Abstract
The La Crosse Virus (LACV) M segment encodes two glycoproteins (Gn and Gc), and plays a critical role in the neuropathogenesis of LACV infection as the primary determinant of neuroinvasion. A recent study from our group demonstrated that the region comprising the membrane proximal two-thirds of Gc, amino acids 860–1442, is critical in mediating LACV fusion and entry. Furthermore, computational analysis identified structural similarities between a portion of this region, amino acids 970–1350, and the E1 fusion protein of two alphaviruses: Sindbis virus and Semliki Forrest virus (SFV). Within the region 970–1350, a 22 amino acid hydrophobic segment (1066–1087) is predicted to correlate structurally with the fusion peptides of class II fusion proteins. We performed site directed mutagenesis of key amino acids in this 22-amino acid segment and determined the functional consequences of these mutations on fusion and entry. Several mutations within this hydrophobic domain affected glycoprotein expression to some extent, but all mutations either shifted the pH threshold of fusion below that of the wild type protein, reduced fusion efficiency, or abrogated cell-to-cell fusion and pseudotype entry altogether. These results, coupled with the aforementioned computational modeling, suggest that the LACV Gc functions as a class II fusion protein and support a role for the region Gc 1066–1087 as a fusion peptide.
INTRODUCTION
The Bunyaviridae are a diverse family of RNA viruses divided into 5 genera (Orthobunyavirus, Hantavirus, Nairovirus, Phlebovirus, and Tospovirus). With the exception of viruses in the genus Hantavirus, most bunyaviruses are arthropod borne. Members of the Bunyaviridae cause human illnesses ranging from mild, asymptomatic infection to pulmonary disease, hemorrhagic fever, and fatal encephalitis (reviewed in (Soldan and Gonzalez-Scarano, 2005)); specifically, several of the orthobunyaviruses are associated with neurological complications including encephalitis and congenital CNS abnormalities (Soldan and Gonzalez-Scarano, 2005). Among these viruses, the best studied and the focus of this report is La Crosse Virus (LACV), a member of the California serogroup of orthobunyaviruses. LACV is an important cause of pediatric encephalitis and aseptic meningitis in the United States, chiefly in areas where its primary mosquito vector, Ochlerotatus (formerly, Aedes) triseriatus, resides (Thompson, Kalfayan, and Anslow, 1965).
The bunyavirus genome contains three single-stranded RNA segments of negative polarity, designated according to size as small (S), medium (M), and large (L). These segments are encapsidated by a nucleocapsid (N) protein to form helical ribonucleoprotein complexes (RNPs). The L RNA segment encodes the viral polymerase that is responsible for both transcription and replication of the genome (Roberts et al., 1995). The S RNA segment encodes the N protein and, for the orthobunyaviruses, tospoviruses, and phleboviruses, a non-structural protein (NSs) through an overlapping reading frame (orthobunyaviruses) or an ambisense coding mechanism (phleboviruses, tospoviruses). The M RNA segment encodes a single open reading frame (ORF) that is processed into the surface transmembrane glycoproteins Gn (formerly called G2) and Gc (formerly G1) and, for the orthobunyaviruses and some members of the genus Phlebovirus, a non-structural protein (NSm) of unknown function. The two surface transmembrane glycoproteins, Gn and Gc, associate as a heteromultimer after cleavage of the precursor polyprotein (Bupp, Stillmock, and Gonzalez-Scarano, 1996; Eshita and Bishop, 1984; Fazakerley et al., 1988). This Gc/Gn heteromultimer is targeted to the Golgi apparatus, the site of viral assembly and budding, by a localization signal encoded within Gn (Bertolotti-Ciarlet et al., 2005; Bupp, Stillmock, and Gonzalez-Scarano, 1996; Chen, Matsuoka, and Compans, 1991; Haferkamp et al., 2005; Lappin et al., 1994; Ruusala et al., 1992). LACV Gc, the larger of the two glycoproteins, is the exclusive target of neutralizing antibodies and plays a critical role in entry as it is the virus attachment protein (Gonzalez-Scarano et al., 1982; Grady et al., 1983; Pekosz et al., 1995a; Pekosz et al., 1995b). In addition, both Gc and a soluble form of Gc lacking a transmembrane domain form homomultimers and undergo a pH dependent conformational change that is associated with virus-cell or cell-to-cell fusion (Gonzalez-Scarano, 1985; Gonzalez-Scarano, Pobjecky, and Nathanson, 1984; Pekosz and Gonzalez-Scarano, 1996).
Previous studies using wild-type LACV and TAH181/57, a highly neurovirulent brain passaged strain of the closely related Tahyna virus with low neuroinvasiveness, mapped the neuroinvasive phenotype to the M segment. (Gonzalez-Scarano et al., 1982) (Janssen, Gonzalez-Scarano, and Nathanson, 1984). In complementary experiments, LACV monoclonal antibody escape variants V22 and V22F, generated with a monoclonal antibody against Gc (807–22), were associated with a more limited viremia and a decreased ability to mediate cell-to-cell fusion (Gonzalez-Scarano et al., 1985; Sundin et al., 1987). Further, V22/V22F mediated fusion occurred at a lower pH than that routinely observed for wild-type LACV, suggesting that Gc is the primary determinant of neuroinvasion and pH dependent fusion (Gonzalez-Scarano et al., 1985; Sundin et al., 1987). More recently, a panel of recombinant M segment constructs using LACV, TAH818/57, and the V22 variants were used to further delineate the molecular basis of cell-to-cell fusion and entry in California serogroup bunyaviruses (Plassmeyer et al., 2005). This study demonstrated that the region corresponding roughly to the membrane proximal two-thirds of Gc, amino acids 860–1442, is critical in mediating fusion and entry, (Plassmeyer et al., 2005). Computational analysis then identified structural similarities between the LACV Gc amino acid region 970–1350 and the E1 fusion protein of two alphaviruses: Sindbis virus and Semliki Forrest virus (SFV) (Zhang et al., 2002) (Plassmeyer et al., 2005). Collectively, these data suggested that the LACV Gc, like the alphavirus E1 and flavivirus E, functions as a class II fusion protein.
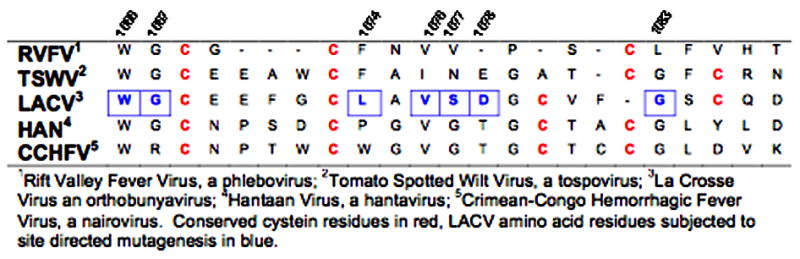
Within the 970–1350 region there is a 22 amino acid hydrophobic segment (1066–1087) that is predicted to correlate structurally with a hydrophobic domain of the Sindbis virus E1 (Garry and Garry, 2004; Plassmeyer et al., 2005). Although the LACV Gc and SFV E1 do not share significant amino acid sequence homology, there is a marked conservation of individual cysteine residues in these hydrophobic regions. The three conserved cysteine residues (Levy-Mintz and Kielian, 1991) of the SFV E1 fusion domain, two of which form disulfide bonds with an adjacent loop that helps maintain the loop structure, are critical for fusion activity (Lescar et al., 2001). The presence of an internal fusion loop within a disulfide-stabilized loop has also been demonstrated for the fusion peptide of Tick-Borne Encephalitis Virus (TBEV) (E) and of avian sarcoma-leukosis virus (ASLV) (Rey et al., 1995) (Delos, Gilbert, and White, 2000; Delos and White, 2000) (Hernandez and White, 1998).
The presence of conserved cysteine residues within the hydrophobic domain of LACV Gc delineated by amino acids 1066–1087 as well as the relative conservation of this region among disparate members of the Bunyaviridae (Table 1) (Garry and Garry, 2004; Plassmeyer et al., 2005), suggest that this hydrophobic region may form a similar disulfide-stablilized loop structure and serve as a fusion peptide. To test this hypothesis, we substituted key amino acids within this putative 22 amino acid fusion peptide. The functional effects of these mutations were then assessed using cell-to-cell fusion and pseudotype transduction assays (Ma et al., 1999; Plassmeyer et al., 2005). Several mutations within this hydrophobic domain affected glycoprotein expression to some extent, but all mutations either shifted the pH threshold of fusion below that of the wild type protein, reduced fusion efficiency, or abrogated cell-to-cell fusion and pseudotype entry altogether. A mutation at position 1066 (W1066A) was particularly informative, since it did not affect glycoprotein expression, yet abolished fusion and entry. Collectively, these results support a role for the region Gc 1066–1087, as the LACV Gc fusion peptide.
Table 1.
Alignment of the putative Gc fusion peptides from representative members of the five genera of the Bunyaviridae (Adopted from Garry and Garry, 2004)
RESULTS
Generation of LACV Gc (1066–1087) and (Gc 761) mutant constructs
We took several approaches to analyzing this putative fusion domain: we (a) substituted several conserved and non-conserved amino acid residues within the LACV Gc (1066–1087), (b) deleted the region in mutant LACV (Δ1066–1087) and (c) prepared an insertion mutant containing the entire fusion peptide of SFV, LACV (SFV-FP). The mutagenesis was accomplished with the QuikChange II XL site directed mutagenesis kit (Stratagene, La Jolla, CA) as indicated in the Methods section. Additionally, we generated constructs with mutations in the tryptic site located at Gc 761. Although there are many potential proteolytic sites in the Gc glycoprotein (Gentsch and Bishop, 1979), the tryptic site at position 761 is uniquely accessible in the whole virion prior to acidification, and its accessibility is affected by the conformational changes that are induced by low pH (González-Scarano, 1985). Therefore, it is of interest to determine whether or not this tryptic site is involved in Gc mediated fusion. All of the mutations were engineered into construct pBluescript II KS(+)–LAC(M), then subcloned into the expression vector pCAGGS (Niwa, Yamamura, and Miyazaki, 1991) and sequenced to verify that there were no additional mutations, as previously described (Plassmeyer et al., 2005).
Expression studies of LACV Gc (1066–1087) mutant constructs
To examine expression as an index of conformational integrity, the LACV wild-type and amino acid point mutant constructs were transiently expressed in QT6 cells. Forty-eight hours after transfection, the cells were stained with conformational monoclonal antibodies (MABs) 807–31 and 807–22 against LACV Gc (50:50, vol/vol), and expression was determined by flow cytometry (Figure 1) (Gonzalez-Scarano et al., 1982). Expression of LACV (D1078A), LACV (V1076A), LACV (G1067A), LACV (W1066A), LACV (S1077A), and LACV (L1074A) was similar to that of the LACV wild-type Gc (Figures 1A and B); expression was not detected with mutant LACV (G1083L) (Figure 1C). In addition, expression of the two tryptic site mutant glycoproteins, LACV (R761A), and LACV (R761H), was also similar to that of wild-type LACV (Figure 1D). Two recombinants, LACV Δ1066–1087 (deletion of the entire putative fusion domain) and LACV SFV FP (insert of the SFV fusion domain, SVF FP), were also examined, but there was no expression above background (Figures 1C and D).
Figure 1. Surface expression of mutant Gc in QT6 cells measured by flow cytometry.
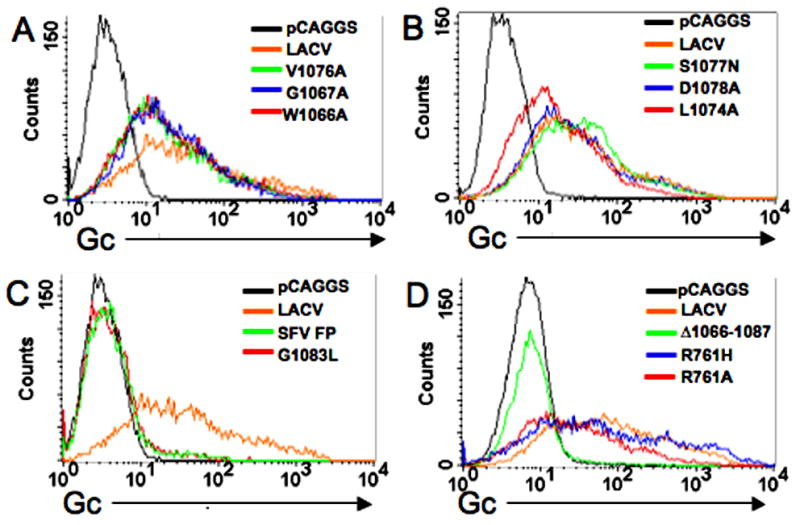
QT6 cells were transfected with M-segment constructs as indicated in the histogram and fixed with 2% paraformaldehyde for 20 minutes. Gc was detected with a 1:100 dilution of a 1:1 (vol/vol) mixture of two conformational mAbs, 807:31 and 807:22. (A) As shown in these representative histograms, surface expression of Gc was consistent between the wild-type LAC Gc and the mutant LAC Gc constructs: W1066A, G1067A, and V1076A, in (B) for constructs L1074A, S1077N and D1078A and in (D) for tryptic site mutant constructs R761H and R761A. (C) Surface expression above background was not detected for G1038L, SFV FP, or (D) LAC (Δ1066–1087) constructs.
To determine if LACV Δ1066–1087 Gc, LACV SFV FP, and LACV (G1083L) were expressed at all, Hela cells transfected with LACV and mutant constructs were examined by IFA at 6, 12, 18, and 48 hours after application of DNA using conformation independent MABs, 807–25 and 807–15 and a MAB specific for LACV Gn, as well as conformation dependent MABs 807–22 and 807–31. Gn and Gc were detected with nonconformational antibodies in Hela cells transfected with LACV wild-type and mutant constructs LACV (V1076A, G1067A, W1066A, S1077N, L1074N, L1074A, D1078A, R761A, R761H, Δ1066–1087, G1083L, and SFV FP) (Figure 2). In contrast to the results with non-conformational antibodies, which detected all of the constructs, three mutants, LACV Δ1066–1087, LACV (G1083L), and LACV (SFV FP) were not detected by staining with the conformational antibodies. Therefore, although deletion of the putative fusion domain or a G to L substitution at position 1083 did not interfere with expression of the glycoprotein, these mutations were not well tolerated as demonstrated by the their altered conformation.
Figure 2. Expression of mutant constructs by Immunofluorescence Assay (IFA).
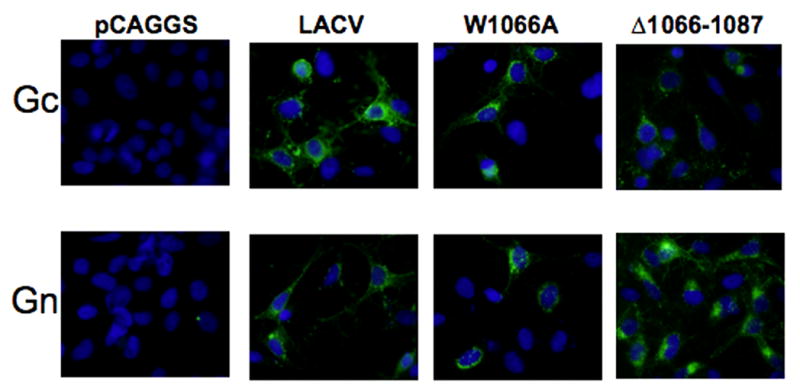
Gc was detected by IFA with a combination of two non-conformational mAbs: 807–15 and 807–25 (1:1, vol/vol). Gn was detected by a monoclonal antibody (Jacoby et al., 1993). Representative images shown here for pCAGGS, LACV, LACV(W1066A), and LACV Δ 1066–1087.
Mutations within the putative fusion domain, LACV Gc (1066–1087), reduce or abrogate cell-to cell fusion
The parental construct and mutants LACV (W1066A), LACV (G1067A), LACV (V1076A), LACV (L1074A), LACV (D1078A), LACV (S1077N), LACV (D1078A), LACV (G1083L), LACV (R761A), LACV (R761H), LACV (Δ 1066–1087), and LACV (SFV FP) were tested in a previously described fusion assay where QT6 served as the effector cells and BSR cells functioned as the target cells (Plassmeyer et al., 2005). In this assay, the pH threshold for cell-to-cell fusion in cells that expressed the LACV wild-type M segment was just below 6.2, similar to the pH previously described as critical for triggering fusion by the native LACV glycoproteins (data not shown) (Pobjecky, Smith, and Gonzalez-Scarano, 1986). Not surprisingly, the three constructs that were not detectable by conformational antibodies, LACV (G1083L), LACV (Δ1066–1087), and LACV (SFV FP) did not mediate cell-to-cell fusion above background (Figure 3C and D). However, under these experimental conditions, all of the constructs that demonstrated expression similar to LACV mediated cell-to-cell fusion with the exception of LACV (W1066A) (Figure 3A and B). The other fusion peptide mutant constructs, LACV (G1067A), LACV (V1076A) LACV (L1074A), LACV (S1077N) and LACV (D1078A), demonstrated decreased fusion efficiency (at least 10-fold lower signal) in comparison to the wild-type LACV. Additionally, substitutions G1067A, V1076A, L1074A, and D1078A appeared to have lower pH thresholds for initiating membrane fusion (Figure 3A and B). Point mutations in the tryptic site located at 761, LACV (R761A) and LACV (R761H), did not affect the magnitude or pH threshold of cell-to-cell mediated fusion (Figure 3E).
Figure 3. Substitutions within the LAC Gc amino acid region 1066–1087 affect pH-dependent cell-to-cell fusion.
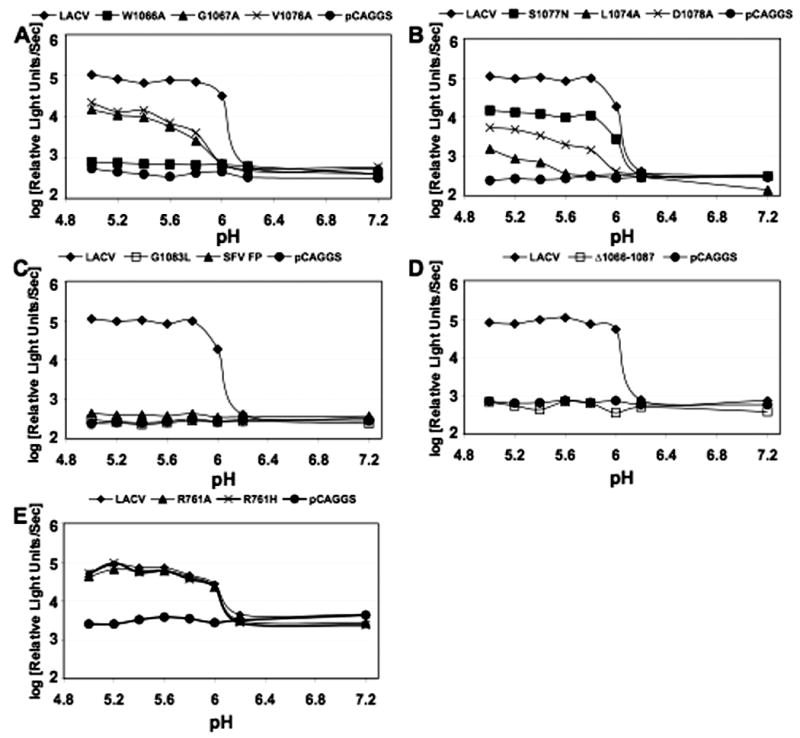
Effector cells (QT6) were transfected with two plasmids: a pCAGGS-M segment construct and a plasmid encoding the Luciferase gene under a T7 promoter. BSR target cells were infected with a vaccinia virus encoding the T7 RNA polymerase (vTF1.1) as indicated in Materials and Methods. Following an overnight incubation, the effector cells were overlaid onto the target cells, and the pH dropped briefly using HEPES-MES buffers. Luciferase activity, which indicates fusion of cells expressing the T7 polymerase and cells expressing M-segment constructs, was then measured in the cell lysates obtained 5 h later. (A) Amino acid substitutions that either decreased fusion or abrogated fusion: G1066A, V1076A, and W1066A. (B) Amino acid substitutions decreased fusion: L1074A, D1078A, S1077N (C and D) Amino acid changes that abolished cell surface expression and also eliminated fusion activity: G1083L, SFV FP, and Δ 1066–1087. (E) Point mutations in the tryptic site located at 761, LACV (R761A) and LACV (R761H) did not affect cell-to-cell fusion.
The percent maximum fusion for each construct at each pH was determined for LAC and mutants G1067A, V1076A, L1074A, and D1078A, then analyzed by non-linear regression with a sigmoidal dose-response model (variable slopes) using GraphPad Prism software, version 4.02 (GraphPad Software, San Diego, Ca) (Martin-Garcia et al., 2005). The pH50 values (at which there is 50% fusion signal) were obtained from the nonlinear regression for each construct and compared by using a t test. The pH50 values for the four mutant constructs were significantly (p<0.005) different from wild-type LACV, further indicating that these amino acid substitutions alter Gc mediated fusion. Importantly, the effect of these mutations on fusion was not the result of altered cellular conformation or reduced expression of the mutant glycoprotein, as their expression profiles were equivalent to those of the wild type construct (Figures 1 and 2).
Transduction of CHME-5 cells by LACV wild-type and mutant M segment pseudotyped MLV particles
To assess the effects of the mutations within this putative fusion domain on entry, we prepared murine leukemia virus (MLV) pseudotype particles incorporating either wild-type or point mutant LAC M segment constructs using a previously described three-plasmid system (Soneoka et al., 1995). Concentrated pseudotypes were analyzed by Western blot for the presence of Gc (Plassmeyer et al., 2005) and MLV Gag p30 to confirm that pseudoviral particles had been produced (Figure 4A). The pseudotypes were then transduced into CHME-5 cells and titrated using the expression of beta-galactosidase to quantify the efficiency of entry (Figure 4B). While pseudotype particles containing with mutations in the tryptic site at 761,LACV (R761A and R761H), were able to mediate entry to the same extent as wild type LACV Gc (data not shown), pseudotype viruses with mutations elsewhere in Gc demonstrated either reduced or abrogated ability to mediate entry. Consistent with the fusion assays, of the Gc constructs that were detectable by conformational antibodies, only LACV (G1067A) and LACV (V1076A) mediated pseudotype virus entry into cells. Pseudotype virus incorporating mutations G1067A and V1076A transduced CHME-5 cells significantly less well that wild-type LACV Gc (p < 0.02). As in the fusion assay, mutant W1066A was unable to mediate entry in spite of normal expression.
Figure 4. MLV pseudotypes of LACV with mutations in the putative fusion peptide demonstrate reduced or abrogated ability to transduce CHME cells.
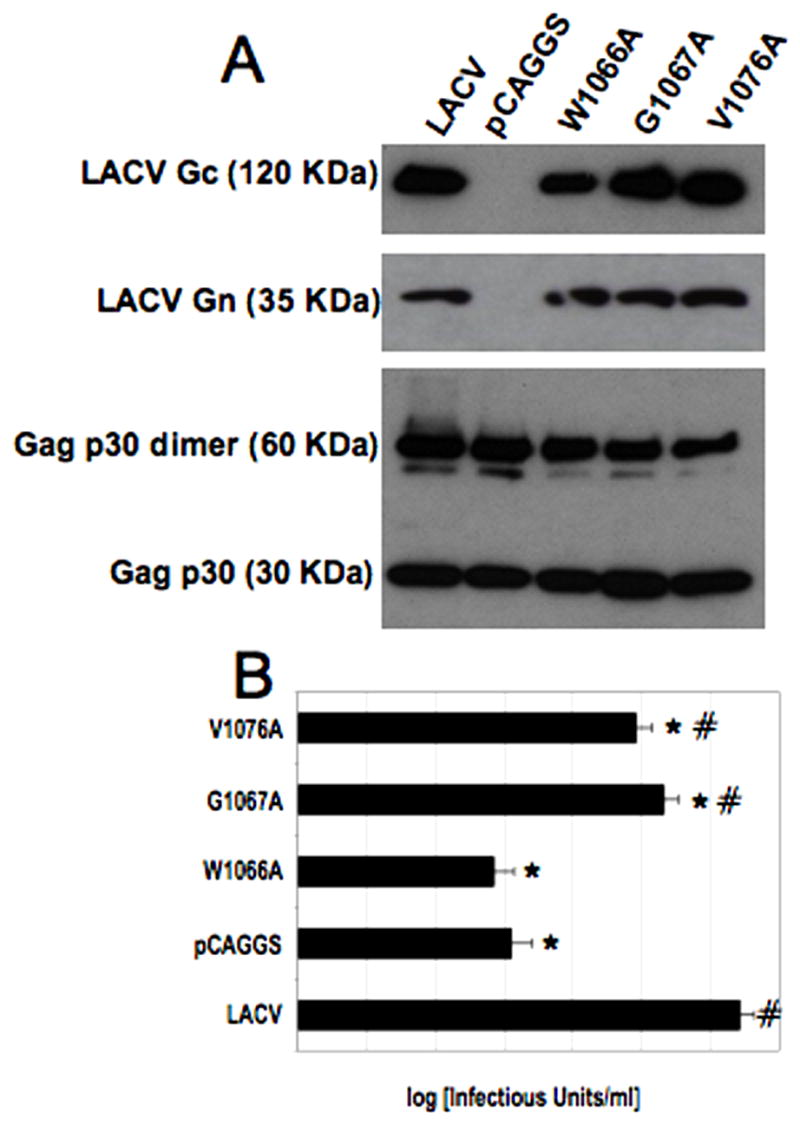
(A) Western blot of MLV pseudotyped particles incorporating LACV wild-type and mutant Gc. Pseudotypes were prepared as described in Materials and Methods. After resolution in an 8–16% tris-glycine polyacrylamide gel, the samples were transferred to a 0.2 μm pore-size nitrocellulose membrane. Top: Wild-type and mutant Gc (as indicated) was detected with mAb 807:31 at a 1:300 dilution. Bottom: MLV Gag was detected using a monoclonal goat antibody recognizing MLV p30. (B) M segment pseudotypes were spinoculated onto CHME-5 cells at 1250 x g for 2 hours at 4°C. Data are presented as the mean plus/minus standard deviation of multiple experiments and were compared by Student’s t test). Constructs significantly different from LACV (p < 0.009) are indicated with an asterisk (*) and constructs significantly (p < 0.05) different from control (MLV-pCAGGS) are indicated with a number sign (#). Pseudotype viruses with mutations in the putative fusion peptide of Gc (1066–1087) demonstrated either reduced or abrogated cell entry. Of the fusion peptide mutant constructs, only LACV (G1067A) and LACV (V1076A) mediated pseudotype virus entry into cells, although their ability to mediate entry was significantly reduced compared to wild-type LACV (p<0.02).
DISCUSSION
We substituted several conserved and non-conserved amino acid residues within the LACV Gc (1066–1087) to examine the contribution of this putative fusion domain. The data from the Gc (1066–1087) mutants demonstrate that multiple mutations within the hydrophobic, putative fusion peptide (1066–1087) of LACV Gc affect expression, shift the pH threshold, and reduce or abrogate total fusion (Summarized in Table 2). All of the mutant constructs expressed Gc and Gn, as detected by non-conformational antibodies. However we were unable to detect three of the mutant Gc constructs [LACV Δ1066–1087, LACV (G1083L), and LACV (SFV FP)] with conformational antibodies. These mutations may have destabilized the hydrophobic domain resulting in altered conformation of Gc. In contrast, amino acid substitutions that did not alter expression may have affected the intra- and intermolecular salt bridges and hydrogen bonds, ultimately affecting fusion efficiency and pH threshold. The reduced pH threshold, decreased fusion efficiency and entry observed for the Gc (1066–1087) point mutant constructs may represent a difference in the overall ability of mutant Gc (1077–1087) to undergo a pH-induced conformational change into a fusion active structure. Alternatively, wild-type and mutant Gc may undergo a conformational change into a fusion active structure at a similar efficiency, but the mutations may result in a Gc glycoprotein that is less fusogenic. The ability to detect LACV (G1067A), (V1076A), (W1066A), (S1077N), (D1078A), and (L1074A) mutant glycoproteins with conformational antibodies suggests that these mutations do not adversely affect the processing, intracellular transport, and overall conformation of these mutants and support the latter hypothesis. Importantly, abrogation of cell-to-cell fusion by the amino acid substitution W1066A, which maintained surface expression, supports a direct role for LACV Gc (1066–1087) in LACV Gc mediated fusion and entry.
Table 2.
Expression and fusion phenotype for LACV Gc (1066–1087) amino acid deletion and substitution mutant constructs and LACV Gc 761 tryptic site mutant constructs.
| CONSTRUCT | EXPRESSION* (Nonconform. Abs) | EXPRESSION (Conform Abs) | Fusion |
|---|---|---|---|
| W1066A | YES | YES | − |
| G1067A | YES | YES | ++ |
| L1074A | YES | YES | ++ |
| V1076A | YES | YES | + |
| S1077N | YES | YES | ++ |
| D1078A | YES | YES | + |
| R761A | YES | YES | ++++ |
| R761H | YES | YES | ++++ |
| G1083L | YES | NO | − |
| Δ1066–1087 | YES | NO | − |
| SFV FP | YES | NO | − |
See results for details of determination of expression using microscopy and FACS.
Since minor amino acid substitutions within LACV Gc (1066–1087) have a significant effect on fusion activity, a specific sequence may be necessary for its full activity. It is helpful to consider these results in comparison with point mutation studies of putative fusion domains for several viruses containing internal fusion peptides such as VSV, ASLV, TBE and SFV (Allison et al., 2001) (Durrer et al., 1995) (Hernandez and White, 1998) (Levy-Mintz and Kielian, 1991) (Whitt et al., 1990). Substitutions of amino acids within the fusion peptides of these viruses have similar phenotypic consequences, including altered fusion protein expression, reduced/abrogated fusogenicity and, in some cases, modification of oligomerization and virus assembly (Allison et al., 2001; Fredericksen and Whitt, 1995; Fredericksen and Whitt, 1996; Fredericksen and Whitt, 1998; Gomara et al., 2004; Russell, Jardetzky, and Lamb, 2004; Ma et al., 1999). Direct interaction between LACV Gc (1066–1087) and the target membrane during fusion remains to be demonstrated. However, in conjunction with computational modeling suggesting that LACV Gc (970–1350) shares structural similarity with class II fusion proteins (Plassmeyer et al., 2005), the results presented in this study support a role for this hydrophobic region as the LACV Gc fusion peptide.
Two classes of viral fusion proteins have been established. Class I fusion proteins are represented by the influenza haemagglutinin protein and by HIV gp41, but also include fusion proteins from many unrelated virus families; class II fusion proteins are represented primarily by members of the Flaviviridae and the Togaviridae (Jardetzky and Lamb, 2004; Kielian and Rey, 2006). Both class I and class II fusion proteins experience a tightly regulated conformational change during fusion. Class I fusion proteins undergo a drastic refolding between the pre-fusion and post-fusion conformations, and are arranged as trimers in both the pre-fusion and post-fusion complex. In contrast, the class II fusion proteins reorganize rather than refold (Jardetzky and Lamb, 2004) and form trimers subsequent to dissociation of the homodimers (flaviviruses) or heterodimers (alphaviruses) by exposure to low pH in the endosome following internalization. During this reorganization, the internal fusion loop is exposed and projected towards the cellular membrane, creating a rod-like structure analogous to the class I fusion complex.
While this study supports a role for the hydrophobic region, Gc (1066–1087) as the fusion peptide, absent three-dimensional structural analysis we cannot demonstrate whether this putative fusion domain directly inserts into the endosomal membrane like the alphavirus E1 or the flavivirus E (Modis et al., 2004). It is also premature to speculate on the potential Gn-Gc arrangements that could be analogous to other class II fusion proteins. Nevertheless, a thorough characterization of the relative contribution of the amino acid residues within the fusion domain can help elucidate the mechanisms of conformational change and virus entry for the LACV Gc and other class II fusion proteins. For example, identification of the flexible “hinge” region, a site of a number of mutations that affect the pH dependence of fusion in other class II fusion proteins may lay the foundation for studies targeting the fusion domain as a possible antiviral strategy for this medically important virus (Kielian, 2006; Kielian and Rey, 2006).
In addition to examining the effect of mutations within the putative fusion domain, we examined the contribution of the tryptic site at Gc position 761 on LACV mediated fusion and entry. While there are many potential proteolytic sites throughout the Gc glycoprotein (Gentsch and Bishop, 1979), the tryptic site at position 761 is easily accessible prior to acidification (González-Scarano, 1985). Therefore, it was of particular interest to determine whether or not this tryptic site is involved in Gc mediated fusion, since other fusion proteins depend on trypsin cleavage for activation. Our results suggest that mutations in the Gc 761 tryptic site do not affect expression, cellular localization, pH mediated cell-to-cell fusion, or pseudotype particle transduction. Therefore, it is unlikely this site plays a direct role in LACV fusion and entry in mammalian cells. However, it is still unknown whether the tryptic site at Gc 761, or for that matter the amino acid region 860–1442, or the LACV Gc putative fusion peptide (1066–1087), are critical for fusion in the mosquitoes. In this regard, it was previously shown that the M segment is a major determinant of oral transmission of LACV by mosquitoes (Beaty et al., 1981; Sundin et al., 1987). Proteolytic enzymes found in the mosquito midgut, including the large quantities of trypsin produced upon blood feeding (Noriega and Wells, 1999), increase virus affinity for mosquito cells. Therefore, while the LACV Gc761 tryptic site does not appear to play a role in mammalian cells, it may play a critical role in mediating fusion and entry in mosquito cells. In addition, it has also been demonstrated that the proteolytic enzymes of the mosquito midgut remove Gc while leaving Gn intact (Ludwig et al., 1989). These data suggest that processing of LACV glycoproteins in the mosquito midgut may facilitate attachment of virus protein and suggests that the role of LACV Gc in fusion and entry in mosquito cells may be different from that in mammalian cells (Beaty et al., 1981; Sundin et al., 1987). It is interesting that mutation of a potential tryptic site (R at 859) affected fusion, although we do not know if this site is used (cleaved) in the process of fusion. Future studies will focus on the contributions of LACV Gc (1066–1087) and the tryptic site at Gc 761 in mediating fusion and entry in the virus’s mosquito vector.
MATERIALS AND METHODS
Cell culture
293T (human fetal kidney), QT6 (quail fibroblast), BSR (baby hamster kidney), and CHME-5 (human fetal microglia) cells were cultured at 37°C/5% CO2 in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal calf serum (FCS) Atlanta Biologicals, Lawrenceville, GA), penicillin/streptomycin (P/S) (Invitrogen), and L-glutamine (Invitrogen).
LACV M segment constructs and site directed mutagenesis
The LACV M segment ORF was subcloned from pcDNA3.1 into the expression vector pCAGGS (Niwa, Yamamura, and Miyazaki, 1991) to employ the CMV enhancer and strong chicken β actin promoter to express the polyprotein, as previously described (Plassmeyer et al., 2005). Briefly, the LACV M ORF was amplified using rTth DNA Polymerase XL (PE Biosystems, Foster City, CA) with a primer set that introduced restriction cut sites ClaI and XhoI at the 5′ and 3′ ends, respectively. The PCR cycle consisted of a 93°C 2 minute denaturing step followed by 35 cycles of (93°C 30 seconds, 48°C 1 minute, 72°C 6 minutes) and an extension of 72°C 10 minutes. The resulting fragment was digested with both ClaI and XhoI and inserted into the linearized pCAGGS expression vector digested with ClaI and XhoI.
To engineer mutations (point, deletion and insertion of SFV FP) in the putative fusion domain of LAC Gc and the Gc ((761–772) tryptic site, the LACV M segment was subcloned into pBluescript II KS+ using ClaI and XhoI (Plassmeyer et al., 2005). The point mutations were engineered into the M segment using the QuikChange II XL site directed mutagenesis kit following manufacturer’s instructions (Stratagene, La Jolla, CA). The forward primers used into engineer the substitutions are listed:
W1066A (5′-GAGAGGACAAGCTCAGCGGGATGCGAAGAGTTTGG);
G1067A (5′-AGGACAAGCTCATGGGCATGCGAAGAGTTTGGTTGC);
L1074A (5′-GAGTTTGGTTGCGCGGCTGTAAGTGATGGG);
V1076A (5′-GGTTGCCTGGCTGCAAGTGATGGGTGTGTATTTGG);
S1077N (5′-GGTTGCCTGGCTGTAAATGATGGGTGTGTATTTGGA);
D1078A (5′-TGCCTGGCTGTAAGTGCTGGGTGTGTATTTGGA);
G1083L (5′-GATGGGTGTGTATTTCTATCATGCCAAGATATA);
R761A (5′-GCCTGCTTATCGCCAGCCTCAGGTGCTATATATGC);
R761H (5′-GCCTGCTTATCGCCACACTCAGGTGCTATATATGC);
Δ1066–1087 (5′GAGAGGACAAGCTCAGGCGTGTACCCGTTCATGTGGGGAGGGGCATATTGCTTCTGCGACT CAAGAAAACATAATAAAAGAAGAACTATCTGTCTATAGG);
SFV (GAGAGGACAAGCTCAATAATAAAAGAAGAACTATCTGTCTATAGG)
For all constructs, the reverse primer is the reverse complement of the respective sequence. For LACV (Δ1066–1087) primer: bold sequence is 5′ to the fusion peptide while the underlined sequence is 3′ to the putative fusion peptide. For LACV Insert SFV primer: underlined sequence is complementary/identical to LACV sequence flanking the putative fusion peptide and the bold sequence is the sequence of the SFV fusion peptide. The point mutant constructs were then cloned into the expression vector pCAGGS. These M segment clones were sequenced to verify that there were no additional mutations.
Expression of wild-type and mutant LAC Gc in QT6 cells: detection by flow cytometry
QT6 cells were plated at a density of 5x105 cells/well in 6 well plates and cultured overnight. Cells were transfected using the ProFection Mammalian Transfection System-Calcium Phosphate (Promega) with 6μg/well of the M segment constructs. 48 hr after application of DNA, the cells were removed using 0.5 mM EDTA and washed twice in PBS before fixation using 2% paraformaldehyde at 4°C. LACV Gc was detected at 4°C with a 1:100 dilution of either individual monoclonal antibodies 807–31 and 807–22 (Gonzalez-Scarano et al., 1982) or a cocktail of the two mAb’s, 807–31 and 807–22 (vol/vol) and goat anti-mouse FITC conjugated mAb (Sigma) at a 1:75 dilution.
Expression of wild-type and mutant Gc in QT6 Cells: detection by IFA and confocal microscopy
Individual German glass cover slips (Electron Microscopy Sciences, Hatfield, PA) plated in 24 well plates were coated with poly-D-lysine overnight then washed with tissue culture grade water and dried. QT6 cells (Invitrogen) were plated on cover slips at a density of 8x104 cells/well and cultured overnight at 37°C/5% CO2 in culture media. Cells were transfected for 6 hours using the ProFection Mammalian Transfection System-Calcium Phosphate (Promega, Madison, WI) with 1μg/well of each plasmid (M segment constructs cloned into pCAGGS). Zero, 6, 12, 18 and 42 hr after removal of the DNA, cells were fixed in 2% paraformaldehyde and permeabilized with 0.5% Triton X-100 at 4°C. Gn was detected with a 1:100 dilution of a Gn specific monoclonal antibody. Gc was detected with a 1:100 dilution of a mix of two non-conformational mAbs, 807:15 and 807:25 (vol/vol), and goat anti-mouse Fluorescein (FITC) conjugated mAb (Sigma, St. Louis, MO) at a 1:75 dilution. Cells were counterstained with a 1:500 dilution of Hoechst. Cells were viewed at 40X, and images were captured using a Leica DC 500 digital camera (Leica, Northvalle, NJ).
Cell-to-cell fusion assay
Cell-to-cell fusion assays were performed as described (Plassmeyer et al., 2005). Briefly, effector cells (QT6) were plated on 6-well plates at 8 X 105 to 1 X 106 cells/well and incubated overnight at 37°C. Cells were transfected using the ProFection Mammalian Transfection System-Calcium Phosphate (Promega) with 6 μg/well of each plasmid, the M segment construct plus the T7LUC vector. The transfected cells were maintained at 37°C for 48 h. Target cells, either QT6 or BSR cells, were plated and incubated overnight at 37°C in a 24-well plate at a density of 8 X 104 cells/well. The target cells were infected with activated recombinant vaccinia expressing T7 RNA pol (vTF1.1) at a MOI of 2–3 pfu/cell using 0.5–1 ml/well for 1–2 h at 37°C with gentle rocking every 15–20 min. vTF1.1 was activated by incubation with trypsin 0.25% [1:1] for 30 min. at 37 °C, then diluted at least 1:10 with Opti-MEM (Invitrogen). Following infection with vTF1.1, the target cells were washed with 0.5 ml PBS, cultured in 2 ml of DMEM supplemented with 10% FBS and 100 μg/ml rifampicin, then incubated overnight at 32°C. Effector cells were collected with 0.5 mM EDTA, pelleted, washed with PBS, and overlaid onto the target cells for 90 min. Cells were exposed to fusion medium (DMEM, 10% FBS, 1XP/S, 10mM HEPES, and 10 mM MES; pH 7.2-5.0) for 30s. Following exposure to fusion medium, the cells were cultured at 37°C in culture medium containing rifampicin for 5–8 h. Cells were lysed with Promega’s reporter lysis buffer and Luciferase activity was evaluated and measured in a LumiCount Luminometer (Packard).
Pseudotype production
A previously described transient three-plasmid system was used to produce Gn/Gc-pseudotyped murine leukemia virus (MLV) particles (Ma et al., 1999; Soneoka et al., 1995); for these experiments the glycoproteins were expressed in pCAGGS, as described above. Supernatants containing pseudotypes were concentrated through 20% sucrose in an SW41 rotor at (150,000) X g) at 4°C for 1 h 45 min (Burns et al., 1993; Ma et al., 1999). The pellets were resuspended overnight in TNE (50 mM Tris-Hcl, pH 8.0, 130 mM NaCl, and 1 mM EDTA).
Western Blots for pseudotype particles
Western blot analysis was performed on concentrated pseudotype particles. For non-reducing conditions, 20μl pseudotype particles were mixed with 4x loading buffer (0.2 M Tris, 0.4% sodium dodecyl sulfate, 40% [vol/vol] glycerol, 0.04% [wt/vol] bromophenol blue) and boiled for 4min. Samples were resolved on 8–16% tris-glycine polyacrylamide gels (Cambrex, East Rutherford, NJ) then transferred to 0.2μm pore-size nitrocellulose membrane (BioRad, Hercules, CA). MLV Gag was detected using monoclonal goat antibody that recognizes p30. Primary antibody was detected with a secondary mouse anti-goat horseradish peroxidase (HPR) conjugated mAb (Sigma). Pseudotype particles containing the wild-type LACV M and LACV mutant constructs were analyzed with an anti-Gn and an anti-Gc (807:31) mAb at a 1:500 dilution. Primary antibody was detected with a sheep anti-mouse HRP conjugated mAb (Sigma) at 1:10,000, respectively. Detection of HRP was performed with the SuperSignal West Pico Chemiluminescence substrate (Pierce, Rockford, IL) following manufacturer’s instructions.
Spinoculation and beta-gal detection
Cells were plated in a 96 well plate at a density of 1 X 104 cells/well and incubated overnight at 37°C. Serial 10-fold dilutions of the pseudotyped viruses in culture medium equilibrated to a final volume of 100 μl were then added to the cells. The cells were centrifugated/spinoculated at 1250 X g for 2h at 4°C to facilitate transduction. Following the centrifugation step, the cells were cultured at 37°C for 48h, then stained for beta-gal expression as previously described (Ma et al., 1999).
Acknowledgments
Supported by NIH grants NS-30606, AI-07325 (training grant for MLP), and NS-007180 (training grant for SSS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001;75(9):4268–75. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty BJ, Holterman M, Tabachnick W, Shope RE, Rozhon EJ, Bishop DH. Molecular basis of bunyavirus transmission by mosquitoes: role of the middle-sized RNA segment. Science. 1981;211(4489):1433–5. doi: 10.1126/science.6781068. [DOI] [PubMed] [Google Scholar]
- Bertolotti-Ciarlet A, Smith J, Strecker K, Paragas J, Altamura LA, McFalls JM, Frias-Staheli N, Garcia-Sastre A, Schmaljohn CS, Doms RW. Cellular localization and antigenic characterization of crimean-congo hemorrhagic fever virus glycoproteins. J Virol. 2005;79(10):6152–61. doi: 10.1128/JVI.79.10.6152-6161.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp K, Stillmock K, Gonzalez-Scarano F. Analysis of the intracellular transport properties of recombinant La Crosse virus glycoproteins. Virology. 1996;220(2):485–90. doi: 10.1006/viro.1996.0336. [DOI] [PubMed] [Google Scholar]
- Burns JC, Friedmann T, Driever W, Burrascano M, Yee JK. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci U S A. 1993;90(17):8033–7. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Matsuoka Y, Compans RW. Golgi complex localization of the Punta Toro virus G2 protein requires its association with the G1 protein. Virology. 1991;183(1):351–65. doi: 10.1016/0042-6822(91)90148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delos SE, Gilbert JM, White JM. The central proline of an internal viral fusion peptide serves two important roles. J Virol. 2000;74(4):1686–93. doi: 10.1128/jvi.74.4.1686-1693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delos SE, White JM. Critical role for the cysteines flanking the internal fusion peptide of avian sarcoma/leukosis virus envelope glycoprotein. J Virol. 2000;74(20):9738–41. doi: 10.1128/jvi.74.20.9738-9741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrer P, Gaudin Y, Ruigrok RW, Graf R, Brunner J. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis viruses. J Biol Chem. 1995;270(29):17575–81. doi: 10.1074/jbc.270.29.17575. [DOI] [PubMed] [Google Scholar]
- Eshita Y, Bishop DH. The complete sequence of the M RNA of snowshoe hare bunyavirus reveals the presence of internal hydrophobic domains in the viral glycoprotein. Virology. 1984;137(2):227–40. doi: 10.1016/0042-6822(84)90215-0. [DOI] [PubMed] [Google Scholar]
- Fazakerley JK, Gonzalez-Scarano F, Strickler J, Dietzschold B, Karush F, Nathanson N. Organization of the middle RNA segment of snowshoe hare Bunyavirus. Virology. 1988;167(2):422–32. [PubMed] [Google Scholar]
- Fredericksen BL, Whitt MA. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J Virol. 1995;69(3):1435–43. doi: 10.1128/jvi.69.3.1435-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Whitt MA. Mutations at two conserved acidic amino acids in the glycoprotein of vesicular stomatitis virus affect pH-dependent conformational changes and reduce the pH threshold for membrane fusion. Virology. 1996;217(1):49–57. doi: 10.1006/viro.1996.0092. [DOI] [PubMed] [Google Scholar]
- Fredericksen BL, Whitt MA. Attenuation of recombinant vesicular stomatitis viruses encoding mutant glycoproteins demonstrate a critical role for maintaining a high pH threshold for membrane fusion in viral fitness. Virology. 1998;240(2):349–58. doi: 10.1006/viro.1997.8921. [DOI] [PubMed] [Google Scholar]
- Garry CE, Garry RF. Proteomics computational analyses suggest that the carboxyl terminal glycoproteins of Bunyaviruses are class II viral fusion protein (beta-penetrenes) Theor Biol Med Model. 2004;1(1):10. doi: 10.1186/1742-4682-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch JR, Bishop DL. M viral RNA segment of bunyaviruses codes for two glycoproteins, G1 and G2. J Virol. 1979;30(3):767–70. doi: 10.1128/jvi.30.3.767-770.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomara MJ, Mora P, Mingarro I, Nieva JL. Roles of a conserved proline in the internal fusion peptide of Ebola glycoprotein. FEBS Lett. 2004;569(1–3):261–6. doi: 10.1016/j.febslet.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Scarano F. La Crosse virus G1 glycoprotein undergoes a conformational change at the pH of fusion. Virology. 1985;140(2):209–16. doi: 10.1016/0042-6822(85)90359-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Scarano F, Janssen RS, Najjar JA, Pobjecky N, Nathanson N. An avirulent G1 glycoprotein variant of La Crosse bunyavirus with defective fusion function. J Virol. 1985;54(3):757–63. doi: 10.1128/jvi.54.3.757-763.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Scarano F, Pobjecky N, Nathanson N. La Crosse bunyavirus can mediate pH-dependent fusion from without. Virology. 1984;132(1):222–5. doi: 10.1016/0042-6822(84)90107-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Scarano F, Shope RE, Calisher CE, Nathanson N. Characterization of monoclonal antibodies against the G1 and N proteins of LaCrosse and Tahyna, two California serogroup bunyaviruses. Virology. 1982;120(1):42–53. doi: 10.1016/0042-6822(82)90005-8. [DOI] [PubMed] [Google Scholar]
- Grady LJ, Srihongse S, Grayson MA, Deibel R. Monoclonal antibodies against La Crosse virus. J Gen Virol. 1983;64 (Pt 8):1699–704. doi: 10.1099/0022-1317-64-8-1699. [DOI] [PubMed] [Google Scholar]
- Haferkamp S, Fernando L, Schwarz TF, Feldmann H, Flick R. Intracellular localization of Crimean-Congo Hemorrhagic Fever (CCHF) virus glycoproteins. Virol J. 2005;2:42. doi: 10.1186/1743-422X-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez LD, White JM. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J Virol. 1998;72(4):3259–67. doi: 10.1128/jvi.72.4.3259-3267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby DR, Cooke C, Prabakaran I, Boland J, Nathanson N, González-Scarano F. Expression of the La Crosse M Segment proteins in a recombinant vaccinia expression system mediates pH dependent cellular fusion. Virology. 1993;193:993–996. doi: 10.1006/viro.1993.1213. [DOI] [PubMed] [Google Scholar]
- Janssen R, Gonzalez-Scarano F, Nathanson N. Mechanisms of bunyavirus virulence. Comparative pathogenesis of a virulent strain of La Crosse and an avirulent strain of Tahyna virus. Lab Invest. 1984;50(4):447–55. [PubMed] [Google Scholar]
- Jardetzky TS, Lamb RA. Virology: a class act. Nature. 2004;427(6972):307–8. doi: 10.1038/427307a. [DOI] [PubMed] [Google Scholar]
- Kielian M. Class II virus membrane fusion proteins. Virology. 2006;344(1):38–47. doi: 10.1016/j.virol.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol. 2006;4(1):67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappin DF, Nakitare GW, Palfreyman JW, Elliott RM. Localization of Bunyamwera bunyavirus G1 glycoprotein to the Golgi requires association with G2 but not with NSm. J Gen Virol. 1994;75 (Pt 12):3441–51. doi: 10.1099/0022-1317-75-12-3441. [DOI] [PubMed] [Google Scholar]
- Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Rey FA. The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105(1):137–48. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- Levy-Mintz P, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J Virol. 1991;65(8):4292–300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig GV, Christensen BM, Yuill TM, Schultz KT. Enzyme processing of La Crosse virus glycoprotein G1: a bunyavirus-vector infection model. Virology. 1989;171(1):108–13. doi: 10.1016/0042-6822(89)90516-3. [DOI] [PubMed] [Google Scholar]
- Ma M, Kersten DB, Kamrud KI, Wool-Lewis RJ, Schmaljohn C, Gonzalez-Scarano F. Murine leukemia virus pseudotypes of La Crosse and Hantaan Bunyaviruses: a system for analysis of cell tropism. Virus Res. 1999;64(1):23–32. doi: 10.1016/s0168-1702(99)00070-2. [DOI] [PubMed] [Google Scholar]
- Martin-Garcia J, Cocklin S, Chaiken IM, Gonzalez-Scarano F. Interaction with CD4 and antibodies to CD4-induced epitopes of the envelope gp120 from a microglial cell-adapted human immunodeficiency virus type 1 isolate. J Virol. 2005;79(11):6703–13. doi: 10.1128/JVI.79.11.6703-6713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–9. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–9. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Noriega FG, Wells MA. A molecular view of trypsin synthesis in the midgut of Aedes aegypti. J Insect Physiol. 1999;45(7):613–620. doi: 10.1016/s0022-1910(99)00052-9. [DOI] [PubMed] [Google Scholar]
- Pekosz A, Gonzalez-Scarano F. The extracellular domain of La Crosse virus G1 forms oligomers and undergoes pH-dependent conformational changes. Virology. 1996;225(1):243–7. doi: 10.1006/viro.1996.0596. [DOI] [PubMed] [Google Scholar]
- Pekosz A, Griot C, Nathanson N, Gonzalez-Scarano F. Tropism of bunyaviruses: evidence for a G1 glycoprotein-mediated entry pathway common to the California serogroup. Virology. 1995a;214(2):339–48. doi: 10.1006/viro.1995.0043. [DOI] [PubMed] [Google Scholar]
- Pekosz A, Griot C, Stillmock K, Nathanson N, Gonzalez-Scarano F. Protection from La Crosse virus encephalitis with recombinant glycoproteins: role of neutralizing anti-G1 antibodies. J Virol. 1995b;69(6):3475–81. doi: 10.1128/jvi.69.6.3475-3481.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassmeyer ML, Soldan SS, Stachelek KM, Martin-Garcia J, Gonzalez-Scarano F. California serogroup Gc (G1) glycoprotein is the principal determinant of pH-dependent cell fusion and entry. Virology. 2005;338(1):121–32. doi: 10.1016/j.virol.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Pobjecky N, Smith J, Gonzalez-Scarano F. Biological studies of the fusion function of California serogroup Bunyaviruses. Microb Pathog. 1986;1(5):491–501. doi: 10.1016/0882-4010(86)90011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375(6529):291–8. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Roberts A, Rossier C, Kolakofsky D, Nathanson N, Gonzalez-Scarano F. Completion of the La Crosse virus genome sequence and genetic comparisons of the L proteins of the Bunyaviridae. Virology. 1995;206(1):742–5. doi: 10.1016/s0042-6822(95)80001-8. [DOI] [PubMed] [Google Scholar]
- Russell CJ, Jardetzky TS, Lamb RA. Conserved glycine residues in the fusion peptide of the paramyxovirus fusion protein regulate activation of the native state. J Virol. 2004;78(24):13727–42. doi: 10.1128/JVI.78.24.13727-13742.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruusala A, Persson R, Schmaljohn CS, Pettersson RF. Coexpression of the membrane glycoproteins G1 and G2 of Hantaan virus is required for targeting to the Golgi complex. Virology. 1992;186(1):53–64. doi: 10.1016/0042-6822(92)90060-3. [DOI] [PubMed] [Google Scholar]
- Soldan SS, Gonzalez-Scarano F. Emerging infectious diseases: the Bunyaviridae. J Neurovirol. 2005;11(5):412–23. doi: 10.1080/13550280591002496. [DOI] [PubMed] [Google Scholar]
- Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, Kingsman AJ. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23(4):628–33. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin DR, Beaty BJ, Nathanson N, Gonzalez-Scarano F. A G1 glycoprotein epitope of La Crosse virus: a determinant of infection of Aedes triseriatus. Science. 1987;235(4788):591–3. doi: 10.1126/science.3810159. [DOI] [PubMed] [Google Scholar]
- Thompson WH, Kalfayan B, Anslow RO. Isolation of California Encephalitis Group Virus from a Fatal Human Illness. Am J Epidemiol. 1965;81:245–53. doi: 10.1093/oxfordjournals.aje.a120512. [DOI] [PubMed] [Google Scholar]
- Whitt MA, Zagouras P, Crise B, Rose JK. A fusion-defective mutant of the vesicular stomatitis virus glycoprotein. J Virol. 1990;64(10):4907–13. doi: 10.1128/jvi.64.10.4907-4913.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, Rossmann MG. Placement of the structural proteins in Sindbis virus. J Virol. 2002;76(22):11645–58. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


