vector with or without HA-Vpr. Two days post transfection, DDB2 was immunoprecipitated and DDB1 that coimmunoprecipitated was detected on an immunoblot. In the absence of Vpr, DDB1 and DDB2 coimmunoprecipitated. The expression of Vpr prevented the coimmunoprecipitation (Fig. 4E). These data suggest that Vpr interferes with the interaction of DDB1 with DDB2.
The Interaction of Vpr with DDB1 May Cause G2 Arrest and Apoptosis.
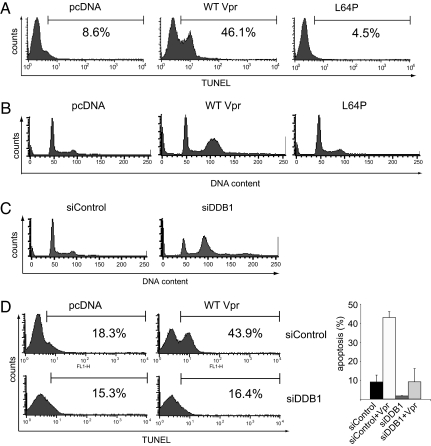
To determine whether Vpr induces apoptosis via DDB1, we tested the proapoptotic activity of L64P Vpr. HeLa cells were transfected with expression vector encoding wild-type or L64P HA-Vpr. Three days later, the extent of apoptosis was determined by TUNEL and G2 arrest was measured by DNA content. Wild-type Vpr induced G2 arrest and induced apoptosis in 40–50% of cells. In contrast, L64P Vpr did not induce apoptosis (Fig. 5A) or G2 arrest (Fig. 5B). To further support these findings, we determined the effect of DDB1 knockdown on cell-cycle progression and apoptosis. DDB1 was knocked down in HeLa cells and the cells were retransfected with Vpr expression vector. In the absence of Vpr, knockdown of DDB1 itself caused the cells to arrest in G2/M (Fig. 5C). In addition, knockdown of Vpr largely prevented Vpr-induced apoptosis (Fig. 5D). Microscopic inspection of the culture supported the TUNEL results. Cells that expressed Vpr were apoptotic in appearance. Apoptosis was almost completely prevented by knockdown of DDB1 (data not shown).
Fig. 5.
Knockdown of DDB1 relieves Vpr-induced apoptosis. (A) HeLa cells were transfected with wild-type or L64P Vpr expression vector or empty vector. Apoptosis was detected 3 days later by TUNEL. (B) Cells were fixed and stained with propidium iodide and analyzed by FACS. (C) HeLa cells were transfected with control siRNA or siRNA against DDB1. Three days posttransfection, the cells were fixed and stained with propidium iodide. The cell-cycle profiles were analyzed by FACS. (D) HeLa cells were transfected with control siRNA or DDB1-specific siRNA and, 24 h later, with wild-type Vpr. Three days later, apoptosis was measured by TUNEL.
Discussion
Several lines of evidence suggest that the interaction with DDB1 is physiologically significant. First, a Vpr point mutant that failed to bind DDB1 failed to induce G2 arrest and apoptosis. Second, knockdown of DDB1 prevented Vpr-induced UNG2 degradation. Third, Vpr binding to DDB1 interfered with the repair of UV-damaged DNA. Fourth, the interaction with DDB1 was conserved in SIV Vpr. Fifth, knockdown of DDB1 results in G2 arrest, a phenotype that is also induced by Vpr. Our findings suggest a model in which Vpr activates the degradation of cellular substrates by hijacking an E3 ubiquitin ligase containing DDB1-Cul4A-Roc1 (SI Fig. 9).
Our results present a potential explanation for several of the known cellular functions of Vpr. In the cell, Vpr induces a response that mimics the response to DNA damage in which ATR is activated to phosphorylate Chk1 and nuclear γH2AX and 53BP1 foci form (9–12). The interaction of Vpr with DDB1 could cause the accumulation of damaged DNA by preventing DNA repair. The damaged DNA could trigger activation of ATR and the subsequent downstream effects. The failure of DDB1 to bind DNA and mediate the repair of DNA lesions generated during S phase might be sensed by the cell as DNA damage and would activate ATR, resulting in G2 arrest followed by apoptosis.
Interestingly, a viral connection to DDB1 has been described. Paramyxoviruses such as SV5, mumps and human parainfluenza virus type 2 encode regulatory proteins that interact with DDB1 (16–18). The paramyxovirus V protein hijacks DDB1-Cul4A to target STAT2 for proteasomal degradation, interfering with the induction of IFN response genes (27–30). Vpr is also similar to the X protein of hepatitis B virus (HBX) and V, both of which slow or arrest the cell cycle, induce a DNA damage response and cause global alterations in cellular transcription (31–33). HBX, like Vpr, induces apoptosis through its interaction with DDB1 (32, 34–36). Like Vpr, HBX and SV5-V disrupt the UV-DDB complex (31, 32). Whether the interaction of Vpr with DDB1 causes the degradation of STAT proteins remains to be determined, although in a preliminary experiment, we did not detect decreased steady-state expression of STATs upon Vpr expression in HeLa cells (data not shown).
Our findings also present a second potential explanation for G2 cell-cycle arrest. The DDB1-Cul4A ubiquitin ligase targets several proteins involved in cell-cycle regulation for proteasomal degradation including Cdt1 and p27kip (20, 21). Vpr could cause G2 arrest by interfering with the degradation of proteins that regulate cell-cycle progression. This possibility would be consistent with our finding that DDB1 knockdown caused G2 arrest. To test this model, we overexpressed DDB1 in cells that expressed limiting amounts of Vpr. However, Vpr overexpression did not abrogate Vpr-induced G2 arrest (data not shown).
In addition to DDB1, the TAP analysis identified VprBP (19). Interestingly, VprBP was recently identified as a binding partner for DDB1. VprBP was proposed to be a member of a novel class of DDB1-Cul4A associated WD40 domain proteins (DCAF) that may be substrates for the DDB1-Cul4A E3 ubiquitin ligase (21, 37). Our findings do not distinguish between direct binding of DDB1 to Vpr or bridging through VprBP.
The findings presented here provide an important clue toward understanding the role of Vpr in HIV-1 replication. They suggest that Vpr mediates some, if not all of its biological activities through its interaction with DDB1. How these functions promote HIV-1 replication remains to be determined. These findings raise interesting possibilities regarding the role of Vpr in HIV-1 replication that can be experimentally addressed.
Materials and Methods
pcHA-Vpr expresses an N-terminal HA-tagged NL4-3 Vpr in the cytomegalovirus promoter driven vector pcDNAI/amp (Invitrogen). The L64P mutation was introduced by site-directed mutagenesis. 3X-Flag-DDB2 expression vector was a gift from Hitoshi Endo (38). The T7-DDB1 expression vector was provided by P. R. Raychaudhuri. The T7 tag was replaced with a myc-tag by PCR, and the W561A mutation was introduced by site-directed mutagenesis. NTAP-Vpr was generated by subcloning Vpr from pcHA-Vpr into the pNTAP (Stratagene) at the Bam-HI and HindIII sites. UNG2 and SMUG1 expression vectors have been reported (8). pNL43(HA-Vpr) was constructed by inserting a HindIII site at the start codon of vpr of pNL43. pNL43 was than cleaved with HindIII and at the unique EcoRI site in vpr, and the resulting fragment was replaced with a HindIII/EcoRI fragment from pcHA-Vpr. The modification does not interfere with virus replication in T cell lines.
Additional experimental methods are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Hitoshi Endo (Jichi Medical School, Tochigi, Japan) and Pradip Raychaudhuri (University of Illinois at Chicago, Chicago, IL) for reagents; Wolfgang Fischer for mass spectrometry; Esther Francisco, Jody Chou, and Dimas Espinola for technical assistance; and Hui Chen and Erica Dhuey for critical reading of the manuscript. This work was supported by National Institutes of Health Grants AI058864-03 and DA014494-05, the University of California at San Diego Center for AIDS Research, and the American Foundation for AIDS Research (AmfAR). N.R.L. is an Elizabeth Glaser Fellow of the Pediatric AIDS Foundation.
Abbreviations
- DDB1
DNA-binding protein 1
- ATM
ataxia telangiectasia-mutated
- ATR
ATM and Rad3-related protein
- TAP
tandem affinity purification
- CPD
cyclobutane pyrimidine dimer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610167104/DC1.
References
- 1.Lang SM, Weeger M, Stahl-Hennig C, Coulibaly C, Hunsmann G, Muller J, Muller-Hermelink H, Fuchs D, Wachter H, Daniel MM, et al. J Virol. 1993;67:902–912. doi: 10.1128/jvi.67.2.902-912.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibbs JS, Lackner AA, Lang SM, Simon MA, Sehgal PK, Daniel MD, Desrosiers RC. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherman MP, De Noronha CM, Williams SA, Greene WC. DNA Cell Biol. 2002;21:679–688. doi: 10.1089/104454902760330228. [DOI] [PubMed] [Google Scholar]
- 4.Muthumani K, Desai BM, Hwang DS, Choo AY, Laddy DJ, Thieu KP, Rao RG, Weiner DB. DNA Cell Biol. 2004;23:239–247. doi: 10.1089/104454904773819824. [DOI] [PubMed] [Google Scholar]
- 5.Goh WC, Rogel ME, Kinsey CM, Michael SF, Fultz PN, Nowak MA, Hahn BH, Emerman M. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 6.Heinzinger NK, Bukinsky MI, Haggerty SA, Ragland AM, Kewalramani V, Lee MA, Gendelman HE, Ratner L, Stevenson M, Emerman M. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. J Virol. 2000;74:7039–7047. doi: 10.1128/jvi.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrofelbauer B, Yu Q, Zeitlin SG, Landau NR. J Virol. 2005;79:10978–10987. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai M, Zimmerman ES, Planelles V, Chen J. J Virol. 2005;79:15443–15451. doi: 10.1128/JVI.79.24.15443-15451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roshal M, Kim B, Zhu Y, Nghiem P, Planelles V. J Biol Chem. 2003;278:25879–25886. doi: 10.1074/jbc.M303948200. [DOI] [PubMed] [Google Scholar]
- 11.Zimmerman ES, Chen J, Andersen JL, Ardon O, Dehart JL, Blackett J, Choudhary SK, Camerini D, Nghiem P, Planelles V. Mol Cell Biol. 2004;24:9286–9294. doi: 10.1128/MCB.24.21.9286-9294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmerman ES, Sherman MP, Blackett JL, Neidleman JA, Kreis C, Mundt P, Williams SA, Warmerdam M, Kahn J, Hecht FM, et al. J Virol. 2006;80:10407–10418. doi: 10.1128/JVI.01212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakai-Murakami C, Shimura M, Kinomoto M, Takizawa Y, Tokunaga K, Taguchi T, Hoshino S, Miyagawa K, Sata T, Kurumizaka H, et al. Oncogene. 2006;26:477–486. doi: 10.1038/sj.onc.1209831. [DOI] [PubMed] [Google Scholar]
- 14.Andersen JL, Zimmerman ES, DeHart JL, Murala S, Ardon O, Blackett J, Chen J, Planelles V. Cell Death Differ. 2005;12:326–334. doi: 10.1038/sj.cdd.4401565. [DOI] [PubMed] [Google Scholar]
- 15.Kino T, Pavlakis GN. DNA Cell Biol. 2004;23:193–205. doi: 10.1089/104454904773819789. [DOI] [PubMed] [Google Scholar]
- 16.Barry M, Fruh K. Sci STKE. 2006:pe21. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- 17.Tang J, Chu G. DNA Repair (Amsterdam) 2002;1:601–616. doi: 10.1016/s1568-7864(02)00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittschieben BB, Wood RD. DNA Repair (Amsterdam) 2003;2:1065–1069. doi: 10.1016/s1568-7864(03)00113-7. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S, Feng Y, Narayan O, Zhao LJ. Gene. 2001;263:131–140. doi: 10.1016/s0378-1119(00)00583-7. [DOI] [PubMed] [Google Scholar]
- 20.Bondar T, Kalinina A, Khair L, Kopanja D, Nag A, Bagchi S, Raychaudhuri P. Mol Cell Biol. 2006;26:2531–2539. doi: 10.1128/MCB.26.7.2531-2539.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J, McCall CM, Ohta T, Xiong Y. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 22.Jian H, Zhao LJ. J Biol Chem. 2003;278:44326–44330. doi: 10.1074/jbc.C300378200. [DOI] [PubMed] [Google Scholar]
- 23.Selig L, Benichou S, Rogel ME, Wu LI, Vodicka MA, Sire J, Benarous R, Emerman M. J Virol. 1997;71:4842–4846. doi: 10.1128/jvi.71.6.4842-4846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li T, Chen X, Garbutt KC, Zhou P, Zheng N. Cell. 2006;124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Nichols AF, Graham JA, Dualan R, Abbas A, Linn S. J Biol Chem. 2000;275:21429–21434. doi: 10.1074/jbc.M000961200. [DOI] [PubMed] [Google Scholar]
- 26.Wakasugi M, Kawashima A, Morioka H, Linn S, Sancar A, Mori T, Nikaido O, Matsunaga T. J Biol Chem. 2002;277:1637–1640. doi: 10.1074/jbc.C100610200. [DOI] [PubMed] [Google Scholar]
- 27.Ulane CM, Rodriguez JJ, Parisien JP, Horvath CM. J Virol. 2003;77:6385–6393. doi: 10.1128/JVI.77.11.6385-6393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin GY, Paterson RG, Richardson CD, Lamb RA. Virology. 1998;249:189–200. doi: 10.1006/viro.1998.9317. [DOI] [PubMed] [Google Scholar]
- 29.Ulane CM, Kentsis A, Cruz CD, Parisien JP, Schneider KL, Horvath CM. J Virol. 2005;79:10180–10189. doi: 10.1128/JVI.79.16.10180-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palosaari H, Parisien JP, Rodriguez JJ, Ulane CM, Horvath CM. J Virol. 2003;77:7635–7644. doi: 10.1128/JVI.77.13.7635-7644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leupin O, Bontron S, Strubin M. J Virol. 2003;77:6274–6283. doi: 10.1128/JVI.77.11.6274-6283.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bontron S, Lin-Marq N, Strubin M. J Biol Chem. 2002;277:38847–38854. doi: 10.1074/jbc.M205722200. [DOI] [PubMed] [Google Scholar]
- 33.Lin-Marq N, Bontron S, Leupin O, Strubin M. Virology. 2001;287:266–274. doi: 10.1006/viro.2001.1036. [DOI] [PubMed] [Google Scholar]
- 34.Becker SA, Lee TH, Butel JS, Slagle BL. J Virol. 1998;72:266–272. doi: 10.1128/jvi.72.1.266-272.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin GY, Lamb RA. J Virol. 2000;74:9152–9166. doi: 10.1128/jvi.74.19.9152-9166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sitterlin D, Bergametti F, Transy C. Oncogene. 2000;19:4417–4426. doi: 10.1038/sj.onc.1203771. [DOI] [PubMed] [Google Scholar]
- 37.Angers S, Li T, Yi X, Maccoss MJ, Moon RT, Zheng N. Nature. 2006;493:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 38.Inoki T, Yamagami S, Inoki Y, Tsuru T, Hamamoto T, Kagawa Y, Mori T, Endo H. Biochem Biophys Res Commun. 2004;314:1036–1043. doi: 10.1016/j.bbrc.2004.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.