Abstract
The OAT (organic anion transporter) family mediates the absorption, distribution and excretion of a diverse array of environmental toxins and clinically important drugs. OAT dysfunction significantly contributes to renal, hepatic, neurological and fetal toxicity and disease. As a first step to establish the topological model of hOAT1 (human OAT1), we investigated the external loops and the cellular orientation of the N- and the C-termini of this transporter. Combined approaches of immunofluorescence studies and site-directed chemical labelling were used for such purpose. Immunofluorescence microscopy of Myc-tagged hOAT1 expressed in cultured cells identified that both the N- and the C-termini of the transporter were located in the cytoplasm. Replacement of Lys59 in the predicted extracellular loop I with arginine resulted in a mutant (K59R), which was largely inaccessible for labelling by membrane-impermeable NHS (N-hydroxysuccinimido)-SS (dithio)-biotin present in the extracellular medium. This result suggests that loop I faces outside of the cell membrane. A single lysine residue introduced into putative extracellular loops III, V and VI of mutant K59R, which is devoid of extracellular lysine, reacted readily with membrane-impermeable NHS-SS-biotin, suggesting that these putative extracellular loops are in the extracellular domains of the protein. These studies provided the first experimental evidence on the extracellular loops and the cellular orientation of the N- and the C-termini of hOAT1.
Keywords: p-aminohippuric acid, biotinylation, human organic anion transporter (hOAT), major facilitator superfamily (MFS), organic anion transporter, topological model
Abbreviations: DMEM, Dulbecco's modified Eagle's medium; LC, long chain; NHS, N-hydroxysuccinimido; OAT, organic anion transporter; hOAT, human OAT; MFS, major facilitator superfamily; OCT, organic cation transporter; PAH, p-aminohippuric acid; SS, dithio
INTRODUCTION
The OATs (organic anion transporters) play essential roles in the body's disposition of clinically important anionic drugs including anti-HIV therapeutics, anti-tumour drugs, antibiotics, anti-hypertensives and anti-inflammatories [1–3]. Six OATs (OAT1–6) have been identified by different laboratory groups [4–15]. These OATs have distinct organ and cellular localizations and different substrate specificities. OAT1 and OAT3 are predominantly expressed at the basolateral membrane of kidney proximal tubule cells and the apical membrane of brain choroid plexus. OAT4 is expressed at the apical membrane of kidney proximal tubule cells and the basolateral membrane of placental trophoblasts. OAT2 is expressed at the basolateral membrane of hepatocytes and is expressed in the kidney. The cellular localization of OAT2 in the kidney is still controversial. OAT5 is expressed only in the kidney. OAT6 is expressed in the olfactory mucosa. The subcellular localization of OAT5 and OAT6 has not been defined.
In the kidney, OAT1 and OAT3 utilize a tertiary transport mechanism to move organic anions across the basolateral membrane into the proximal tubule cells for subsequent exit across the apical membrane into the urine for elimination [1–3]. Through this tertiary transport mechanism, Na+/K+-ATPase maintains an inwardly directed (blood-to-cell) Na+ gradient. The Na+ gradient then drives a sodium dicarboxylate co-transporter, sustaining an outwardly directed dicarboxylate gradient that is utilized by a dicarboxylate/organic anion exchanger (termed OAT) to move the organic anion substrate into the cell. This cascade of events indirectly links organic anion transport to metabolic energy and the Na+ gradient, allowing the entry of a negatively charged substrate against both its chemical concentration gradient and the electrical potential of the cell.
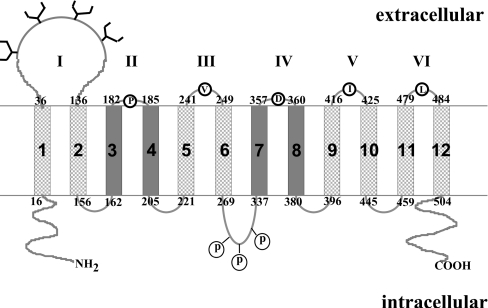
OATs were predicted to have 12 transmembrane segments (Figure 1, [9] and topology prediction program TopPred). Based on this topological model, all members of this family share the following common structural features: 12 transmembrane segments are interrupted by alternating intra- and extra-cellular loops; a cluster of potential glycosylation sites located in the large, putative extra-cellular loop between the first and the second transmembrane segments; and multiple presumed phosphorylation sites. N- and C-termini of the proteins are proposed to be located intracellularly. We have recently shown [16–18] that removal of potential glycosylation sites in the predicted first extracellular loop of hOAT1 (human OAT1) and of hOAT4 resulted in an increased electrophoretic mobility, indicating that these glycosylation sites are utilized and therefore in the extracellular domains of the protein. Despite this initial finding, there has been no direct biochemical evidence to support any of the computer-generated topological models. As a first step to mapping the topology of hOAT1, we investigated the extracellular loops and cellular orientation of the N- and the C-termini of this transporter.
Figure 1. Predicted transmembrane topology of OAT family.
Twelve transmembrane segments are numbered from 1 to 12. Putative extracellular loops are numbered from I to VI. The boundaries of extracellular loops are also numbered. Residues replaced by lysine are indicated inside the open circles. Potential glycosylation sites are denoted by tree-like structures. Potential phosphorylation sites are labelled as ‘P’. Transmembrane domain helices 3, 4, 7 and 8 are shown in dark grey to indicate the uncertainty of the prediction.
MATERIALS AND METHODS
Materials
[14C]PAH (p-aminohippuric acid) was from NEN Life Science Products (Hercules, CA, U.S.A.). Membrane-impermeable biotinylation reagents NHS (N-hydroxysuccinimido)-SS (dithio)-biotin and biotin-LC (long chain)-hydrazide and streptavidin–agarose beads were purchased from Pierce (Rockford, IL, U.S.A.). Protein A–agarose beads were purchased from Invitrogen (Carlsbad, CA, U.S.A.). COS-7 cells were purchased from American Type Culture Collection (Manassas, VA, U.S.A.). All other reagents were purchased from Sigma (St. Louis, MO, U.S.A.).
Construction of epitope-tagged hOAT1
A 10-amino-acid epitope Myc tag (EQKLISEEDL) was inserted into the N-terminus of hOAT1 (hOAT1–Myc-N), immediately after the first methionine residue, using a QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) following the manufacturer's instructions. The mutant sequences were confirmed by the dideoxy chain termination method.
Mutagenesis to make lysine mutants
Predicted extracellular Lys59 in hOAT1 was removed by replacement with arginine. The resulting mutant K59R was then used as a template into which single lysine residue was introduced into each of the predicted extracellular loops. The mutant sequences were confirmed by the dideoxy chain termination method.
Transport measurements
Uptake of [14C]PAH was initiated by adding uptake solution. The uptake solution consisted of PBS/CM (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 0.1 mM CaCl2 and 1 mM MgCl2, pH 7.4) and PAH (20 μM: 5 μM 14C-labelled and 15 μM unlabelled). At times as indicated in the Figure legends, the uptake was stopped by rapidly washing the cells with ice-cold PBS. The cells were then solubilized in 0.2 M NaOH, neutralized in 0.2 M HCl and divided into aliquots for liquid-scintillation counting. The uptake count in each well was standardized by the respective amount of protein in that well.
For efflux studies, cells expressing wild-type hOAT1 or its mutants were preloaded with [14C]PAH (100 μM) for 1 h, followed by an exposure to PBS (control group) or PBS containing α-oxoglutarate (α-ketoglutarate) (200 μM, experimental group) for 10 min. The uptake was then stopped by rapidly washing the cells with ice-cold PBS. The cells were then solubilized in 0.2 M NaOH, neutralized in 0.2 M HCl and divided into aliquots for liquid-scintillation counting. The uptake count in each well was standardized by the respective amount of protein in that well.
Transfection of cells with different constructs
COS-7 cells were grown at 37 °C and 5% CO2 in DMEM (Dulbecco's modified Eagle's medium; Invitrogen) supplemented with 10% (v/v) fetal bovine serum. Confluent cells were transfected with DNA plasmids using Lipofectamine™ 2000 reagent (Invitrogen). For each well of 12-well plates, 2 μg of plasmid DNA was added into 125 μl of OPTI medium (Invitrogen) for 5 min and then mixed with another 125 μl of OPTI medium containing 5 μl of Lipofectamine™ 2000 reagent. The mixture was incubated at room temperature (23 °C) for 20 min and diluted with 1250 μl of DMEM before addition to the cell monolayer. As for cells plated on 48-well plates for uptake assays, 0.32 μg of plasmid DNA and 0.8 μl of Lipofectamine™ 2000 reagent were used for the transfection. Transfected cells were incubated for 48 h at 37 °C and then used for transport assay and cell-surface biotinylation.
Immunofluorescence of transfected cells
Forty-eight hours after transfection, cells were washed three times in PBS, fixed for 15 min at room temperature in 4% (w/v) paraformaldehyde in PBS and rewashed in PBS. The fixed cells were then permeabilized with or without 0.1% Triton X-100 for 10 min. The cells were incubated for 30 min at room temperature in PBS containing 5% (v/v) goat serum and then incubated for 1 h in the same medium containing anti-Myc antibody (1:100; Mount Sinai School of Medicine, Hybridoma Center, New York, NY, U.S.A.) at room temperature. The cells were washed and bound primary antibodies were detected by reaction with FITC-coupled goat anti-mouse IgG (Chemicon), diluted 1:200, for 1 h. Cells were thoroughly washed and the coverglasses were mounted in GEL/MOUNT™ (Biomeda, Foster City, CA, U.S.A.). Samples were examined on a fluorescence microscope.
Electrophoresis and immunoblotting
Protein samples (30 μg) were resolved on SDS/7.5% PAGE minigels and electroblotted on to PVDF membranes. The blots were blocked for 1 h with 5% (w/v) non-fat dried milk powder in PBS/0.05% Tween 20, washed and incubated overnight at 4 °C with appropriate primary antibodies followed by horseradish-peroxidase-conjugated secondary antibodies incubation for 1 h at room temperature. The signals were detected by SuperSignal West Dura Extended Duration Substrate kit (Pierce, Rockford, IL, U.S.A.).
Cell-surface biotinylation
Cell-surface biotinylation was performed using the membrane-impermeable biotinylation reagents, NHS-SS-biotin or biotin-LC-hydrazide. For labelling with NHS-SS-biotin, COS-7 cells expressing hOAT1–Myc-C and its mutants were incubated on ice with NHS-SS-biotin (0.5 mg/ml in PBS, pH 8.0/0.01 mM Ca2+/1 mM Mg2+) for two successive 20 min periods with very gentle shaking. The reagent was freshly prepared before incubation. After biotinylation, cells were briefly rinsed with 3 ml of PBS/Ca2+/Mg2+ containing 100 mM glycine and then incubated on ice with the same solution for 20 min to ensure complete quenching of the unchanged NHS-SS-biotin. The cells were then dissolved in 400 μl of lysis buffer A [10 mM Tris/HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100 with protease inhibitor cocktail] for 1 h at 4 °C. Cell pellets were removed by centrifugation at 11000 g for 20 min at 4 °C. Supernatants were transferred to clean Eppendorf tubes and 50 μl streptavidin–agarose beads were added and incubated for 1 h at 4 °C with end-over-end mixing. Beads were then washed three times with lysis buffer and once with PBS. Proteins were released from the beads by incubation with Laemmli buffer for 30 min at 50 °C. The proteins were then resolved on SDS/7.5% PAGE minigels and electroblotted on to PVDF membranes. The membranes were probed with anti-Myc antibody and signals were detected by SuperSignal West Dura Extended Duration Substrate kit (Pierce).
For labelling with biotin-LC-hydrazide, COS-7 cells expressing hOAT1–Myc-C and its mutants were incubated on ice with sodium periodate (10 mM in PBS, pH 7.0/0.1 mM Ca2+/1 mM Mg2+) for 30 min. The cells were washed three times with PBS and once with sodium acetate (pH 5.5). Then 1 ml of biotin-LC-hydrazide (2 mM in sodium acetate, pH 5.5/0.1 mM Ca2+/1 mM Mg2+) was added to each well and incubated on ice for 30 min. The unchanged biotin-LC-hydrazide was removed by washing the cells five times with PBS (pH 7.0). The cells were then dissolved in 200 μl of lysis buffer B [10 mM Tris/HCl (pH 7.5), 10 mM NaCl, 2 mM EDTA, 10% glycerol, 0.5% Triton X-100 with protease inhibitor cocktail] for 1 h at 4 °C. Cell pellets were removed by centrifugation at 11000 g for 20 min at 4 °C. Supernatants were transferred to clean Eppendorf tubes and precleaned with 50 μl protein A–agarose beads for 1 h at 4 °C. Anti-Myc antibody (5 μg) was then added to the supernatant to immunoprecipitate hOAT1–Myc. The immunoprecipitates were analysed by Western blotting with horseradish-peroxidase-conjugated streptavidin and signals were detected by SuperSignal West Dura Extended Duration Substrate kit (Pierce). To ensure that biotin was only labelling surface proteins, the integrity of the cell membrane during biotinylation was tested in each experiment by immunoblotting with an anti-actin antibody as previously described [19]. In all experiments, actin immunoreactivity was only detected when cell membranes were permeabilized with 0.1% Nonidet P40, confirming the impermeability of the biotinylation reagent.
Data analysis
Each experiment was repeated a minimum of three times. The statistical analysis given was from multiple experiments. Statistical analysis was performed using Student's paired t tests. A P value of <0.05 was considered significant.
RESULTS
Functional analysis of Myc-tagged hOAT1
We have previously shown that Myc epitope tagged at the C-terminus of hOAT1 retained the functional properties of its native structure [17]. To facilitate the immunolocalization of N-terminus of hOAT1, we engineered Myc tag to the N-terminus of this protein (hOAT1–Myc-N). The expressed protein could be detected using an anti-Myc antibody. To ensure that the epitope tag had little effect on the transport function, we measured the [14C]PAH uptake into both wild-type hOAT1 and hOAT1–Myc-N-transfected cells. As shown in Table 1, hOAT1–Myc-N exhibited approx. 70% transport activity as compared with that of the wild-type OAT1. This 30% decrease in transport activity of hOAT1–Myc-N mainly resulted from a decreased cell-surface expression of the transporter (results not shown). Wild-type OAT1 is known to function as an exchanger [1–3] with one organic anion transported into the cells to be exchanged for another organic anion effluxed from the cells. To determine whether the epitope tag affects such functional characteristics, cells expressing wild-type hOAT1 or hOAT1–Myc-N were preloaded with [14C]PAH followed by exposure to the medium with or without α-oxoglutarate, another substrate for OAT1. As shown in Table 1, similar efflux of intracellular [14C]PAH was observed with both wild-type hOAT1 and hOAT1–Myc-N when cells were exposed to a medium containing an exchangeable substrate α-oxoglutarate, as compared with a medium without α-oxoglutarate. hOAT1–Myc-N was also equally sensitive to the inhibition by probenecid, an inhibitor for OAT, as compared with that of wild-type hOAT1. These results are consistent with our previous observation [19] and suggest that cell-surface-expressed hOAT1–Myc-N preserves the basic functional characteristics of the unmodified transporter.
Table 1. Functional analysis of wild-type (Wt) hOAT1 and hOAT1–Myc-N.
For PAH uptake, transport of [14C]PAH (20 μM, 10 min) in COS-7 cells expressing Wt hOAT1 or hOAT1–Myc-N was measured. Uptake activity was expressed as a percentage of the uptake measured in Wt. For efflux studies, cells expressing Wt hOAT1 or hOAT1–Myc-N were preloaded with [14C]PAH (100 μM) for 1 h, followed by an exposure to PBS (control group) or PBS containing α-ketoglutarate (experimental group). The amount of intracellular [14C]PAH in the experimental group was expressed as a percentage of that in the control group. For inhibition by probenecid, transport of PAH in COS-7 cells expressing Wt hOAT1 or hOAT1–Myc-N was measured in the presence (experimental group) and absence (control group) of 0.5 mM probenecid. Uptake activity remaining in the experimental group was expressed as a percentage of that measured in the control group. The results represent data from three experiments, with triplicate measurements for each transporter construct. The uptake value from pcDNA vector-transfected cell (mock cells) was subtracted from all other uptake values.
| Mutant | Uptake of PAH | Efflux of PAH | Inhibition of PAH uptake by probenecid |
|---|---|---|---|
| Wt-hOAT1 | 100±8.0 | 78.6±4.5 | 33.8±6.9 |
| hOAT1–Myc-N | 69.6±6.9 | 85.0±5.9 | 43.4±4.1 |
Immunofluorescence staining of Myc-tagged hOAT1-transfected cells
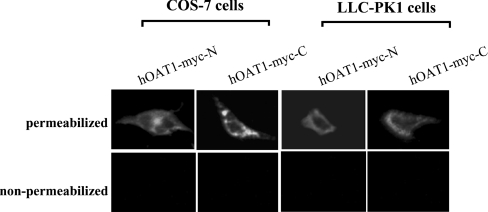
To determine the cellular orientation of both N- and C-termini of hOAT1, we expressed Myc-tagged hOAT1 protein in both COS-7 and LLC-PK1 cells. Myc epitope was tagged at either the N-terminus (hOAT1–Myc-N) or the C-terminus (hOAT1–Myc-C). The transfected cells were then used for indirect immunofluorescent labelling with anti-Myc antibody in conjunction with FITC-conjugated secondary antibody. The cells were incubated in the presence or absence of Triton X-100 to compare intracellular staining and cell-surface staining patterns. As shown in Figure 2, in both hOAT1–Myc-N- and hOAT1–Myc-C-transfected cells, plasma membrane staining could only be observed under permeabilized conditions (incubation with Triton X-100) but not under non-permeabilized conditions, consistent with both the N- and the C-termini being localized intracellularly.
Figure 2. Immunofluorescent localization of Myc-tagged hOAT1 in COS-7 cells and LLC-PK1 cells.
Cells transfected with cDNAs encoding hOAT1–Myc-N or hOAT1–Myc-C were visualized with indirect immunofluorescent labelling. Permeabilized cells were generated by treatment with 0.1% Triton X-100. Proteins were detected with anti-Myc antibody as primary antibody and FITC-conjugated goat anti-mouse IgG as secondary antibody. Specific immunostaining appears as green fluorescence.
Labelling of external lysine residues
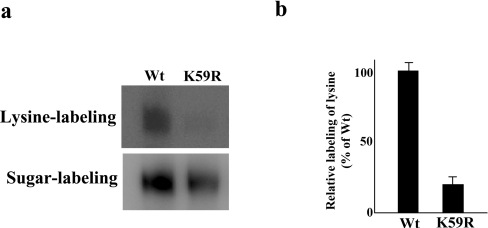
hOAT1 has one predicted extracellular lysine residue (Lys59), which lies in the first extracellular loop. This lysine was removed from hOAT1–Myc-C by substitution with arginine. The resulting mutant K59R has normal transport properties as compared with that of wild-type hOAT1 (Table 2), suggesting that this mutant is properly folded in the cell membrane. To analyse the accessibility of this mutant to membrane-impermeable reagents present in the extracellular medium, we performed two sets of experiments in parallel. In one set of the experiments, we labelled hOAT1–Myc-C- and K59R-transfected cells with membrane-impermeable biotinylation reagent NHS-SS-biotin, which modifies lysine. This experiment was designed to test the presence of external lysine residues. In another set of the experiments, we labelled hOAT1–Myc-C- and K59R-transfected cells with membrane-impermeable biotinylation reagent biotin-LC-hydrazide, which modifies sugar moieties. Since hOAT1 is a glycoprotein, the experiment was designed to determine the total cell-surface expression of this glycoprotein. Western blotting was performed to reveal the results of such labelling. Figure 3(a) showed the labelling of lysine with NHS-SS-biotin and the labelling of sugar moieties with biotin-LC-hydrazide. When the amount of lysine labelled in the hOAT1–Myc-C and in K59R was normalized to their respective total cell-surface expression (Figure 3b), it was apparent that hOAT1–Myc-C was much more reactive to NHS-SS-biotin than that of K59R, indicating that Lys59 is responsible for the reactivity of hOAT1 towards NHS-SS-biotin. This result is consistent with our previous observation [16–18] that the predicted extracellular loop I indeed faces extracellularly.
Table 2. Functional analysis of wild-type (Wt) hOAT1 and K59R.
For PAH uptake, transport of [14C]PAH (20 μM, 10 min) in COS-7 cells expressing Wt hOAT1 or K59R was measured. Uptake activity was expressed as a percentage of the uptake measured in Wt. For efflux studies, cells expressing Wt hOAT1 or K59R were preloaded with [14C]PAH (100 μM) for 1 h, followed by an exposure to PBS (control group) or PBS containing α-oxoglutarate (experimental group). The amount of intracellular [14C]PAH in the experimental group was expressed as a percentage of that in the control group. For inhibition by probenecid, transport of PAH in COS-7 cells expressing Wt hOAT1 or K59R was measured in the presence (experimental group) and absence (control group) of 0.5 mM probenecid. Uptake activity remaining in the experimental group was expressed as a percentage of that measured in the control group. The results represent data from three experiments, with triplicate measurements for each transporter construct. The uptake value from pcDNA vector-transfected cell (mock cells) was subtracted from all other uptake values.
| OAT | Uptake of PAH | Efflux of [14C]PAH | Inhibition of PAH uptake by probenecid |
|---|---|---|---|
| Wt hOAT1 | 100.0±15.6 | 72.8±4.5 | 10.9±1.7 |
| K59R | 112.4±8.0 | 72.4±3.5 | 12.7±1.3 |
Figure 3. Chemical labelling of hOAT1–Myc-C and K59R mutant.
(a) Cells expressing hOAT1–Myc-C or K59R mutant were treated with NHS-SS-biotin to label lysine. Biotinylated proteins were extracted with immobilized streptavidin and subjected to SDS/PAGE and Western blotting with anti-Myc antibody. Cells expressing hOAT1–Myc-C or K59R mutant were treated with biotin-LC-hydrazide to label sugar moieties. Biotinylated proteins were extracted with anti-Myc antibody and subjected to SDS/PAGE and Western blotting with horseradish-peroxidase-conjugated streptavidin. (b) Densitometry analysis of the ratio of labelled lysine to labelled sugar from the experiment shown in (a). The data were quantified relative to the wild-type (Wt) intensity. Each column shows the mean for three experiments and the error bars show the range of observations.
Labelling of introduced lysine residues
By now, we have generated a mutant (K59R) devoid of exposed lysine. Into this background construct, we introduced single lysine into each of the remaining five putative extracellular loops (II, III, IV, V and VI) (Figure 1). Each new lysine substitution mutant was first tested for the transport function. All of the mutants with lysine substitution retained transport activity comparable with that of the wild-type (Table 3), indicating that they are folded properly at the cell surface.
Table 3. Functional analysis of lysine mutants.
Transport of [14C]PAH (20 μM, 10 min) in COS-7 cells expressing wild-type (Wt) hOAT1 or lysine mutant was measured. Uptake activity was expressed as a percentage of the uptake measured in Wt. The results represent data from three experiments, with triplicate measurements for each transporter construct. The uptake value from pcDNA vector-transfected cell (mock cells) was subtracted from all other uptake values.
| OAT | Extracellular loop | Uptake of PAH (% of control) |
|---|---|---|
| Wt hOAT1 | 100.0±15.6 | |
| P183K | II | 64.0±5.2 |
| V244K | III | 75.6±9.5 |
| D359K | IV | 74.0±4.1 |
| I420K | V | 82.4±9.7 |
| L481K | VI | 101.1±9.5 |
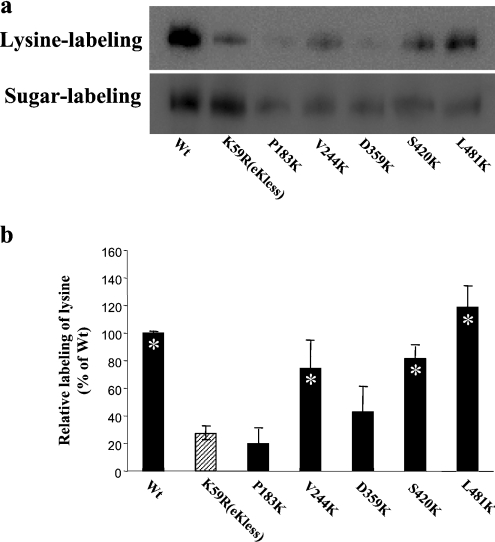
Each of these lysine substitution mutants was then examined for its reactivity to extracellular NHS-SS-biotin (Figure 4). Again, the amount of lysine labelled for each mutant was normalized to its total cell-surface expression to allow evaluation for the different expression levels of the mutants. For V244K in extracellular loop III, S420K in extracellular loop V and L481K in extracellular loop VI, the average labelling in a series of three experiments was similar to that of hOAT1 wild-type. These results suggested that loops III, V and VI are accessible to external NHS-SS-biotin. In contrast, the labelling efficiency of P183K in extracellular loop II as well as D359K in the extracellular loop IV was almost the same as that of K59R, suggesting that the lysine residue introduced into these two loops was not accessible to extracellular NHS-SS-biotin. Lysine was also introduced to three additional positions (L360K, Q361K and G362K) in loop IV. Again, the substituted lysine was not accessible to the extracellular NHS-SS-biotin (results not shown). Replacement of lysine by Val357 and Met358 in this loop resulted in non-functional protein and therefore was not studied further.
Figure 4. Chemical labelling of introduced lysine residues.
(a) Cells expressing hOAT1–Myc-C or lysine mutants were treated with NHS-SS-biotin to label lysine. Biotinylated proteins were extracted with immobilized streptavidin and subjected to SDS/PAGE and Western blotting with anti-Myc antibody. Cells expressing hOAT1–Myc-C or lysine mutant were treated with biotin-LC-hydrazide to label sugar moieties. Biotinylated proteins were extracted with anti-Myc antibody and subjected to SDS/PAGE and Western blotting with horseradish-peroxidase-conjugated streptavidin. (b) Densitometry analysis of the ratio of labelled lysine to labelled sugar from the experiment shown in (a). The data were quantified relative to the wild-type (Wt) intensity. Each column shows the mean for three experiments and the error bars show the range of observations. Asterisks indicate values significantly different (P<0.05) from that of K59R.
DISCUSSION
OATs play essential roles in the body's disposition of clinically important anionic drugs including anti-HIV therapeutics, anti-tumour drugs, antibiotics, anti-hypertensives and anti-inflammatories [1–3]. The OAT family and another closely related OCT (organic cation transporter) family belong to a large group of related proteins, the MFS (major facilitator superfamily). The members of MFS share common structural features, including 12 putative transmembrane-spanning segments and intracellular C- and N-termini. Recent elucidation of high-resolution crystal structures of two other MFS members, LacY [20] and GlpT [21], suggests that all MFS members may share a common fold. Based on such an assumption, a three-dimensional structure model of rat OCT1 has been developed using the template structure of LacY [22]. Such a model revealed a complex 12-transmembrane structure with transmembrane segments 1, 2, 4, 5, 7, 8, 10 and 11 forming a large hydrophilic cleft for substrate binding. However, there has been no experimental data supporting such a topology.
Experimentally establishing the topology of OATs will provide the groundwork that is essential for experimental design and interpretation of results for future structure–function studies. As an initial step to map the topology of OATs, we provided the first experimental evidence on the extracellular loops and the cellular orientation of the N- and the C-termini of hOAT1. All the mutants used for our study had similar functional properties as those of the wild-type, ensuring that our conclusion was derived from the studies of active molecules.
We first showed that both the N- and the C-termini are on the cytoplasmic side of the plasma membrane. This conclusion was supported by immunofluorescent localization of Myc-tagged hOAT1 in permeabilized but not in non-permeabilized cells. Our results are consistent with a transporter with even numbers of transmembrane segments and therefore reject any computer-generated model consisting of odd number of transmembrane segments. It is important to note that our Myc-tagged construct hOAT1–Myc-N showed approx. 30% decrease in transport activity as compared with that of the wild-type OAT1 (Table 1) mainly resulting from a decreased cell-surface expression. However, in the case of studying membrane orientation, our main concern was whether those cell-surface-expressed transporters (not those in the intracellular compartments) had correct membrane conformation. Our results showed that the Myc-tagged hOAT1 has similar behaviour as that of wild-type hOAT1 in terms of efflux and inhibition (Table 1), strongly supporting that the cell-surface-expressed transporters retain the same conformation as that of the wild-type and therefore are suitable for the studies of membrane orientation. Several published papers by other investigators on mapping the topology of the transporters used epitope-tagged transporters, which have lower transport activity as compared with that of the wild-type. For example, the HA (haemagglutinin)-tagged vacuolar Na+/H+ antiporter has approx. 78% transport activity as that of the wild-type [23] and the FLAG-tagged Na+/dicarboxylate co-transporter has only approx. 50% transport activity as that of the wild-type [24].
However, our initial attempt to insert Myc tag into those predicted extracellular loops was unsuccessful. Some of the loops consist of as little as two amino acid residues. Insertion of a ten-amino-acid Myc tag resulted in non-functional proteins, presumably by perturbing the native structure of the transporter. We also considered other methods used in the studies of the membrane topology of other transporters. For example, a reporter group can be fused to the C-terminus or N-terminus of the truncated transporters, and the orientation of the reporter in the cell membrane can then be determined [25,26]. However, if this method is used, usually functional activity is lost so that no reliable test can be made to demonstrate that the truncated polypeptide is processed or folded in the same way as the intact native protein. To circumvent these difficulties, we took a different strategy of site-directed chemical labelling. In such a strategy, a lysine residue was introduced into each predicted extracellular loop of an hOAT1 ‘null’ mutant, in which the exposed lysine residue had been removed. The accessibility of the introduced lysine to membrane-impermeable biotinylation reagent was then analysed. This approach has been successfully used previously to determine the extracellular loops of several transporter proteins [27,28]. The advantages of using such an approach are that small impermeable biotinylation reagents can reach and detect residues in small loops, which might not be accessible to macromolecular reagents such as antibodies and proteases, and that substitution of a single amino acid residue causes only small changes in the protein sequence. Therefore the resulting mutants used in the present study retain most or all of the transport activity of the wild-type transporter.
The predicted external loop I of hOAT1 contains multiple consensus sites for glycosylation. We previously demonstrated [16–18] that these sites were utilized and therefore were part of the external domain. Substitution of lysine residue Lys59 in this loop resulted in a ‘null’ mutant protein (K59R), which was largely inaccessible for labelling by membrane-impermeable NHS-SS-biotin, suggesting that Lys59 is the only exposed lysine residue in hOAT1 responsible for the reactivity to NHS-SS-biotin. The residual labelling of K59R might have resulted from broken or leaky cells in the preparation, which allowed NHS-SS-biotin to react with intracellular lysine residues. The elevated pH required for NHS-SS-biotin labelling may have promoted this effect. Using this null protein (K59R) as a starting point, we introduced one lysine residue into each of the putative extracellular loops so the accessibility of these loops to NHS-SS-biotin could be determined. The lysine residue introduced into predicted extracellular loops III, V and VI had significantly higher reactivity to NHS-SS-biotin as compared with that of the null mutant, establishing that these loops face outside of the cell. The low reactivity of lysine introduced to the predicted extracellular loops II and IV as compared with that of lysine introduced to the predicted extracellular loops III, V and VI is consistent with the relatively smaller size of these two loops (only two amino acid residues in loops II and IV) as compared with that of loops I, III, V and VI. Moreover, introducing lysine residues into several additional locations in the predicted extracellular loop IV also failed to elevate the reactivity of these mutants to NHS-SS-biotin as compared with that of the null mutant. The most likely explanation for our result is that the loops II and IV are too small to be accessible for NHS-SS-biotin, although we cannot absolutely exclude the possibility that the predicted topology model shown in Figure 1 is incorrect. The transmembrane domain helices 3, 4, 7 and 8 may not fully cross the membrane. Further studies are needed to differentiate between these possibilities.
In conclusion, the current studies provided first experimental evidence that both the N- and the C-termini of hOAT1 are localized intracellularly, consistent with a transporter with an even number of transmembrane segments and therefore rejecting any computer-generated model consisting of an odd number of transmembrane segments. We also provided the first experimental evidence that the putative extracellular loops I, III, V and VI are in the extracellular domains of hOAT1. Establishing the membrane topology of OATs is crucial for understanding its structure–function relationship. The studies described here provide the groundwork for future investigations of the membrane topology of these transporters.
Acknowledgments
This work was supported by grant R01-DK 60034 from the National Institutes of Health (to G. Y.).
References
- 1.You G. Structure, function and regulation of renal organic anion transporters. Med. Res. Rev. 2002;22:602–616. doi: 10.1002/med.10019. [DOI] [PubMed] [Google Scholar]
- 2.You G. The role of organic ion transporters in drug disposition: an update. Curr. Drug Metab. 2004;5:55–62. doi: 10.2174/1389200043489207. [DOI] [PubMed] [Google Scholar]
- 3.You G. Towards an understanding of organic anion transporters: structure–function relationships. Med. Res. Rev. 2004;24:762–774. doi: 10.1002/med.20014. [DOI] [PubMed] [Google Scholar]
- 4.Sweet D. H., Wolff N. A., Pritchard J. B. Expression cloning and characterization of ROAT1: the basolateral organic anion transporter in rat kidney. J. Biol. Chem. 1997;272:30088–30095. doi: 10.1074/jbc.272.48.30088. [DOI] [PubMed] [Google Scholar]
- 5.Sekine T., Watanabe N., Hosoyamada M., Kanai Y., Endou H. Expression cloning and characterization of a novel multispecific organic anion transporter. J. Biol. Chem. 1997;272:18526–18529. doi: 10.1074/jbc.272.30.18526. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Nieto C. E., You G., Bush K. T., Barros E. J., Beier D. R., Nigam S. K. Molecular cloning and characterization of NKT, a gene product related to the organic cation transporter family that is almost exclusively expressed in the kidney. J. Biol. Chem. 1997;272:6471–6478. doi: 10.1074/jbc.272.10.6471. [DOI] [PubMed] [Google Scholar]
- 7.Wolff N. A., Werner A., Burkhardt S., Burckhardt G. Expression cloning and characterization of a renal organic anion transporter from winter flounder. FEBS Lett. 1997;417:287–291. doi: 10.1016/s0014-5793(97)01304-5. [DOI] [PubMed] [Google Scholar]
- 8.Sekine T., Cha S. H., Tsuda M., Apiwattanakul N., Nakajima N., Kanai Y., Endou H. Identification of multispecific organic anion transporter 2 expressed predominantly in the liver. FEBS Lett. 1998;429:179–182. doi: 10.1016/s0014-5793(98)00585-7. [DOI] [PubMed] [Google Scholar]
- 9.Cihlar T., Lin D. C., Pritchard J. B., Fuller M. D., Mendel D. B., Sweet D. H. The antiviral nucleotide analogs cidofovir and adefovir are novel substrates for human and rat renal organic anion transporter 1. Mol. Pharmacol. 1999;56:570–580. doi: 10.1124/mol.56.3.570. [DOI] [PubMed] [Google Scholar]
- 10.Lu R., Chan B. S., Schuster V. L. Cloning of the human kidney PAH transporter: narrow substrate specificity and regulation by protein kinase C. Am. J. Physiol. 1999;276:F295–F303. doi: 10.1152/ajprenal.1999.276.2.F295. [DOI] [PubMed] [Google Scholar]
- 11.Kusuhara H., Sekine T., Utsunomiya-Tata N., Tsuda M., Kojima R., Cha S. H., Sugiyama Y., Kanai Y., Endou H. Molecular cloning and characterization of a new multispecific organic anion transporter from rat brain. J. Biol. Chem. 1999;274:13675–13680. doi: 10.1074/jbc.274.19.13675. [DOI] [PubMed] [Google Scholar]
- 12.Cha S. H., Sekine T., Kusuhara H., Yu E., Kim J. Y., Kim D. K., Sugiyama Y., Kanai Y., Endou H. Molecular cloning and characterization of multispecific organic anion transporter 4 expressed in the placenta. J. Biol. Chem. 2000;275:4507–4512. doi: 10.1074/jbc.275.6.4507. [DOI] [PubMed] [Google Scholar]
- 13.Youngblood G. L., Sweet D. H. Identification and functional assessment of the novel murine organic anion transporter Oat5 (Slc22a19) expressed in kidney. Am. J. Physiol. Renal Physiol. 2004;287:F236–F244. doi: 10.1152/ajprenal.00012.2004. [DOI] [PubMed] [Google Scholar]
- 14.Monte J. C., Nagle M. A., Eraly S. A., Nigam S. K. Identification of a novel murine organic anion transporter family member, OAT6, expressed in olfactory mucosa. Biochem. Biophys. Res. Commun. 2004;323:429–436. doi: 10.1016/j.bbrc.2004.08.112. [DOI] [PubMed] [Google Scholar]
- 15.Ekaratanawong S., Anzai N., Jutabha P., Miyazaki H., Noshiro R., Takeda M., Kanai Y., Sophasan S., Endou H. Human organic anion transporter 4 is a renal apical organic anion/dicarboxylate exchanger in the proximal tubules. J. Pharmacol. Sci. 2004;94:297–304. doi: 10.1254/jphs.94.297. [DOI] [PubMed] [Google Scholar]
- 16.Kuze K., Greves P., Leahy A., Wilson P., Stuhlmann H., You G. Heterologous expression and functional characterization of a mouse renal organic anion transporter in mammalian cells. J. Biol. Chem. 1999;274:1519–1524. doi: 10.1074/jbc.274.3.1519. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka K., Xu W., Zhou F., You G. Role of glycosylation in the organic anion transporter OAT1. J. Biol. Chem. 2004;279:14961–14966. doi: 10.1074/jbc.M400197200. [DOI] [PubMed] [Google Scholar]
- 18.Zhou F., Xu W., Hong M., Pan Z., Sinko P. J., Ma J., You G. The role of N-linked glycosylation in protein folding, membrane targeting, and substrate binding of human organic anion transporter hOAT4. Mol. Pharmacol. 2005;67:868–876. doi: 10.1124/mol.104.007583. [DOI] [PubMed] [Google Scholar]
- 19.Xu W., Tanaka K., Sun A. Q., You G. The functional role of the C terminus of human organic anion transporter hOAT1. J. Biol. Chem. 2006;281:31178–31183. doi: 10.1074/jbc.M605664200. [DOI] [PubMed] [Google Scholar]
- 20.Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 22.Popp C., Gorboulev V., Muller T. D., Gorbunov D., Shatskaya N., Koepsell H. Amino acids critical for substrate affinity of rat organic cation transporter 1 line the substrate binding region in a model derived from the tertiary structure of lactose permease. Mol. Pharmacol. 2005;67:1600–1611. doi: 10.1124/mol.104.008839. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi T., Apse M. P., Shi H., Blumwald E. Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12510–12515. doi: 10.1073/pnas.2034966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F. F., Pajor A. M. Topology of the Na+/dicarboxylate cotransporter: the N-terminus and hydrophilic loop 4 are located intracellularly. Biochim. Biophys. Acta. 2001;1511:80–89. doi: 10.1016/s0005-2736(00)00385-0. [DOI] [PubMed] [Google Scholar]
- 25.Clark J. A. Analysis of the transmembrane topology and membrane assembly of the GAT-1 γ-aminobutyric acid transporter. J. Biol. Chem. 1997;272:14695–14704. doi: 10.1074/jbc.272.23.14695. [DOI] [PubMed] [Google Scholar]
- 26.Olivares L., Aragon C., Gimenez C., Zafra F. Analysis of the transmembrane topology of the glycine transporter GLYT1. J. Biol. Chem. 1997;272:1211–1217. doi: 10.1074/jbc.272.2.1211. [DOI] [PubMed] [Google Scholar]
- 27.Chen J. G., Liu-Chen S., Rudnick G. The third transmembrane domain of the serotonin transporter contains residues associated with substrate and cocaine binding. J. Biol. Chem. 1998;273:12675–12681. doi: 10.1074/jbc.272.45.28321. [DOI] [PubMed] [Google Scholar]
- 28.Hu Y. K., Kaplan J. H. Site-directed chemical labelling of extracellular loops in a membrane protein. The topology of the Na,K-ATPase α-subunit. J. Biol. Chem. 2000;275:19185–19191. doi: 10.1074/jbc.M000641200. [DOI] [PubMed] [Google Scholar]