Abstract
Alzheimer's disease (AD) is associated with accumulation of the neurotoxic peptide amyloid-β (Aβ), which is produced by sequential cleavage of amyloid precursor protein (APP) by the aspartyl protease β-secretase and the presenilin-dependent protease γ-secretase. An increase of casein kinase 1 (CK1) expression has been described in the human AD brain. We show, by using in silico analysis, that APP, β-secretase, and γ-secretase subunits contain, in their intracellular regions, multiple CK1 consensus phosphorylation sites, many of which are conserved among human, rat, and mouse species. Overexpression of constitutively active CK1ε, one of the CK1 isoforms expressed in brain, leads to an increase in Aβ peptide production. Conversely, three structurally dissimilar CK1-specific inhibitors significantly reduced endogenous Aβ peptide production. By using mammalian cells expressing the β C-terminal fragment of APP, it was possible to demonstrate that CK1 inhibitors act at the level of γ-secretase cleavage. Importantly, Notch cleavage was not affected. Our results indicate that CK1 represents a therapeutic target for prevention of Aβ formation in AD.
Keywords: γ-cleavage, neurodegenerative, amyloid precursor protein
Several lines of evidence led us to evaluate the possible role of casein kinase 1 (CK1) in the production of amyloid-β (Aβ)-40 and Aβ-42 peptides. CK1δ mRNA has been shown to be up-regulated in brain samples from subjects with Alzheimer's disease (AD) (1) and may have a pathological association with tau (2). It is also interesting to note that glycogen synthase kinase 3 (GSK3), one of the most studied kinases in the AD field (3), can phosphorylate its substrates only if they are prephosphorylated by a priming kinase; furthermore CK1 is one of the few known GSK3-priming kinases, which also include PKA, CK2, Cdk5, and DYRK1A (for a review see ref. 4). It has also been shown that CK1 is an upstream regulator of Cdk5, another protein kinase implicated in AD (5, 6). Moreover, CK1 phosphorylates β-secretase (BACE) and regulates its subcellular location (7). CK1 has also been shown to phosphorylate presenilin 2 (8).
CK1 consists of a family of eight genes in mice, which appear to function as monomeric enzymes: α, γ1, γ2, γ3, δ, ε1, ε2, and ε3. Family members contain a highly conserved N-terminal catalytic domain of ≈290 residues coupled to a variable C-terminal region that ranges in size from 40 to 180 aa. It is possible that the different isoforms are expressed in different neuronal populations and/or are targeted to different regions of the neuron and thus may have access to different substrates. Little is known about the regulation of CK1. CK1 is basally active, but certain isoforms (particularly CK1δ and CK1ε) are regulated by inhibitory autophosphorylation at their C-terminal regions (9, 10). Notably, it has been shown that the C-terminal region of CK1ε is phosphorylated at multiple sites and that enzyme activity can be increased after dephosphorylation by a signaling pathway involving activation of the serine/threonine phosphatase, calcineurin (also termed PP2B), in response to stimulation of metabotropic glutamate receptors (5, 11).
CK1 is present in both the cytosol and the nucleus; the C-terminal region of CK1 has been shown to promote differential subcellular localization of individual isoforms (e.g., cellular membrane versus cytoplasm) (12, 13). A number of proteins have been found to interact with CK1 isoforms in nonneuronal tissues resulting in its targeting to specific signaling pathways (14–16). Association of CK1 with the plasma membrane and the cytoskeleton has also been reported (12, 17, 18). In neurons, CK1 phosphorylates a variety of proteins including transcription factors, as well as certain synaptic vesicle proteins (19, 20). CK1δ and CK1ε, both expressed predominantly in the brain (21), have been implicated in several important brain processes, including but not limited to: dopamine signaling (phosphorylation of dopamine- and cAMP-regulated phosphoprotein of 32 kDa), circadian rhythm (phosphorylation of protein Period), and neurotransmitter receptor phosphorylation (22–24).
Our analysis of consensus phosphorylation sites for CK1 conserved in mammalian proteins related to AD has revealed potential regulation of these proteins by CK1 (Fig. 1). Here we show that three different structurally unrelated, CK1-specific inhibitors, significantly reduce endogenous Aβ peptide production by selectively interfering with amyloid precursor protein (APP) γ-cleavage. Conversely, overexpression of constitutively active CK1 leads to an increase of Aβ peptide production. This effect is specific for one of the brain CK1 isoforms (CK1ε). Importantly, Notch cleavage is not affected. Taken together, these results strongly suggest that CK1 represents an attractive target for development of novel therapies for the treatment of AD.
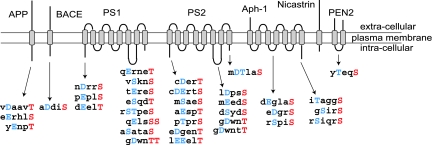
Fig. 1.
Conservation of CK1 consensus phosphorylation sites in APP, BACE, and γ-secretase. Consensus sites were determined by using different computational tools (ELM-motifs, NetPhos 2.0). Only intracellular regions were considered for this analysis. The amino acid sequences of the consensus phosphorylation sites are listed below the regions of the proteins in which they are found. The sites indicated are conserved in human, rat, and mouse genes. The residue potentially phosphorylated by CK1 (Ser or Thr) is indicated in red, and the acidic residue (Asp/Glu or phosphorylated Ser/Thr) required two or three residues N-terminal is indicated in blue. Transmembrane domains are indicated by gray rectangles.
Results
In Silico Analysis of Putative CK1 Phosphorylation Sites in AD-Related Proteins.
The human, rat, and mouse amino acid sequences corresponding to amyloid-β related proteins (APP, BACE, PS1, PS2, Aph-1, PEN2, and nicastrin) were screened for the presence of putative CK1 phosphorylation sites by using different computational tools (e.g., ELM-motifs; http://elm.eu.org, NetPhos 2.0) (Fig. 1). Only the putative phosphorylation sites conserved between human, mouse, and rat species are indicated. This in silico analysis of intracellular domains in APP, BACE, PS1, PS2, Aph-1, PEN2, and nicastrin has revealed the presence of numerous putative phosphorylation sites, particularly in PS1 and PS2. Moreover, the same in silico analysis of the two kinases that have been implicated in AD (Cdk5/p35 and GSK3-β) has revealed the presence of numerous putative phosphorylation sites for CK1, conserved in human, rat, and mouse sequences. Indeed, we have found 7 putative sites in Cdk5, 15 in p35, and 24 in GSK3-β.
Overexpression of Constitutively Active CK1ε Leads to an Increase in Aβ Peptide Production.
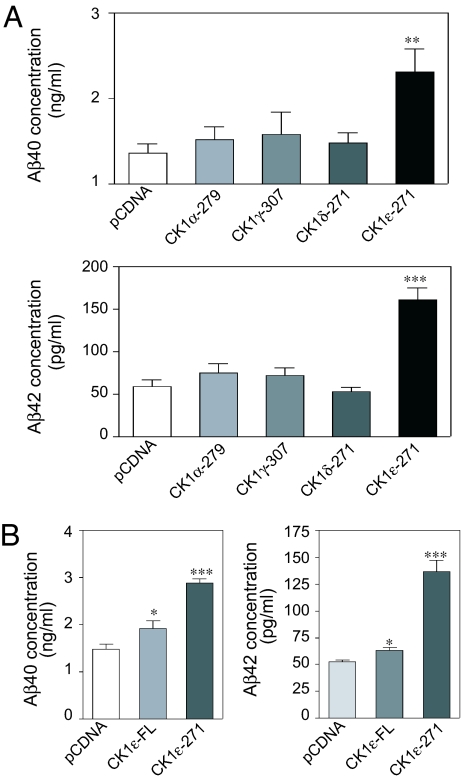
We tested the capacity of the four isoforms of CK1, namely CK1α, CK1γ, CK1δ, and CK1ε, to regulate APP processing by overexpressing each isoform in N2A cells stably expressing APP (N2A-APP695 cells). All of the isoforms are basally active, but CK1γ, CK1δ, and CK1ε are autoinhibited by their respective C-terminal domains. We therefore generated constitutively active mutants of CK1γ, CK1δ, and CK1ε by truncating the C-terminal autoinhibitory region of each isoform. We measured the level of expression of each isoform after transient transfection in N2A cells and normalized the results based on these levels. As shown in Fig. 2A, only CK1ε-271 was able to induce a substantial increase in either Aβ40 or Aβ42 peptide production.
Fig. 2.
Constitutively active CK1ε selectively increases Aβ formation. N2A cells stably expressing APP-695 were transiently transfected with plasmids containing different isoforms of truncated (constitutively active) or full-length (not constitutively active) CK1. Forty-eight hours after transfection, cell supernatants were collected and subjected to Aβ40/Aβ42 sandwich ELISAs. The same amount of plasmid DNA was used for each of the transfections, and the level of expression was verified and normalized by Western blot analysis. (A) Comparison of the capacity of the four constitutively active CK1 isoforms to increase Aβ40 peptide production. The CK1 numbers indicate the last amino acid contained in the different truncation mutants. (B) Comparison of full-length CK1ε (CK1ε-FL) and constitutively active CK1ε (CK1ε-271) to regulate Aβ40/Aβ42 peptide regulation. Data for at least three experiments (mean ± SEM) were compared (GraphPad Prism ver. 4.0, GraphPad Software, San Diego, CA) with the control conditions (pcDNA). ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; one-tailed Student's t test, 95% significance level. pcDNA; empty pcDNA-3.1 plasmid.
We next compared the effect of overexpressing constitutively active CK1ε (CK1–271) versus full-length CK1ε in N2A cells stably expressing APP. CK1ε-271 induced an increase of Aβ40 and Aβ42 levels to 194% and 261% of control, respectively (Fig. 2B). In contrast, full-length CK1ε was weakly active, producing an increase of Aβ40 and Aβ42 peptide levels to 129% and 121%, respectively (Fig. 2B). These results, obtained by ELISA and confirmed by Aβ peptide immunoprecipitation and Western blotting analysis (data not shown), indicate that CK1ε activity is capable of regulating Aβ peptide levels.
Effect of CK1 Inhibitors on Aβ Peptide Production After Overexpression of Constitutively Active CK1ε.
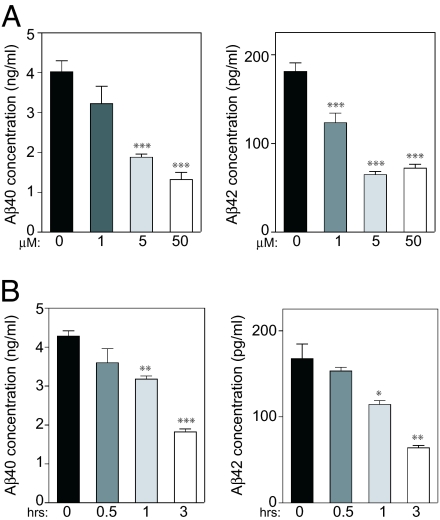
We next examined the effect of the selective CK1 inhibitor IC261 (25) on Aβ40 and Aβ42 peptide production in N2A cells transiently expressing CK1ε-271. We varied the concentration of IC261 (1–50 μM) and the time of incubation (0.5–3 h). IC261 treatment reduced the generation of Aβ40 and Aβ42, with a maximal effect being obtained with 5–50 μM IC261 after 3 h (Fig. 3). To exclude nonspecific effects of IC261, we tested two other CK1 inhibitors, CKI-7 (26) and D4476 (27). Both inhibitors caused a significant and dose-dependent reduction of Aβ40 and Aβ42 production in cells overexpressing CK1ε-271 (data not shown).
Fig. 3.
CK1 inhibitor IC261 abolishes CK1ε-271-dependent Aβ40/Aβ42 increase in a dose- and time-dependent manner. N2A cells stably expressing APP-695 were transiently transfected with CK1ε-271. (A) IC261 dose response (3 h). Forty-eight hours after transfection, cells were incubated in the absence or presence of various concentrations of CK1 inhibitor IC261. Cell supernatants were collected after 3 h of incubation and subjected to Aβ40 (Left)/Aβ42 (Right) ELISAs. (B) IC261 time response (50 μM). Forty-eight hours after transfection, CK1 inhibitor IC261 (50 μM) was added. Cell supernatants were collected at different times of incubation and subjected to Aβ40 (Left)/Aβ42 (Right) sandwich ELISAs. Data for at least three experiments (mean ± SEM.) were compared (GraphPad Prism ver. 4.0) with the control conditions (no drug or zero time point). ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; one-tailed Student's t test, 95% significance level.
Regulation of Aβ Peptide Production by Endogenous CK1.
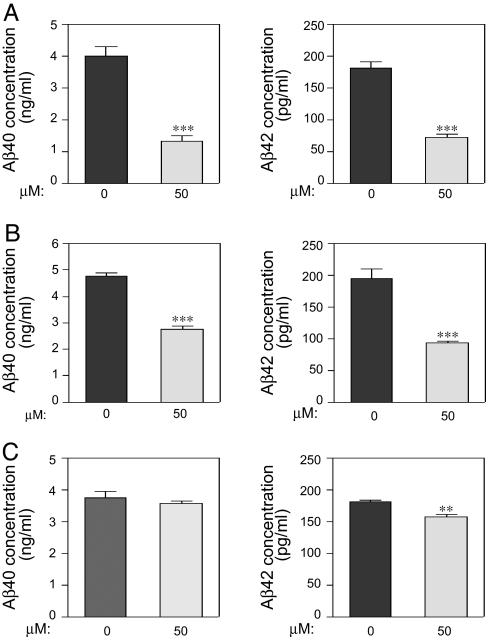
We next compared the effect of the three different classes of CK1 inhibitors, IC-261, D4476, and CKI-7 on Aβ peptide production in N2A cells expressing APP-695, assuming that these three inhibitors will block endogenous CK1ε activity. Aβ40 peptide production was significantly reduced after 3 h of incubation with IC261 or D4476, and Aβ42 peptide production was significantly reduced after 3 h of incubation with each of the three inhibitors (Fig. 4). The reduction was shown by using two different methods: sandwich ELISA (Fig. 4) and Western blot analysis after immunoprecipitation (data not shown). We observed the following decreases after incubation for 3 h: IC261 (50 μM), 67.9% and 60.8% decrease for Aβ40 and Aβ42 peptides, respectively (Fig. 4A); D4476 (50 μM), 42.8% and 52.7% decrease for Aβ40 and Aβ42 peptides, respectively (Fig. 4B), and CKI-7 (50 μM), 4.6% and 13.7% decrease for Aβ40 and Aβ42 peptides, respectively (Fig. 4C). These results demonstrate that endogenous CK1, under basal conditions, participates in regulation of Aβ peptide production. In contrast to all three CK1 inhibitors for which a maximal effect was observed at 3 h, other kinase inhibitors (e.g., lithium chloride, Gleevec) required at least 24 h of incubation to induce a significant effect on Aβ generation (3, 28).
Fig. 4.
Capacity of three different CK1 inhibitors to lower endogenous Aβ peptide production. N2A cells stably expressing APP-695 were incubated for 3 h in the absence or presence of CK1 inhibitors (50 μM). Cell supernatants were collected and subjected to Aβ40/Aβ42 sandwich ELISAs. (A) IC261. (B) D4476. (C) CKI-7. Data for at least three experiments (mean ± SEM) were compared (GraphPad Prism ver. 4.0) with the control conditions (no drug). ∗∗, P < 0.01; ∗∗∗, P <0.001; one-tailed Student's t test, 95% significance level.
To exclude the possibility that the inhibitory effects on Aβ peptide production were because of cellular toxicity, we investigated the effects of the drugs on cell viability. Media were collected and tested for the level of lactate dehydrogenase, a stable cytosolic enzyme released after cell lysis, with a nonradioactive CytoTox96 kit (Promega, Madison, WI). Values obtained from drug-treated cells were compared with values obtained with DMSO and nontreated cells. No toxicity was observed for CKI-7, even after 24 h of incubation, and for the highest concentration tested (50 μM). A slight toxicity (less than doubling) was observed for D4476, only for the higher dose tested (50 μM) and only after 24 h of incubation. No effect on toxicity was observed with IC261 at 3 h of incubation.
CK1 Modulation of γ-Secretase Activity in N2A Cells.
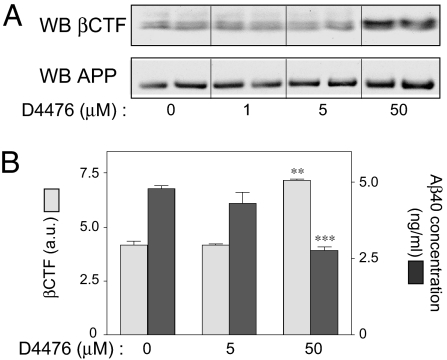
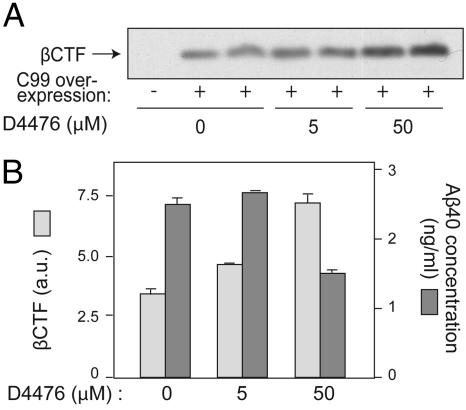
We next compared the effects of D4476 on APP, the C-terminal fragment of βAPP cleaved by BACE (βCTF), and Aβ40 peptide levels in N2A cells. Inhibition of CK1 with D4476 (50 μM) significantly increased βCTF levels and considerably reduced Aβ peptide production under conditions in which it had no effect on APP expression level (Fig. 5). These results strongly suggest that CK1 activity regulates Aβ peptide formation at the level of the γ-secretase cleavage site of APP. This conclusion received direct support from experiments in which an APP fragment corresponding to the membrane-bound C-terminal region produced by BACE (APP-C99) was stably expressed in N2A cells instead of the full-length APP protein. D4476 inhibited the breakdown of C99 and consequently decreased Aβ peptide production in a dose-dependent manner (Fig. 6).
Fig. 5.
Effect of CK1 inhibitor D4476 on APP, βCTF, and Aβ levels. N2A cells stably expressing APP-695 were incubated for 3 h with the indicated concentrations of D4476. Cellular extracts were subjected to Western blot analysis by using βCTF or APP antibodies or to Aβ40 ELISAs. (A) Western blot analysis of βCTF and APP in one representative experiment carried out in duplicate. (B) Means ± SEM for βCTF and Aβ40 in at least three independent experiments analyzed in duplicate. Data for at least three experiments (mean ± SEM) were compared (GraphPad Prism ver. 4.0) with the control conditions (no drug). ∗∗, P < 0.01; ∗∗∗, P < 0.001; one-tailed Student's t test, 95% significance level.
Fig. 6.
Effect of CK1 inhibitor D4476 on APP-C99 and Aβ levels. N2A cells transiently expressing APP-C99 for 24 h were incubated with various concentrations of D4476 for 3 h. Cell lysates were analyzed by Bis·Tris acrylamide gel electrophoresis and Western blotting by using βCTF antibodies (CT695) or by Aβ40 ELISA. (A) Western blot analysis of one representative experiment carried out in duplicate. (B) βCTF Western blot analysis shown in A was quantified by using the program NIH image (ver. 1.63) and is shown as a histogram (light gray bars). The same samples were also subjected to Aβ40 ELISA (dark gray bars). Data for at least three experiments analyzed in duplicate (mean ± SEM) were compared (GraphPad Prism ver. 4.0) with the control condition (no drug). ∗, P < 0.05; ∗∗ P < 0.01; ∗∗∗, P < 0.001; one-tailed Student's t test, 95% significance level.
Notch Cleavage Is Not Affected by CK1 Inhibitors.
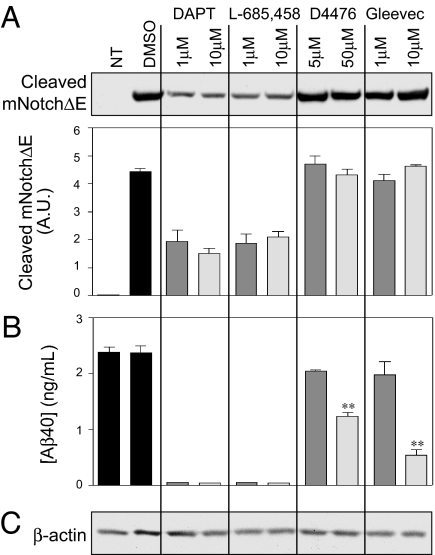
Major side-effects of γ-secretase inhibitors that have been developed for treatment of AD include important organ dysfunctions (e.g., gastrointestinal tract, spleen, and thymus) most likely caused by Notch cleavage inhibition, a type I transmembrane protein that serves as a γ-secretase substrate (for a review see ref. 29). We therefore investigated the effect of CK1 inhibitors on Notch processing. In N2A cells overexpressing Notch and APP-695, we confirmed a strong inhibition of Notch cleavage after only 3 h of incubation with DAPT and L-685,458, two typical γ-secretase inhibitors (Fig. 7A). We also confirmed the results of earlier studies showing that Gleevec does not inhibit Notch cleavage (28) (Fig. 7A). Under the same experimental conditions, none of the CK1 inhibitors tested, D4476 (Fig. 7A), IC261, or CKI-7 (data not shown), inhibited Notch cleavage. The same samples were also analyzed by sandwich ELISA for Aβ40 peptide production levels and gave results that were comparable to those described above (Fig. 7B) and in previous reports (28).
Fig. 7.
CK1 inhibitor D4476 does not inhibit Notch cleavage. N2A cells transiently expressing N-terminally truncated mNotchΔE-myc were incubated with the indicated concentrations of DAPT, L-685,458, D4476, or Gleevec for 3 h. Cell lysates (20 μg) were analyzed by Bis·Tris acrylamide gel electrophoresis and Western blotting by using anti-cleaved Notch (A) and anti-actin antibodies (C), or by Aβ40 ELISA (B). DAPT and L-685,458 are γ-secretase inhibitors known to inhibit βCTF and Notch cleavage; Gleevec is known to inhibit βCTF but not Notch cleavage; D4476 is a CK1 inhibitor. Data for at least three experiments (mean ± SEM) were compared (GraphPad Prism ver. 4.0) with the control condition (no drug). NT, nontransfected. ∗∗, P < 0.01; one-tailed Student's t test, 95% significance level.
Discussion
It is widely accepted that Aβ is a causative agent in the development of AD. It is also well established that Aβ is produced from amyloid precursor protein by sequential cleavage by BACE and γ-secretase. Major efforts to develop selective inhibitors of these enzymes have met with only limited success. Most γ-secretase inhibitors suffer from the drawback that they inhibit the cleavage of Notch, a receptor essential not only for normal development but also for cell fate decisions (for a review, see ref. 29).
We have demonstrated in the present study that constitutively active CK1ε increases the formation of Aβ. Conversely, three distinct classes of CK1 inhibitors reduce the formation of β-amyloid peptide produced in N2A cells either expressing endogenous levels of CK1 or overexpressing constitutively active CK1ε. D4476 has recently been described as being a CK1 inhibitor and is reported to be more potent and more selective in vitro than the two others (27). However, a 50 μM concentration was required to observe an effect on CK1 activity in cell lines (27), suggesting that cellular permeability may be limiting. The 50 μM concentration effective in other experiments (27) is comparable to our results obtained by using N2A cells. Interestingly, by using the Simplified Molecular Input Line Entry System (or SMILES string; Daylight Chemical Information Systems, Aliso Viejo, CA), we searched for compounds similar to the known CK1 inhibitors. We have identified a compound very similar to the compound D4476 which is commercially available, namely SB-431542 (Sigma–Aldrich, St. Louis, MO), and we have shown comparable properties on Aβ peptide formation for D4476 and SB-431542 (data not shown).
Under conditions in which inhibition of CK1 activity reduces Aβ formation, no inhibition of Notch cleavage was observed. Experiments using N2A cells demonstrate that the effect of the CK1 inhibitors is expressed at the level of γ-secretase activity. This result was further confirmed in a different cellular system by examination of the effect of CKI-7 and D4476 on Aβ formation in the yeast Pichia pastoris, which had been stably transfected with the four mammalian γ-secretase subunits and with APP-C99 (Y. Chungiang and S. Sisodia, unpublished data). CKI-7 and D4476 significantly reduced Aβ peptide production at low-micromolar concentrations (data not shown). No effect on expression of any of the γ-secretase subunits was observed, nor was any toxicity observed at concentrations up to 50 μM. These studies confirm the regulatory role of CK1 in APP processing and support the idea that a component of the γ-secretase complex, or a modulator thereof, is regulated by CK1.
The observations that three structurally unrelated CK1 inhibitors significantly decrease Aβ peptide production greatly increases the likelihood that the three inhibitors act through a common target, namely CK1. This conclusion is further supported by the fact that constitutively active CK1ε leads to increased Aβ peptide formation. Altogether, these results suggest the involvement of CK1ε activity in the regulation of the APP γ-secretase cleavage. The results presented here, and the fact that Notch cleavage is not affected by any of the CK1 inhibitors tested, indicate that CK1 acts directly or indirectly on the γ-secretase activity to selectively interfere with the APP γ-cleavage site but not at the Notch/APP ε-cleavage site.
The mechanism by which CK1ε activity leads to the regulation of Aβ formation is an important topic for future investigation. Further studies will be required to identify and characterize the CK1 substrate(s) involved in regulation of APP processing. It is surprising that no effect was observed after overexpression of the constitutively active CK1δ isoform because CK1δ and CK1ε constitutively active isoforms are 97% identical. Presumably, differences exist in the substrates phosphorylated or in the subcellular localization of these two isoforms. Interestingly, CK1δ mRNA is elevated in postmortem human AD brain (1). No comparable studies appear to have been carried out for CK1ε.
Another important question concerns the mechanism by which CK1ε activity is regulated in vivo. It has been shown that phosphorylation of CK1ε in its C-terminal region is associated with inhibition of enzymatic activity (9). One known mechanism involved in activation of CK1ε is the activation by glutamate of group I metabotropic mGlu receptors via a Gq/PLC-IP3-Ca2+cascade (11). Through this signaling cascade, Ca2+ activates calcineurin, leading to a transient dephosphorylation of autoinhibitory C-terminal phosphorylation sites and activation of CK1ε. CK1ε can also be dephosphorylated and activated by other protein phosphatases 1 and 2A (PP1 and PP2A) (9), which are themselves subject to regulation by various physiologically relevant mechanisms. Aβ production may therefore be modulated by a variety of signaling pathways that regulate CK1ε activity. It will be of interest to further study the specific regulation of the three different CK1ε isoforms, namely CK1ε1, CK1ε2, and CK1ε3, each resulting from the expression of a distinct gene. Additional studies with each CK1ε isoform would help to clarify the mechanism by which CK1ε regulates Aβ peptide production. The region used to test the impact of CK1ε activity on Aβ peptide formation (the N-terminal 271 amino acid) is 100% identical among the three isoforms; for this reason, we only tested one of the three isoforms (CK1ε1). However, differences exist in their C-terminal regulatory regions, and these could result in differential regulation of kinase activity.
Several protein kinases and protein phosphatases have been implicated in the progression of AD, especially in the regulation of β-amyloid formation. These include the protein kinases PKA, PKC, GSK3-β, and Cdk5 (3, 30–32), and the protein phosphatases PP1/PP2A and PP2B (30, 33). Early studies demonstrated that activation of PKA or PKC or inhibition of PP1/PP2A or PP2B causes a dramatic reduction in the formation of Aβ and/or an increase in the amount of the α-secretase cleavage product sAPPα (30). Unfortunately, some of the drugs used in these studies had issues related to toxicity, and others could inhibit Notch cleavage. Indeed, by using the same cellular conditions as described above for the CK1 inhibitors, we have found that roscovitine (a Cdk5 inhibitor) and kenpaullone (a GSK3 inhibitor) were at least partially associated with inhibition of Notch cleavage (data not shown).
Reducing selectively and specifically the biogenesis of Aβ peptide has been a major focus in AD studies. γ-Secretase is a key enzyme in the rate-limiting step in Aβ peptide production. So far, numerous efforts have been directed toward development of potent inhibitors for γ-secretase. However, the success of such strategies has been limited, most likely because of the essential nature of this enzyme in multiple pathways implicated in developmental regulation as well as adult homeostasis (for a review, see ref. 34). For example, it has long been recognized that γ-secretase is needed in many signaling cascades, especially in Notch activation, which play critical roles in development and in the immune response (35). γ-Secretase inhibitors have shown significant side-effects in animal studies (29). Therefore, it will be essential to develop reagents that can inhibit Aβ generation and minimize side-effects, especially in regard to the Notch signaling cascade. From our current study, because CK1 has an impact on γ- but not β-cleavage of APP, because Notch cleavage is not inhibited, and because certain γ-secretase-related proteins are candidates for CK1 phosphorylation, CK1 represents a potential AD therapeutic target.
Methods
Plasmids.
Full-length cDNAs corresponding to the four CK1 isoforms were subcloned from a rat oligonucleotide-dT cDNA library, by using standard PCR and molecular techniques, into the pcDNA3.1/Myc-His6 A+ plasmid (Invitrogen, Carlsbad, CA). Constitutively active CK1 constructs were derived from full-length cDNAs by PCR and cloned into pcDNA3.1/Myc-His6 A+ plasmid (Invitrogen). Subcloned DNA fragments were systematically checked by sequencing. CK1α was truncated at amino acid 279; CK1δ was truncated at amino acid 271; CK1ε was truncated at amino acid 271; and CK1γ was truncated at amino acid 307. The construct encoding the fragment of APP corresponding to the membrane-bound C-terminal region (APP-C99) produced by BACE was a kind gift from Y. Li (Merck Research Laboratories, West Point, PA) (36).
Antibodies and Western Blotting.
Several antibodies used in this study were commercially available: Myc 9E10 (Covance, Princeton, NJ), cleaved Notch 1 (Val-1744) (Cell Signaling Technology, Beverly, MA), βCTF and APP (polyclonal CT695; Zymed Laboratories, South San Francisco, CA), and β-actin (Cell Signaling Technology). Western blotting analysis was performed by using 4–12% Bis-Tris acrylamide gels (Invitrogen) with Mes or Mops buffer, and proteins were transferred onto nitrocellulose membranes. Antigens were detected by using the ECL system (Amersham Pharmacia Biosciences, Piscataway, NJ).
Kinase Inhibitors.
The following inhibitors were commercially available: CKI-7 (Seikagatu, Tokyo, Japan), IC-261 (Calbiochem, San Diego, CA), D4476 (Calbiochem), DAPT (Calbiochem), and Gleevec (Sequoia Research Products, Oxford, U.K.). L-685,458 was a kind gift from Y. Li. Stocks were prepared in dimethylsulfoxide (Sigma–Aldrich) at 5 mM.
Cell Culture.
Transfected N2a cells were grown as reported in ref. 37. Briefly, N2A-APP695 cells were plated in 12-well dishes at a density of 3 × 105 cells per well. Cells were grown to 60% confluence and then transfected with relevant plasmid constructs by using FuGENE6 (Roche Applied Science, Indianapolis, IN). After transfection, cells were incubated under the conditions described for each experiment. Drugs were added to cell cultures in fresh medium (0.5% FBS). After incubation, cells were recovered with 100 μl of RIPA buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS), incubated for 30 min on ice, and centrifuged at 10,000 × g for 20 min at 4°C. The supernatants and/or cells were collected and subjected to bicinchoninic acid (Pierce Chemical, Rockford, IL) quantification. Equal amounts ofprotein samples were subjected to Western blot analysis by using appropriate antibodies as described above. For immu-noprecipitation assays, media were incubated with antibody 4G8 (Covance) to detect Aβ and full-length βAPP.
Aβ Quantification.
After immobilization of total Aβ (coated monoclonal antibody specific for the N terminus of Aβ) from media, Aβ40/42 peptide determinations were made by sandwich ELISA (BioSource International, Camarillo, CA) by using rabbit polyclonal antibodies specific for the C terminus of Aβ40 or Aβ42. Aβ levels were normalized to total protein levels. Aβ40 and Aβ42 standard curves were plotted as a sigmoidal dose-response curve (variable slope) by using GraphPad Prism ver. 4.0. Data presented are the results of at least three independent experiments done in triplicate. Results were analyzed with GraphPad Prism ver. 4.0.
Immunoprecipitation/Western Blot Analysis.
Immunodetection and Western blot analysis were carried out as reported in ref. 30 by using standard methods.
mNotchΔE Transfection and Notch-1 Cleavage Assays in N2A Cells.
N2A cells stably overexpressing APP-695 were transiently transfected with mNotchΔE (truncated Notch-1, lacking most of the Notch extracellular domain) (38). Cultures were preincubated with the various inhibitors for 3 h. mNotchΔE was detected in cell lysates by Western blot analysis by using the cleaved Notch 1 antibody (Val-1744).
Acknowledgments
We thank Ameet Talwalkar and Maria Metcalfe for their technical help, and Heike Rebholz for helpful comments on the manuscript. This project was supported by National Institutes of Health Grant AG09464, Administration on Aging/Department of Health and Human Services Grant 90AZ2791, and the F. M. Kirby Foundation.
Abbreviations
- CK1
casein kinase 1
- AD
Alzheimer's disease
- APP
amyloid precursor protein
- BACE
β-secretase
- βCTF
C-terminal fragment of βAPP cleaved by β-secretase
- Aβ
APP fragment cleaved by the β- and γ-secretases
- APP-C99
APP fragment corresponding to the membrane-bound C-terminal region produced by β-secretase
- GSK3
glycogen synthase kinase 3.
Footnotes
The authors declare no conflict of interest.
References
- 1.Yasojima K, Kuret J, DeMaggio AJ, McGeer E, McGeer PL. Brain Res. 2000;865:116–120. doi: 10.1016/s0006-8993(00)02200-9. [DOI] [PubMed] [Google Scholar]
- 2.Schwab C, DeMaggio AJ, Ghoshal N, Binder LI, Kuret J, McGeer PL. Neurobiol Aging. 2000;21:503–510. doi: 10.1016/s0197-4580(00)00110-x. [DOI] [PubMed] [Google Scholar]
- 3.Phiel CJ, Wilson CA, Lee VM, Klein PS. Nature. 2003;423:435–439. doi: 10.1038/nature01640. [DOI] [PubMed] [Google Scholar]
- 4.Meijer L, Flajolet M, Greengard P. Trends Pharmacol Sci. 2004;25:471–480. doi: 10.1016/j.tips.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Liu F, Ma XH, Ule J, Bibb JA, Nishi A, DeMaggio AJ, Yan Z, Nairn AC, Greengard P. Proc Natl Acad Sci USA. 2001;98:11062–11068. doi: 10.1073/pnas.191353898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walter J, Fluhrer R, Hartung B, Willem M, Kaether C, Capell A, Lammich S, Multhaup G, Haass C. J Biol Chem. 2001;276:14634–14641. doi: 10.1074/jbc.M011116200. [DOI] [PubMed] [Google Scholar]
- 7.Pastorino L, Ikin AF, Nairn AC, Pursnani A, Buxbaum JD. Mol Cell Neurosci. 2002;19:175–185. doi: 10.1006/mcne.2001.1065. [DOI] [PubMed] [Google Scholar]
- 8.Walter J, Grunberg J, Schindzielorz A, Haass C. Biochemistry. 1998;37:5961–5967. doi: 10.1021/bi971763a. [DOI] [PubMed] [Google Scholar]
- 9.Gietzen KF, Virshup DM. J Biol Chem. 1999;274:32063–32070. doi: 10.1074/jbc.274.45.32063. [DOI] [PubMed] [Google Scholar]
- 10.Graves PR, Roach PJ. J Biol Chem. 1995;270:21689–21694. doi: 10.1074/jbc.270.37.21689. [DOI] [PubMed] [Google Scholar]
- 11.Liu F, Virshup DM, Nairn AC, Greengard P. J Biol Chem. 2002;277:45393–45399. doi: 10.1074/jbc.M204499200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vancura A, Sessler A, Leichus B, Kuret J. J Biol Chem. 1994;269:19271–19278. [PubMed] [Google Scholar]
- 13.Babu P, Bryan JD, Panek HR, Jordan SL, Forbrich BM, Kelley SC, Colvin RT, Robinson LC. J Cell Sci. 2002;115:4957–4968. doi: 10.1242/jcs.00203. [DOI] [PubMed] [Google Scholar]
- 14.Amit S, Hatzubai A, Birman Y, Andersen JS, Ben-Shushan E, Mann M, Ben-Neriah Y, Alkalay I. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cong F, Schweizer L, Varmus H. Mol Cell Biol. 2004;24:2000–2011. doi: 10.1128/MCB.24.5.2000-2011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, Glinka A, Niehrs C. Nature. 2005;438:867–872. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed K. Cell Mol Biol Res. 1994;40:1–11. [PubMed] [Google Scholar]
- 18.Walter J, Schnolzer M, Pyerin W, Kinzel V, Kubler D. J Biol Chem. 1996;271:111–119. doi: 10.1074/jbc.271.1.111. [DOI] [PubMed] [Google Scholar]
- 19.Issinger OG. Pharmacol Ther. 1993;59:1–30. doi: 10.1016/0163-7258(93)90039-g. [DOI] [PubMed] [Google Scholar]
- 20.Gross SD, Hoffman DP, Fisette PL, Baas P, Anderson RA. J Cell Biol. 1995;130:711–724. doi: 10.1083/jcb.130.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross SD, Anderson RA. Cell Signalling. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- 22.Desdouits F, Siciliano JC, Greengard P, Girault JA. Proc Natl Acad Sci USA. 1995;92:2682–2685. doi: 10.1073/pnas.92.7.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh TJ, Grundke-Iqbal I, Iqbal K. J Neurochem. 1995;64:1420–1423. doi: 10.1046/j.1471-4159.1995.64031420.x. [DOI] [PubMed] [Google Scholar]
- 24.Kloss B, Rothenfluh A, Young MW, Saez L. Neuron. 2001;30:699–706. doi: 10.1016/s0896-6273(01)00320-8. [DOI] [PubMed] [Google Scholar]
- 25.Mashhoon N, DeMaggio AJ, Tereshko V, Bergmeier SC, Egli M, Hoekstra MF, Kuret J. J Biol Chem. 2000;275:20052–20060. doi: 10.1074/jbc.M001713200. [DOI] [PubMed] [Google Scholar]
- 26.Chijiwa T, Hagiwara M, Hidaka H. J Biol Chem. 1989;264:4924–4927. [PubMed] [Google Scholar]
- 27.Rena G, Bain J, Elliott M, Cohen P. EMBO Rep. 2004;5:60–65. doi: 10.1038/sj.embor.7400048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Netzer WJ, Dou F, Cai D, Veach D, Jean S, Li Y, Bornmann WG, Clarkson B, Xu H, Greengard P. Proc Natl Acad Sci USA. 2003;100:12444–12449. doi: 10.1073/pnas.1534745100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barten DM, Meredith JE, Jr, Zaczek R, Houston JG, Albright CF. Drugs R D. 2006;7:87–97. doi: 10.2165/00126839-200607020-00003. [DOI] [PubMed] [Google Scholar]
- 30.Buxbaum JD, Gandy SE, Cicchetti P, Ehrlich ME, Czernik AJ, Fracasso RP, Ramabhadran TV, Unterbeck AJ, Greengard P. Proc Natl Acad Sci USA. 1990;87:6003–6006. doi: 10.1073/pnas.87.15.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desdouits-Magnen J, Desdouits F, Takeda S, Syu LJ, Saltiel AR, Buxbaum JD, Czernik AJ, Nairn AC, Greengard P. J Neurochem. 1998;70:524–530. doi: 10.1046/j.1471-4159.1998.70020524.x. [DOI] [PubMed] [Google Scholar]
- 32.Cruz JC, Kim D, Moy LY, Dobbin MM, Sun X, Bronson RT, Tsai LH. J Neurosci. 2006;26:10536–10541. doi: 10.1523/JNEUROSCI.3133-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desdouits F, Buxbaum JD, Desdouits-Magnen J, Nairn AC, Greengard P. J Biol Chem. 1996;271:24670–24674. doi: 10.1074/jbc.271.40.24670. [DOI] [PubMed] [Google Scholar]
- 34.Wolfe MS. Curr Top Med Chem. 2002;2:371–383. doi: 10.2174/1568026024607535. [DOI] [PubMed] [Google Scholar]
- 35.Jenkinson EJ, Jenkinson WE, Rossi SW, Anderson G. Nat Rev Immunol. 2006;6:551–555. doi: 10.1038/nri1883. [DOI] [PubMed] [Google Scholar]
- 36.Lai MT, et al. J Biol Chem. 2003;278:22475–22481. doi: 10.1074/jbc.M300974200. [DOI] [PubMed] [Google Scholar]
- 37.Xu H, Gouras GK, Greenfield JP, Vincent B, Naslund J, Mazzarelli L, Fried G, Jovanovic JN, Seeger M, Relkin NR, Liao F, Checler F, Buxbaum JD, Chait BT, Thinakaran G, Sisodia SS, Wang R, Greengard P, Gandy S. Nat Med. 1998;4:447–451. doi: 10.1038/nm0498-447. [DOI] [PubMed] [Google Scholar]
- 38.Kopan R, Schroeter EH, Weintraub H, Nye JS. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]