Abstract
ATF3 (activating transcription factor 3) gene encodes a member of the ATF/CREB (cAMP-response-element-binding protein) family of transcription factors. Its expression is induced by a wide range of signals, including stress signals and signals that promote cell proliferation and motility. Thus the ATF3 gene can be characterized as an ‘adaptive response’ gene for the cells to cope with extra- and/or intra-cellular changes. In the present study, we demonstrate that the p38 signalling pathway is involved in the induction of ATF3 by stress signals. Ectopic expression of CA (constitutively active) MKK6 [MAPK (mitogen-activated protein kinase) kinase 6], a kinase upstream of p38, indicated that activation of the p38 pathway is sufficient to induce the expression of the ATF3 gene. Inhibition of the pathway indicated that the p38 pathway is necessary for various signals to induce ATF3, including anisomycin, IL-1β (interleukin 1β), TNFα (tumour necrosis factor α) and H2O2. Analysis of the endogenous ATF3 gene indicates that the regulation is at least in part at the transcription level. Specifically, CREB, a transcription factor known to be phosphorylated by p38, plays a role in this induction. Interestingly, the ERK (extracellular-signal-regulated kinase) and JNK (c-Jun N-terminal kinase)/SAPK (stress-activated protein kinase) signalling pathways are neither necessary nor sufficient to induce ATF3 in the anisomycin stress paradigm. Furthermore, analysis of caspase 3 activation indicated that knocking down ATF3 reduced the ability of MKK6(CA) to exert its pro-apoptotic effect. Taken together, our results indicate that a major signalling pathway, the p38 pathway, plays a critical role in the induction of ATF3 by stress signals, and that ATF3 is functionally important to mediate the pro-apoptotic effects of p38.
Keywords: activating transcription factor 3 (ATF3), mitogen activated protein kinase (MAPK), MAPK kinase (MKK), p38, stress kinase, stress response
Abbreviations: ATF, activating transcription factor; C/EBP, CCAAT/enhancer-binding protein; CA, constitutively active; Chop10, C/EBP-homologous protein 10; CRE, cAMP-response element; CREB, CRE-binding protein; DMEM, Dulbecco's modified Eagle's medium; DN, dominant negative; DTT, dithiothreitol; ERK, extracellular-signal-regulated kinase; FBS, fetal bovine serum; gadd153, growth-arrest and DNA-damage-inducible protein 153; β-Gal, β-galactosidase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathione S-transferase; HA, haemagglutinin; HEK-293 cells, human embryonic kidney 293 cells; IL-1β, interleukin 1β; IP–kinase, immunoprecipitation coupled with kinase; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; MEF, mouse embryonic fibroblast; MEK1, MAPK/ERK kinase 1; MKK, MAPK kinase; NF-κB, nuclear factor κB; RT, reverse transcriptase; SAPK, stress-activated protein kinase; shRNA, small-hairpin RNA; tetO, tet operator; TGF-β, transforming growth factor-β; TNFα, tumour necrosis factor α
INTRODUCTION
Cellular responses to extracellular stress signals play an important role in the maintenance of homoeostasis under adverse conditions. An early stress response is to activate cascades of phosphorylation events that, in many cases, increase the expression of the immediate early genes. The immediate early genes encode transcription factors, which in turn regulate downstream genes, initiating a network of transcriptional regulation. Overwhelming evidence indicates that the ATF3 gene, which encodes a member of the ATF (activating transcription factor)/CREB [CRE (cAMP-response element)-binding protein] family of transcription factors, is an immediate early gene induced by a variety of stress signals in different cell types (reviewed in [1,2]). The wide use of the DNA microarray technique has added to the long list of signals that can induce the expression of ATF3 (see [3] for a list of signals). One feature of ATF3 induction is that it is neither tissue-specific nor stimulus-specific. As a few examples, ATF3 is induced in the heart by ischaemia coupled with reperfusion, in the liver by chemical toxicity [4], in the pancreatic β-cells by inflammatory cytokines [5] and in the endothelium by endoplasmic reticulum stress [6]. Interestingly, several other immediate early genes, such as c-Jun and Erg, are often induced in the same cluster as ATF3 in the microarray analysis. Therefore the initial genome response to extra- and/or intra-cellular stress signals appears to turn on a set of common genes, irrespective of the nature of the signals or the cell types exposed to the signals. The diversity in the final readouts is most likely to be determined by the context of the cells.
In addition to induction by stress signals, ATF3 expression is induced under conditions that are not usually considered as stresses. As an example, ATF3 is induced in the MCF-7 breast cancer cells by adipokines [7], the secreted factors from adipocytes. Since adipokines promote the cell survival and motility of MCF-7 cells, they do not fit the conventional definition of stress signals. Furthermore, ATF3 expression is induced in S-phase [8,9]. Thus the characterization of ATF3 as a stress-inducible gene is overly simplistic. We suggest to characterize ATF3 as an ‘adaptive response’ gene that participates in cellular processes to adapt to extra- and/or intra-cellular changes. We note that, similar to the microarray studies of stress responses, immediate early genes such as Jun-B and Erg are induced in the same cluster as ATF3 in the S-phase [8,9]. The involvement of these genes in apparently unrelated processes – cell-cycle regulation and stress response – indicates that a common subset of genes may be required for different cellular processes. Although this may appear to be counter-intuitive, it is not a unique phenomenon. In yeast, a common set of genes was found to play a role in widely divergent biological processes [10]. Thus ATF3 is one of the common subset of ‘adaptive response’ genes that plays a role in a variety of cellular processes.
The induction of ATF3 under such a wide range of conditions suggests that many signalling pathways may be involved in the induction of ATF3. Dissecting the signalling pathways will probably provide insights for future designs to dampen or enhance ATF3 induction using either pharmacological or genetic means. Although several signalling pathways have been demonstrated to be involved in the induction of ATF3 by stress signals, many gaps exist. In the present study, we examined three main MAPK (mitogen-activated protein kinase) pathways – the ERK (extracellular-signal-regulated kinase), JNK (c-Jun N-terminal kinase)/SAPK (stress-activated protein kinase) and p38 pathways – for their potential involvement in the induction of ATF3 by stress signals. For the convenience of discussion, we will refer to the JNK/SAPK pathway as the JNK pathway in the rest of the report. We focused on the MAPK pathways for the following reasons. First, MAPKs are a group of kinases that play an important role in cellular response to extracellular signals [11–13], and many of these signals are known to induce ATF3 expression (reviewed in [1,2]). Secondly, the ATF3 promoter was shown to be activated by ATF2 and c-Jun [14], two transcription factors that are the downstream phosphorylation targets of MAPKs [15–18]. Despite significant cross-talks between the pathways in vitro, there appears to be a reasonable specificity in vivo presumably by the formation of protein–protein complexes through scaffold proteins [19,20]. Therefore it should be possible to distinguish the selective (if not specific) roles of each pathway in the induction of ATF3.
Since all the work on ATF3 induction indicated an increase in the steady-state mRNA level of ATF3, the induction could be due to the increase in ATF3 gene transcription, or the increase in ATF3 mRNA stability, or both. The presence of binding sites for transcription factors known to be phosphorylated (and thus activated) by MAPKs on the ATF3 promoter suggests that the induction is at least in part at the transcription level. Therefore, in addition to the signalling pathways, we addressed the issue of transcription. In the present study, we demonstrate that the p38 pathway is necessary and sufficient to up-regulate the transcription of the ATF3 gene. We also demonstrate for the first time that ATF3 is a functionally important mediator for the pro-apoptotic effects of p38.
MATERIALS AND METHODS
Cell culture
HeLa cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) FBS (fetal bovine serum). COS-1 cells were maintained in MEM (minimum essential medium) supplemented with 10% FBS. Primary MEFs (mouse embryonic fibroblasts) and immortalized MEFs derived from wild-type or ATF3-deficient mice were detailed previously [21] and maintained in DMEM supplemented with 10% FBS, 2 mM glutamine, 0.1 mM non-essential amino acid and 55 μM 2-mercaptoethanol. All cells were maintained in the growing medium in a humidified 5% CO2 atmosphere at 37 °C; no prior serum starvation was included in any experiment.
Plasmid DNAs and reagents
Plasmid DNAs expressing different proteins were kindly provided by various investigators: β-Gal (β-galactosidase) by Dr A. Young (Ohio State University), MEK1 (MAPK/ERK kinase 1)–ERK2 by Dr M. Cobb (University of Texas Southwestern Medical Center at Dallas), MKK7(CA) (where MKK7 is MAPK kinase 7 and CA is constitutively active) by Dr M. Kracht (Medical School Hannover, Germany), JNK1 by Dr J. Woodgett (Ontario Cancer Institute, and Samuel Lunenfeld Research Institute, Ontario, Canada), MKK6(CA) by Dr J. Han (The Scripps Research Institute, La Jolla, CA, U.S.A.), C/EBPβ (CCAAT/enhancer-binding protein) by Dr J. DeWille (Ohio State University), A-CREB by Dr C. Vinson (National Cancer Institute, Bethesda, MD, U.S.A.), MEF2A, MEF2C, MEF2C(R24L) and MEF2C(R3T) by Dr J. D. Molkentin (Cincinnati Children's Hospital Medical Center, University of Cincinnati, Cincinnati, OH, U.S.A.). DNA expressing gadd153/Chop10 (growth-arrest and DNA-damage-inducible protein 153/C/EBP-homologous protein 10) was described previously [4]. pCG-CREB was generated by inserting the CREB open reading frame (from pCREB, a gift of Dr R. Goodman, Vollum Institute, Oregon Health and Science University, Portland, OR, U.S.A.) into the pCG vector. DN (dominant negative) MKK6 construct was generated by site-directed mutagenesis to mutate Lys82 to Ala (‘AAG’ to ‘GCG’). The ATF3 shRNA (small-hairpin RNA) construct targeting at the sense sequence 5′-GAAUAAACACCUCUGCCAUCGGAUG-3′ was generated in pENTR/D-TOPO (Invitrogen) under the control of the U6 promoter (pGEM-U6, a gift from Dr N. Hernandez, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, U.S.A.). Anisomycin (Sigma) was used at 50 ng/ml for all experiments in the present study; SB203580, PD98059 (Calbiochem), IL-1β (interleukin 1β), TNFα (tumour necrosis factor α), TGF-β (transforming growth factor-β; R&D Systems), PMA, adriamycin, camptothecin (Sigma) and H2O2 (Fisher Scientific) were used at the concentrations indicated in the Figure legends. JNKi peptide, a cell-permeable peptide that inhibits the activity of the JNK pathway [22], was custom-made by the Cleveland Clinic Foundation (Cleveland, OH, U.S.A.) and used at the indicated concentrations.
Adenovirus
Adenoviral construct expressing HA (haemagglutinin)-tagged ATF3 was generated using the Invitrogen Gateway® technology. A cassette expressing HA–ATF3 under the control of the tetO (tet operator) was inserted in pENTR/D-TOPO (Invitrogen). The leaky expression from the tetO promoter (in the absence of the tet activator) provided a sufficient level of HA–ATF3 for this study. The tetO-HA–ATF3 insert in the pENTR-TOPO vector was swapped into the destination vector pAd/PL-DEST (Invitrogen). To generate adenovirus, the resulting DNA was transfected into the HEK-293A (human embryonic kidney 293A) cells using Lipofectamine™ 2000 (Invitrogen) according to the instructions from the manufacturers. All viruses were purified following a two-step caesium chloride gradient ultracentrifugation protocol [23]. The ratio of A260/A280 (where A is absorbance) was used to estimate the purity of the virus (expected to be ∼1.2). The titres were calculated as A260 1=1×1012 pfu (plaque-forming units)/ml. Cells were infected at MOI (multiplicity of infection) 50.
RT (reverse transcriptase)–PCR and real-time PCR
Total RNAs were isolated using the TRIzol® method (Invitrogen) and converted into cDNAs using the oligo-dT primer. Five per cent of the cDNAs were used for PCR or real-time PCR as described previously [24] using the following primers. Regular PCR: ATF3 upstream, 5′-GCTGCAAAGTGCCGAAACAAG3′, downstream, 5′-TCTCCAATGGCTTCAGGGTT-3′; GAPDH (glyceraldehyde-3-phosphate dehydrogenase) upstream, 5′-CCGGATCCTGGGAAGCTTGTCATCAACGG-3′, downstream, 5′-GGCTCGAGGCAGTGATGGCATGGACTG-3′. Real-time PCR: ATF3 upstream, 5′-AGCCTGGAGCAAAATGATGCTT-3′, downstream, 5′-AGGTTAGCAAAATCCTCAAACAC-3′, probe, 5′-CACCCAGGCCAGGTCTCTGCCTC-3′; GAPDH upstream, 5′-TCATCAATGGAAATCCCATCA-3′, downstream, 5′-GCCAGCATCGCCCCACTT-3′, probe, 5′-TCTTCCAGGAGCGAGATCCCTCCAAA-3′.
Immunoblot analysis
Whole cell lysates were prepared with 2% (v/v) Triton lysis buffer [4 mM EDTA, 40 mM Tris/HCl, pH 7.5, 20% (v/v) glycerol, 2% Triton X-100, 1 mM DTT (dithiothreitol) and 275 mM NaCl] containing inhibitors for proteases and phosphatases. An equal amount of total protein (30–50 μg) was resolved by SDS/PAGE, and the fractionated proteins were transferred on to polyvinylidene fluoride membranes (Immobilon-P; Millipore). After being blocked with 5% (w/v) non-fat dry milk or 5% (w/v) BSA in TBST (Tris-buffered saline/Tween; 50 mM Tris/HCl, pH 7.5, 50 mM NaCl and 0.1% Tween 20), the membranes were incubated with the indicated primary antibodies diluted (1:1000) in the blocking buffer: ATF3, ERK, p-ERK (phosphorylated ERK), JNK, p38 (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), p-JNK (phosphorylated JNK), p-p38 (phosphorylated p38), cleaved caspase 3 (Cell Signaling Technology, Beverly, MA), and actin (Sigma, St Louis, MO, U.S.A.). Bound primary antibodies were detected using the appropriate horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology) at 1:3000 dilution and Lumi-Light Western Blotting substrate (Roche).
IP–kinase (immunoprecipitation coupled with kinase) reaction for JNK
Whole cell lysates were prepared with 0.1% Triton lysis buffer (0.2 mM EDTA, pH 8.0, 25 mM Hepes, pH 7.6, 1.5 mM MgCl2, 0.1% Triton X-100, 1 mM DTT and 0.3 M NaCl) containing protease and phosphatase inhibitors. Lysates (250 μg) were immunoprecipitated with 0.4 μg of pan-JNK antibody (Santa Cruz Biotechnology) and 20 μl of 50% (v/v) slurry of Protein A–Sepharose (Sigma) for 2 h at 4 °C. Immunoprecipitates were washed twice with lysis buffer and once with kinase reaction buffer (20 mM Hepes, pH 7.6, and 20 mM MgCl), before incubation with 20 μl of kinase buffer containing 20 μM unlabelled ATP, 5 μCi [γ-32P]ATP (at 3000 Ci/mmol) and 5 μg of GST (glutathione S-transferase)–c-Jun-(1–79) substrate in the presence or absence of JNKi peptide for 20 min at 30 °C. The reaction was stopped by the addition of an equal volume of 2×Lamini buffer. The samples were boiled for 5 min before fractionation on an SDS/10% PAGE, and analysed by autoradiography using an X-ray film (Kodak).
Transcription assay of endogenous ATF3 gene
To isolate nuclei, the cell pellet was resuspended in lysis buffer (5 mM Pipes, pH 8.0, 85 mM KCl and 0.5% Nonidet P40) followed by incubation on ice for 10 min. The nuclei were collected by centrifugation at 2000 g for 5 min at 4 °C. Nuclear RNAs were extracted using the TRIzol® method (Invitrogen). ATF3 pre-mRNAs were measured by RT–PCR using an intron–exon primer set: upstream targeted at intron 1, 5′-AGAGCTTCAGCAATGGTTTGC-3′, and downstream targeted at exon B, 5′-CCGCTCGAGACCTGGCCAGGATGTTGAAGC-3′. The control primers for mature ATF3 mRNA are the same as the ones described above for regular RT–PCR. Reactions without RT were included to confirm the lack of genomic DNA contamination. The PCR reaction was carried out as follows: 94 °C for 3 min (one cycle), 94 °C for 45 s followed by 56 °C for 45 s and 72 °C for 2 min (40 cycles), 72 °C for 10 min (one cycle).
RESULTS AND DISCUSSION
Anisomycin activates the MAPK pathways and increases the expression of ATF3
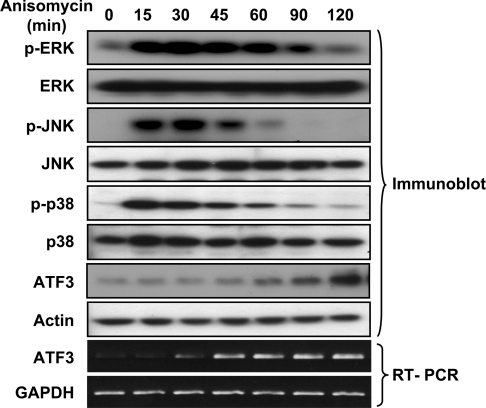
As a stress paradigm to activate the MAPK pathways, we used anisomycin, a well-characterized activator of the JNK and p38 pathways [25–27]. Whether anisomycin activates the ERK pathway has been a subject of debate [25,28], and we found that it also activated the ERK pathway under the condition we examined. Immunoblot analyses using phospho-specific antibodies indicated that anisomycin treatment of COS-1 cells activated all three main MAPK pathways: ERK, JNK and p38 (Figure 1). Control antibodies against the respective kinases confirmed that equivalent amounts of kinases were loaded in each lane. Importantly, anisomycin induced the expression of ATF3 and the time-course analysis indicated that the increase of ATF3 steady-state protein level ensued after the activation of MAPK kinases (Figure 1). Thus anisomycin represents a good model system to delineate the importance of MAPK pathways in the induction of ATF3. Although anisomycin is also a protein synthesis inhibitor, the concentration we used (50 ng/ml) is a sub-inhibitory concentration that does not inhibit translation but activates the stress kinase pathways [25]. RT–PCR confirmed that the steady-state mRNA level of ATF3 also increased upon anisomycin treatment. This increase was observed in a variety of cell lines examined so far, including COS-1, HeLa, MEFs and HEK-293 cells (Figures 1–8 and results not shown).
Figure 1. Anisomycin activates MAPKs and induces ATF3 expression.
COS-1 cells were treated with anisomycin (50 ng/ml) for the indicated times and analysed by immunoblot with the indicated antibodies or by RT–PCR with primers specific to ATF3 or GAPDH.
The ERK pathway is not necessary or sufficient for the induction of ATF3
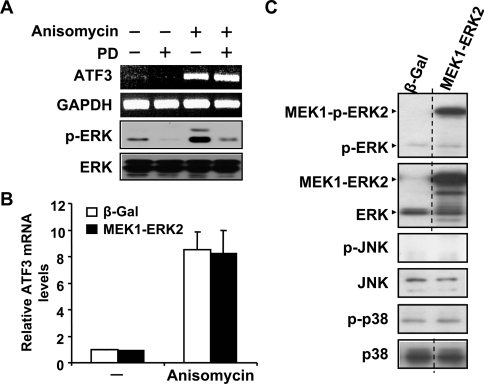
To address whether the ERK pathway is required for the induction of ATF3 expression by anisomycin, we examined whether PD98059, an MEK1 inhibitor [29], affects the induction of ATF3. As shown in Figure 2(A), this inhibitor efficiently blocked the phosphorylation of ERK, but did not decrease the level of ATF3 mRNA, suggesting that ERK is not necessary for anisomycin to induce ATF3. To address whether activation of the ERK pathway is sufficient to induce ATF3, we used a novel CA ERK, which is a fusion protein between ERK2 and its upstream kinase MEK1 [30]. As shown by real-time PCR, MEK1–ERK2 did not increase the steady-state ATF3 mRNA level either in the absence or presence of anisomycin treatment (Figure 2B). Control immunoblot analysis using phospho-specific antibodies confirmed that MEK1–ERK2 activated the ERK pathway but not the JNK or p38 pathway (Figure 2C). Taken together, these results indicate that activation of the ERK pathway is not sufficient to induce ATF3, and is not necessary for the induction of ATF3 by anisomycin.
Figure 2. The ERK pathway is not necessary or sufficient for ATF3 induction.
(A) HeLa cells were pretreated with DMSO or 25 μM PD98059 (PD) for 30 min, followed by anisomycin or buffer treatment (i.e. control) for 1 h, and analysed by RT–PCR with primers specific to ATF3 or GAPDH (top two panels). The effectiveness of the PD98059 pretreatment was determined by the analysis of phospho-ERK at 30 min after anisomycin treatment: immunoblot with antibody against p-ERK or ERK (bottom two panels). (B) COS-1 cells were transfected with DNA expressing β-Gal or MEK1–ERK2 for 36 h, treated with anisomycin or buffer (–) for 1 h and analysed by real-time RT–PCR. ATF3 mRNA level in the β-Gal-transfected untreated cells was arbitrarily defined as 1. Results shown represent the means±S.E.M. for three experiments. (C) The specificity of the MEK1–ERK2 construct to activate the ERK but not the JNK or p38 pathway was demonstrated by phospho-kinase blot. COS-1 cells were transfected with DNA expressing β-Gal or MEK1–ERK2 for 36 h and analysed by immunoblot with the indicated antibodies. The dotted line indicates the grouping of images from different parts of the same gel.
The JNK pathway is not necessary or sufficient for the induction of ATF3
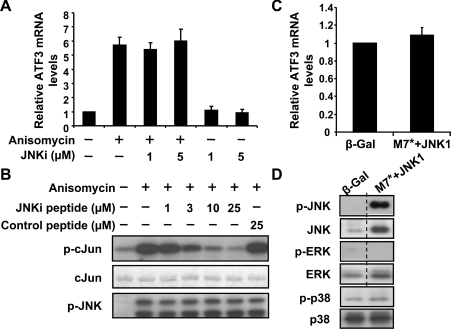
To address whether the JNK pathway is required for the induction of ATF3 by anisomycin, we used a cell-permeable peptide inhibitor JNKi [22] to inhibit the JNK pathway. This peptide contains the minimal inhibitory region derived from the human homologue of JIP (JNK-interacting protein), linked to a ten-amino-acid HIV-Tat (HIV transactivator of transcription) sequence that allows rapid penetration of the peptide into the cytoplasm and nucleus [31]. Real-time PCR indicated that JNKi did not reduce the steady-state ATF3 mRNA level in the presence of anisomycin (Figure 3A), suggesting that the JNK pathway is not necessary for the induction of ATF3 by anisomycin. The effectiveness of this peptide was confirmed by IP–kinase assay: JNK from COS-1 cells treated with anisomycin was immunoprecipitated and assayed for its ability to phosphorylate GST–c-Jun in the absence or presence of JNKi. As shown in Figure 3(B), the JNKi peptide efficiently inhibited the JNK activity (top panel). Coomassie Blue staining indicated that an equivalent amount of substrate (GST–c-Jun) was used in each reaction (middle panel), and immunoblot assay of the extracts confirmed that the same amount of activated JNK (phospho-JNK) was used (bottom panel).
Figure 3. The JNK pathway is not necessary or sufficient for ATF3 induction.
(A) COS-1 cells were pretreated with the indicated amount of JNKi peptide for 30 min, followed by anisomycin or buffer treatment for 1 h and analysed by real-time RT–PCR. ATF3 mRNA level in untreated cells was defined as 1 and means±S.E.M. for four experiments are shown. (B) COS-1 cells were treated with anisomycin for 30 min and analysed by IP–kinase assay for JNK activity in the absence or presence of the indicated inhibitors (top panel). The bottom two panels indicate that comparable amounts of substrate and active kinase were used in each reaction: Coomassie Blue stain of GST–c-Jun (middle panel) and immunoblot of p-JNK in the immunoprecipitate (bottom panel). (C, D) COS-1 cells were transfected with DNA expressing β-Gal or DNAs expressing CA MKK7 (M7*) and JNK1 for 36 h, and analysed by real-time RT–PCR (C) or immunoblot (D). The ATF3 mRNA level in the β-Gal-transfected cells was arbitrarily defined as 1 and means±S.E.M. for five experiments are shown in (C). The dotted line in (D) indicates the grouping of images from different parts of the same gel.
To address the issue of sufficiency, we transfected COS-1 cells with plasmids expressing JNK1 and a CA form of MKK7, an upstream kinase of JNK that has been demonstrated to activate JNK but not ERK or p38 [32]. As shown in Figure 3(C), real-time PCR indicated that co-transfection of JNK1 and MKK7(CA) did not induce ATF3 gene expression. The selectivity of JNK1+MKK7(CA) was demonstrated by the activation of the JNK pathway but not the ERK or p38 pathway (Figure 3D).
The p38 pathway is necessary and sufficient for the induction of ATF3
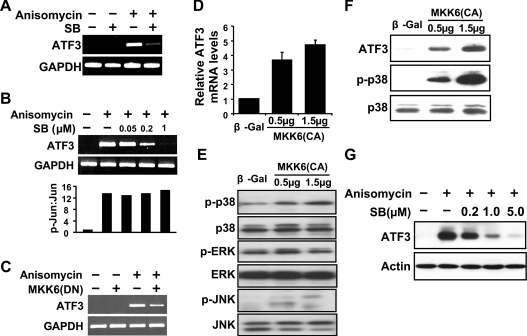
To examine the involvement of the p38 pathway in the induction of ATF3 by stress signals, we treated the cells with SB203580, a p38 kinase inhibitor [33], before the induction of ATF3 by anisomycin. ATF3 mRNA levels were reduced in the presence of SB203580 (Figure 4A) in a dose-dependent manner within the range from 0.05 to 1 μM (Figure 4B, top panel). At high concentrations, SB203580 was demonstrated to also inhibit JNK activity [34]. To determine whether SB203580 inhibited JNK at the concentrations used, we examined the phosphorylation of endogenous c-Jun, a substrate of JNK. As shown in Figure 4(B) (bottom panel), c-Jun phosphorylation was not affected by SB203580 at the doses examined, indicating that the conditions we used did not inhibit JNK activity. As another method to inhibit the p38 pathway, we used a DN inhibitor of MKK6, a p38 upstream kinase. Consistent with SB203580, MKK6(DN) reduced the induction of ATF3 by anisomycin (Figure 4C). Taken together, these results indicate that the p38 pathway is required for the induction of ATF3 by anisomycin.
Figure 4. The p38 pathway is necessary and sufficient for ATF3 induction.
(A) HeLa cells were pretreated with DMSO or 1 μM SB203580 (SB) for 30 min, followed by anisomycin or buffer treatment for 1 h and analysed by RT–PCR. (B) Same as (A), except that the indicated doses of SB203580 were used in the pretreatment (top panel). The lack of JNK inhibition by SB203580 at the indicated doses was demonstrated by the p-Jun/Jun ratio in the cell extracts at 30 min after anisomycin treatment (bottom panel). The p-Jun/Jun ratio in untreated cells was arbitrarily defined as 1. (C) HeLa cells were transfected with DNA expressing β-Gal or MKK6 (DN) for 36 h followed by anisomycin treatment for 1 h, and analysed by RT–PCR. (D, E) COS-1 cells were transfected with DNA expressing β-Gal or MKK6 (CA) for 36 h and analysed by real-time RT–PCR (D) or immunoblot (E). ATF3 mRNA level in the β-Gal-transfected cells was arbitrarily defined as 1 and means±S.E.M. for three experiments are shown in (D). (F) Immortalized MEFs were transfected with DNA expressing β-Gal or MKK6(CA) for 36 h and analysed by immunoblot. (G) Immortalized MEFs were pretreated with the indicated amounts of SB203580 (SB), followed by anisomycin treatment for 3 h and analysed by immunoblot.
To determine whether activation of the p38 pathway is sufficient to induce ATF3, we transfected the cells with DNA expressing MKK6(CA) and examined ATF3 mRNA by real-time PCR. As shown in Figure 4(D), the MKK6(CA) construct induced ATF3 expression. Immunoblot using phospho-specific antibodies indicated that MKK6(CA) activated the p38 but not the ERK pathway (Figure 4E). It also activated the JNK pathway, but only slightly (Figure 4E). Therefore our results suggest that the p38 pathway is necessary and sufficient for ATF3 induction. We have repeated these experiments in several other cells and found similar results. Figures 4(F) and 4(G) show representative immunoblots from immortalized MEFs, confirming the induction of ATF3 by MKK6(CA) (Figure 4F) and inhibition by SB203580 (Figure 4G). Results from HEK-293 cells and primary MEFs are not shown.
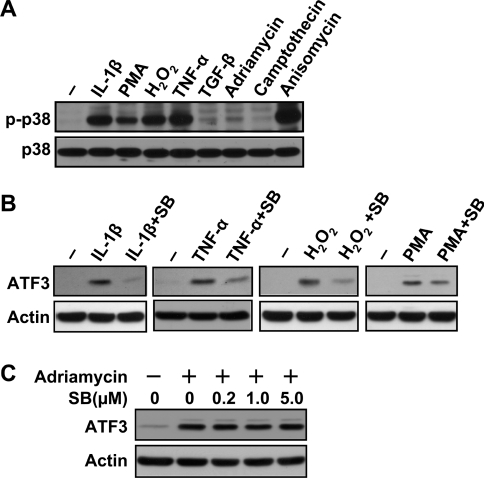
Since ATF3 is induced by various signals, we asked whether the p38 pathway plays a necessary role in treatments other than anisomycin. We first examined several signals to determine whether they can activate the p38 pathway. Among those examined, IL-1β, PMA, H2O2 and TNFα activated the p38 pathway, but TGF-β, adriamycin and camptothecin did not (slightly if any) (Figure 5A). We then examined the role of p38 by pretreating the cells with SB203580 before inducing ATF3 by these agents. As shown in Figure 5(B), SB203580 clearly reduced the induction of ATF3 by IL-1β, TNF-α and H2O2. Although it also reduced the induction of ATF3 by PMA, the effect was less obvious. Thus the p38 pathway plays a necessary role in the induction of ATF3 by various signals (that activate p38), not just anisomycin. As a control, we also pretreated the cells with SB203580 and examined its effect on ATF3 induction by adriamycin, camptothecin and TGF-β. As expected, SB203580 did not reduce the induction of ATF3 to any appreciable degree (Figure 5C and results not shown).
Figure 5. p38 pathway is necessary for ATF3 induction by various stress signals.
(A) HeLa cells were treated with the indicated stresses for 30 min (10 ng/ml IL-1β, 0.5 μg/ml PMA, 100 μM H2O2, 60 ng/ml TNFα, 5 ng/ml TGF-β, 0.5 μg/ml adriamycin, 10 μM camptothecin and 50 ng/ml anisomycin) and analysed for p38 activation by immunoblot using the antibody against p-p38 or p38. (B) HeLa cells were pretreated with 1 μM SB203580 (SB) for 30 min, followed by the treatment with IL-1β, TNFα, H2O2 or PMA at the above indicated concentrations for 3 h, and analysed by immunoblot using the indicated antibodies. Shown is a representative result of three repeats. (C) HeLa cells were pretreated with the indicated amounts of SB203580 (SB) for 30 min, followed by adriamycin treatment (0.5 μg/ml) for 3 h and analysed by immunoblot.
Previously, several studies demonstrated the involvement of MAPKs in the induction of ATF3 by stress signals. As an example, ERK was shown to be necessary for the induction of ATF3 by sulindac sulfide and troglitazone in the HCT-116 human colorectal cancer cells [35]. JNK was shown to be necessary for the induction of ATF3 by homocysteine in endothelial cells [6], by pro-inflammatory cytokines in pancreatic β-cells [5], and by injury in neurons [36]. Both JNK and p38 were shown to be necessary for ionizing radiation to induce ATF3 in fibroblasts [37]. However, these studies did not systematically compare all three major MAPK pathways side-by-side. Thus it is not clear whether the pathway(s) that were not addressed in the reports play(s) a role in the given stress paradigms. During the course of the present study, Inoue et al. [38] reported that inhibition of the p38 pathway reduced the ability of TNFα to induce ATF3. However, the effect was less obvious than the one we observed (Figure 5B). A potential explanation is the differences in cell types – epithelium in our studies versus endothelium in their studies. Furthermore, they described that the ERK pathway inhibits but the JNK pathway promotes the induction of ATF3 by TNFα in the endothelium. This is in contrast with our findings that neither ERK nor JNK played a role using the anisomycin paradigm. These differences again highlight the importance of cellular and stress contexts in signalling events.
Anisomycin increased the transcription of the endogenous ATF3 gene
To address whether the increase in ATF3 steady-state mRNA level is at least in part due to the increase in ATF3 promoter activity, we analysed the transcriptional activity of the endogenous ATF3 gene. An established method to detect endogenous gene transcription is to measure the primary transcripts (pre-mRNAs) [39]. We thus isolated the nuclear RNAs from anisomycin-treated or untreated cells, and examined their ATF3 pre-mRNA levels by RT–PCR using an upstream primer targeted at intron 1 and a downstream primer targeted at exon B of the ATF3 gene. As shown in Figure 6(A), the ATF3 pre-mRNA level was easily detectable in anisomycin-treated cells (lane 5) but not in untreated cells (lane 4). The lack of RT–PCR signals in the absence of the RT (lanes 2 and 3) confirmed that the signals were not derived from the genomic DNAs. That the primer set (intron–exon primers) only detected pre-mRNAs but not mature mRNAs was confirmed by the lack of signals when oligo-dT was used to generate the cDNAs from the total RNAs (lanes 6 and 7). The lack of signals from oligo-dT-derived cDNAs was not due to the degradation of the RNAs, since the control primer set targeted against the mature ATF3 mRNAs detected strong signals (lanes 8 and 9). Using this assay, we demonstrated that SB203580 inhibited anisomycin-induced transcription of the ATF3 gene (Figure 6B) and MKK6(CA) was sufficient to increase the transcription (Figure 6C), indicating that the regulation of ATF3 expression by the p38 pathway is at least in part at the transcriptional level.
Figure 6. The p38 pathway increases the transcription of ATF3 gene.
(A) Immortalized MEFs were treated with anisomycin for 40 min. Nuclear RNAs (for pre-mRNA detection, lanes 1–5) and total RNAs (for mature mRNA detection, lanes 6–9) were isolated and analysed by RT–PCR using the indicated primer sets. The intron–exon primers hybridize to intron 1 and exon B; they detect the ATF3 pre-mRNA and genomic DNA (G. DNA) but not mature mRNA. The control primers hybridize to exon C and exon E; they detect the mature ATF3 mRNA. (B) Immortalized MEFs were pretreated with SB203580 (SB) for 30 min, followed by anisomycin treatment for 40 min and analysed by RT–PCR using the intron–exon primers to detect ATF3 pre-mRNA (as indication of transcription). (C) Immortalized MEFs were transfected with DNAs expressing β-Gal or MKK6(CA) for 36 h and analysed by RT–PCR as in (B).
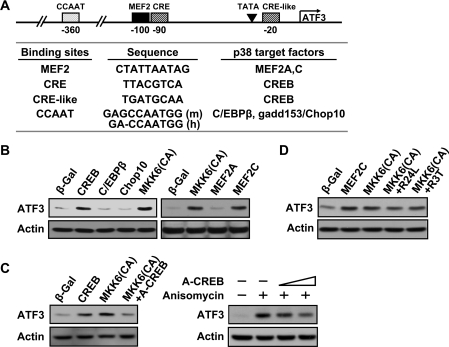
To determine the specific transcription factor(s) mediating the effects of p38, we used a candidate search approach. We inspected the conserved regions of human and mouse ATF3 promoters for binding sites of transcription factors that are known to be the phosphorylation targets of the p38 pathway (reviewed in [40]). Figure 7(A) listed several potential candidates: CREB, C/EBPβ, gadd153/Chop10, MEF2A and MEF2C. As a first step to test these candidates, we transiently transfected DNAs expressing these transcription factors and examined ATF3 expression by immunoblot. As shown in Figure 7(B), ectopic expression of CREB and MEF2C increased the expression of the endogenous ATF3 gene, but C/EBPβ, gadd153/Chop10 and MEF2A did not. In this experiment, β-Gal was used as a negative control and MKK6(CA) as a positive control. To examine whether CREB plays a necessary role, we used A-CREB, a DN CREB [41]. As shown in Figure 7(C), A-CREB reduced the ability of MKK6(CA) (left panel) and anisomycin (right panel) to induce ATF3. The effect of A-CREB on anisomycin is less efficient than that on MKK6(CA). One explanation is that not all cells were transfected by pA-CREB but they all received anisomycin treatment. This was not an issue for the experiment using MKK6(CA), since pMKK6(CA) and pA-CREB were delivered by co-transfection. We also examined the effects of two DN MEF2C constructs: one expressing the R24L mutation and the other R3T [42]. As shown in Figure 7(D), neither DN construct reduced the induction of ATF3 by MKK6(CA) to any appreciable degree. MEF2C is selectively expressed in brain, muscle and spleen [43]. Thus the lack of abundant MEF2C levels in fibroblasts may provide an explanation for the results we observed. We note that many binding sites have been identified in the ATF3 promoter, including the AP-1, CRE/ATF, E2F, p53, NF-κB (nuclear factor κB), SBE [STAT (signal transducer and activator of transcription)-binding element], C/EBP, MEF2 and Egr-1 (early-growth-response-gene product 1) sites [14,35,44–46]. The above result identified CREB as a necessary transcription factor for the p38 pathway to induce ATF3, but is by no means a comprehensive study. Other factors are likely to play a role. In addition, it is possible that the transcription factors required to induce ATF3 under various stress conditions are context-dependent.
Figure 7. The p38 pathway induces ATF3 gene expression at least in part via the transcription factor CREB.
(A) The conserved regions of the human (h) and mouse (m) ATF3 promoter contain potential binding sites for several transcription factors known to be phosphorylated by p38: CREB, C/EBPβ, gadd153/Chop10, MEF2A and MEF2C. (B) Immortalized MEFs were transfected with DNA expressing β-Gal, CREB, C/EBPβ, gadd153/Chop10, MEF2A, MEF2C or MKK6(CA) as indicated for 36 h and analysed by immunoblot. (C) Left panel: immortalized MEFs were transfected with DNA expressing β-Gal, CREB, MKK6(CA), or MKK6(CA) plus A-CREB (a DN CREB) for 36 h and analysed by immunoblot. Right panel: immortalized MEFs were transfected with DNA expressing β-Gal or increasing amounts of A-CREB for 36 h, followed by anisomycin treatment for 3 h and analysed by immunoblot. (D) Immortalized MEFs were transfected with DNA expressing β-Gal, MEF2C, MKK6(CA), or MKK6(CA) plus DN MEF2C (R24L or R3T) for 36 h and analysed by immunoblot. Shown in (B–D) are representative results of three repeats.
The functional relevance of ATF3 induction by the p38 pathway
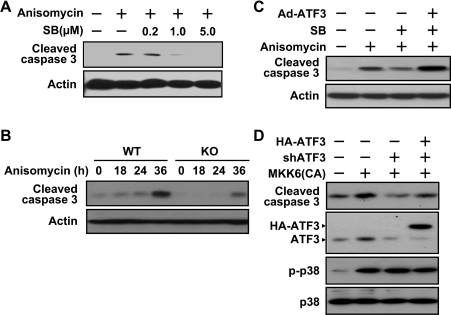
To address the functional significance of ATF3 induction in the context of p38 activation, we first examined the importance of p38 activation upon anisomycin treatment. Activation of the p38 pathway has been implicated to either enhance or reduce apoptosis [47], depending on the cell types and stimuli used in the studies. We found that anisomycin treatment activated caspase 3 in MEFs, and inhibition of p38 by SB203580 reduced anisomycin-induced caspase 3 cleavage (Figure 8A), indicating that p38 is pro-apoptotic in this stress paradigm. Previously, ATF3 was demonstrated to be pro-apoptotic in various stress paradigms using different cells types, including fibroblasts [21], epithelium [5] and endothelium [48]. We found that, in the context of anisomycin treatment, ATF3 was also pro-apoptotic in the MEFs. As shown in Figure 8(B), MEFs derived from the ATF3 knockout (ATF3−/−) mice had reduced caspase 3 activation upon anisomycin treatment compared with those from ATF3+/+ mice. Thus both p38 and ATF3 are pro-apoptotic in the context of anisomycin treatment in MEFs. Although the p38 pathway induces the expression of the ATF3 gene (shown above), it undoubtedly elicits other downstream events. To dissect the functional importance of ATF3, we designed the following experiment. We ectopically expressed ATF3 in the presence of SB203580 to restore only the expression of ATF3 without restoring other p38 functions. As shown in Figure 8(C), ATF3 restored the activation of caspase 3 by anisomycin under the condition that p38 was inhibited. Thus, in the context of anisomycin stress condition, ATF3 is sufficient to activate caspase 3 in the absence of other downstream events elicited by p38. We then asked whether ATF3 is necessary for the p38 pathway to induce apoptosis. As shown in Figure 8(D), ectopic expression of MKK6(CA) activated caspase 3, and reduction of ATF3 by shRNA knockdown diminished the ability of MKK6(CA) to activate caspase 3, supporting a necessary role of ATF3 in the pro-apoptotic function of p38. This effect was reversed by the expression of shRNA-resistant ATF3 (Figure 8D, lane 4), excluding the possibility that the effect of the ATF3 shRNA is due to its non-specific inhibition of other genes.
Figure 8. ATF3 is a downstream effector to mediate the pro-apoptotic effect of p38.
(A) Immortalized MEFs were pretreated with SB203580 for 30 min, followed by anisomycin or buffer treatment for 36 h and analysed by immunoblot with antibody against cleaved caspase 3 or actin. (B) Immortalized wild-type (WT) or ATF3 knockout (KO) MEFs were treated with anisomycin for the indicated times and analysed for caspase 3 activation by immunoblot. (C) Immortalized MEFs were infected with adenovirus expressing HA–ATF3 or a control vector for 6 h, pretreated with 5 μM of SB203580 (SB) for 30 min, followed by anisomycin treatment for 36 h and analysed for caspase 3 activation by immunoblot. (D) Immortalized MEFs were co-transfected with the indicated DNAs expressing the following products: MKK6 (CA), shRNA targeted at mouse ATF3 (shATF3), and HA-tagged human ATF3 that is resistant to the above shATF3. Cells were transfected for 36 h before immunoblot analysis using the indicated antibodies.
Taken together, our results indicate that the p38 pathway plays a critical role in the induction of ATF3 by anisomycin and are the first to show that ATF3 is a functionally important downstream effector for p38 to exert its pro-apoptotic effect. The pro-apoptotic role of ATF3 reported here is consistent with our recent finding that ATF3 represses Ras-mediated tumorigenesis of MEFs in part through promoting apoptosis [21]. Interestingly, ATF3 was shown to be induced by anticancer agents such as curcumin and LY294002, and this induction contributes to the pro-apoptotic effects of these anticancer compounds [49,50]. Similarly, the p38 pathway was demonstrated to mediate the pro-apoptotic effects of anticancer drugs [51], including cannabinoids [52] and 8-chloro-cAMP [53]. Thus our study identifying ATF3 as a functionally important target of the p38 pathway may provide a new mechanistic understanding for p38-mediated apoptosis during chemotherapy.
Acknowledgments
We thank Dr J. Han for pMKK6(CA), Dr M. Kracht for pMKK7(CA), Dr M. Cobb for pMEK1-ERK2, Dr J. Woodgett for pJNK1, Dr N. Hernandez for pGEM-U6, Dr J. DeWille for pC/EBPβ, Dr A. Young for pβ-Gal, Dr R. Goodman for pCREB, Dr C. Vinson for pA-CREB and Dr J. Molkentin for pMEF2A, pMEF2C, pMEF2C(R24L) and pMEF2C(R3T). We acknowledge the past and current members of our laboratory for their contributions: M. Kim for ATF3 shRNA construct and D. Li for the adenoviruses. This work was supported by the National Institutes of Health grants DK59605 and DK064938 (to T. H.) and American Diabetes Association research grant 7-05-RA-52 (to T. H.).
References
- 1.Hai T., Wolfgang C. D., Marsee D. K., Allen A. E., Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7:321–335. [PMC free article] [PubMed] [Google Scholar]
- 2.Hai T., Hartman M. G. The molecular biology and nomenclature of the ATF/CREB family of transcription factors: ATF proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 3.Hai T. The ATF transcription factors in cellular adaptive responses. In: Ma J., editor. Gene Expression and Regulation, a Current Scientific Frontiers Book. People's Republic of China and Springer-Verlag, New York: Higher Education PressBeijing; 2006. pp. 322–333. [Google Scholar]
- 4.Chen B. P. C., Wolfgang C. D., Hai T. Analysis of ATF3: a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol. Cell. Biol. 1996;16:1157–1168. doi: 10.1128/mcb.16.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartman M. G., Lu D., Kim M. L., Kociba G. J., Shukri T., Buteau J., Wang X., Frankel W. L., Guttridge D., Prentki M., et al. Role for activating transcription factor 3 in stress-induced beta-cell apoptosis. Mol. Cell. Biol. 2004;24:5721–5732. doi: 10.1128/MCB.24.13.5721-5732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai Y., Zhang C., Nawa T., Aso T., Tanaka M., Oshiro S., Ichijo H., Kitajima S. Homocysteine-responsive ATF3 gene expression in human vascular endothelial cells: activation of c-Jun NH2-terminal kinase and promoter response element. Blood. 2000;96:2140–2148. [PubMed] [Google Scholar]
- 7.Iyengar P., Combs T. P., Shah S. J., Gouon-Evans V., Pollard J. W., Albanese C., Flanagan L., Tenniswood M. P., Guha C., Lisanti M. P., et al. Adipocyte-secreted factors synergistically promote mammary tumorigenesis through induction of anti-apoptotic transcriptional programs and proto-oncogene stabilization. Oncogene. 2003;22:6408–6423. doi: 10.1038/sj.onc.1206737. [DOI] [PubMed] [Google Scholar]
- 8.Cho R. J., Huang M., Campbell M. J., Dong H., Steinmetz L., Sapinoso L., Hampton G., Elledge S. J., Davis R. W., Lockhart D. J. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 2001;27:48–54. doi: 10.1038/83751. [DOI] [PubMed] [Google Scholar]
- 9.van der Meijden C. M., Lapointe D. S., Luong M. X., Peric-Hupkes D., Cho B., Stein J. L., van Wijnen A. J., Stein G. S. Gene profiling of cell cycle progression through S-phase reveals sequential expression of genes required for DNA replication and nucleosome assembly. Cancer Res. 2002;62:3233–3243. [PubMed] [Google Scholar]
- 10.Ross-Macdonald P., Coelho P. S., Roemer T., Agarwal S., Kumar A., Jansen R., Cheung K. H., Sheehan A., Symoniatis D., Umansky L., et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- 11.Cowan K. J., Storey K. B. Mitogen-activated protein kinases: new signaling pathways functioning in cellular responses to environmental stress. J. Exp. Biol. 2003;206:1107–1115. doi: 10.1242/jeb.00220. [DOI] [PubMed] [Google Scholar]
- 12.Whitmarsh A. J., Davis R. J. Signal transduction by MAP kinases: regulation by phosphorylation-dependent switches. Science STKE 1999. 1999:PE1. doi: 10.1126/stke.1999.1.pe1. [DOI] [PubMed] [Google Scholar]
- 13.Chang L., Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 14.Liang G., Wolfgang C. D., Chen B. P. C., Chen T. H., Hai T. ATF3 gene: genome organization, promoter and regulation. J. Biol. Chem. 1996;271:1695–1701. doi: 10.1074/jbc.271.3.1695. [DOI] [PubMed] [Google Scholar]
- 15.Derijard B., Hibi M., Wu I.-H., Barrett T., Su B., Deng T., Karin M., Davis R. J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 16.Gupta S., Campbell D., Derijard B., Davis R. J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 17.Kallunki T., Su B., Tsigelny I., Sluss H. K., Derijard B., Moore G., Davis R. J., Karin M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 18.Kyriakis J. M., Banerjee P., Nikolakaki E., Dai T., Rubie E. A., Ahmad M. F., Avruch J., Woodgett J. R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 19.Morrison D. K., Davis R. J. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 20.Zanke B. W., Elizabeth A. R., Winnet E., Chan J., Randall S., Parsons M., Bordreau K., McInnis M., Yan M., Templeton D., et al. Mammalian mitogen-activated protein kinase pathways are regulated through formation of specific kinase-activator complexes. J. Biol. Chem. 1996;271:29876–29881. doi: 10.1074/jbc.271.47.29876. [DOI] [PubMed] [Google Scholar]
- 21.Lu D., Wolfgang C. D., Hai T. ATF3: a stress-inducible gene, suppresses Ras-stimulated tumorigenesis. J. Biol. Chem. 2006;281:10473–10481. doi: 10.1074/jbc.M509278200. [DOI] [PubMed] [Google Scholar]
- 22.Bonny C., Oberson A., Negri S., Sauser C., Schorderet D. F. Cell-permeable peptide inhibitors of JNK. Diabetes. 2001;50:77–82. doi: 10.2337/diabetes.50.1.77. [DOI] [PubMed] [Google Scholar]
- 23.Southgate T. D., Kingston P. A., Castro M. G. Current Protocols in Neuroscience. New York: John Wiley and Sons; 2000. Gene transfer into neural cells in vitro using adenoviral vectors; pp. 4.23.21–24.23.40. [DOI] [PubMed] [Google Scholar]
- 24.Allen-Jennings A. E., Hartman M. G., Kociba G. J., Hai T. The roles of ATF3 in glucose homeostasis: a transgenic mouse model with liver dysfunction and defects in endocrine pancreas. J. Biol. Chem. 2001;276:29507–29514. doi: 10.1074/jbc.M100986200. [DOI] [PubMed] [Google Scholar]
- 25.Cano E., Hazzalin C. A., Mahadevan L. C. Anisomycin-activated protein kinases p45 and p55 but not mitogen-activated protein kinases ERK-1 and -2 are implicated in the induction of c-fos and c-jun. Mol. Cell. Biol. 1994;14:7352–7362. doi: 10.1128/mcb.14.11.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cano, Mahadevan Parallel signal processing among mammalian MAPKs. Trends Biochem. Sci. 1995;20:117–122. doi: 10.1016/s0968-0004(00)88978-1. [DOI] [PubMed] [Google Scholar]
- 27.Nahas N., Molski T. F., Fernandez G. A., Sha'afi R. I. Tyrosine phosphorylation and activation of a new mitogen-activated protein (MAP)-kinase cascade in human neutrophils stimulated with various agonists. Biochem. J. 1996;318:247–253. doi: 10.1042/bj3180247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhawan P., Bell A., Kumar A., Golden C., Mehta K. D. Critical role of p42/44(MAPK) activation in anisomycin and hepatocyte growth factor-induced LDL receptor expression: activation of Raf-1/Mek-1/p42/44(MAPK) cascade alone is sufficient to induce LDL receptor expression. J. Lipid Res. 1999;40:1911–1919. [PubMed] [Google Scholar]
- 29.Dudley D. T., Pang L., Decker S. J., Bridges A. J., Saltiel A. R. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson M. J., Stippec S. A., Goldsmith E., White M. A., Cobb M. H. A constitutively active and nuclear form of the MAP kinase ERK2 is sufficient for neurite outgrowth and cell transformation. Curr. Biol. 1998;8:1141–1150. doi: 10.1016/s0960-9822(07)00485-x. [DOI] [PubMed] [Google Scholar]
- 31.Schwarze S. R., Ho A., Vocero-Akbani A., Dowdy S. F. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 32.Tournier C., Whitmarsh A. J., Cavanagh J., Barrett T., Davis R. J. Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7337–7342. doi: 10.1073/pnas.94.14.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuenda A., Rouse J., Doza Y. N., Meier R., Cohen P., Gallagher T. F., Young P. R., Lee J. C. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 34.Clerk A., Sugden P. H. The p38-MAPK inhibitor, SB203580, inhibits cardiac stress-activated protein kinases/c-Jun N-terminal kinases (SAPKs/JNKs) FEBS Lett. 1998;426:93–96. doi: 10.1016/s0014-5793(98)00324-x. [DOI] [PubMed] [Google Scholar]
- 35.Bottone F. G., Jr, Moon Y., Alston-Mills B., Eling T. E. Transcriptional regulation of activating transcription factor 3 involves the early growth response-1 gene. J. Pharmacol. Exp. Ther. 2005;315:668–677. doi: 10.1124/jpet.105.089607. [DOI] [PubMed] [Google Scholar]
- 36.Lindwall C., Dahlin L., Lundborg G., Kanje M. Inhibition of c-Jun phosphorylation reduces axonal outgrowth of adult rat nodose ganglia and dorsal root ganglia sensory neurons. Mol. Cell. Neurosci. 2004;27:267–279. doi: 10.1016/j.mcn.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Kool J., Hamdi M., Cornelissen-Steijger P., van der Eb A. J., Terleth C., van Dam H. Induction of ATF3 by ionizing radiation is mediated via a signaling pathway that includes ATM, Nibrin1, stress-induced MAP kinases and ATF-2. Oncogene. 2003;22:4235–4242. doi: 10.1038/sj.onc.1206611. [DOI] [PubMed] [Google Scholar]
- 38.Inoue K., Zama T., Kamimoto T., Aoki R., Ikeda Y., Kimura H., Hagiwara M. TNFalpha-induced ATF3 expression is bidirectionally regulated by the JNK and ERK pathways in vascular endothelial cells. Genes Cells. 2004;9:59–70. doi: 10.1111/j.1356-9597.2004.00707.x. [DOI] [PubMed] [Google Scholar]
- 39.Lipson K. E., Baserga R. Transcriptional activity of the human thymidine kinase gene determined by a method using the polymerase chain reaction and an intron-specific probe. Proc. Natl. Acad. Sci. U.S.A. 1989;86:9774–9777. doi: 10.1073/pnas.86.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y., Gaestel M. In the cellular garden of forking paths: how p38 MAPKs signal for downstream assistance. Biol. Chem. 2002;383:1519–1536. doi: 10.1515/BC.2002.173. [DOI] [PubMed] [Google Scholar]
- 41.Ahn S., Olive M., Aggarwal S., Krylov D., Ginty D. D., Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol. Cell. Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molkentin J. D., Black B. L., Martin J. F., Olson E. N. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin J. F., Schwarz J. J., Olson E. N. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5282–5286. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang Y., Chen C. R., Massague J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell. 2003;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 45.Zhang C., Gao C., Kawauchi J., Hashimoto Y., Tsuchida N., Kitajima S. Transcriptional activation of the human stress-inducible transcriptional repressor ATF3 gene promoter by p53. Biochem. Biophys. Res. Commun. 2002;297:1302–1310. doi: 10.1016/s0006-291x(02)02382-3. [DOI] [PubMed] [Google Scholar]
- 46.Huo J. S., McEachin R. C., Cui T. X., Duggal N. K., Hai T., States D. J., Schwartz J. Profiles of growth hormone (GH)-regulated genes reveal time-dependent responses and identify a mechanism for regulation of activating transcription factor 3 by GH. J. Biol. Chem. 2006;281:4132–4141. doi: 10.1074/jbc.M508492200. [DOI] [PubMed] [Google Scholar]
- 47.Wada T., Penninger J. M. Mitogen-activated protein kinases in apoptosis regulation. Oncogene. 2004;23:2838–2849. doi: 10.1038/sj.onc.1207556. [DOI] [PubMed] [Google Scholar]
- 48.Nawa T., Nawa M. T., Adachi M. T., Uchimura I., Shimokawa R., Fujisawa K., Tanaka A., Numano F., Kitajima S. Expression of transcriptional repressor ATF3/LRF1 in human atherosclerosis: colocalization and possible involvement in cell death of vascular endothelial cells. Atherosclerosis. 2002;161:281–291. doi: 10.1016/s0021-9150(01)00639-6. [DOI] [PubMed] [Google Scholar]
- 49.Yan C., Jamaluddin M. S., Aggarwal B., Myers J., Boyd D. D. Gene expression profiling identifies activating transcription factor 3 as a novel contributor to the proapoptotic effect of curcumin. Mol. Cancer Ther. 2005;4:233–241. [PubMed] [Google Scholar]
- 50.Yamaguchi K., Lee S. H., Kim J. S., Wimalasena J., Kitajima S., Baek S. J. Activating transcription factor 3 and early growth response 1 are the novel targets of LY294002 in a phosphatidylinositol 3-kinase-independent pathway. Cancer Res. 2006;66:2376–2384. doi: 10.1158/0008-5472.CAN-05-1987. [DOI] [PubMed] [Google Scholar]
- 51.Olson J. M., Hallahan A. R. p38 MAP kinase: a convergence point in cancer therapy. Trends Mol. Med. 2004;10:125–129. doi: 10.1016/j.molmed.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 52.Herrera B., Carracedo A., Diez-Zaera M., Guzman M., Velasco G. p38 MAPK is involved in CB2 receptor-induced apoptosis of human leukaemia cells. FEBS Lett. 2005;579:5084–5088. doi: 10.1016/j.febslet.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 53.Ahn Y. H., Jung J. M., Hong S. H. 8-Chloro-cyclic AMP-induced growth inhibition and apoptosis is mediated by p38 mitogen-activated protein kinase activation in HL60 cells. Cancer Res. 2005;65:4896–4901. doi: 10.1158/0008-5472.CAN-04-3122. [DOI] [PubMed] [Google Scholar]