Abstract
X-ray structures of bovine heart cytochrome c oxidase have suggested that the enzyme, which reduces O2 in a process coupled with a proton pumping process, contains a proton pumping pathway (H-pathway) composed of a hydrogen bond network and a water channel located in tandem across the enzyme. The hydrogen bond network includes the peptide bond between Tyr-440 and Ser-441, which could facilitate unidirectional proton transfer. Replacement of a possible proton-ejecting aspartate (Asp-51) at one end of the H-pathway with asparagine, using a stable bovine gene expression system, abolishes the proton pumping activity without influencing the O2 reduction function. Blockage of either the water channel by a double mutation (Val386Leu and Met390Trp) or proton transfer through the peptide by a Ser441Pro mutation was found to abolish the proton pumping activity without impairment of the O2 reduction activity. These results significantly strengthen the proposal that H-pathway is involved in proton pumping.
Keywords: mutagenesis, mitochondrial import, HeLa cell, peptide bond, keto-enol tautomerism
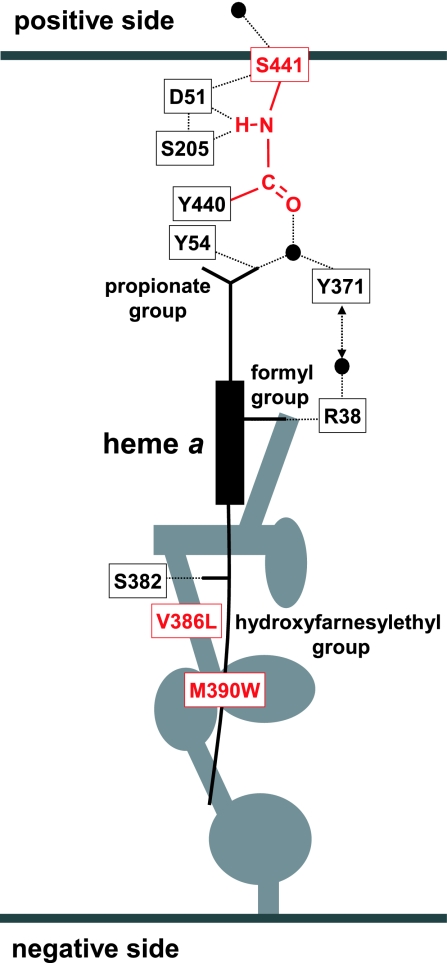
The mechanism of the proton pumping process of cytochrome c oxidase (CcO) is one of the most intriguing research subjects in the field of bioenergetics. Three possible proton transfer pathways (K-, D-, and H-pathways) have been identified in the x-ray structures of bovine and bacterial CcOs at high resolution (1–7). The K- and D-pathways connect the O2 reduction site with the inner space of the mitochondrial membrane (negative side space), whereas the H-pathway, which is composed of a hydrogen bond network and a water channel, extends across the enzyme from the negative side surface to the surface facing the outside space of mitochondrial membrane (positive side space) (Fig. 1).
Fig. 1.
Schematic representation of the H-pathway of oxidized bovine heart CcO. Hydrogen bond network extends from Arg-38 to Asp-51 including a peptide bond between Tyr-440 and Ser-441. The dotted line represents the hydrogen bond. The water channel (represented by the gray area) allows access of water molecules in the negative side space to the formyl group of heme a, which is hydrogen bonded to Arg-38. Heme a also interacts with the H-pathway via the other hydrogen bond between the propionate group and water (represented by the black sphere). Mutation sites are highlighted in red.
X-ray structures of bovine heart CcO have been determined at 1.8 Å resolution for the oxidized state and 1.9 Å for the reduced state (6). These structures indicate that the H-pathway contains structural elements that are sufficient to drive the proton pumping function (6). Asp-51 undergoes a redox-coupled conformational change near the positive side surface of the enzyme which could eject protons transferred from the negative side surface via the hydrogen bond network. The peptide bond between Tyr-440 and Ser-441 in the hydrogen bond network could induce unidirectionality to the process of proton transfer through the hydrogen bond network (6). Protons could be transferred through a peptide bond via an imidic acid intermediate ( C(OH)
C(OH) N+H
N+H ) formed upon protonation of the peptide bond (8). The imidic acid intermediate provides the enol form of the peptide (
) formed upon protonation of the peptide bond (8). The imidic acid intermediate provides the enol form of the peptide ( C(OH)
C(OH) N
N ) after removal of the proton at the nitrogen atom. The enol form is then tautomerized to the keto form (
) after removal of the proton at the nitrogen atom. The enol form is then tautomerized to the keto form ( CO
CO NH
NH ) because the enol form is less stable than the keto form. The difference in the stability of the two tautomers provides unidirectionality to the process of proton transfer through the peptide bond. The low spin heme (heme a) participates in the hydrogen bond network via two peripheral substituents, the propionate group and the formyl group (Fig. 1). In this arrangement, protons from the water channel could be actively transported through the hydrogen bond network by the net positive charge created upon oxidation of heme a and delocalized to these peripheral groups (6). A redox-coupled conformational change in the water-channel could promote collection of protons from the negative side space (6).
) because the enol form is less stable than the keto form. The difference in the stability of the two tautomers provides unidirectionality to the process of proton transfer through the peptide bond. The low spin heme (heme a) participates in the hydrogen bond network via two peripheral substituents, the propionate group and the formyl group (Fig. 1). In this arrangement, protons from the water channel could be actively transported through the hydrogen bond network by the net positive charge created upon oxidation of heme a and delocalized to these peripheral groups (6). A redox-coupled conformational change in the water-channel could promote collection of protons from the negative side space (6).
In our previous paper, the function of one of the key residues, Asp-51, was examined by investigation of an Asp51Asn mutant. The mutation was found to abolish the proton pumping function without impairment of the O2 reduction activity (6). However, we cannot exclude the possibility that the mutation disrupts the conformation of an alternative proton pumping pathway to abolish the proton pumping function. To evaluate this possibility, we examined two additional key structural features of the H-pathway; the Tyr-440–Ser-441 peptide bond and the water channel. The mutant enzymes had no proton pumping activity and retained full electron transfer activity, demonstrating the functions of these key structures, proposed by the x-ray structural analyses (6).
Results
Evaluation of Potential Conformational Changes Induced by H-Pathway Mutations.
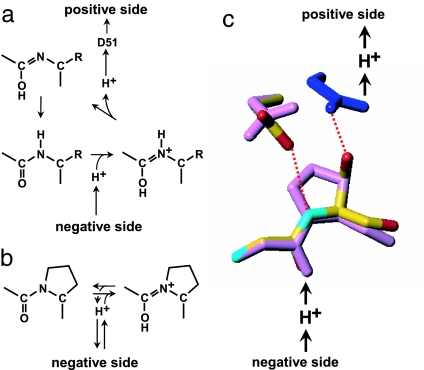
The Ser441Pro mutation was designed to prevent formation of an imidic acid intermediate with a dissociable proton at the peptide nitrogen. This mutation provides a means to evaluate the likelihood of proton transfer occurring through the peptide bond (Fig. 2 a and b). However, proline substitution often induces additional conformational changes that complicate the interpretation of the effect of the substitutions.
Fig. 2.
Proton transfer through the peptide bond between Tyr-440 and Ser-441. (a) Mechanism of proton transfer through the peptide bond. Protonation of the oxygen of the peptide bond by proton from the negative side yields the imidic acid intermediate ( C(OH)
C(OH) N+H
N+H ) (8), followed by removal of the imidic acid proton by D51 to form the enol tautomer of the peptide (
) (8), followed by removal of the imidic acid proton by D51 to form the enol tautomer of the peptide ( C(OH)
C(OH) N
N ) (6). The enol form is then tautomerized back to the keto form (
) (6). The enol form is then tautomerized back to the keto form ( CO-NH
CO-NH ) because the latter is more stable. (b) Protonation of the peptide bond by proton from the negative side in the Ser441Pro mutant. (c) Prediction of the conformation of the Ser441Pro mutant. The purple structures show the conformational changes of the oxidized form induced by the Ser441Pro mutation as predicted by X-PLOR analysis. The dark blue structure denotes the conformational change occurring upon reduction of the enzyme, wherein no influence by the mutation is detectable. The red dotted lines indicate hydrogen bonds, and the red, yellow, and light blue structures denote oxygen, carbon, and nitrogen atoms, respectively.
) because the latter is more stable. (b) Protonation of the peptide bond by proton from the negative side in the Ser441Pro mutant. (c) Prediction of the conformation of the Ser441Pro mutant. The purple structures show the conformational changes of the oxidized form induced by the Ser441Pro mutation as predicted by X-PLOR analysis. The dark blue structure denotes the conformational change occurring upon reduction of the enzyme, wherein no influence by the mutation is detectable. The red dotted lines indicate hydrogen bonds, and the red, yellow, and light blue structures denote oxygen, carbon, and nitrogen atoms, respectively.
A prediction of the structural effects arising from the Ser441Pro mutation was performed based on x-ray structures of bovine heart CcO. The dihedral angle, φ, of Ser-441 was found to be −84° in the x-ray structure of the oxidized form at 1.8 Å resolution (6), which is within the allowable angle of a proline residue (−100° < φ < −50°) (9). The mutant structure was predicted by energy minimization using the program X-PLOR (Fig. 2c). The main chain structure of Pro-441 converged to (φ = −64° and ϕ = −31°) in the favored region (9). The side chain of Asp-51 in the oxidized form was found to be rotated about the Cβ Cγ bond to reduce the repulsive energy between Oδ1 and Oδ2 of Asp-51 and Cγ and Cδ of Pro-441, whereas the conformation of the rest of the protein environment was found to be identical to that of the native structure. Additional significant structural changes arising as a result of the mutation are not predicted for either the oxidized or reduced forms (Fig. 2c).
Cγ bond to reduce the repulsive energy between Oδ1 and Oδ2 of Asp-51 and Cγ and Cδ of Pro-441, whereas the conformation of the rest of the protein environment was found to be identical to that of the native structure. Additional significant structural changes arising as a result of the mutation are not predicted for either the oxidized or reduced forms (Fig. 2c).
An energy minimization analysis for a double mutation of selected residues in the water channel (Val386Leu/Met390Trp) (Fig. 1) indicated the incidence of complete blockage of the water channel without the occurrence of significant conformational changes in the rest of the protein.
Expression and Function of the Mutant Enzymes.
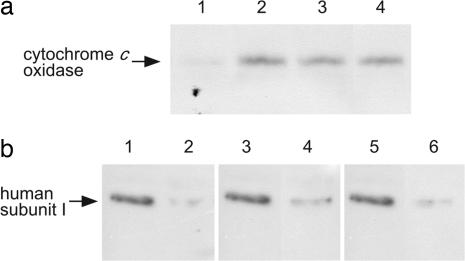
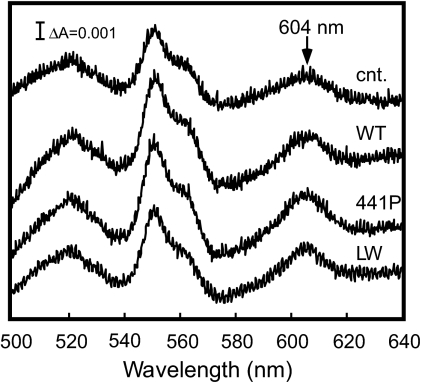
Dodecylmaltoside-solubilized mitochondrial proteins from stable transfectants of HeLa cells expressing bovine subunit I were fractionated by blue native PAGE (10). The presence of bovine subunit I in the CcO fractions was demonstrated at 210 kDa by immunoblot analyses with antibody specific to bovine subunit I (Fig. 3a). The analyses clearly indicated that the wild type as well as the mutant subunit I formed hybrid enzymes with the remaining 12 human subunits. Immunoblot analyses of the mitochondrial proteins fractionated by SDS/PAGE using an antibody specific to the human subunit I showed that subunit I fraction from the HeLa cells transfected with the gene encoding subunit I of bovine wild type or mutant contained ≈20% of the residual human subunit I (Fig. 3b). These results indicate that the hybrid enzymes were dominantly expressed over the endogenous human enzyme. The observation of dominant expression of the hybrid enzymes is consistent with previous results (6). Both mutant enzymes exhibited normal absorption spectra (Fig. 4), indicating that the mutations have minimal effects on the enzyme conformation. This work confirmed the predictions made on the basis of energy minimization analyses of the mutant described in the previous section.
Fig. 3.
Immunoblot analyses of CcO of HeLa cells. (a) Immunoblot analyses of CcO fractionated by blue-native PAGE of solubilized mitochondrial fractions. Solubilized mitochondria (70 μg) from HeLa cells harboring the expression vector without the bovine subunit I gene (lane 1) and with that of wild type (lane 2), Ser441Pro (lane 3), and Val386Leu/Met390Trp (lane 4) were fractionated on blue native PAGE (10). CcO complex electrophoresed at 210 kDa was demonstrated by immunoblot analysis to be positive for the bovine subunit I specific antibody (6), indicating that the bovine subunit I formed the hybrid enzyme with the other human subunits. (b) Immunoblot analysis of human subunit I of CcO. Dodecylmaltoside (1.4%)-solubilized CcO (2.5 pmol) from mitochondrial preparations was fractionated by SDS/PAGE. Mitochondrial samples were obtained from HeLa cells harboring the expression vector without the bovine subunit I gene (lanes 1, 3, and 5), and with that of wild type (lane 2), Ser441Pro (lane 4) and Val386Leu/Met390Trp (lane 6). Human subunit I was detected by immunoblot analysis using the specific antibody (6). When bovine subunit I was expressed simultaneously, the intensity of the subunit I band detected by human subunit I antibody was ≈20% that of the pure human subunit I, indicating that ≈80% of CcO contained bovine subunit I.
Fig. 4.
Absorption spectra of mitochondrial preparations. Visible difference absorption spectra of samples before and after treatment with a slight excess of dithionite. The mitochondrial preparations were isolated from HeLa cells harboring the expression vectors without the bovine heart cytochrome c subunit I gene (cnt.) and with that of the wild type (wt), Ser441Pro (441P), and Val386Leu/Met390Trp double mutant (LW). The mitochondrial preparations were treated with 1.4% dodecylmaltoside before obtaining absorption measurements. The protein concentration for each measurement was ≈1 mg/ml in 100 mM HEPES-KOH buffer, pH 7.0, containing 0.14% dodecylmaltoside in a 1-cm cuvette.
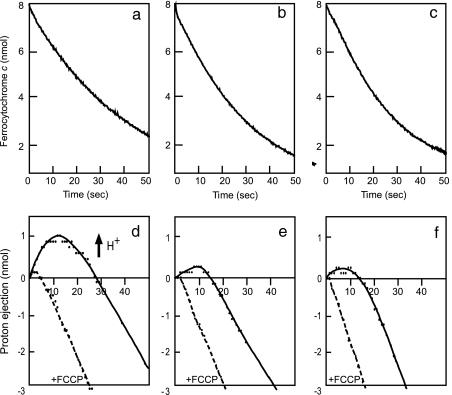
The wild-type enzyme exhibited rapid proton ejection after initiation of the reaction (Fig. 5d), which was quenched by addition of carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), a proton ionophore. The initial rates of proton ejection and ferrocytochrome c oxidation measured in four to five independent experiments showed that the ratio of pumped protons to electrons transferred (H+/e−) is 0.6–0.8. This finding indicates efficient coupling of proton ejection with electron transfer in the wild-type enzyme. The mutants exhibited normal ferrocytochrome c oxidation (Fig. 5 b and c), with inefficient proton ejection (Fig. 5 e and f). The observation of weak proton ejections for both enzymes is most likely due to the presence of residual endogenous human enzyme in the mitochondria, because the proton ejection rate relative to that of the wild-type enzyme is consistent with the amount of residual endogenous enzyme relative to the total enzyme content determined by the immunoblot analyses as described above. These results indicate that these two mutants have essentially no proton pumping activity. No significant differences in the specific electron transfer activities (electron transfer rate/enzyme molecule, each averaged for several assays) were detected between the mutants and the wild-type enzyme, suggesting that the mutations have an inhibitory effect on electron transfer activity.
Fig. 5.
Ferrocytochrome c oxidation and proton pumping of bovine/human hybrid CcO. The experimental conditions are described in Materials and Methods. (a–c) Ferrocytochrome c oxidation monitored at 550 nm, the absorption peak characteristics of cytochrome c in the ferrous state. (d–f) Proton ejection from mitoplasts by CcO after addition of ferrocytochrome c. The hybrid enzymes investigated were wild type (a and d), Ser441Pro mutant (b and e), and Val386Leu/Met390Trp double mutant (c and f). Proton ejection by the wild-type hybrid enzyme (d) in the initial 15 s after addition of ferrocytochrome c is followed by a decrease in proton concentration due to the proton back-leak into the matrix space. Weak acidification in the initial 10 s for the Ser441Pro mutant (e) and the double mutant (f) is induced by 20% residual human CcO. +FCCP denotes the proton ejection trace obtained in the presence of FCCP, a proton ionophore. The results of the two mutant enzymes in (b, c, e, and f) were obtained at the enzyme concentration 20% higher than those of the wild-type enzyme.
The mutants exhibit proton uptake rates (due to the O2 reduction by the mutant enzymes after the initial burst of the proton ejection by the residual human enzymes) that are significantly slower than the proton uptake rates in the presence of FCCP (Fig. 5 e and f). This acceleration of the proton uptake by FCCP suggests that, for the mutants, the abolishment of the proton pumping process is not due to the enhancement of passive proton leakage caused by the artificial subunit assembly conditions, if any. Furthermore, if the increase in passive proton conductivity occurred for the mutants, similar conductivity increase would also occur in the subunit assembly that includes the wild-type subunit I, resulting in elimination of the proton ejection.
Discussion
A Stable Expression System for the Subunit I Gene.
Although the hybrid enzyme has not been isolated from the mitochondrial preparation, the formation and integrity of the hybrid enzyme complex have been well established by various methods including immunochemical analyses, proton pumping, and electron transfer activity measurements and absorption spectral analyses (6). The possibility of occurrence of homologous recombination during the stable transfection of the bovine subunit I gene to HeLa cells is negligible [see supporting information (SI) Text].
The mitochondrial codons for Met and Trp (ATA and TGA) in the bovine heart subunit I gene were exchanged for the universal codons (ATG and TGG), respectively, before incorporation of the gene into the vector. Transfection of HeLa cells with the wild-type subunit I gene produced the hybrid enzyme which was indistinguishable from the human nonhybrid enzyme in terms of the enzymatic activities and the absorption spectra (6). This result shows that the codon exchange procedure does not influence the function of the expressed enzyme.
Proton Pumping Mechanism of Bovine Heart CcO.
Examination of x-ray structures of the fully oxidized and reduced states of bovine heart CcO has led to the proposal that protons are pumped through the H-pathway driven by heme a (6). This proposal has been confirmed by analyses of three mutants. The Ser441Pro mutant blocks irreversible proton transfer through the peptide bond. The Val386Leu/Met390Trp double mutant blocks the water transfer through the water channel. The Asp51Asn mutant blocks the proton pumping site, as reported previously. It is unlikely that each of these three different mutations destroys an alternative proton pumping system other than the H-pathway to give the identical phenotype (i.e., complete abolishment of the proton pumping without having a significant inhibitory influence on the electron transfer activity). These analyses demonstrate that animal CcOs that include the H-pathway with Asp-51 pump protons by H-pathway, whereas O2 is reduced at the heme a3 CuB dinuclear O2 reduction site. The K- and D-pathways connecting the negative side surface with the O2 reduction site are only used to transfer protons used for water formation.
CuB dinuclear O2 reduction site. The K- and D-pathways connecting the negative side surface with the O2 reduction site are only used to transfer protons used for water formation.
It should be noted that the proton pumping pathway is separated from the system for O2 reduction. This separation is indispensable for avoiding leakage of protons used in the proton pumping into the pathway used to transfer protons used for formation of water. Such leakage would result in dissipation of the energy for proton pumping (11).
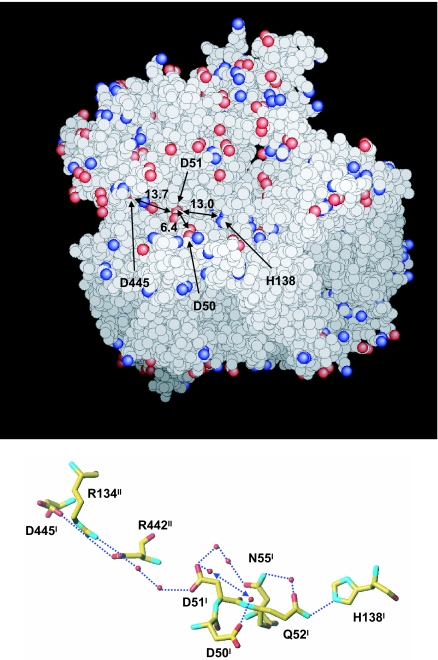
It has been reported that, upon oxidation of heme a, protons are released from the bovine heart CcO to the positive side, inconsistent with the redox coupled x-ray structural changes in Asp-51 (12–14). However, x-ray structures of the bovine heart enzyme in the oxidized and reduced states at 1.8 and 1.9 Å resolutions suggest the existence of reversible proton accepting system (Fig. 6). Two proton-accepting groups, His-138 and Asp-445, are located 13–14 Å apart from Asp-51 in the reduced state, each connected with Asp-51 with a hydrogen bond network. The carboxyl group of Asp-445 is partially buried inside the protein, where its proton affinity would be higher than in aqueous solution. These two functional groups would be expected to retain protons under physiological conditions. Asp-50, which is exposed to the bulk water phase, is also connected to Asp-51 via two fixed water molecules as shown in Fig. 6. These two negatively charged and closely located (≈6 Å distance) carboxyl groups are capable of trapping protons. These high-resolution x-ray structures near Asp-51 site suggest that the protons released from Asp-51 upon reduction are not readily released to the bulk water phase of the external phase, but are trapped and distributed over these amino acids and the hydrogen bond networks under the negative charge influence of the deprotonated Asp-51. Elimination of the negative charge influence from Asp-51 upon oxidation would decrease the overall proton affinity of the surface structure given in Fig. 6 to release protons to the external phase. Extensive infrared analyses for these structures would evaluate this proposal based on the x-ray structures.
Fig. 6.
Possible proton acceptor sites near Asp-51 in bovine heart CcO. (Upper) Space filling model of bovine heart CcO at 1.9 Å resolution in the reduced state. Red and blue spheres represent oxygen and nitrogen atoms of acidic and basic amino acid side chains. Asp-51 and three possible proton acceptors (H138, D50, and D445) from Asp-51 are indicated by arrows. The distances from the Asp-51 carboxyl group are indicated by the digits in Å unit on the double-headed arrows. (Lower) Hydrogen bond network connecting Asp-51 with the possible proton accepting sites (H138, D50, and D445). Dotted lines denote hydrogen bonds. The double headed dotted arrow shows a possible pathway where either one of the two water molecules can migrate to the other water molecule within the hydrogen-bond distance for proton transfer.
Experimental Results in Support of Involvement of the D-Pathway in the Proton Pumping Mechanism.
An alternative proton pumping mechanism in which D-pathway transfers both pumping and water-forming protons has been proposed (15). All of the critical experimental results supporting this mechanism should be discussed in light of the currently proposed H-pathway mechanism.
Effects of K- and D-pathway mutations on the F-to-O transitions.
During the complete oxidation process of the fully reduced CcO (R) with the excess molecular oxygen, three intermediate species, denoted A, P, and F, are detectable before the final fully oxidized species, O, appears. The initial intermediate, A, is the oxygenated species (Fea3 O2), whereas P and F represent two types of ferryl species (Fea3
O2), whereas P and F represent two types of ferryl species (Fea3 O). One electron equivalent is necessary for the transition from F to O.
O). One electron equivalent is necessary for the transition from F to O.
The effects of K- and D-pathway mutations on the F-to-O transition were examined by a combined technique of the flash-induced reduction and time-resolved electrometric monitoring of the membrane potential. A D-pathway mutant, Asp132Asn of Rhodobacter sphaeroides shows no significant electrogenic proton transfer, whereas K-pathway mutations (Lys362Met and Thr359Ala) have virtually no effect on the transition (16). It has been established that the F-to-O transition is coupled with transfer of one equivalent each of electrons and protons used in the proton pumping process and for water-formation (17). These results have been often referred as experimental evidence showing that both protons used in the pumping and water-formation protons are transferred through the D-pathway. However, the results simply indicate that the three processes, (i) the proton transfer through the D-pathway, (ii) the electron transfer to heme a3, and (iii) the proton pumping are tightly coupled with each other. These results do not disprove the presence of the proton pump system apart from the D-pathway.
Effects of D-pathway mutations on the steady state turnover reactions of CcO.
A D-pathway mutant, Asn139Asp (R. sphaeroides) has no proton pumping activity, but exhibits significantly higher electron transfer activity than that of the wild type enzyme (18). Similar results were reported for CcO of Paracoccus denitrificans (19). The proton pumping efficiency (H+/e−) is restored by introduction of the Asp132Asn mutation into the Asn139Asp mutant enzyme, although, its electron transfer activity is ≈20% of that of the wild type enzyme (20). The Asp132Asn/Asn139Asp double mutant enzyme pumps four protons per molecule of O2 reduced. The decoupling and recoupling by these mutations are correlated with the change in the pKa of Glu-286 (20). It has been proposed that Glu-286 (R. sphaeroides), located at the end of D-pathway, is the branching point for the pathways to the O2 reduction site and to the proton pumping site including the propionates of the two hemes (17). The pKa of Glu-286 was assigned as the pKa value determined from the pH dependency of the P-to-F transition rate, assuming that the P-to-F transition is controlled by the protonation state of Glu-286 (21). The pKa of the P-to-F transition of Asn139Asp mutant is ≈1.6 pH units higher than that of the wild type (pKa = 9.4) (21). The double mutant exhibits about the same pKa value for the P-to-F transition as that of the wild type enzyme. Assuming that the pKa values of the proton pumping site and the O2 reduction site are ≈10 and 12, respectively, Glu-286 of the Asn139Asp mutant enzyme, which has a pKa of 11, would be unable to protonate the proton pumping site but could transfer protons to the O2 reduction site, which has a pKa higher than that of Glu-286 (21). Thus, proton pump is decoupled from the electron transfer.
However, the identity of the functional group, which controls the P-to-F transition rate to provide the pKa value of 9.4, has not been deduced from experimental results. Thus, the following alternative interpretation is equally possible for these results: the P-to-F transition is controlled by Arg-52 (R. sphaeroides number, equivalent to bovine Arg-38), which is located at one end of the hydrogen bond network in the H-pathway and forms a hydrogen bond with the formyl group of heme a. This is one of the trigger sites for active proton transport through the hydrogen bond network. The proton affinity of a guanidino group is lower within a hydrophobic environment. Thus, assignment of a pKa of 9.4 for Arg-52 is reasonable, considering the environment of Arg-38 of bovine CcO indicated in the x-ray structure (6). Thus, a conformational change in the guanidino group of Arg-52 arising from the Asn139Asp mutation could weaken the hydrogen bond between the formyl group and the guanidino group to increase the pKa value. Both the increased pKa and the weakened hydrogen bond between the formyl group and Arg-52 would significantly decrease the efficiency of the coupling between the proton pumping through the H-pathway and the electron transfer through heme a. Therefore, these mutational results are not in conflict with the H-pathway mechanism. Further discussions on the K-pathway mutation and the possibility of the proton transfer through a water exit pathway from the O2 reduction site are provided in SI Text.
Mutational analyses of bacterial H-pathway.
CcOs from P. denitrificans and R. sphaeroides have possible proton transfer pathways that are very similar to the H-pathway of bovine CcO (22, 23). One notable exception is that the analogous location of Asp-51 of the bovine CcO has a glycine residue and one or two water molecules. Mutations of residues of the H-pathways of the bacterial CcOs have been examined to evaluate the function of the H-pathway (22, 23). The Arg52Met mutant exhibits only a small percentage of the electron transfer activity of the wild-type enzyme while retaining proton pumping efficiency (H+/e−) of approximate unity (24). It has been proposed that the normal proton pumping efficiency of the mutant enzyme, in which proton transfer through the H-pathway appears to be blocked, argues against the mechanism where transfer of the pumping protons occurs through the H-pathway. However, the methionine thioether is capable of forming hydrogen bonds with two hydrogen donor groups to provide a proton transfer pathway. In fact, both the K- and D-pathways of bovine heart CcO include methionine thioesters of Met-273 and Met-71, respectively (6). Therefore, the mutation would not disrupt the hydrogen bond network of the H-pathway. Methionine cannot form a hydrogen bond with the formyl group of heme a. However, if the methionine is located close enough to the formyl group, proton transfer through the methionine would be accelerated by the delocalized positive charge on the formyl group (i.e., the net electron density decrease in the formyl group). Therefore, these results do not disprove the proposal that proton pumping occurs through the H-pathway.
Tyr-414 (R. sphaeroides, equivalent to bovine Tyr-371) of the hydrogen bond network of the H-pathway (Fig. 1) can be replaced by Phe without causing significant effects on electron transfer and proton transfer activities (23). However, X-PLOR analyses of this mutant indicated that a fixed water molecule is introduced at the site of the phenol OH group of Tyr-371 and thus an essentially identical hydrogen bond network is retained. Several amino acid residues of water channels were replaced with less bulky amino acids. These mutations would not be expected to block transfer of water molecules through the channel and this interpretation is consistent with the finding that H+/e− ratios of these mutant CcOs are normal (22, 23).
A double mutation of amino acid residues contributing to the water channel of the bacterial enzyme (Val421Leu/Phe425Trp) did not significantly affect the proton pumping efficiency (25). The mutation points in the amino acid sequence correspond to those of the double mutation of the bovine enzyme as given in Results. X-ray structures of the bacterial water channel indicate that the Phe425Trp mutation would expand the channel instead of blocking it. The x-ray structures of the bovine and bacterial enzymes indicate that the structure of the water channel has significant diversity.
The investigation of mutations of the bacterial H-pathway thus far does not provide conclusive evidence against the H-pathway mechanism.
Diversity of the H-pathway.
Asp-51 is not conserved. Plant and bacterial CcOs do not have it. Some bacterial CcOs do not have the H-pathway. Furthermore, some bacterial CcOs have heme b instead of heme a (26). The diversity of the H-pathway clearly indicates that the proton pumping pathways of plant and bacterial CcOs are not identical to that of the animal CcOs. However, the complete conservation of the low spin heme suggests that proton pump is driven by the low spin heme also in the plant and bacterial CcOs, although not in the same fashion as that of the animal CcOs. In fact, it has been well known that different sets of amino acids could drive an identical physiological function (26). On the other hand, the O2 reduction system, which includes the O2 reduction site and the two proton transfer pathways (D and K), is well conserved. It seems unlikely that the O2 reduction system pumps protons in plant and bacterial CcOs but not in the animal CcOs. Therefore, the possibility of the proton pumping driven by the low spin heme needs to be reexamined for bacterial CcOs. A more detailed discussion is given on physiological relevance of the diversity of H-pathway in SI Text.
Many theoretical analyses have been conducted to examine how the structures of the D-pathway and the O2 reduction site in the enzyme are able to pump protons (27–32). Experimental evaluations of these theoretical proposals have not yet been provided.
Conclusion
The three mutations in the H-pathway of bovine heart CcO clearly show that proton pumping is driven by heme a through H-pathway in animal CcOs. Although the low spin heme is completely conserved, further structural and functional studies on the bacterial enzymes, especially x-ray structural analyses at the resolution at least as high as those of the bovine enzyme, are required for evaluation of the proposal for the proton pumping system separated from the O2 reduction system and driven by the low spin heme. However, we cannot exclude the possibility that the proton pumping driven by the low spin heme occurs only in the animal enzyme. Direct observation of proton transfer through the key structures of these pathways by time-resolved infrared spectroscopy is expected to provide the means of conclusively identifying the pathway. The possibility that the low spin heme functions as the driving element of the proton pumping process has been proposed on several occasions back to 1978 (33–36).
Materials and Methods
Stable Transfectants of HeLa Cells.
The subunit I gene was modified to encode subunit I protein with an N-terminal presequence identical to that of the nuclear encoded subunit IV of the bovine enzyme, which is targeted to mitochondria. The processes of transfection of HeLa cells with the expression vectors and selection of the stable transfectants were also performed according to previously described procedures (6).
Measurements of Enzymatic Activities.
The catalytic activities of wild type and mutant hybrid enzymes were measured by using mitoplast samples prepared in hypotonic buffer. Ferrocytochrome c oxidation by CcO was measured at 15°C, monitoring absorbance changes at 550 nm on a PerkinElmer UV-Visible spectrophotometer lambda 18 (Shelton, CT), equipped with a temperature controlled cell holder. Measurements of proton ejection and uptake by the mitoplast samples were taken at 15°C according to described procedures (6).
Supplementary Material
Acknowledgments
This work was supported by Grant-in-Aid for Scientific Research on Priority Areas 16087206 (to T.T.), 16087208 (to S.Y.), and 18054030 (to H.S.), and 21st Century Center of Excellence Program (to T.T. and S.Y.) from MEXT; and Creative Scientific Research 17GS0419 (to M.S.) from JSPS; Japan Biological Informatics Consortium (to T.T. and H.S); CREST (to S.Y.); and Keio University (to H.S). H.M. is a research fellow supported by Leading Project for Biosimulation. S.Y. is a Senior Visiting Scientist in RIKEN Harima Institute.
Abbreviations
- CcO
cytochrome c oxidase
- FCCP
carbonyl cyanide p-trifluoromethoxyphenylhydrazone.
Footnotes
The authors declare no conflict of interest.
Preliminary results were presented at the 13th European Bioenergetics Conference, Pisa, Italy, August 2004.
Preliminary results were presented at the 13th European Bioenergetics Conference, Pisa, Italy, August 2004.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611627104/DC1.
References
- 1.Iwata S, Ostermeier C, Ludwig B, Michel H. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 2.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 3.Ostermeier C, Harrenga A, Ermler U, Michel H. Proc Natl Acad Sci USA. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshikawa S, Shinzawa-Itoh K, Nakashima R, Yaono R, Yamashita E, Inoue N, Yao M, Fei MJ, Libeu CP, Mizushima T, et al. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 5.Svensson-Ek M, Abramson J, Larsson G, Tornroth S, Brzezinski P, Iwata S. J Mol Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 6.Tsukihara T, Shimokata K, Katayama Y, Shimada H, Muramoto K, Aoyama H, Mochizuki M, Shinzawa-Itoh K, Yamashita E, Yao M, et al. Proc Natl Acad Sci USA. 2003;100:15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Proc Natl Acad Sci USA. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perrin CL. Acc Chem Res. 1989;22:268–275. [Google Scholar]
- 9.Lovell SC, Davis IW, Arendall WB, III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 10.Schagger H, von Jagow G. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 11.Williams RJP. Nature. 1995;376:643. doi: 10.1038/376643a0. [DOI] [PubMed] [Google Scholar]
- 12.Capitanio N, Capitanio G, Minuto M, De Nitto E, Palese LL, Nicholls P, Papa S. Biochemistry. 2000;39:6373–6379. doi: 10.1021/bi0003137. [DOI] [PubMed] [Google Scholar]
- 13.Capitanio N, Capitanio G, Boffoli D, Papa S. Biochemistry. 2000;39:15454–15461. doi: 10.1021/bi001940z. [DOI] [PubMed] [Google Scholar]
- 14.Forte E, Barone MC, Brunori M, Sarti P, Giuffre A. Biochemistry. 2002;41:13046–13052. doi: 10.1021/bi025917k. [DOI] [PubMed] [Google Scholar]
- 15.Gennis RB. Front Biosci. 2004;9:581–591. doi: 10.2741/1237. [DOI] [PubMed] [Google Scholar]
- 16.Konstantinov AA, Siletsky S, Mitchell D, Kaulen A, Gennis RB. Proc Natl Acad Sci USA. 1997;94:9085–9090. doi: 10.1073/pnas.94.17.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brzezinski P, Larsson G. Biochim Biophys Acta. 2003;1605:1–13. doi: 10.1016/s0005-2728(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 18.Pawate AS, Morgan J, Namslauer A, Mills D, Brzezinski P, Ferguson-Miller S, Gennis RB. Biochemistry. 2002;41:13417–13423. doi: 10.1021/bi026582+. [DOI] [PubMed] [Google Scholar]
- 19.Pfitzner U, Hoffmeier K, Harrenga A, Kannt A, Michel H, Bamberg E, Richter OM, Ludwig B. Biochemistry. 2000;39:6756–6762. doi: 10.1021/bi992235x. [DOI] [PubMed] [Google Scholar]
- 20.Branden G, Pawate AS, Gennis RB, Brzezinski P. Proc Natl Acad Sci USA. 2006;103:317–322. doi: 10.1073/pnas.0507734103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Namslauer A, Pawate AS, Gennis RB, Brzezinski P. Proc Natl Acad Sci USA. 2003;100:15543–15547. doi: 10.1073/pnas.2432106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfitzner U, Odenwald A, Ostermann T, Weingard L, Ludwig B, Richter OM. J Bionenerg Biomembr. 1998;30:89–97. doi: 10.1023/a:1020515713103. [DOI] [PubMed] [Google Scholar]
- 23.Lee H-M, Das TK, Rousseau DL, Mills D, Ferguson-Miller S, Gennis RB. Biochemistry. 2000;39:2989–2996. doi: 10.1021/bi9924821. [DOI] [PubMed] [Google Scholar]
- 24.Jasaitis A, Backgren C, Morgan JE, Puustinen A, Verkhovsky MI, Wikström M. Biochemistry. 2001;40:5269–5274. doi: 10.1021/bi002948b. [DOI] [PubMed] [Google Scholar]
- 25.Salje J, Ludwig B, Richter OM. Biochem Soc Trans. 2005;33:829–831. doi: 10.1042/BST0330829. [DOI] [PubMed] [Google Scholar]
- 26.Pereira MM, Santana M, Teixeira M. Biochim Biophys Acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 27.Siegbahn PEM, Blomberg MRA, Blomberg ML. J Phys Chem B. 2003;107:10946–10955. [Google Scholar]
- 28.Popovic DM, Stuchebrukhov AA. J Am Chem Soc. 2004;126:1858–1871. doi: 10.1021/ja038267w. [DOI] [PubMed] [Google Scholar]
- 29.Olkhova E, Hutter MC, Lill MA, Helms V, Michel H. Biophys J. 2004;86:1873–1889. doi: 10.1016/S0006-3495(04)74254-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Voth GA. Proc Natl Acad Sci USA. 2005;102:6795–6800. doi: 10.1073/pnas.0408117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wikström M, Ribacka C, Molin M, Laakkonen L, Verkhovsky M, Puustinen A. Proc Natl Acad Sci USA. 2005;102:10478–10481. doi: 10.1073/pnas.0502873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olkhova E, Helms V, Michel H. Biophys J. 2005;89:2324–2331. doi: 10.1529/biophysj.105.062091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Artzatbanov VY, Konstantinov AA, Skulachev VP. FEBS Lett. 1978;87:180–185. doi: 10.1016/0014-5793(78)80327-5. [DOI] [PubMed] [Google Scholar]
- 34.Babcock GT, Callahan PM. Biochemistry. 1983;22:2314–2319. doi: 10.1021/bi00279a002. [DOI] [PubMed] [Google Scholar]
- 35.Rousseau DL, Sassaroli M, Ching YC, Dasgupta S. Ann NY Acad Sci. 1988;550:223–237. doi: 10.1111/j.1749-6632.1988.tb35338.x. [DOI] [PubMed] [Google Scholar]
- 36.Papa S, Capitanio N, Villani G. FEBS Lett. 1998;439:1–8. doi: 10.1016/s0014-5793(98)01305-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.