Abstract
TorsinA is a widely expressed AAA+ (ATPases associated with various cellular activities) ATPase of unknown function. Previous studies have described torsinA as a type II protein with a cleavable signal sequence, a single membrane spanning domain, and its C-terminus located in the ER (endoplasmic reticulum) lumen. However, in the present study we show that torsinA is not in fact an integral membrane protein. Instead we find that the mature protein associates peripherally with the ER membrane, most likely through an interaction with an integral membrane protein. Consistent with this model, we provide evidence that the signal peptidase complex cleaves the signal sequence of torsinA, and we show that the region previously suggested to form a transmembrane domain is translocated into the lumen of the ER. The finding that torsinA is a peripheral, and not an integral membrane protein as previously thought, has important implications for understanding the function of this novel ATPase.
Keywords: AAA+ ATPase, early onset torsion dystonia, endoplasmic reticulum, membrane protein, signal peptide, torsinA
Abbreviations: AAA+, ATPases associated with various cellular activities; CAPSO, 3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid; endo H, endoglycosidase H; ER, endoplasmic reticulum; PDI, protein disulfide-isomerase; SPC, signal peptidase complex; TCA, tricarboxylic acid; TMD, transmembrane domain
INTRODUCTION
TorsinA is the founding member of a novel family of four related proteins that belong to the AAA+ (ATPases associated with various cellular activities) superfamily of ATPases. TorsinA was first identified as the protein mutated in early onset torsion dystonia, an inherited neurological disorder characterized by uncontrollable movements and twisted postures [1]. Although its biological role is unknown, torsinA is expressed in most tissues [1] and knockout mice die within 48 h of birth [2], suggesting that it performs an essential and ubiquitous cellular function. Interestingly, torsinA and its relative torsinB are the only AAA+ ATPases known to be located inside the ER (endoplasmic reticulum) of eukaryotic cells. Experiments in cultured cells have established that torsinA is membrane-associated and faces the lumen of the ER [3–6]. However, the mechanisms that mediate the biosynthesis of torsinA and its association with the ER membrane remain unclear, and current evidence indicates that these may differ from conventional mechanisms of signal sequence cleavage and membrane integration. Hence, torsinA contains a 40 amino acid hydrophobic region at its N-terminus (Figure 1A), and recent studies have shown that the first 20 residues function as a signal sequence and are removed from the mature protein by cleavage of the polypeptide after Ala20 [3,5]. The hydrophobic residues 21–40 that remain after cleavage are proposed to anchor torsinA in the ER membrane [1,7]. Consistent with this hypothesis, full-length torsinA has been shown to behave as an integral membrane protein upon alkaline sodium carbonate extraction, whereas a version lacking residues 24–40 behaves as a soluble ER luminal protein [3]. These data have lead to the conclusion that residues 21–40 form a membrane-spanning domain that anchors torsinA in the ER membrane in a type II orientation, with its C-terminus containing the AAA+ domain located in the ER lumen. However, this model of torsinA biosynthesis appears to create a paradox, as cleavable signal sequences usually impose a type I orientation on a mature membrane protein [8]. Since signal sequence cleavage by the SPC (signal peptidase complex) generates a new N-terminus that resides in the lumen of the ER, it is difficult to envisage how a protein with a cleavable signal sequence could become integrated into the ER membrane with a type II orientation.
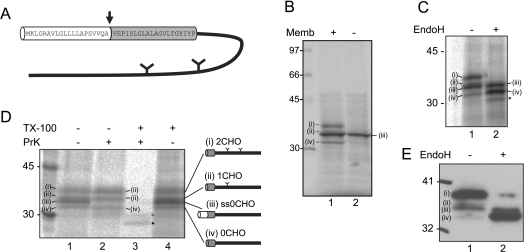
Figure 1. TorsinA undergoes membrane-dependent processing.
(A) Schematic of torsinA showing the sequence of the N-terminal hydrophobic region. The signal sequence is white and the putative TMD is grey. The signal sequence cleavage site is shown with an arrow and the two glycosylation sites are indicated. (B) TorsinA mRNA was translated in the presence or absence of canine pancreatic microsomes, membrane-associated material was isolated by centrifugation and analysed by SDS/PAGE and phosphorimaging. (C) Membrane-associated torsinA was treated with endo H or mock treated. Additional product seen after endo H treatment is labelled with *. (D) Membrane-associated torsinA was treated with proteinase K (PrK), proteinase K and 1% Triton X-100 (TX-100), or 1% Triton X-100. Two limited digestion products are labelled ● and ○. Schematics illustrate the deduced identity of each product. (E) HeLa cells were transiently transfected with torsinA and lysates analysed by Western blotting with affinity purified anti-torsinA.
In order to address these issues, we have examined the signal sequence cleavage and membrane association of torsinA in mammalian cells. We provide evidence that the SPC is responsible for the N-terminal cleavage of torsinA, and demonstrate that the remaining hydrophobic region is fully translocated into the lumen of the ER. In contrast with previous work, the present study shows that torsinA is not a bona fide integral membrane protein. Instead, we find that the mature protein is peripherally associated with the ER membrane, most likely via binding to integral ER protein(s). This novel finding that torsinA is a peripheral and not an integral membrane protein as previously thought has important implications for our understanding of the function of torsinA, and will help to inform future studies aimed at defining the properties of this widely expressed AAA+ ATPase.
EXPERIMENTAL
Reagents and antibodies
T7 RNA polymerase, transcription reagents and rabbit reticulocyte lysate were from Promega, endo H (endoglycosidase H) was from New England Biolabs, and Easytag [35S] protein labelling mix was from PerkinElmer. All other chemicals were from Sigma or BDH/Merck. TorsinA antiserum was obtained from rabbits immunized with a GST (glutathione S-transferase)-tagged protein corresponding to residues 191–332 of human torsinA. Rabbit anti-PDI (protein disulfide-isomerase) was a gift from Professor Neil Bulleid (Faculty of Life Sciences, University of Manchester, Manchester, U.K.), rabbit anti-calnexin was from Stressgen and ribophorin I antiserum was from rabbits immunized with a synthetic peptide KSAVEAERLVAGKLKKD (single letter amino acid codes are used). Horseradish peroxidase-conjugated secondary antibodies were from Dako.
Molecular cloning and DNA manipulations
An expressed sequence tag IMAGE clone (5504660) containing the 5′ end of torsinA was obtained from the HGMP Resource Centre (Human Genome Mapping Project, Hinxton Hall, Cambridge, U.K.) and sequenced to confirm it contained the correct full-length coding sequence. The coding sequence was amplified by PCR and cloned into the EcoRI/BamHI sites of pcDNA3.1(−) (Invitrogen). QuikChange® mutagenesis (Stratagene) was used to generate the torsinA point mutations. A phenylalanine residue was chosen to replace Val18 and Ala20 in the cleavage site mutants since its bulky hydrophobic side chain should not disrupt the hydrophobic signal sequence or introduce a charge that might influence the orientation of the protein.
Cell culture and transient transfection
HeLa cells were grown in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% foetal calf serum. Cells were transiently transfected using Lipofectamine™ 2000 (Invitrogen) and analysed after 18–24 h.
In vitro transcription and translation
Transcription was performed with T7 RNA polymerase as described by the manufacturer and the mRNA obtained was purified with an RNeasy Mini Kit (Qiagen). TorsinA mRNA was translated in rabbit reticulocyte lysate (Promega) for 20 min at 30 °C in the presence of 0.825 μCi/μl [35S] protein labelling mix (NEN) and canine pancreatic microsomes as a source of ER membrane [9]. Aurintricarboxylic acid (0.1 mM) was then added to inhibit translation initiation and samples were incubated at 30 °C for a further 5 min. Membrane-associated products were isolated by centrifugation through a high salt sucrose cushion [250 mM sucrose, 500 mM KOAc, 5 mM Mg(OAc)2, 50 mM Hepes/KOH (pH 7.9)] at 130000 g for 10 min at 4 °C. The resulting membrane pellet was analysed by SDS/PAGE and phosphorimaging, or further processed as described below.
Proteinase K treatment
Protease protection was used to identify in vitro translation products that were not protected within the lumen of the ER. Membranes were resuspended in low salt sucrose (as high salt sucrose with 100 mM KOAc) with or without 1% (w/v) Triton X-100, then incubated with 7.5 μg of proteinase K for 45 min on ice. Reactions were terminated by the addition of 2 mM PMSF, and samples were precipitated in a final concentration of 10% TCA (tricarboxylic acid) and 25% acetone on ice for 1 h. Samples were centrifuged at approx. 16000 g for 20 min and the resulting pellet washed with acetone and analysed by SDS/PAGE.
Alkaline extraction
HeLa cells were detached from culture flasks with trypsin, resuspended in 100 μl KHM [20 mM Hepes (pH 7.2), 110 mM potassium acetate and 2 mM magnesium acetate) and passed twenty times through a ball bearing cell cracker. The resulting homogenate was resuspended in 900 μl of either 25 mM CAPSO [3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid] buffer (pH 9.5) or 100 mM Na2CO3 (pH 11.3), and incubated on ice for 1 h with intermittent vortexing. Samples were centrifuged at 130000 g for 10 min at 4 °C, and the resulting membrane pellet re-extracted with the appropriate buffer. Samples were centrifuged as above to pellet non-extractable proteins, and the two supernatant fractions containing the alkaline extractable material were pooled and precipitated with TCA. Samples of the total cell homogenate, supernatant and pellet fractions were analysed by SDS/PAGE and Western blotting.
Saponin permeabilization and salt washing
HeLa cells were detached from culture flasks with trypsin, resuspended in Hepes buffer [20 mM Hepes (pH 7.4), 100 mM NaCl and 2 mM EDTA) containing 0.2% saponin, and incubated on ice for 10 min. Permeabilized cells were pelleted by centrifugation at approx. 14000 g for 30, and the resulting cell pellet resuspended in Hepes buffer containing 0.1–1 M NaCl or 1–4 M urea. After 10 min incubation on ice, the washed cells were pelleted by centrifugation at 25000 g for 10 min at 4 °C. The cell pellets were resuspended in SDS/PAGE sample buffer, and the supernatant was collected and precipitated with TCA as described above prior to SDS/PAGE and Western blotting.
RESULTS
TorsinA undergoes membrane-dependent processing
Previous studies have found that multiple forms of torsinA are present in mammalian cells [4,5,10,11]. In order to characterize these, we first examined the biosynthesis of torsinA in a cell-free translation system. Four distinct membrane-associated products were generated when translation was carried out in the presence of ER-derived membranes (Figure 1B, lane 1). These were denoted (i)–(iv) on the basis of their migration upon SDS/PAGE. In the absence of membranes, a single torsinA species was produced, which appeared to correspond to product (iii) obtained in the presence of membranes (Figure 1B, compare lanes 1 and 2). The presence of this product in the absence of ER membranes suggests that it is a non-glycosylated, uncleaved form of torsinA. TorsinA contains two N-glycosylation sites [4,12], and treatment with endo H caused a decrease in the intensity of products (i) and (ii), with a corresponding increase in the intensity of product (iv) (Figure 1C, compare lanes 1 and 2). Thus we conclude that products (i) and (ii) represent doubly and singly glycosylated forms of torsinA respectively. The mobility of products (iii) and (iv) were not altered by endo H, confirming that they are not glycosylated. A faint product migrating at approx. 30 kDa was occasionally seen in untreated samples and became more prevalent following endo H treatment (Figure 1C, lane 2, marked with *). This may represent an alternatively processed form or a degradation product of torsinA. In order to examine the orientation of the torsinA species with respect to the ER membrane, samples were treated with proteinase K. In the presence of intact membranes, product (iii) disappeared following proteinase K treatment (Figure 1D, lane 2), confirming that this species is exposed to the cytosol. In contrast, the other forms of torsinA were resistant to protease digestion (Figure 1D, lane 2), consistent with them residing in the lumen of the ER. All of the torsinA products were digested in the presence of detergent (Figure 1D, lane 3). Similarly, when torsinA was transiently expressed in HeLa cells, several forms of the protein were seen (Figure 1E, lane 1). As found in vitro, products (iv) and (iii) remain following endo H treatment, with product (iv) increasing in intensity (Figure 1E, lane 2).
Together these results show that products (i) and (ii) are glycosylated forms of torsinA located in the ER lumen, product (iii) is not glycosylated and is exposed to the cytosol, whilst product (iv) is a non-glycosylated form located in the ER lumen. The fact that the luminal product (iv) migrates more rapidly than the cytosolically oriented product (iii), suggests that it has been subject to cleavage of its N-terminal signal sequence upon translocation across the ER membrane. The difference in apparent molecular mass between products (iii) and (iv) is consistent with this explanation. Furthermore, it is product (iv) that increases in intensity following endo H treatment (Figure 1B, compare lanes 1 and 2), suggesting that the glycosylated products in the ER lumen have undergone signal sequence cleavage as would be predicted.
The simplest interpretation of these results is that product (i) represents the doubly glycosylated, signal sequence-cleaved ‘mature’ form of torsinA, product (ii) is a singly glycosylated, signal sequence-cleaved intermediate, product (iii) is a non-glycosylated, uncleaved form of torsinA associated with the cytoplasmic face of the ER membrane, and product (iv) is a signal sequence-cleaved but non-glycosylated form of the protein. The nature of these four torsinA-derived products is summarized in Figure 1(D).
It is clear that the proportion of torsinA which has undergone signal sequence cleavage and glycosylation is greater in vivo than in vitro [product (i), Figure 1C, lane 1 compared with Figure 1E, lane 1], suggesting that the maturation of torsinA is more efficient in vivo. Therefore an in vivo expression system was used to examine torsinA biogenesis.
The SPC mediates cleavage of the torsinA signal sequence
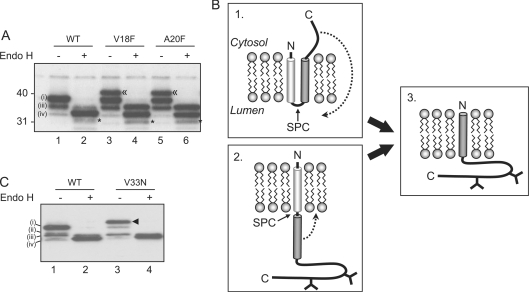
Having established that we could distinguish torsinA that had undergone signal sequence cleavage from the full-length uncleaved protein, we examined this cleavage event in more detail. To determine whether the SPC is responsible for the cleavage of torsinA, amino acid substitutions were made in the cleavage site (Figure 1A). Val18 and Ala20 were replaced with a phenylalanine residue (V18F and A20F respectively) since these occupy the −1 and −3 positions relative to the cleavage site known to be critical for cleavage by SPC [13]. Wild-type and SPC cleavage site mutants were transiently expressed in HeLa cells and the extent of signal sequence cleavage was assessed following endo H treatment. This reduced the complexity of the torsinA species present, allowing us to differentiate between the full-length protein and that which had undergone signal sequence cleavage. Following endo H treatment of wild-type torsinA, two major products were visible, representing uncleaved and cleaved torsinA [Figure 2A, lane 2, (iii) and (iv) respectively]. It is clear from this analysis that the vast majority of wild-type torsinA had undergone signal sequence cleavage. In marked contrast, a significant amount of uncleaved torsinA was seen following endo H treatment of the cleavage site mutants [Figure 2A, lanes 3 and 5, (iii)]. Thus, whilst wild-type torsinA was almost completely cleaved, both V18F and A20F torsinA exhibited a dramatic reduction in signal sequence cleavage. Expression of the mutant proteins also generated a distinct higher molecular mass form of torsinA (Figure 2A, lanes 3 and 5, indicated by ≪). This product was sensitive to endo H (Figure 2A, lanes 4 and 6), suggesting that it represents a glycosylated form. Its increased size [Figure 2A, ≪ compared with (i)] is consistent with the signal sequence still being present. Similar results were obtained with both mutants, and no additive effect was seen with a double V18F/A20F mutation (results not shown). Together these data reveal that altering the putative SPC recognition site of torsinA reduces the extent of signal sequence cleavage. This strongly suggests that the SPC is responsible for cleavage of the N-terminal signal peptide in torsinA.
Figure 2. The SPC mediates cleavage of the torsinA signal sequence.
(A) Wild-type (WT) torsinA or cleavage site mutants (V18F and A20F) were transiently transfected into HeLa cells. Cell lysates were treated with endo H or mock treated and analysed by Western blotting with affinity purified anti-torsinA. A higher molecular mass species generated by the cleavage site mutants is labelled ≪. An additional product seen after endo H treatment is labelled *. A singly glycosylated intermediate (ii) was not apparent in this experiment. (B) Potential mechanisms for torsinA to adopt a type II orientation. (C) Wild-type torsinA (WT) or a form containing a third glycosylation site (V33N) was transiently transfected into HeLa cells, and lysates treated as in (A). An additional species representing triply glycosylated torsinA V33N is labelled ◀.
The N-terminal hydrophobic region is fully translocated into the ER lumen
We next addressed the question of how torsinA adopts the correct final type II orientation (Figure 2B, box 3) following cleavage of the N-terminal signal sequence. One possibility is that the polypeptide is first inserted with its N-terminus towards the ER lumen and subsequently re-orientates following cleavage (Figure 2B, box 1). Alternatively, the putative TMD (transmembrane domain) could be fully translocated into the lumen of the ER before being inserted into the lipid bilayer (Figure 2B, box 2). In order to determine whether the latter scenario occurs, a third glycosylation site was introduced into the putative TMD [14] by mutating Val33 to an asparagine residue (V33N). Glycosylation of this site would indicate that the hydrophobic region is completely translocated into the ER lumen, and does not exit the translocon laterally as is conventional for TMDs. When V33N was expressed in HeLa cells, an additional product migrating more slowly than the wild-type torsinA products was seen (Figure 2C, lane 3 indicated by ◀). This abundant product was sensitive to endo H (Figure 2C, lane 4), and its increased size is consistent with the presence of a third N-linked glycan. Thus the third glycosylation site of V33N is efficiently modified, suggesting that the putative TMD of torsinA is fully translocated into the lumen of the ER. Therefore in order for this region to act as a TMD, it would have to be integrated into the membrane from the ER lumen post-translationally (Figure 2B, box 2). This would represent extremely unusual behaviour as TMDs usually enter the lipid bilayer laterally from the Sec61 translocon [15].
TorsinA is not an integral membrane protein
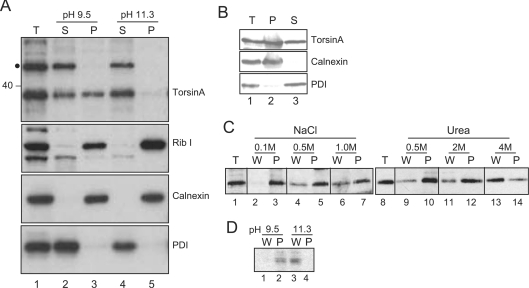
The surprising finding that the putative TMD enters the ER lumen, raised the possibility that torsinA may not be a bona fide integral membrane protein. In order to address this issue, alkaline extractions were performed at two levels of stringency using buffers at pH 9.5 and 11.3 [16]. HeLa cells were disrupted using a cell cracker and extracted twice with the appropriate buffer to completely remove any soluble or peripheral proteins from the ER membrane. Total cell extract, and the supernatant and membrane fractions were analysed by SDS/PAGE and Western blotting (Figure 3). At the less stringent pH 9.5, endogenous torsinA was distributed equally between the supernatant and pellet fractions (Figure 3A, top panel, lanes 2 and 3). Unexpectedly, at pH 11.3, torsinA was found exclusively in the supernatant (Figure 3A, top panel, lanes 4 and 5). The authentic single spanning integral membrane proteins calnexin and ribophorin I remained entirely in the membrane pellet at either pH (Figure 3A, middle panels, lanes 3 and 5), whereas the soluble ER luminal protein PDI was located exclusively in the supernatant at both pH 9.5 and 11.3 (Figure 3A, bottom panel, lanes 2 and 4). These results clearly demonstrate that torsinA does not behave as a typical integral membrane protein, since extraction at pH 11.3 is sufficient to completely remove the protein from the ER membrane. However, torsinA does not behave as a completely soluble protein either, since approximately half of the protein remains membrane associated following extraction at pH 9.5, even though this treatment is sufficient to completely extract PDI. On the basis of these results, we conclude that torsinA is peripherally associated with the inner face of the ER membrane.
Figure 3. TorsinA is peripherally associated with the ER membrane.
(A) HeLa cells were mechanically cracked and extracted twice with CAPSO buffer (pH 9.5) or sodium carbonate (pH 11.3). Membrane-associated material was isolated by centrifugation, and proteins in the supernatant precipitated with TCA. Samples of the total lysate (T), supernatant (S) and membrane pellet (P) were analysed by Western blotting with affinity purified anti-torsinA, or antibodies to ribophorin I (Rib I), calnexin or PDI. ● shows an approx. 50 kDa protein that cross-reacts with the torsinA antisera. (B) HeLa cells were permeabilized with 0.2% saponin and isolated by centrifugation. Samples of total cells (T), supernatant (S) and cell pellet (P) were analysed by Western blotting with anti-torsinA. (C and D) Saponin-permeabilized cells were washed with buffer containing NaCl, urea, CAPSO buffer (pH 9.5) or sodium carbonate (pH 11.3). Cells were reisolated by centrifugation. Samples of the total cells (T), wash (W) and cell pellet (P) were analysed as in (B).
In contrast, Lui et al. [3] found that overexpressed torsinA remained in the membrane pellet following extraction at pH 11.3. However, in their study, extraction was carried out by mixing alkaline sodium carbonate buffer with an equal volume of cell lysate. This may have reduced the effective pH of the buffer, resulting in a lower stringency analysis. Consistent with this interpretation, we found that lowering the pH of the extraction buffer significantly increased the proportion of torsinA that remained membrane associated.
In order to examine the membrane association of torsinA in more detail, HeLa cells were permeabilized with saponin to release soluble ER luminal proteins. PDI was efficiently released into the supernatant following permeabilization (Figure 3B, bottom panel), whilst calnexin remained associated with the cell pellet (Figure 3B, middle panel). TorsinA was distributed equally between the supernatant and pellet (Figure 3B, top panel), suggesting that only a proportion of the endogenous protein is associated with the ER membrane under normal conditions. To provide information on the nature of the association between torsinA and the ER membrane, permeabilized cells were washed with buffers containing increasing concentrations of salt or urea. When cells were washed with low salt buffer in the absence of urea, torsinA remained associated with the cell pellet (Figure 3C, lane 3). In contrast, when cells were washed with buffer containing 1.0 M NaCl or 4 M urea, torsinA was released into the wash fraction (Figure 3C, lanes 6 and 13), suggesting that these conditions were sufficient to disrupt the interaction between torsinA and the ER membrane. Consistent with the results of the alkaline extractions, the membrane-associated torsinA recovered after saponin treatment was released into the wash fraction at pH 11.3 (Figure 3D, lane 3), but remained associated with the cell pellet at pH 9.5 (Figure 3D, lane 2). These results show that the association of torsinA with the ER membrane is sensitive to salt, pH and urea, and therefore most likely reflects binding of torsinA to integral ER membrane protein(s). In this respect it is noteworthy that two of the three torsinA binding partners identified to date are integral membrane proteins; LAP1 in the nuclear envelope and LULL1 in the ER [10,17].
DISCUSSION
Despite recent progress in understanding the biology of torsinA, several of its fundamental biochemical properties remain undefined. Previous studies have described torsinA as a type II protein with a single membrane spanning domain and its C-terminus located in the ER lumen [1,3–5]. This posed the question of how torsinA is both N-terminally cleaved and integrated into the ER membrane in the correct orientation. However, we now show that endogenous torsinA does not behave as a typical integral membrane protein, since it can be fully extracted from the ER membrane at pH 11.3. Under less stringent conditions, a proportion of torsinA does remain associated with the membrane, but we clearly show that the membrane-associated torsinA can be removed by a high-salt or urea wash. Based on these results, we conclude that torsinA is peripherally associated with the ER membrane, most likely via binding to an integral membrane protein. Consistent with this notion, we provide evidence that the SPC mediates the cleavage of torsinA after Ala20, as would be predicted for an ER luminal protein with a cleavable N-terminal signal sequence. Moreover, in the present study we show that the remaining hydrophobic residues are translocated into the ER lumen and available for N-glycosylation, and are not integrated laterally into the ER membrane as would be expected for a typical TMD.
The surprising finding that torsinA is not an integral membrane protein has some important implications. Although torsinA does not have an obvious ER retention motif, the wild-type protein is located primarily in the ER. It has been suggested that TMD-based signals may be responsible for its ER localization, as has been shown for a number of ER membrane proteins [18,19]. However, the lack of a TMD indicates that an alternative mechanism, such as binding to an ER resident protein, is responsible for retaining torsinA in the ER. The absence of a TMD, also raises the possibility that torsinA may interact dynamically with the ER membrane. Such behaviour is important during the catalytic cycle of several other AAA+ ATPases, including NSF (N-ethylmaleimide-sensitive fusion protein) and p97/VCP (97 kDa valosin-containing protein), which catalyse conformational changes in membrane-localized substrates [20]. This idea is supported by our finding that a significant pool of endogenous torsinA is not tightly associated with the ER membrane.
Based on the results detailed above, we propose a model for torsinA biogenesis whereby the nascent polypeptide is targeted to the ER via the hydrophobic sequence at its N-terminus. The first 20 amino acids of this sequence are then removed by the SPC, and the rest of the polypeptide is completely translocated into the ER lumen where it associates peripherally with the luminal face of the ER membrane, most likely via binding to an integral membrane protein. Future research will be aimed at identifying the protein(s) that torsinA binds to at the ER membrane, and establishing whether any such interaction is regulated by ATP binding and/or hydrolysis.
Acknowledgments
This work was supported by the BBSRC (Biotechnology and Biological Sciences Research Council) and co-operative group funding from the MRC (Medical Research Council). S.H. is a BBSRC Professorial Fellow.
References
- 1.Ozelius L. J., Hewett J. W., Page C. E., Bressman S. B., Kramer P. L., Shalish C., de Leon D., Brin M. F., Raymond D., Corey D. P., et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- 2.Goodchild R. E., Kim C. E., Dauer W. T. Loss of the dystonia-associated protein torsina selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z., Zolkiewska A., Zolkiewski M. Characterization of human torsinA and its dystonia-associated mutant form. Biochem. J. 2003;374:117–122. doi: 10.1042/BJ20030258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kustedjo K., Bracey M. H., Cravatt B. F. TorsinA and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J. Biol. Chem. 2000;275:27933–27939. doi: 10.1074/jbc.M910025199. [DOI] [PubMed] [Google Scholar]
- 5.Hewett J., Ziefer P., Bergeron D., Naismith T., Boston H., Slater D., Wilbur J., Schuback D., Kamm C., Smith N., et al. TorsinA in PC12 cells: localization in the endoplasmic reticulum and response to stress. J. Neurosci. Res. 2003;72:158–168. doi: 10.1002/jnr.10567. [DOI] [PubMed] [Google Scholar]
- 6.Hewett J., Gonzalez-Agosti C., Slater D., Ziefer P., Li S., Bergeron D., Jacoby D. J., Ozelius L. J., Ramesh V., Breakefield X. O. Mutant torsinA, responsible for early-onset torsion dystonia, forms membrane inclusions in cultured neural cells. Hum. Mol. Genet. 2000;9:1403–1413. doi: 10.1093/hmg/9.9.1403. [DOI] [PubMed] [Google Scholar]
- 7.Breakefield X. O., Kamm C., Hanson P. I. TorsinA: movement at many levels. Neuron. 2001;31:9–12. doi: 10.1016/s0896-6273(01)00350-6. [DOI] [PubMed] [Google Scholar]
- 8.High S., Dobberstein B. Mechanisms that determine the transmembrane disposition of proteins. Curr. Opin. Cell Biol. 1992;4:581–586. doi: 10.1016/0955-0674(92)90075-n. [DOI] [PubMed] [Google Scholar]
- 9.Walter P., Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- 10.Kamm C., Boston H., Hewett J., Wilbur J., Corey D. P., Hanson P. I., Ramesh V., Breakefield X. O. The early onset dystonia protein torsinA interacts with kinesin light chain 1. J. Biol. Chem. 2004;279:19882–19892. doi: 10.1074/jbc.M401332200. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Alegre P., Paulson H. L. Aberrant cellular behavior of mutant torsinA implicates nuclear envelope dysfunction in DYT1 dystonia. J. Neurosci. 2004;24:2593–2601. doi: 10.1523/JNEUROSCI.4461-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bragg D. C., Kaufman C. A., Kock N., Breakefield X. O. Inhibition of N-linked glycosylation prevents inclusion formation by the dystonia-related mutant form of torsinA. Mol. Cell. Neurosci. 2004;27:417–426. doi: 10.1016/j.mcn.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 13.von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 14.Dalley J. A., Bulleid N. J. The endoplasmic reticulum (ER) translocon can differentiate between hydrophobic sequences allowing signals for glycosylphosphatidylinositol anchor addition to be fully translocated into the ER lumen. J. Biol. Chem. 2003;278:51749–51757. doi: 10.1074/jbc.M303978200. [DOI] [PubMed] [Google Scholar]
- 15.Rapoport T. A., Goder V., Heinrich S. U., Matlack K. E. S. Membrane–protein integration and the role of the translocation channel. Trends Cell Biol. 2004;14:568–575. doi: 10.1016/j.tcb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Nicchitta C. V., Blobel G. Lumenal proteins of the mammalian endoplasmic reticulum are required to complete protein translocation. Cell. 1993;73:989–998. doi: 10.1016/0092-8674(93)90276-v. [DOI] [PubMed] [Google Scholar]
- 17.Goodchild R. E., Dauer W. T. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J. Cell. Biol. 2005;168:855–862. doi: 10.1083/jcb.200411026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato K., Sato M., Nakano A. Rer1p, a retrieval receptor for ER membrane proteins, recognizes transmembrane domains in multiple modes. Mol. Biol. Cell. 2003;14:3605–3616. doi: 10.1091/mbc.E02-12-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker A. K. T., Gergely F. V., Taylor C. W. Targeting of inositol 1,4,5-trisphosphate receptors to the endoplasmic reticulum by multiple signals within their transmembrane domains. J. Biol. Chem. 2004;279:23797–23805. doi: 10.1074/jbc.M402098200. [DOI] [PubMed] [Google Scholar]
- 20.Hanson P. I., Whiteheart S. W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]