Abstract
The 369-residue glycoprotein D (gD) is the entry, receptor-binding protein of herpes simplex virus 1. The common receptors for viral entry are nectin-1, HveA, and a specific O-linked sulfated proteoglycan. The major receptor-binding sites of gD are at the N terminus, whereas the domain required for fusion of viral envelope with the plasma membrane is at the C terminus of the ectodomain (residues 260–310). In the course of retargeting gD to the urokinase plasminogen activator (uPA) receptor for potential therapeutic applications, we obtained a genetically engineered infectious virus in which the receptor-binding domain consisting of the N-terminal domain of uPA fused to residues 33–60 of gD was separated from an independently expressed C-terminal domain of gD containing residues 219–369. The intervening sequences (residues 62–218) were replaced by a stop codon and a promoter for the C-terminal domain of gD. The physical interaction of the two components was reconstructed by coimmunoprecipitation of the N-terminal domain of uPA with the C-terminal domain of gD. These results indicate that codons 61–218 of gD do not encode executable functions required for viral entry into cells and suggest that the receptor-binding ligand must interact with but need not alter the structure of the residual portion of gD to effect virus entry. This finding opens the way for the development of a family of recombinant viruses in which the profusion domain of gD and independently furnished, interacting receptor-binding domains effect entry of the virus via a range of receptors.
Keywords: targeting, urokinase plasminogen activator, virus entry
Herpes simplex virus 1 (HSV-1) encodes five glycoproteins that play key roles in virus entry into susceptible cells (reviewed in refs. 1–3). Thus, glycoprotein B (gB) and glycoprotein C interact with heparan sulfate proteoglycan to enable initial attachment to cell surfaces. In the next step in the entry cascade, gD then binds a specific surface receptor such as nectin-1 or HveA (1, 4, 5). The interaction of glycoprotein D (gD) with the receptor alters the conformation of gD to enable the activation of gB, glycoprotein H, and glycoprotein L, a trio of glycoproteins that execute the fusion of the viral envelope with the plasma membrane (6). In this sequence of events, gD emerges as the surface protein that imparts to the virion the specificity of entry into cells. This 369-residue glycoprotein has been the subject of numerous structural and functional studies (6–8). The protein appears to function as a dimer (9). At least some of the interactions of gD with its receptors have been mapped to the N terminus of gD, whereas the fusogenic determinant maps in the C-terminal portion of the ectodomain (residues 260–310). These residues (260–310) become actively engaged in membrane fusion after the interaction of the N-terminal domain with one of its cognate receptors (6–8, 10–14). gD also blocks apoptosis resulting from endocytosis of virus particles lacking gD. Thus, virus particles may also enter cells by endocytosis. In the absence of gD, however, these virions are degraded, viral replication does not ensue, and a discharge of lysosomal enzymes causes cell death (15). Transduction of cells with specific domains of gD, treatment with chloroquine, or overexpression of cation-independent mannose 6-phosphate receptor prevents cell death (15–17). The domains required for blocking cell death have been mapped by insertional mutagenesis at several sites of gD (17).
In this article, we show that residues 61–218 of gD do not perform functions required for entry of virus into cells. As detailed elsewhere, we constructed two series of recombinant viruses designed to target receptors that are uniquely present on the surface of tumor cells (18–20). In one recombinant (R5111) designed to target the IL-13α2 receptor enriched in malignant glioma cells, 140 residues encoding IL-13 were inserted between residues 24 and 25 of gD. This recombinant virus can infect cells expressing the IL-13α2 receptor, but it also retained the capacity to interact with nectin-1 and HveA receptors (18). In the second recombinant virus (R5141), the IL-13 ligand was fused to residue 33 of gD. In addition, valine 34 was replaced with serine. This recombinant entered cells solely via the IL-13α2 receptor. IL-13 added to the medium blocked entry of the recombinant virus expressing the IL-13α2 receptor (20). The second target selected for these studies was urokinase plasminogen activator receptor (uPAR), commonly found on the surface of most cells but highly enriched on the surface of cancer cells (19). In the first of this series of recombinant viruses, R5181, the N-terminal 155 residues of urokinase plasminogen activator (uPA) were inserted after residue 24 of gD. This recombinant virus infected cells expressing nectin-1 or HveA or even hamster cells that lacked known HSV receptors but were subsequently shown to express hamster-specific uPAR mRNA. In this instance, removal of uPAR by digestion of the glucosyl-inositol anchor with phosphatidyl inositol-specific phospholipase C blocked virus entry (19). The results reported below emerged from an attempt to construct a chimeric gD modeled after the recombinant virus R5141 in that uPA sequences were fused to residue 33 of gD and valine 34 was replaced with serine. Although the genetically engineered virus designated R5322 replicated as expected in all cell lines tested, sequencing of the chimeric gD present in the recombinant virus revealed that it contained two frameshift mutations, one each after residues 60 and 201. The frameshifts introduced four stop codons between residues 60 and 201. The first available potential initiator methionine codon was at position 219. In light of the evidence that the R5322 recombinant virus was infectious and produced viral progeny, we examined in more detail the role of the gD peptides so propitiously created by frameshifts.
Results
Residues 61–218 of gD Are Dispensable in the Genetically Engineered Virus R5322.
The design of the R5322 recombinant virus involved replacement of residues 1–32 with the N-terminal 155 residues of uPA and substitution of valine 34 with serine, but it retained the signal sequence of wild-type gD. R5322 was generated by cotransfecting R6 cells with intact R5110 DNA, which lacks the gD gene, and a plasmid encoding the desired chimeric gD. R6 cells were derived by transduction of rabbit skin cells with plasmid pEA102 containing the HSV-1 gD coding sequence under the UL26.5 promoter. They express gD ectopically and enable viruses lacking gD to replicate and spread (15). The infectious virus was plaque purified, and initial stocks were produced in R6 cells. To verify the presence of the N-terminal domain of uPA, the recombinant gD was amplified by PCR and sequenced. Repeated sequencing concurrently with appropriate wild-type controls revealed that in gD of recombinant R5322 one cytosine was inserted after codon 60 and one was deleted after codon 201 (Fig. 1). The frameshifts resulting from the deletion and insertion of the cytosines introduced stop codons at residues 139, 148, 159, and 190. The first potential initiator methionine is at position 219. Further tests showed that the virus infected and replicated in four test cell lines: i.e., in J1.1, a hamster cell line that lacks receptors for wild-type virus; in J-HveA, a hamster cell line that expresses the HveA receptor; in J-nectin, a hamster cell line that expresses nectin 1; and in Vero-13R, a Vero-derived cell line that expresses IL-13α2 receptor in addition to the natural receptors for HSV-1 (data not shown). The observation that it replicated in J1.1 cells is consistent with earlier results showing that these cells accumulate uPAR mRNA (19).
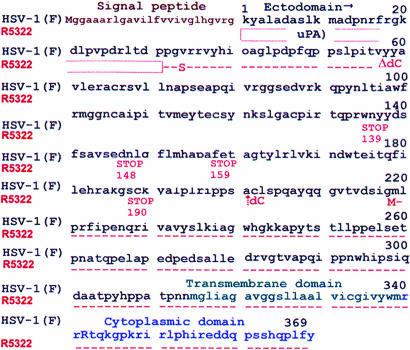
Fig. 1.
Amino acid sequence of wild-type and R5322 gD. The 155 N-terminal residues of uPA were inserted between the gD signal peptide and residue 33 of gD. In addition, valine 34 was substituted with serine. The frameshifts resulting from insertion of a cytosine after codon 60 and deletion of a cytosine after codon 201 introduced in-frame new codons including four stop codons at the positions shown. The wild-type gD sequences resume at codon 202. Residue 219 is the first methionine of the C-terminal portion of gD.
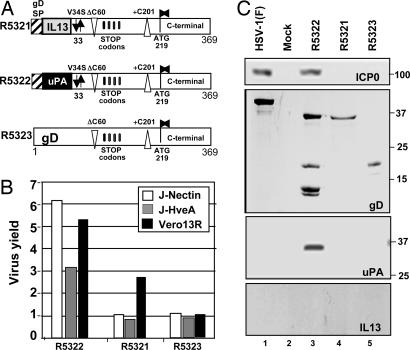
These results raised the question of whether residues 60–218 of gD are dispensable for entry just in this virus or whether they play no essential role in wild-type virus as well. To answer this question, we attempted to construct two additional viruses with designations R5321 and R5323. To produce R5321, R6 cells were transfected with R5110 DNA and a chimeric gD construct that was identical to that of R5322 except that the N-terminal domain of uPA was replaced with that of IL-13 (19). In the chimeric plasmid designed to generate R5323, the residues uPAR–gD33–60 of R5322 were replaced with residues 1–60 of wild-type gD. Schematic representations of the chimeric gDs are shown in Fig. 2A. The stocks were made and characterized as described above and tested by exposing replicate 25-cm2 cultures of J-HveA, J-nectin, or Vero-13R cells to 0.1 pfu per cell. The progeny viruses were titered in Vero-13R cells. gD ectopically expressed in R6 cells enabled the amplification of recombinants produced in these cells. The spread and replication of recombinant viruses in J-HveA, J-nectin, or Vero-13R cells were dependent on the presence of appropriate receptor-binding domains in the chimeric gD. As shown in Fig. 2B, R5322 replicated in all cell lines to approximately the same extent as the R5181 recombinant virus in which the chimeric gD consisted of the uPA sequences inserted after codon 24 of gD (19). The recombinant R5321 replicated to a very low level and only in Vero-13R cells (Fig. 2B). The products of transfection to generate R5323 did not recombine to form an infectious virus.
Fig. 2.
Structure of chimeric gDs present in R5322 or designed for construction of recombinant R5321 or R5323 viruses and the properties of the recombinant viruses. (A) The sequence arrangement of gD present in R5322 or constructed for other recombinant viruses. (B) Replication of recombinant viruses in J-nectin, J-HveA, or Vero-13R cell lines. Cells grown in 25-cm2 flasks were exposed to 0.1 pfu of the recombinant virus per cell and harvested 24 h after infection. Progeny virus was titrated on Vero-13R cells. (C) Photograph of electrophoretically separated proteins from lysates of cells infected with R5321, R5322, and R5323 recombinant viruses. Vero-13R cells grown in 25-cm2 flasks were exposed to 1.0 pfu of R5321, R5322, and R5323 virus per cell. The cells were harvested 24 h after infection, solubilized, subjected to electrophoresis in 10% denaturing polyacrylamide gels, electrically transferred onto a nitrocellulose sheet, and reacted with a monoclonal antibody against ICP0, gD (ZC25), uPA, and IL-13, respectively.
To determine whether the recombinant viruses express the appropriate protein, Vero-13R cells were exposed to 1.0 pfu of wild-type HSV-1(F), R5321, R5322, or R5323 virus. As shown in Fig. 2C, the only protein detected in lysates of R5323-infected cells was a truncated form of gD. Both wild-type virus- and the R5322 mutant virus-infected cells produced ICP0, a major α (immediate–early) regulatory protein of the virus. As could be expected from the presence of in-frame stop codons, the major product of the R5322 virus reacting with either gD or uPA antibody was significantly smaller than the wild-type gD made in wild-type virus-infected cells, consistent with the hypothesis that the N-terminal and C-terminal domains were produced independently.
HSV-1-infected cells can suppress one stop codon at a very low level (21). To make a single protein based on the sequence of the chimeric gD in R5322, all four stop codons would have to be suppressed. An alternative explanation is that a promoter domain within the gD ORF enables the synthesis of mRNA that encodes a functional protein starting with methionine 219. Implicit in this hypothesis is the notion that residues 61–218 simply link the N terminus of gD to the C-terminal domain, but that the sequence itself is important only in the sense that it maintains the two ends in the proper relationship. To test these hypotheses, two sets of viruses were constructed (Fig. 3).
Fig. 3.
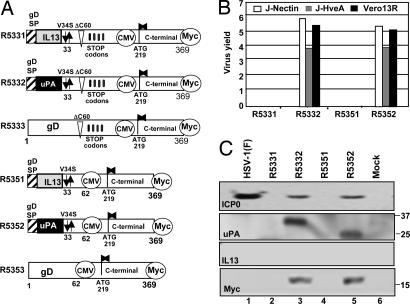
Structure of chimeric gD and the properties of the recombinant viruses incorporating these glycoproteins. (A) Sequence arrangement of the chimeric gD designed for construction of recombinant viruses. (B) Yields of R5332 and R5352 from cells infected with 0.1 pfu of virus per cell. The recombinant viruses R5331 and R5351 did not infect these cells or produce plaques in Vero-IL13 cells. (C) Accumulation of ICP0, the N-terminal polypeptide of gD identified by the antibody against uPA or the C-terminal polypeptide of gD identified by its reactivity with the anti-Myc antibody.
The viruses predicted to be formed by recombinant gD constructs designated R5331, R5332, and R5333 differed from the corresponding recombinant gD constructs in R5321, R5322, and R5323 in that a human cytomegalovirus (CMV) immediate–early promoter was inserted upstream of residue 219. In recombinant gD constructs designed to make R5351, R5352, and R5353, residues 63–218 were deleted, and a stop codon and the CMV immediate–early promoter were inserted between residues 62 and 219. In addition, a Myc epitope tag was inserted at the C terminus of gD to enable identification of the protein made from the corresponding domain of chimeric gD. The results of our attempts to construct these viruses were as follows.
Recombinant viruses R5332 and R5352 were produced and titered in R6 cells and were used to infect other cell lines at ratios of 0.1 pfu per cell. As shown in Fig. 3B, these recombinant viruses replicated in J-nectin, J-HveA, and Vero-13R cell lines to the same extent as R5322 (Fig. 2B). R6 cells transfected with chimeric gD constructs designed to produce the recombinant viruses R5331 or R5351 exhibited extensive cytopathic effects. However, the lysates of these cells did not replicate in the three test lines (J-HveA, J-nectin, or Vero-13R). No infectious virus progeny of the transfection of R6 cells with R5333 or R5353 were detected, and these were not tested further.
Lastly, analyses of key proteins made by the recombinant viruses constructed in this series of experiments are shown in Fig. 3C. Thus, Vero-13R cells infected with either R5332 or R3552 accumulated two sets of comigrating bands, one that reacted with antibody to ICP0 and one that reacted with anti-Myc antibody. The anti-uPA antibody reacted with a protein band in R5332 mutant virus-infected cells that migrated more slowly than the protein detected by the anti-uPA antibody in R5352 mutant virus-infected cells. We conclude that (i) the tagged gD proteins containing the residues 219–369 were expressed as separate entities from the proteins reactive with the anti-uPA antibody, and (ii) the differences in electrophoretic mobility of proteins reactive with the anti-uPA antibody reflect their sizes. Thus, in R5352 mutant virus-infected cells, the N-terminal polypeptide would comprise uPA and gD residues 33–60 only. In R5332 mutant virus-infected cells, translation of the mRNA would proceed beyond codon 60 to at least the first stop codon at position 139.
We conclude that (i) the residues 61–218 of gD are not required for entry into cells, (ii) the essential N-terminal and C-terminal domains of gD can be produced independently of each other, and (iii) infectivity depends on the nature of the ligand attached to the N-terminal domain of gD.
The N-Terminal Domain of gD Linked to uPA Coprecipitates with the C-Terminal Domain of gD.
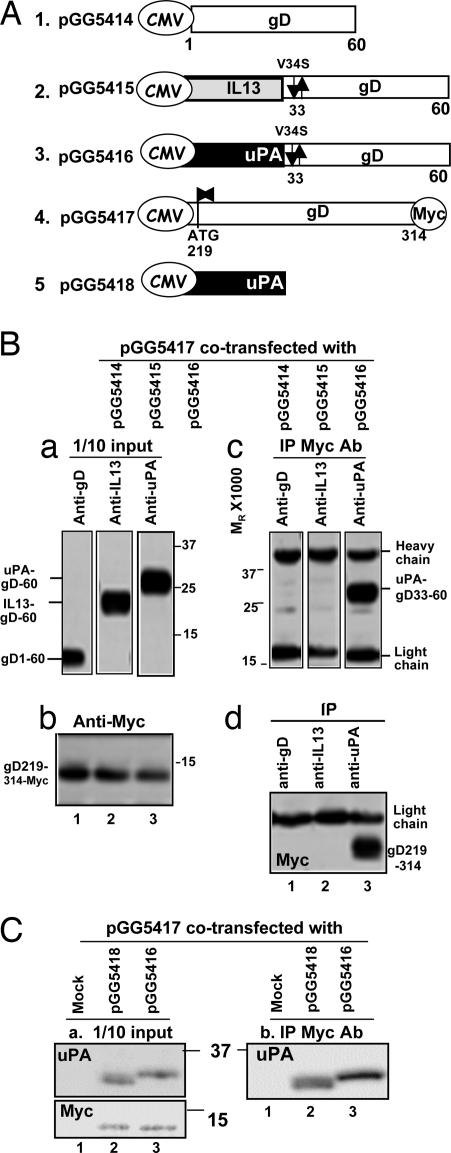
The results shown in Fig. 3 indicated that in R5332- or R5352-infected cells the amino acid stretch comprising the N-terminal domain of uPA linked to residues 33–60 of gD were in a different polypeptide (uPA–gD33–60) than the C-terminal domain of gD consisting of residues 219–369 (gD219–369). To infect cells, however, the two functional domains of the chimeric gD had to interact. To determine whether the two polypeptides interact, we constructed four plasmids schematically illustrated in Fig. 4A. Plasmid pGG5414 contained the residues 1–60 of gD driven by the CMV promoter. Plasmids pGG5415 and pGG5416 consisted of uPA–gD33–60 and IL-13–gD33–60 driven by the CMV promoter, respectively. In each of the latter two plasmids, the valine codon 34 was substituted with that of serine. Finally, plasmid R5417 contained the gD residues 219–314, that is, without the transmembrane and cytoplasmic domains tagged at the C terminus with the Myc epitope and driven by the CMV promoter. The plasmids were transfected into HEK293 cells in pairwise fashion, i.e., pGG5417 with either pGG5414, pGG5415, or pGG5416. The cells were harvested 40 h after transfection, lysed, and reacted with antibodies to either gD (transfection pGG5417 + pGG5414), IL-13 (pGG5417 + pGG5415), uPA (pGG5417 + pGG5416), or Myc. As shown in Fig. 4 Bc, anti-Myc antibody precipitated a protein reactive with the anti-uPA antibody. Conversely, as shown in Fig. 4 Bd, the anti-uPA antibody precipitated a protein reactive with Myc. Anti-Myc antibody did not pull down proteins reactive with the gD219–314 polypeptide from cells transfected with pGG5417 + pGG5414 or pGG5417 + pGG5415. The results show that the uPA–gD33–60 polypeptide physically interacts with the gD219–314 polypeptide. We have not found evidence that either IL-13–gD33–60 or wild-type gD1–60 interacts physically with the gD219–360 polypeptide. One explanation for the failure to detect infectious R5333 or R5353 virus is the failure of the two key components of gD to interact with each other.
Fig. 4.
Coprecipitation of the polypeptides consisting of the N-terminal uPA–gD33–60 and the C-terminal gD219–314 domains. (A) Structure of the plasmids transfected in a pairwise manner into HEK293 cells. The transfected cells were harvested 40 h after transfection. (B and C) Portions comprising 10% of the total cell lysates were solubilized, subjected to electrophoresis in a denaturing gel, and probed with antibody to uPA (Ba and C Left), IL-13 (Ba), or gD (Ba) or with antibody to Myc (Bb and C Right). Other aliquots of the lysates were reacted with antibodies to gD, uPA, or IL-13. The precipitates were collected, solubilized, subjected to electrophoresis in denaturing gels, and reacted with antibody to uPA (Bc) or Myc (Bd).
One hypothesis that would explain our results is that the coprecipitation of uPA–gD33–60 with gD219–369 reflects the interaction of the kringle domain (residues 50–132) of the N-terminal domain of uPA with lysines in the C-terminal domain of gD. To test this hypothesis, HEK293 cells were transfected with mixtures of pGG5417 and a plasmid (pGG5418) encoding the N-terminal domain of uPA or pGG5417 and pGG5416. The lysates of the transfected cells were reacted with antibody to Myc. The immunoprecipitates were collected, electrophoretically separated in a denaturing gel, transferred to a nitrocellulose membrane, and reacted with anti-uPA antibody. As shown in Fig. 4 C Left, anti-Myc antibody coprecipitated uPA from lysates of cells cotransfected with pGG5417 and pGG5418 (lane 2) and uPA119–314 from lysates of cells cotransfected with pGG5417 and pGG5416 (lane 3).
Discussion
HSV-1 gD serves as the entry receptor-binding protein. The domains of gD critical for its functions may be summarized as follows. The interaction of gD with HveA is abolished by deletion of N-terminal 32 residues (12). The interactions of gD with nectin-1 are abolished by mutations at residues 34, 38, 215, 222, and 223 (13, 14, 20). A profusion domain, essential for activation of fusogenic glycoproteins B, H, and L, has been mapped to residues 260–310 in close proximity to the transmembrane domain beginning at residue 314 (6). gD suppresses apoptosis induced by mutants lacking gD (17). Linker insertion analyses have identified several sites that abolish this function; some are located between residues 61 and 218 (17). Although failure of many linker insertion mutants to inactivate infectivity was attributed to the flexibility of the gD structure, the data could have presaged the conclusion that at least a portion of gD plays no sequence-specific role in virus entry other than to hold the receptor-binding domain linked to the profusion domain.
The initial objective of the studies described in this paper was to enhance the yield of a virus designed to target uPAR. In the course of constructing the recombinant viruses, we obtained an infectious virus with two frameshifts, one after codon 60 and the second after codon 201. The frameshifts replaced codons 61–201 with a sequence that contained four stop codons. The salient feature of the recombinant virus that emerged from these studies is that gD consisting of polypeptide A containing the N-terminal domain of uPA fused to residues 33–60 of gD and polypeptide B containing the C-terminal domain of gD was infectious. Attempts to construct recombinant viruses in which the A polypeptides consisted solely of gD residues 1–60 or of the 132 residues encoding IL-13 fused to gD residues 33–60 were not infectious. The significance of these findings stems from three conclusions:
(i) Residues 61–218 do not execute a function required for HSV-1 entry into cells. They appear to serve as linkers between the N-terminal domain and the C-terminal profusion domains of gD. It is of interest to note that residues 61–218 coincide almost entirely with the Ig-like core of gD located between residues 56 and 184 (7). In light of our results, its primary function appears to be to hold the key N-terminal and C-terminal domains in proper orientation. This domain may also be required to block cell death as a consequence of a discharge of lysosomal enzymes (17).
(ii) The fundamental difference between the constructs that yielded an infectious virus and those that failed is that polypeptide A consisting of uPA33–60 interacted with the C-terminal domain of gD, whereas those containing gD1–60 or IL-13–gD33–60 did not. Although it may be tempting to suggest that the N-terminal domain of uPA conferred a conformation to gD33–60 that enabled it to interact with the C-terminal domain of gD, the evidence supports the conclusion that uPA itself can interact with gD219–314. As reported here, the results support the conclusion that physical interaction of the domain capable of binding a cell surface receptor with the C-terminal domain of gD may lead to successful virus entry into cells, whereas lack of physical interaction results in failure. The results reinforce the conclusion reported earlier that entry requires the physical rapprochement of gD to the cell surface receptor (22).
(iii) Current studies favor the hypothesis that interaction of gD with its receptors alters its conformation and exposes the C terminal of gD to fusogenic glycoproteins B, H, and L (23–25). It is unlikely that each of the three ligands selected by chance alters the conformation of the remaining portions of gD in an identical manner to enable virus entry. One hypothesis that remains to be explored is that gD is “armed” by a conformational change resulting from replacement of residues 1–32 by any of the three ligands and that cell membrane–envelope fusion is executed when the ligands interact with their cognate receptors. A corollary of this hypothesis is that, although the interaction of the receptor-binding domain with its natural cell surface receptor may alter the conformation of the distal portions of gD, such changes are not obligatory consequences of the interaction of chimeric gD molecules with cognate receptors.
Lastly, with respect to the initial objective of these studies, it has not escaped our attention that the uPA ligand might be replaced by any of a number of alternative ligands in trans, provided that they can associate with the profusogenic domain to extend the host range of recombinant viruses in useful ways.
Materials and Methods
Cells.
Vero and HEK293 were obtained from the American Type Culture Collection (Manassas, VA). R6 cells were derived by transduction of rabbit skin cells with plasmid pEA102 containing the HSV-1 gD coding sequence under the UL26.5 promoter (15). R6 cells express gD and enable ΔgD viruses to replicate and spread from cell to cell by complementing the virions with gD made ectopically. The thymidine kinase-minus J1.1 cell line lacking all receptors for wild-type virus, J-HveA, and J-nectin cell lines were the kind gifts of G. Campadelli-Fiume (University of Bologna, Bologna, Italy) (18). The properties of the Vero-13R cell line expressing the IL-13α2 receptor have been described previously (26).
Viruses.
HSV-1(F) is the prototype wild-type virus used in this laboratory. The recombinant viruses described in this report were generated by cotransfection of R6 cells with a plasmid containing the desired construct of chimeric gD and intact DNA of the ΔgD virus R5110 as described elsewhere (18). The progeny of transfection was then plated on R6 cells for further plaque purification and virus amplification. The gD contained in the virion was analyzed to ensure that it contained the sequence of the desired gD construct. The plasmids encoding gD constructs and used for production of recombinant viruses are described in Results and were made by standard procedures described elsewhere (18).
Virus Replication in Cell Lines.
Replicate cultures of J-HveA, J-nectin, or Vero-13R cells were exposed to 0.1 pfu of recombinant viruses or HSV-1(F) per cell. After 24 h at 36°C, the cells were harvested and disrupted by sonication. Viral progeny was titered on Vero-13R cells.
Transfection Plasmids.
Transfer vector pAc-CMV, which contains the CMV immediate–early promoter-enhancer sequences in the XhoI–BamHI sites of pAc-SG2, was described elsewhere (15). To construct pGG5414, pGG5415, and pGG5416, an EcoRI–PstI fragment was derived by PCR from R5321, R5322, and R5323, respectively, with primers CGGAATTCATGGGGGGGGCTGCCGCCAG (for 5414), CGGAATTCATGGCGCTTTTGTTGACCACGG (for 5415), and CGGAATTCATGAGAGCCCTGCTGGCGCGCC (for 5416), along with AACTGCAGCTAGGCGTAGTAAACCGTGATCGGG, and then inserted into the EcoRI–PstI sites of pAc-CMV transfer vector. To construct the transfer vector expressing 219–314 residues of gD with the Myc tag, an EcoRI–PstI fragment was amplified by PCR from gD-A (16) with primers CGGAATTCATGCTGCCCCGCTTCATCCCCGAG and AACTGCAGTTACAGGTCCTCCTCTGAGATCAGCTTCTGCATTGATGCGTTGTTCGGGGTGGCCGGGGG and then inserted into the EcoRI–PstI sites of pAc-CMV. Transfer vector pGG5418 was constructed by inserting the uPA N-terminal fragment into the EcoRI–PstI sites of pAc-CMV transfer vector by PCR amplification with the primers CGGAATTCATGAGAGCCCTGCTGGCGCGCC along with AACTGCAGCTATTTTCCATCTGCGCAGTCATGC.
Antibodies.
The ZC25 antibody to the C-terminal domain of gD was the kind gift of G. H. Cohen and R. J. Eisenberg (University of Pennsylvania, Philadelphia, PA) (20). Monoclonal antibodies against gD (clone H170) and ICP0 (clone H1083) were from the Goodwin Institute (Plantation, FL). Monoclonal antibodies against IL-13 protein were described previously (18). Anti-Myc antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal antibodies against the human N-terminal fragment of uPA (19) were purchased from American Diagnostica (Stamford, CT).
Coimmunoprecipitation Assays.
Subconfluent cultures of HEK293 cells in 25-cm2 flasks were cotransfected with 1 μg of pGG5417 and 1 μg of pGG5414, pGG5415, or pGG5416. The cells were harvested 40 h after transfection, collected by centrifugation, rinsed twice with 5 ml of PBS, resuspended in 200 μl of lysis buffer [20 mM Tris (pH 8.0), 1 mM EDTA, 1% Nonidet P-40, 400 mM NaCl, 2 mM DTT, 0.1 mM NaVO4, 10 mM NaF, and 1× protease inhibitor mixture (Sigma, St. Louis, MO)], and chilled on ice for 40 min. The supernatant fluid (150 μl) collected after centrifugation at 100 × g for 2 min (Eppendorf 5415C; Avant, St. Louis, MO) was diluted with 150 μl of low-salt lysis buffer [20 mM Tris (pH 8.0), 1 mM EDTA, 1% Nonidet P-40, 16 mM NaCl, and 2 mM DTT] and incubated with 5% rabbit preimmune serum at 4°C for 1 h, followed by incubation with 50 μl of protein A-Sepharose for 1 h at 4°C and centrifugation at 900 × g for 3 min to remove nonspecifically bound proteins. The supernatant fluids were reacted with anti-Myc monoclonal antibody at 4°C for 16 h and then reacted with 20 μl of protein A-Sepharose at 4°C for 1 h. Immune complexes bound to protein A-Sepharose were rinsed three times with rinse buffer [50 mM Tris (pH 7.4), 10 mM MgCl2, and 5 mM DTT], collected by centrifugation, disrupted by boiling in sample buffer for 5 min, subjected to electrophoresis on denaturing gels, transferred to a nitrocellulose sheet, and probed with monoclonal antibodies to gD (H170), IL-13, or uPA. In-parallel aliquots of the supernatant fluids were reacted with antibodies to gD (H170), IL-13, or uPA. The collected immunoprecipitates were subjected to electrophoresis in denaturing gels, transferred to a nitrocellulose sheet, and probed with antibody to the Myc epitope.
Acknowledgments
We thank Haidong Gu (University of Chicago) for the pCCM plasmid, which contains CMV promoter and Myc epitope tag, and Robert Haselkorn and Anthony Kosiakoff for helpful discussions. This work was supported by National Cancer Institute Grants CA115662, CA83939, CA71933, CA78766, and CA88860.
Abbreviations
- CMV
cytomegalovirus
- gB
glycoprotein B
- gD
glycoprotein D
- HSV
herpes simplex virus
- uPA
urokinase plasminogen activator
- uPAR
urokinase plasminogen activator receptor.
Footnotes
The authors declare no conflict of interest.
References
- 1.Campadelli-Fiume G, Cocchi F, Menotti L, Lopez M. Rev Med Virol. 2000;10:305–319. doi: 10.1002/1099-1654(200009/10)10:5<305::aid-rmv286>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 2.Spear PG. Cell Microbiol. 2004;6:401–410. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 3.Spear PG, Longnecker R. J Virol. 2003;77:10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montgomery RI, Warner MS, Lum BJ, Spear PG. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 5.Spear PG, Eisenberg RJ, Cohen GH. Virology. 2000;275:1–9. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- 6.Fusco D, Forghieri C, Campadelli-Fiume G. Proc Natl Acad Sci USA. 2005;102:9323–9328. doi: 10.1073/pnas.0503907102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krummenacher C, Supekar VM, Whitneck JC, Connolly SA, Eisenberg RJ, Cohen G, Wiley DC, Carfi A. EMBO J. 2005;24:4144–4153. doi: 10.1038/sj.emboj.7600875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocchi F, Fusco D, Menotti L, Gianni T, Eisenberg RJ, Cohen GH, Campadelli-Fiume G. Proc Natl Acad Sci USA. 2004;101:7445–7450. doi: 10.1073/pnas.0401883101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willis SH, Rux AH, Peng C, Whitbeck JC, Nicola AV, Lou H, Hou W, Salvador L, Eisenberg RJ, Cohen GH. J Virol. 1998;72:5937–5947. doi: 10.1128/jvi.72.7.5937-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connolly SA, Landsburg DJ, Carfi A, Wiley DC, Eisenberg RJ, Cohen GH. J Virol. 2002;76:10894–10904. doi: 10.1128/JVI.76.21.10894-10904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly SA, Landsburg DJ, Carfi A, Wiley DC, Cohen GH, Eisenberg RJ. J Virol. 2003;77:8127–8140. doi: 10.1128/JVI.77.14.8127-8140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon M, Spear PG. Proc Natl Acad Sci USA. 2004;101:17252–17257. doi: 10.1073/pnas.0407892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly SA, Landsburg DJ, Carfi A, Whitbeck JC, Zuo Y, Wiley DC, Cohen GH, Eisenberg RJ. J Virol. 2005;79:1282–1295. doi: 10.1128/JVI.79.2.1282-1295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manoj S, Jogger CR, Myscofski D, Yoon M, Spear PG. Proc Natl Acad Sci USA. 2004;101:12414–12421. doi: 10.1073/pnas.0404211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou G, Galvan V, Campadelli-Fiume G, Roizman B. J Virol. 2000;74:11782–11791. doi: 10.1128/jvi.74.24.11782-11791.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou G, Roizman B. J Virol. 2002;76:6197–6204. doi: 10.1128/JVI.76.12.6197-6204.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou G, Avitabile E, Campadelli-Fiume G, Roizman B. J Virol. 2003;77:3759–3767. doi: 10.1128/JVI.77.6.3759-3767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou G, Ye GJ, Debinski W, Roizman B. Proc Natl Acad Sci USA. 2002;99:15124–15129. doi: 10.1073/pnas.232588699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamiyama H, Zhou G, Roizman B. Gene Ther. 2006;13:621–629. doi: 10.1038/sj.gt.3302685. [DOI] [PubMed] [Google Scholar]
- 20.Zhou G, Roizman B. Proc Natl Acad Sci USA. 2006;103:5508–5513. doi: 10.1073/pnas.0601258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou J, Poon APW, Johnson J, Roizman B. J Virol. 1994;68:8304–8311. doi: 10.1128/jvi.68.12.8304-8311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cocchi F, Menotti L, Mirandola P, Campadelli-Fiume G. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gianni T, Menotti L, Campadelli-Fiume G. J Virol. 2005;79:7042–7049. doi: 10.1128/JVI.79.11.7042-7049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connolly SA, Landsburg DJ, Carfi A, Whitbeck JC, Zuo Y, Wiley DC, Cohen GH, Eisenberg RJ. J Virol. 2005;79:1282–1295. doi: 10.1128/JVI.79.2.1282-1295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez WM, Spear PG. J Virol. 2002;76:7255–7262. doi: 10.1128/JVI.76.14.7255-7262.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou G, Roizman B. J Virol. 2005;79:5272–5277. doi: 10.1128/JVI.79.9.5272-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]