Abstract
Macromolecular signalling complexes that link neurotransmitter receptors to functionally and structurally associated proteins play an important role in the regulation of neurotransmission. Thus the identification of proteins binding to neurotransmitter receptors describes molecular mechanisms of synaptic signal transduction. To identify interacting proteins of GABAC (where GABA is γ-aminobutyric acid) receptors in the retina, we used antibodies specific for GABAC receptor ρ1–3 subunits. Analysis of immunoprecipitated proteins by MALDI–TOF MS (matrix-assisted laser-desorption ionization–time-of-flight MS) identified the liver regeneration-related protein 2 that is identical with amino acids 253–813 of the Tax1BP1 (Tax1-binding protein 1). A C-terminal region of Tax1BP1 bound to an intracellular domain of the ρ1 subunit, but not to other subunits of GABAC, GABAA or glycine receptors. Confocal laser-scanning microscopy demonstrated co-localization of Tax1BP1 and ρ1 in clusters at the cell membrane of transfected cells. Furthermore, Tax1BP1 and GABAC receptors were co-expressed in both synaptic layers of the retina, indicating that Tax1BP1 is a component of GABAC receptor-containing signal complexes.
Keywords: γ-aminobutyric acid (GABA), immunoprecipitation, matrix-assisted laser-desorption ionization–time-of-flight (MALDI–TOF), protein interaction, synaptic signalling complex, Tax1-binding protein 1 (Tax1BP1)
Abbreviations: CNS, central nervous system; CRABP-I, cellular retinoic acid-binding protein-I; DTAF, 5-(4,6-dichlorotriazinyl)aminofluorescein; GABA, γ-aminobutyric acid; GCL, ganglion cell layer; GLYT-1, glycine transporter-1; GST, glutathione S-transferase; HEK-293 cells, human embryonic kidney cells; INL, inner nuclear layer; IPL, inner plexiform layer; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; MAP, microtubule-associated protein; NCBI, National Center for Biotechnology Information; ONL, outer nuclear layer; OPL, outer plexiform layer; PKC, protein kinase C; RhoGAP, Rho GTPase-activating protein; RT, reverse transcriptase; Tax1BP1, Tax1-binding protein 1; TM, transmembrane domain; TRAF-6, tumour-necrosis-factor-receptor-associated factor 6
INTRODUCTION
Neuronal excitability is regulated by a delicate balance between excitatory and inhibitory neurotransmission. GABA (γ-aminobutyric acid) is the most important inhibitory neurotransmitter receptor in the CNS (central nervous system). The molecule binds to three GABA receptor types, termed A, B and C. Although GABAA and GABAC receptors form ligand-gated ion channels, GABAB receptors couple with G-protein-associated signal cascades [1,2]. GABAC receptors are insensitive to the classical GABAA receptor antagonist bicuculline, do not respond to anaesthetics such as barbiturates and benzodiazepines and have a faster kinetic when compared with GABAA receptors. Within the CNS, GABAC receptors are highly expressed in the retina where they contribute to the sharpening of the visual image [3].
Mammalian GABAC receptors are composed of three ρ subunits (ρ1–3) that assemble into pentameric homo- and hetero-oligomeric receptor complexes [4]. In the retina, ρ subunits are concentrated in discrete clusters at bipolar cells axon terminal systems, while they are more diffusively expressed at the dendrites of these neurons [5–7]. In analogy to the nicotinic acetylcholine receptor, it is assumed that each ρ subunit is anchored in the membrane by four transmembrane domains (TM1–4). N- and C-termini are on the extracellular side of the membrane and one extra- and two intracellular loops connect the four TMS. The intracellular loop between TM3 and TM4 contains consensus sequences for enzymes and phosphorylation of this region was demonstrated in [8]. In recent years, several proteins binding to the TM3–4 loop of different ρ subunits were identified, including proteins of the ZIP family that interact with PKC (protein kinase C), GLYT-1 (glycine transporter-1), CRABP-I (cellular retinoic acid-binding protein-I) and MAP-1B (microtubule-associated protein 1B) [9–12]. However, MAP-1B knockout mice had a normal retina [13] and the physiological consequences of the other protein interactions have not been elucidated either.
Using antibodies specific for the extracellular N-termini of ρ subunits [5], we immunoprecipitated GABAC receptors from solubilized retinal proteins and identified the Tax1BP-1 (Tax1-binding protein 1) as a new interactor of this receptor class. Tax1BP1 was identified by different groups in yeast two-hybrid screens against the human T-cell leukaemia virus type I Tax protein, the zinc-finger protein A20 and TRAF-6 (tumour-necrosis-factor-receptor-associated factor 6) [14–18]. Depending on the originally assigned function of Tax1BP1, the protein is also designated T6BP and TXBP151. In the retina, Tax1BP1 is co-expressed with GABAC receptors in synaptic layers and binds specifically to the TM3–4 loop of the ρ1 subunit, but not to other subunits of GABAC, GABAA or glycine receptors. Based on our data we suggest that Tax1BP1 is a component of GABAC receptor-containing signal complexes in the retina.
EXPERIMENTAL
Preparation of membrane proteins and immunoprecipitation
Six retinae of adult rat were homogenized in 1.5 ml of buffer I (150 mM NaCl and 50 mM Tris/HCl, pH 7.5) containing 1% Nonidet P40 (Sigma–Aldrich, Taufkirchen, Germany), 0.5% sodium deoxycholate, 10 mg/ml DNaseI and a cocktail of protease inhibitors (Roche Diagnostics, Mannheim, Germany) using a Teflon glass homogenizer. The sample was sonificated with 3 bursts for 10 s at 300 W and incubated for 30 min under slow agitation. Subsequently, ultracentrifugation was carried out at 100000 g for 1 h at 4 °C. The supernatant was precleared for 2 h with 25 μl of Protein A-conjugated agarose (Roche Diagnostics). After sedimentation of the beads, the supernatant was incubated with 50 μl of GABAC receptor-specific antibodies [5] for 2 h under slow agitation, supplemented with 25 μl of fresh Protein A-conjugated agarose and agitated for another 16 h. Thereafter, the beads were sedimented, washed twice in 1 ml of buffer I and twice in buffer II (500 mM NaCl and 50 mM Tris/HCl, pH 7.5) containing 0.1% Nonidet P40, 0.05% sodium deoxycholate and the protease inhibitor cocktail. Finally, bound proteins were eluted from the beads by boiling in 30 μl of SDS sample buffer for 10 min. All protein preparation steps were carried out on ice or at 4 °C.
Peptide mass ‘fingerprinting’
The identification of immunoprecipitated proteins by MS was described previously [19]. Briefly, precipitated proteins were separated on 10–15% protein gels, visualized with SyproRuby™ and bands of interest were excised. Gel bands were destained, dried and resuspended in 50 μl of 25 mM NH4HCO3 containing 0.2 μg/μl sequencing grade trypsin for the enzymatic digestion of the purified proteins. The generated peptide fragments were eluted from the gel, eluates were collected and dried and peptides were resuspended in 10 μl of water containing 0.1% trifluoroacetic acid. Subsequently, peptide solutions were diluted 1:10 to 1:50 with water and 0.5 μl of diluted samples were mixed with 0.5 μl of 3-hydroxy-α-cyanocinnamic acid matrix, dotted on to a steel target and air-dried. MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) analysis was performed on an Autoflex (Bruker Daltonics, Bremen, Germany) in the reflector mode, using a nitrogen laser (337 nm) for sample desorption and an acceleration with 19 kV after a delay of 3500 ns. Masses obtained by MALDI–TOF MS were used for a peptide mass ‘finger-print’ search using the search program MS-Fit and the NCBI (National Center for Biotechnology Information) protein database.
Pull-down techniques
Coding sequences for the rat TM3–4 loops of the ρ1–3 subunits, of the α1, β2, γ2 subunits of the GABAA receptor and of the α1 subunit of the glycine receptor were ligated in-frame to the coding sequence of GST (glutathione S-transferase) in pET-41. The coding sequences of the liver regeneration-related protein 2 and of Tax1BP1 were tagged with a T7 epitope by cloning in pET-21. DNA sequencing verified the identity of all constructs, as well as of the plasmids used in yeast two-hybrid experiments and for expression in HEK-293 cells (human embryonic kidney cells), as described below. Plasmids were transformed in Escherichia coli BL21(DE3)pLysS and protein expression was induced by adding 1 mM IPTG (isopropyl β-D-thiogalactoside; Sigma–Aldrich). Fusion proteins were purified under native conditions and immobilized to glutathione–Sepharose as described in [20]. Similar concentrations of immobilized fusion proteins were incubated with the cytosolic fraction of E. coli expressing T7-tagged proteins for 1 h at 4 °C under slow agitation, followed by three washes (4.3 mM Na2HPO4, 1.47 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl and 0.1% Triton X-100, pH 7.3). Bound proteins were eluted by boiling in SDS sample buffer, separated by SDS/PAGE and analysed by Western blotting using a monoclonal anti-T7 immune serum and the enhanced chemiluminescence system (ECL®; Amersham, Braunschweig, Germany). Unless otherwise stated, all reagents were purchased from Novagen (Madison, WI, U.S.A.).
To analyse the interaction between GABAC receptors and Tax1BP1 in the retina, rat retinae were homogenized in buffer I as described above and cell debris was removed from the extract by centrifugation at 20000 g for 30 min. The resulting protein preparation was incubated overnight at 4 °C with 30 μl of a 50% (v/v) suspension of Protein A–agarose (Amersham) that was pre-incubated with antibodies specific for GABAC receptors (50 μl; [5]) or for mGluR2 (metabotropic glutamate receptor type 2; 5 μg; Upstate Biotechnology, Lake Placid, NY, U.S.A.) as control. Precipitates were washed four times with 500 μl of 150 mM NaCl, 50 mM Tris/HCl and 0.1% Triton X-100 (pH 7.5), bound proteins were eluted from the beads by boiling in 20 μl of SDS sample buffer and Tax1BP1 was detected on Western blots using a specific immune serum (sc-15274; 1:100; Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.).
Yeast two-hybrid techniques
The coding sequence of the TM3–4 loop of the rat ρ1 subunit was fused to the activation domain of Gal4 in the bait vector pGBKT7-BD. Coding sequences of Tax1BP1 or of N-terminal deletions were fused to the DNA binding domain of Gal4 in the prey vector pGADT7. Yeast AH109 cells were first transformed with the ρ1 bait vector and the resulting bait strain was subsequently transformed with 1 μg of prey vector constructs. Transformed yeast cells were plated on media selecting for His3 reporter gene activation and growing yeast colonies were then transferred to plates containing ONPG (o-nitrophenyl β-D-galactopyranoside; Sigma–Aldrich) to test β-galactosidase reporter gene activation. All reagents including the plasmid for the negative control were parts of the MATCHMAKER GAL4 Two-Hybrid System 3 (Clontech, Palo Alto, CA, U.S.A.).
Cell culture and immunocytochemistry
Coding sequences of the GABAC receptor ρ1 subunit and of the T7-tagged Tax1BP1 were ligated in the eukaryotic expression vectors pRC/CMV and pRK7 to allow protein expression under the control of the CMV (cytomegalovirus) promoter. Human embryonic kidney cells (HEK-293 cells, ATCC no. CRL 1573) were grown in 3 cm dishes on glass coverslips coated with poly(L-lysine) to approx. 80% confluency and transfected with 1 μg of each plasmid using the calcium phosphate method [21]. After 2 days, the cells were washed three times with PBS, fixed in 4% (v/v) formaldehyde/PBS for 10–15 min on ice, washed again as above and blocked with 10% (v/v) fetal calf serum containing 2% (w/v) BSA and 0.1% Triton X-100 for 10 min. Subsequently, cells were incubated for 2 h with GABAC receptor-specific antibodies from rabbit (1:100) [5] and an anti-T7 serum from mouse to detect Tax1BP1 (1:3000; Novagen) diluted in PBS containing 1% fetal calf serum and 0.1% Triton X-100. After washing, Texas Red (1:500; red fluorescence) and DTAF [5-(4,6-dichlorotriazinyl)aminofluorescein; 1:300; green fluorescence] conjugated goat anti-rabbit or goat anti-mouse IgG secondary antibodies (Dianova, Hamburg, Germany) were applied for 45 min to reveal the binding sites of the primary antibodies. The cells were extensively washed and mounted in Mowiol (Sigma–Aldrich). Labelled proteins were examined with a Leica inverted microscope (DMIRB; Bensheim, Germany) equipped with a cooled MicroMax CCD camera (charge-coupled-device camera; Princeton Instruments, Stanford, CA, U.S.A.) or a Leica confocal microscope (TCS SL). Fluorescence profiles were determined with the beta 4.0.2 version of the data acquisition and analysis software from Scion Corporation (Frederick, MD, U.S.A.).
Isolation of RNA and RT (reverse transcriptase)–PCR
Total RNA was extracted from adult rat tissues following a method described in [22], reverse transcribed and amplified as described previously [20]. The following primers were used for PCR amplification: Tax1BP1 sense, 5′-AGGGAGAAGGAACTTACAAAG-3′; Tax1BP1 antisense, 5′-CTAGTCGAAGTTGAGAACATTC-3′; EF1α sense, 5′-GTCTGCCCAGAAAGCTCAG-3′; EF1α antisense, 5′-AATGGTCTCAAAATTCTGTGAC-3′. Five microlitres of each PCR product were separated on a 1.5% agarose gel and stained with ethidium bromide. Controls were treated as above without adding cDNA or RT and showed no PCR products. Both PCR products were identified by DNA sequencing.
Immunohistochemistry
Adult mice were anaesthetized with halothane and decapitated. Retinal cryostat sections with a thickness of 12 μm were prepared as described in [23] and incubated with a mixture of antibodies specifically recognizing Tax1BP1 (1:100; Santa Cruz Biotechnology) and GABAC receptor ρ1–3 subunits (1:100; [5]). The binding sites of the primary antibodies were revealed by the secondary antibodies Alexa Fluor® 594 (red fluorescence) and Alexa Fluor® 488 (green fluorescence) donkey anti-goat or donkey anti-rabbit IgG (H+L) conjugates respectively (1:500; Molecular Probes, Eugene, OR, U.S.A.). The sections were examined as described above.
RESULTS
Identification of GABAC receptor-associated proteins
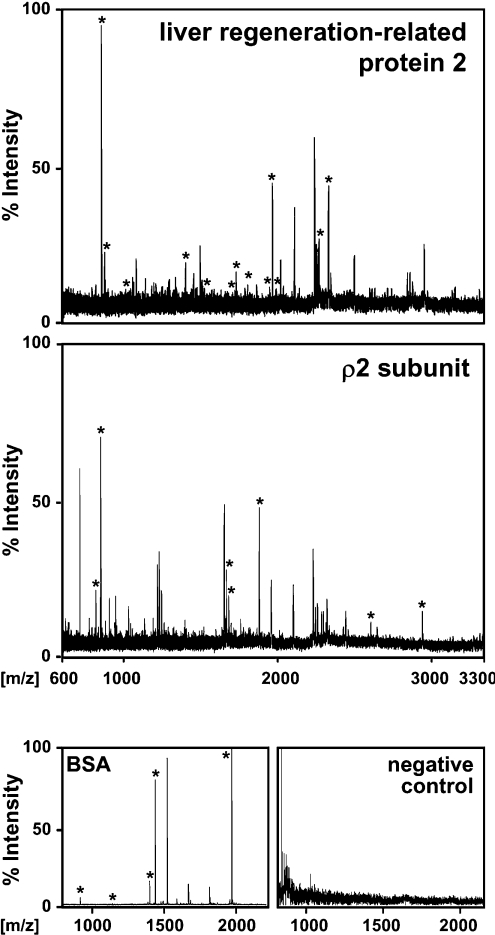
In order to identify new binding partners of GABAC receptors in the retina, detergent extracts from retinal membranes were immunoprecipitated by using antibodies specifically recognizing the extracellular N-termini of the GABAC receptors ρ1–3 subunits [5]. Precipitated proteins were separated on SDS gels, stained with SyproRuby™ (PerkinElmer, Norwalk, CT, U.S.A.) and visualized under UV light. Protein bands were excised from the SDS gel, digested with trypsin and the mass of the generated peptides was determined by MALDI–TOF MS. The resulting peptide mass ‘fingerprints’ (Figure 1) were aligned to theoretical trypsin digests of proteins listed in the NCBI database and yielded several potential GABAC receptor interactors (Table 1). The highest coverage of identified peptides showed the protein sequence of the rat liver regeneration-related protein 2. Importantly, among the precipitated proteins was the GABAC receptor ρ2 subunit, proving that we isolated GABAC receptor-associated protein complexes.
Figure 1. MALDI–TOF analysis of immunoprecipitated proteins using GABAC receptor-specific antibodies.
The spectra show relative peak intensities of peptide masses analysed by MALDI–TOF MS. Peaks derived from peptides that matched the theoretical trypsin digests of proteins in the NCBI protein database are indicated by stars and the identified proteins are labelled in the upper corner of each spectrum. BSA served as a positive control and the indicated negative control was generated from a gel fragment not containing protein.
Table 1. Potential GABAC receptor binding partners identified by immunoprecipitation with GABAC receptor-specific antibodies and subsequent MALDI–TOF analysis.
The liver regeneration-related protein 2 is identical with amino acids 253–813 of Tax1BP1. Two peptides of the liver regeneration-related protein 2 with m/z values larger than 3300 are not labelled in Figure 1(A).
| Candidate | NCBI accession number | Molecular mass (kDa) | Matched peptides | Coverage (%) |
|---|---|---|---|---|
| Liver regeneration-related protein 2 | 21314404 | 64.59 | 15 | 41 |
| GABAC receptor ρ2 subunit | 6679921 | 54.00 | 7 | 17 |
| BCR (B-cell receptor) protein | 29179429 | 44.82 | 5 | 14 |
| Dynamin-1 like protein | 26252094 | 68.70 | 4 | 10 |
| piUS protein | 11067377 | 49.52 | 4 | 9 |
| fez-like protein | 11125691 | 48.94 | 4 | 7 |
Tax1BP1 binds to the TM3–4 loop of the ρ1 subunit in vitro
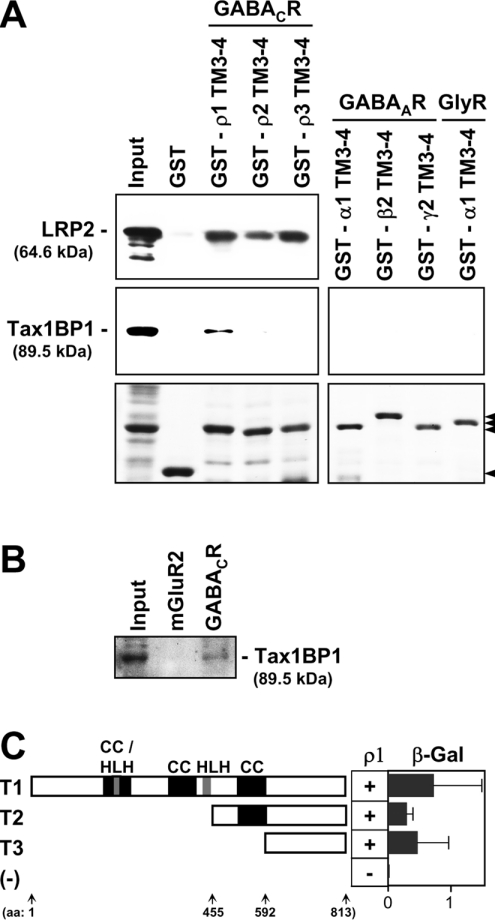
Up to date, all interacting proteins that bind to acetylcholine-type neurotransmitter receptors interact with the intracellular TM3–4 loops of their subunits. Because also GABAC receptors are part of this receptor family, the candidates listed in Table 1 were tested for their binding affinities to the intracellular TM3–4 loops of ρ subunits. In GST pull-down assays, we identified the liver regeneration-related protein 2 as an interactor of these domains (Figure 2A, top panel). The cDNA sequence of the liver regeneration-related protein 2 was cloned from regenerated liver of rat and directly submitted to the NCBI database (accession number AAM46928). A corresponding protein sequence with the mass of 64.6 kDa was translated, but no further data about this protein was available in the literature. In addition, the predicted amino acid sequence does not start with a methionine, suggesting that N-terminal sequences are missing. By searching the NCBI databases we found that amino acids 253–813 of the Tax1BP1 (accession number NP_001004199) are identical with the liver regeneration-related protein 2. Interestingly, the 89.5 kDa protein Tax1BP1 is cleaved by caspases, resulting in a prominent proteolysis product of approx. 65 kDa [14]. Therefore we assumed that cleavage of an N-terminal region of Tax1BP1 generated a 65 kDa proteolytic fragment that we immunoprecipitated together with GABAC receptors. To test this possibility, we analysed Tax1BP1 for its ability to bind the TM3–4 loops of ρ subunits and found a specific interaction with the ρ1 subunit (Figure 2A, middle panel). Intracellular TM3–4 loops of other GABAC receptors ρ subunits, or of the related GABAA and glycine receptors did not bind to Tax1BP1 (Figure 2A, right panel). From these results we concluded that Tax1BP1 specifically interacts with the ρ1 TM3–4 loop and thus we focused on Tax1BP1 for all future experiments.
Figure 2. Specific binding between a C-terminal region of Tax1BP1 and the TM3–4 loop of the GABAC receptor ρ1 subunit.
(A) GST and GST-fusion proteins were immobilized on glutathione–Sepharose and incubated with T7-tagged liver regeneration-related protein 2 (LRP2) or Tax1BP1. Bound proteins were detected on Western blots using a monoclonal anti-T7 immune serum as indicated. Protein concentrations of coated Sepharose beads are shown on Coomassie-stained SDS/PAGE (arrowheads in the lower panels). (B) Immunoprecipitations were performed with antibodies specifically recognizing GABAC receptors or the metabotropic glutamate receptor type 2 as indicated, and bound Tax1BP1 was detected on a Western blot. (C) Schematic representation of the domain structure of Tax1BP1 (T1) and of N-terminal deletion constructs (T2, T3). Encoded amino acids are given as numbers in parentheses. T1–3 constructs were individually tested in yeast cells for their ability to bind the TM3–4 loop of the ρ1 subunit. Interactions were monitored by the ability of transformants to grow on selective media, indicated by ‘+’ or ‘−’. Relative affinities were quantified and visualized as arbitrary β-galactosidase units (β-Gal; horizontal bars). Each value represents the mean for three yeast clones. Error bars are ±S.E. [CC, coiled-coil; HLH, helix–loop–helix; (−), negative control.]
Tax1BP1 and the TM3–4 loop of the ρ1 subunit interact in vivo
To analyse if this interaction also occurs in vivo, we incubated solubilized retinal proteins with Protein A–agarose that was preconjugated with antibodies recognizing GABAC receptors or the metabotropic glutamate receptor type 2 as negative control. Sedimentation of the agarose beads specifically co-precipitated Tax1BP1 with GABAC receptors, indicating that both proteins can interact in retinal cells (Figure 2B).
Furthermore, we transformed the coding sequences of Tax1BP1 and the ρ1 TM3–4 loop, fused to the activation and DNA binding domains of the Gal4 transcription factor, into yeast cells. Binding between Tax1BP1 and the ρ1 TM3–4 loop was demonstrated by the ability of transformants to grow on selective media upon activation of the His3 reporter gene (Figure 2C, construct T1). Tax1BP1 is composed of 813 amino acids and contains three coiled-coil and two helix–loop–helix domains (Figure 2C). To map the ρ1 interacting region in Tax1BP1, we generated the N-terminal deletion constructs T2 and T3 that were tested for their ability to bind the TM3–4 loop of the ρ1 subunit as above. To our surprise, the C-terminal 220 amino acids of construct T3 were sufficient for the binding, while none of the defined protein domains was required for the interaction with the ρ1 TM3–4 loop (Figure 2C).
We estimated the relative binding strengths between the different Tax1BP1 constructs and the ρ1 TM3–4 loop using a quantitative β-galactosidase assay. The results are presented as arbitrary β-galactosidase units (horizontal columns in Figure 2C). Compared with the wild-type Tax1BP1 (T1), construct T3 showed a small, statistically not significant reduction in binding strength, indicating that the C-terminal 220 amino acids of Tax1BP1 are the main region that mediated the binding to the ρ1 TM3–4 loop. These results indicated that Tax1BP1 interacts with the ρ1 TM3–4 loop in vivo and that its C-terminal 220 amino acids play an important role in this interaction.
Tax1BP1 and the ρ1 subunit are co-localized at the cell membrane
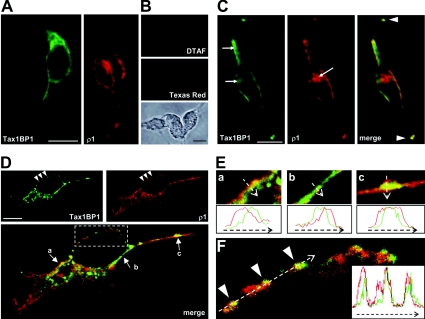
In a next step we analysed the subcellular localization of Tax1BP and the GABAC receptor ρ1 subunit in mammalian cells. First, eukaryotic expression plasmids for TAX1BP1 and the ρ1 subunit were separately transfected in HEK-293 cells. The localization of the expressed proteins was analysed using antibodies recognizing the extracellular ρ1 N-terminus or the T7 epitope fused to the N-terminus of Tax1BP1. Inspection of the subcellular distribution of both proteins by conventional light microscopy demonstrated a comparable localization of Tax1BP1 and the GABAC receptors ρ1 subunit (Figure 3A). Specific signals were detected in cytosolic compartments surrounding the nucleus and in or near the outer cell membrane. Negative controls that were incubated with secondary antibodies only showed no signals (Figure 3B).
Figure 3. Tax1BP1 is present in clusters of the outer cell membrane that contain the GABAC receptor ρ1 subunit.
(A) HEK-293 cells transiently transfected with T7-tagged Tax1BP1 or GABAC receptor ρ1 subunits were incubated with specific primary antibodies, as indicated. Immunoreactivity was detected by secondary antibodies coupled with DTAF (green) or Texas Red (red) using conventional light microscopy. (B) No signals were visible when cells were incubated with secondary antibodies only. The lower panel shows a phase-contrast image of the cells. (C) Tax1BP1 and GABAC receptor ρ1 subunits were co-expressed in the same cell and the distribution of expressed proteins was analysed as in (A). Co-localization of Tax1BP1 and ρ1 at the outer cell membrane can be seen in the merge of the two stainings, resulting in yellow signals. Arrows point to regions of different cytoplasmic localization of both proteins, while arrowheads indicate co-localization in cellular processes. (D) Confocal micrographs of a single z-stack of a HEK-293 cell co-transfected as in (C). The lower panel shows the merge of both upper panels and yellow signals indicate a co-localization of Tax1BP1 and ρ1 subunits. Regions marked by arrows are shown in higher magnification in (E) and the boxed area is enlarged in (F). Arrowheads in (D) and (F) point to clusters at the outer cell membrane containing Tax1BP1 and ρ1 subunits. In (E) and (F), fluorescence profiles were measured along the arrows and relative signal intensities are compared in the diagrams. Scale bars, 10 μm.
To compare the distribution of the binding partners, both proteins were co-expressed in the same cells and analysed as above. Co-localization between Tax1BP1 and the ρ1 subunit was clearly detected as yellow label in several regions of the outer cell membrane of HEK-293 cells (Figure 3C). In addition, specific signals for both proteins were superimposed in cellular processes (arrowheads in Figure 3C). Tax1BP1 and the ρ1 subunit were also observed in the cytoplasm of transfected HEK-293 cells, but mostly at different localizations (arrows in Figure 3C). This is consistent with Tax1BP1 being a cytosolic protein that uses a different machinery for protein biosynthesis than the membrane targeted ρ1 subunit. Indeed, when the same experiment was performed with non-permeabilized cells, the intracellular staining for Tax1BP1 disappeared completely and the ρ1 subunit was detected at the membrane only, proving the cytosolic localization of Tax1BP1 (results not shown).
To verify our findings, we analysed co-transfected HEK-293 cells by confocal laser-scanning microscopy. Signals of a single z-stack specific for Tax1BP1 and the ρ1 subunit demonstrated that both proteins were co-localized in several regions of the outer cell membrane, including cellular processes (Figure 3D). Tax1BP1 and the ρ1 subunit were also present in the cytoplasm. However, as evident from a large amount of red and green label in the lower panel of Figure 3(D), the two proteins showed nearly no intracellular co-localization, consistent with our data from Figures 3(A)–3(C).
The GABAC receptor-specific antibody recognizes the extracellular N-terminus of the ρ1 subunit, while Tax1BP1 binds to its intracellular TM3–4 loop and thus is present within the cytosol. As a result, red label arising from the binding of antibodies to the extracellular ρ1 N-terminus is located at the outer surface of the cell membrane, while green signals represent Tax1BP1 inside the cell. Thus in the event of co-localization, red and green signals will only partially overlap generating a yellow region between both colours. Indeed, we observed the described pattern of red-yellow-green at several regions along the cell membrane (arrows in Figure 3D), which can be best seen in higher power views for the three indicated areas (Figure 3E). The spatial distribution of red and green signals across the outer cell membrane was measured (arrows in Figure 3E) and the fluorescence profiles were compared in the lower panels. Clearly, red and green signals labelling the positions of the ρ1 subunit and Tax1BP1 relative to the membrane overlap in a middle region, visible in yellow.
The ρ1 subunit did not appear homogeneously distributed at the cell surface, but was rather concentrated in clusters along the outer cell membrane (arrowheads in Figure 3D). Importantly, Tax1BP1-specific signals were also present in these clusters. To compare the spatial distribution of these red and green signals in detail, the boxed area of Figure 3(D) was magnified and signal intensities were measured along the arrow drawn in Figure 3(F). The fluorescence profile shown in the inset unequivocally demonstrated that Tax1BP1 is present in clusters of the ρ1 subunit. Based on these findings we concluded that Tax1BP1 and the ρ1 subunit are present at the same subcellular compartments, which is a prerequisite for their physical interaction.
Tax1BP1 is expressed in the mammalian retina and is co-localized with GABAC receptors
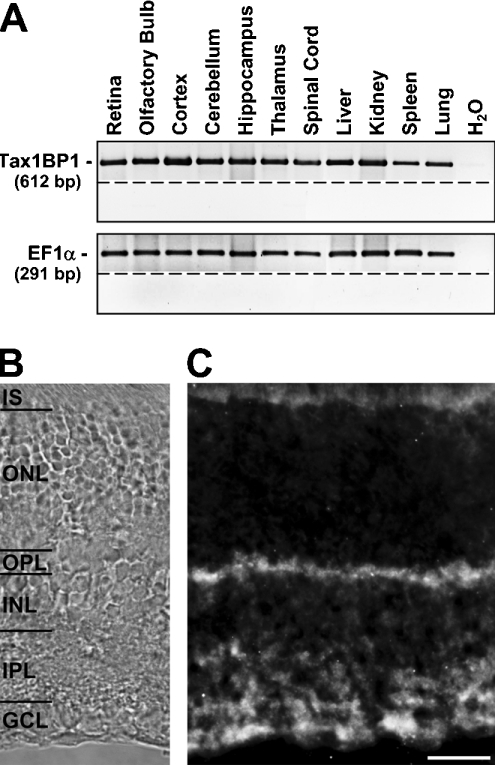
From the results presented so far we postulate a protein interaction between Tax1BP1 and the ρ1 subunit of GABAC receptors. Because GABAC receptors are mainly expressed in the retina, we tested if Tax1BP1 is also present in this tissue. Semi-quantitative RT–PCR experiments detected a robust expression of Tax1BP1 in the retina (Figure 4A). In addition, similar transcript levels were observed for other neuronal or non-neuronal tissues. PCR products of about equal intensity for the housekeeping gene EF1α indicated that comparable amounts of cDNA were used for each sample.
Figure 4. Tax1BP1 is expressed in synaptic layers of the retina.
(A) DNA fragments amplified by RT–PCR from cDNA, obtained after reverse transcription of total RNA isolated from different tissues, as indicated. Amplification of fragments for EF1α ensured similar cDNA concentrations in all samples. Control reactions without the addition of cDNA (H2O), or RT (below dashed lines) resulted in the absence of PCR products. (B) Retinal layers are indicated using phase contrast (IS, inner segments of photoreceptors). (C) Fluorescent micrograph of a vertical cryostat section through the adult mouse retina incubated with Tax1BP1-specific antibodies. Tax1BP1 was strongly expressed in both synaptic layers, the OPL and the IPL and in cell bodies of the GCL. In contrast, weak signals are present in the INL and no label was detected in the ONL. Inner segments of photoreceptors located in the upper part of the micrograph show a faint autofluorescence. Scale bar, 20 μm.
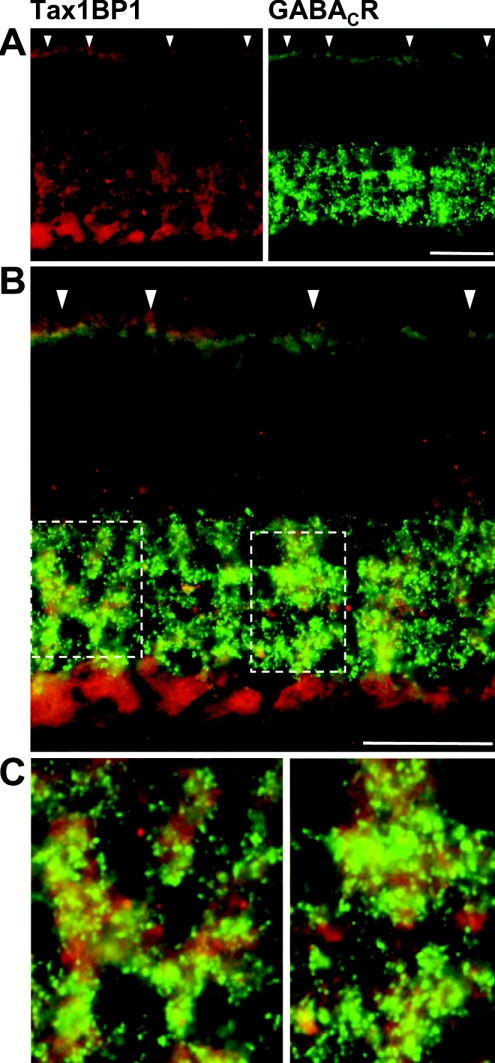
The distribution of GABAC receptor ρ subunits in the retina has been extensively studied [5–7]. Therefore we analysed the retinal localization of Tax1BP1 and compared it with the expression pattern of GABAC receptors. Upon staining vertical cryostat sections of the adult mouse retina with antibodies specific for Tax1BP1, label was present in both synaptic layers, the OPL (outer plexiform layer) and IPL (inner plexiform layer) (Figure 4C). No signals were detected in the ONL (outer nuclear layer) containing cell bodies of photoreceptors, and only faint label was observed in the INL (inner nuclear layer), where the somata of bipolar, horizontal and amacrine cells are located. In contrast, strong staining was present in the GCL (ganglion cell layer). A weak autofluorescence of the inner segments of photoreceptors is visible at the upper border of the micrograph.
Next, we compared the expression of Tax1BP1 and GABAC receptors by double-labelling retinal sections with specific antibodies for both proteins. Tax1BP1 and GABAC receptors showed a comparable expression in both synaptic layers of the retina (Figure 5A). Indeed, the merge of the two panels demonstrated a good co-localization of both proteins in the OPL and in the IPL (Figures 5B and 5C). In contrast, Tax1BP1 expressed in the GCL did not co-localize with GABAC receptors, indicating that Tax1BP1 and GABAC receptors are co-expressed in a subset of retinal neurons.
Figure 5. Tax1BP1 and GABAC receptors are co-expressed in synaptic layers of the retina.
(A) Micrographs of vertical cryostat sections through an adult mouse retina double-immuno-labelled for Tax1BP1 (red) and GABAC receptors (green) demonstrate a similar distribution of both proteins in the synaptic layers of the retina. This can be best seen as yellow colour in the merge of the two stainings shown in higher magnification in (B). In contrast, cell bodies located in the GCL are labelled for Tax1BP1 but do not express GABAC receptors. Regions of extensive co-localization in the OPL are indicated by arrowheads, while the boxed areas in the IPL are enlarged in (C). Scale bars, 20 μm.
DISCUSSION
Macromolecular signalling complexes that physically link neurotransmitter receptors, ion channels, enzymes and scaffold proteins are important regulators for the neurotransmission in the CNS. In the present study, we identified Tax1BP1 as a new binding partner of GABAC receptors. Antibodies that specifically recognize the extracellular N-termini of GABAC receptor ρ subunits immunoprecipitated the liver regeneration-related protein 2 with a size of approx. 65 kDa that is a shorter variant of the 89.5 kDa protein Tax1BP1. The sequence of the NCBI database entry for the liver regeneration-related protein 2 (accession number AAM46928) does not start with a methionine residue, suggesting that N-terminal sequences are missing. Interestingly, Tax1BP1 is cleaved by members of the caspase3-like subfamily, e.g. caspase 6, resulting in a prominent proteolysis product of approx. 65 kDa [14]. Based on these data, we assume that an N-terminal region of Tax1BP1 was cleaved by caspases, generating a 65 kDa proteolytic fragment that bound to the immunoprecipitated GABAC receptors and that we detected by MS. This assumption is supported by our finding that the C-terminal 220 amino acids of Tax1BP1 being identical with the liver regeneration-related protein 2 are sufficient for the interaction with the ρ1 subunit. Furthermore, all peptides of the peptide mass ‘fingerprints’ that led to the identification of the liver regeneration-related protein 2 are also present in Tax1BP1.
GST pull-down and yeast two-hybrid experiments demonstrated a direct interaction between the intracellular TM3–4 loop of the ρ1 subunit with the C-terminal 220 amino acids of Tax1BP1. In contrast, the same domains of the ρ2 and ρ3 subunits did not bind to Tax1BP1. This was surprising, because the mass ‘fingerprints’ of our initial immunoprecipitation identified only the ρ2 subunit. An explanation for this discrepancy could be that (i) native GABAC receptors are hetero-oligomeric protein complexes mostly composed of ρ1 and ρ2 subunits [24] and that (ii) the retinal expression level of the ρ2 subunit is much higher than that of ρ1 [25,26]. Thus, although we identified ρ2 but not ρ1 in our immunoprecipitate, both subunits are co-assembled in hetero-oligomeric GABAC receptor pentamers, enabling ρ1 to bind Tax1BP1, while the higher expressed ρ2 was detected in our mass ‘fingerprints’. Alternatively, the proteolytic 65 kDa fragment of Tax1BP1 could simply fold into a three-dimensional structure different from the 89.5 kDa Tax1BP1, resulting in different binding characteristics for ρ subunits. Indeed, we found that the shorter fragment interacted with all ρ subunits, including ρ2.
Tax1BP1 is an anti-apoptotic protein that is constitutively expressed in human cells and that binds to the Tax protein of the human T-cell leukaemia virus type 1. Tax is localized in the nucleus and in the cytoplasm of infected cells. There, it interacts with different host proteins such as Tax1BP1 in order to regulate viral and cellular transcription, signal transduction pathways of the host cell and apoptosis [27,28]. Tax1BP1 also interacts with A20, a Cys2/Cys2 zinc-finger protein that is induced by inflammatory stimuli and inhibits apoptosis [14]. Binding of Tax1BP1 to the C-terminal zinc-finger domains of A20 mediated its anti-apoptotic activity [29]. Furthermore, Tax1BP1 is associated with the Rho GTPase-activating protein RhoGAP [30].
Because Tax1BP1 is a TRAF-6-binding protein it is also designated T6BP [18]. The interaction between Tax1BP and TRAF-6 is mediated by coiled-coil domains of Tax1BP1 that bind to the N-terminal ring- and zinc-finger domains of TRAF-6 [18]. Because Tax1BP uses its C-terminal 220 amino acids to interact with the ρ1 TM3–4 loop, its coiled-coil domains are free to bind TRAF-6, enabling a simultaneous association of the three proteins. In addition to binding Tax1BP1, TRAF-6 interacts with members of the PKC-ζ interacting protein family that are termed p62 or ZIP1-3 [31]. Interestingly, as Tax1BP1, also ZIP proteins bind to the TM3–4 loops of GABAC receptor ρ subunits and it was proposed that ZIP proteins function as a linker between GABAC receptors and PKC-ζ [11]. Similar to Tax1BP1, ZIP proteins use different domains to bind GABAC receptors, TRAF-6 and PKC-ζ and thus could serve as a scaffold between these proteins [11,31]. This hypothesis is supported by our finding that Tax1BP1 and the ZIP-binding PKC-ζ are co-localized with GABAC receptors in the retina.
Tax1BP1 and its binding partners A20, RhoGAP, TRAF-6 and proteins of the ZIP family are associated with intracellular signal cascades that are regulated by interleukin-1, nerve growth factors, tumour necrosis factors and ubiquitin, effecting e.g. gene expression and apoptosis [18,29,31]. Recently, CRABP was described as a binding partner of the GABAC receptor ρ1 subunit in the retina [12]. The authors suggest that this interaction could serve as a link between the GABA signalling pathway and the control of gene expression in neurons. However, an influence of GABAC receptors-mediated signalling on the above-mentioned factors such as interleukin-1 or tumour necrosis factor is not known. Interestingly, GABAC receptor ρ1 and ρ2 subunits were expressed normally in apoptotic retinae of an animal model for secondary angle closure glaucoma [32].
Besides being expressed in the retina, GABAC receptor ρ subunits were also found in other brain regions such as the hippocampus, cerebellum and the superior colliculus, as well as in the spinal cord [33–36]. Furthermore, ρ subunits were detected in secretory cells of the pituitary, in neurons of the gastrointestinal tract and in a neuroendocrine tumour cell line of the gut [37–40]. These findings suggest that in addition to regulating neurotransmission, GABAC receptors are associated with secretory pathways. This idea is strengthened by the described binding between ρ1 and Tax1BP1, because Tax1 itself interacts with several secretory pathways [41].
The lowest sequence similarity between ρ subunits is found in their intracellular TM3–4 loops. The same holds true when the TM3–4 loops of GABAC receptors are compared with subunits of GABAA and glycine receptors. Because of this sequence heterogeneity, these domains offer an ideal molecular mechanism for the diverse regulation of different receptor types, simply by specifically interacting with a large variety of intracellular proteins. Thus it was not surprising that also the immunoprecipitated Tax1BP1 bound to this domain. Tax1BP used a C-terminal region to interact with the ρ1 subunit, leaving its coiled-coil and helix–loop–helix domains accessible for other protein interactions as well as for dimerization [18]. By this means one can imagine a picture in which the intracellular TM3–4 loop of a GABAC receptor ρ1 subunit binds to a C-terminal region of Tax1BP1. Dimerization of Tax1BP1 via its second coiled-coil domain could cross-link two GABAC receptors. If the cross-linked GABAC receptors contain more than one ρ1 subunit, additional Tax1BP1 proteins can bind and dimerize, forming connections to other GABAC receptors which would generate large clusters at synaptic sites. In addition, the first and third coiled-coil domains and the helix–loop–helix regions of Tax1BP1 are still accessible for other protein interactions, thus enabling the formation of multimeric signal complexes at GABAC receptor-expressing synapses in the retina.
Acknowledgments
We thank Melanie Rose and Angela Graulich for continuous support in staining retinal cryostat sections and Nadja Schröder for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft.
References
- 1.Bormann J. The ‘ABC’ of GABA receptors. Trends Pharmacol. Sci. 2000;21:16–19. doi: 10.1016/s0165-6147(99)01413-3. [DOI] [PubMed] [Google Scholar]
- 2.Rudolph U., Crestani F., Möhler H. GABA(A) receptor subtypes: dissecting their pharmacological functions. Trends Pharmacol. Sci. 2001;22:188–194. doi: 10.1016/s0165-6147(00)01646-1. [DOI] [PubMed] [Google Scholar]
- 3.Roska B., Werblin F. Vertical interactions across ten parallel, stacked representations in the mammalian retina. Nature. 2001;410:583–587. doi: 10.1038/35069068. [DOI] [PubMed] [Google Scholar]
- 4.Enz R. GABAC receptors: a molecular view. Biol. Chem. 2001;382:1111–1122. doi: 10.1515/BC.2001.141. [DOI] [PubMed] [Google Scholar]
- 5.Enz R., Brandstätter J. H., Wässle H., Bormann J. Immunocytochemical localization of the GABAc receptor rho subunits in the mammalian retina. J. Neurosci. 1996;16:4479–4490. doi: 10.1523/JNEUROSCI.16-14-04479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fletcher E. L., Koulen P., Wässle H. GABAA and GABAC receptors on mammalian rod bipolar cells. J. Comp. Neurol. 1998;396:351–365. doi: 10.1002/(sici)1096-9861(19980706)396:3<351::aid-cne6>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 7.Koulen P., Brandstätter J. H., Enz R., Bormann J., Wässle H. Synaptic clustering of GABAC receptor rho-subunits in the rat retina. Eur. J. Neurosci. 1998;10:115–127. doi: 10.1046/j.1460-9568.1998.00005.x. [DOI] [PubMed] [Google Scholar]
- 8.Sedelnikova A., Weiss D. S. Phosphorylation of the recombinant rho1 GABA receptor. Int. J. Dev. Neurosci. 2002;20:237–246. doi: 10.1016/s0736-5748(02)00037-0. [DOI] [PubMed] [Google Scholar]
- 9.Hanley J. G., Jones E. M., Moss S. J. GABA receptor rho1 subunit interacts with a novel splice variant of the glycine transporter, GLYT-1. J. Biol. Chem. 2000;275:840–846. doi: 10.1074/jbc.275.2.840. [DOI] [PubMed] [Google Scholar]
- 10.Hanley J. G., Koulen P., Bedford F., Gordon-Weeks P. R., Moss S. J. The protein MAP-1B links GABAC receptors to the cytoskeleton at retinal synapses. Nature. 1999;397:66–69. doi: 10.1038/16258. [DOI] [PubMed] [Google Scholar]
- 11.Croci C., Brandstätter J. H., Enz R. ZIP3, a new splice variant of the PKC-ξ-interacting protein family, binds to GABAC receptors, PKC-ξ and Kvβ2. J. Biol. Chem. 2003;278:6128–6135. doi: 10.1074/jbc.M205162200. [DOI] [PubMed] [Google Scholar]
- 12.Song X. Q., Meng F., Ramsey D. J., Ripps H., Qian H. The GABA rho1 subunit interacts with a cellular retinoic acid binding protein in mammalian retina. Neuroscience. 2005;136:467–475. doi: 10.1016/j.neuroscience.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 13.Meixner A., Haverkamp S., Wässle H., Fuhrer S., Thalhammer J., Kropf N., Bittner R. E., Lassmann H., Wiche G., Propst F. MAP1B is required for axon guidance and is involved in the development of the central and peripheral nervous system. J. Cell Biol. 2000;151:1169–1178. doi: 10.1083/jcb.151.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Valck D., Jin D. Y., Heyninck K., Van de Craen M., Contreras R., Fiers W., Jeang K. T., Beyaert R. The zinc finger protein A20 interacts with a novel anti-apoptotic protein which is cleaved by specific caspases. Oncogene. 1999;18:4182–4190. doi: 10.1038/sj.onc.1202787. [DOI] [PubMed] [Google Scholar]
- 15.Gachon F., Peleraux A., Thebault S., Dick J., Lemasson I., Devaux C., Mesnard J. M. CREB-2, a cellular CRE-dependent transcription repressor, functions in association with Tax as an activator of the human T-cell leukemia virus type 1 promoter. J. Virol. 1998;72:8332–8337. doi: 10.1128/jvi.72.10.8332-8337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin D. Y., Teramoto H., Giam C. Z., Chun R. F., Gutkind J. S., Jeang K. T. A human suppressor of c-Jun N-terminal kinase 1 activation by tumor necrosis factor alpha. J. Biol. Chem. 1997;272:25816–25823. doi: 10.1074/jbc.272.41.25816. [DOI] [PubMed] [Google Scholar]
- 17.Wu K., Bottazzi M. E., de la Fuente C., Deng L., Gitlin S. D., Maddukuri A., Dadgar S., Li H., Vertes A., Pumfery A., et al. Protein profile of tax-associated complexes. J. Biol. Chem. 2004;279:495–508. doi: 10.1074/jbc.M310069200. [DOI] [PubMed] [Google Scholar]
- 18.Ling L., Goeddel D. V. T6BP, a TRAF-6-interacting protein involved in IL-1 signaling. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9567–9572. doi: 10.1073/pnas.170279097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seeber S., Humeny A., Herkert M., Rau T., Eschenhagen T., Becker C. M. Formation of molecular complexes by N-methyl-D-aspartate receptor subunit NR2B and ryanodine receptor 2 in neonatal rat myocard. J. Biol. Chem. 2004;279:21062–21068. doi: 10.1074/jbc.M313009200. [DOI] [PubMed] [Google Scholar]
- 20.Croci C., Sticht H., Brandstätter J. H., Enz R. Group I metabotropic glutamate receptors bind to protein phosphatase 1C. Mapping and modeling of interacting sequences. J. Biol. Chem. 2003;278:50682–50690. doi: 10.1074/jbc.M305764200. [DOI] [PubMed] [Google Scholar]
- 21.Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 23.Brandstätter J. H., Koulen P., Kuhn R., van der Putten H., Wassle H. Compartmental localization of a metabotropic glutamate receptor (mGluR7): two different active sites at a retinal synapse. J. Neurosci. 1996;16:4749–4756. doi: 10.1523/JNEUROSCI.16-15-04749.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D., Pan Z. H., Zhang X., Brideau A. D., Lipton S. A. Cloning of a gamma-aminobutyric acid type C receptor subunit in rat retina with a methionine residue critical for picrotoxinin channel block. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11756–11760. doi: 10.1073/pnas.92.25.11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enz R., Brandstätter J. H., Hartveit E., Wässle H., Bormann J. Expression of GABA receptor rho 1 and rho 2 subunits in the retina and brain of the rat. Eur. J. Neurosci. 1995;7:1495–1501. doi: 10.1111/j.1460-9568.1995.tb01144.x. [DOI] [PubMed] [Google Scholar]
- 26.Enz R., Cutting G. R. GABAC receptor rho subunits are heterogeneously expressed in the human CNS and form homo- and heterooligomers with distinct physical properties. Eur. J. Neurosci. 1999;11:41–50. doi: 10.1046/j.1460-9568.1999.00423.x. [DOI] [PubMed] [Google Scholar]
- 27.Grassmann R., Aboud M., Jeang K. T. Molecular mechanisms of cellular transformation by HTLV-1 Tax. Oncogene. 2005;24:5976–5985. doi: 10.1038/sj.onc.1208978. [DOI] [PubMed] [Google Scholar]
- 28.Jeang K. T., Giam C. Z., Majone F., Aboud M. Life, death, and tax: role of HTLV-I oncoprotein in genetic instability and cellular transformation. J. Biol. Chem. 2004;279:31991–31994. doi: 10.1074/jbc.R400009200. [DOI] [PubMed] [Google Scholar]
- 29.Klinkenberg M., Van Huffel S., Heyninck K., Beyaert R. Functional redundancy of the zinc fingers of A20 for inhibition of NF-κB activation and protein-protein interactions. FEBS Lett. 2001;498:93–97. doi: 10.1016/s0014-5793(01)02504-2. [DOI] [PubMed] [Google Scholar]
- 30.Nagaraja G. M., Kandpal R. P. Chromosome 13q12 encoded Rho GTPase activating protein suppresses growth of breast carcinoma cells, and yeast two-hybrid screen shows its interaction with several proteins. Biochem. Biophys. Res. Commun. 2004;313:654–665. doi: 10.1016/j.bbrc.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Geetha T., Wooten M. W. Structure and functional properties of the ubiquitin binding protein p62. FEBS Lett. 2002;512:19–24. doi: 10.1016/s0014-5793(02)02286-x. [DOI] [PubMed] [Google Scholar]
- 32.Dyka F. M., May C. A., Enz R. Metabotropic glutamate receptors are differentially regulated under elevated intraocular pressure. J. Neurochem. 2004;90:190–202. doi: 10.1111/j.1471-4159.2004.02474.x. [DOI] [PubMed] [Google Scholar]
- 33.Rozzo A., Armellin M., Franzot J., Chiaruttini C., Nistri A., Tongiorgi E. Expression and dendritic mRNA localization of GABAC receptor rho1 and rho2 subunits in developing rat brain and spinal cord. Eur. J. Neurosci. 2002;15:1747–1758. doi: 10.1046/j.1460-9568.2002.02013.x. [DOI] [PubMed] [Google Scholar]
- 34.Alakuijala A., Palgi M., Wegelius K., Schmidt M., Enz R., Paulin L., Saarma M., Pasternack M. GABA receptor rho subunit expression in the developing rat brain. Brain Res. Dev. Brain Res. 2005;154:15–23. doi: 10.1016/j.devbrainres.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Boue-Grabot E., Roudbaraki M., Bascles L., Tramu G., Bloch B., Garret M. Expression of GABA receptor rho subunits in rat brain. J. Neurochem. 1998;70:899–907. doi: 10.1046/j.1471-4159.1998.70030899.x. [DOI] [PubMed] [Google Scholar]
- 36.Zheng W., Xie W., Zhang J., Strong J. A., Wang L., Yu L., Xu M., Lu L. Function of gamma-aminobutyric acid receptor/channel rho 1 subunits in spinal cord. J. Biol. Chem. 2003;278:48321–48329. doi: 10.1074/jbc.M307930200. [DOI] [PubMed] [Google Scholar]
- 37.Fletcher E. L., Clark M. J., Senior P., Furness J. B. Gene expression and localization of GABAC receptors in neurons of the rat gastrointestinal tract. Neuroscience. 2001;107:181–189. doi: 10.1016/s0306-4522(01)00339-6. [DOI] [PubMed] [Google Scholar]
- 38.Boue-Grabot E., Taupignon A., Tramu G., Garret M. Molecular and electrophysiological evidence for a GABAc receptor in thyrotropin-secreting cells. Endocrinology. 2000;141:1627–1632. doi: 10.1210/endo.141.5.7476. [DOI] [PubMed] [Google Scholar]
- 39.Jansen A., Hoepfner M., Herzig K. H., Riecken E. O., Scherubl H. GABAC receptors in neuroendocrine gut cells: a new GABA-binding site in the gut. Pflugers Arch. 2000;441:294–300. doi: 10.1007/s004240000412. [DOI] [PubMed] [Google Scholar]
- 40.Gamel-Didelon K., Kunz L., Fohr K. J., Gratzl M., Mayerhofer A. Molecular and physiological evidence for functional γ-aminobutyric acid (GABA)-C receptors in growth hormone-secreting cells. J. Biol. Chem. 2003;278:20192–20195. doi: 10.1074/jbc.M301729200. [DOI] [PubMed] [Google Scholar]
- 41.Alefantis T., Jain P., Ahuja J., Mostoller K., Wigdahl B. HTLV-1 Tax nucleocytoplasmic shuttling, interaction with the secretory pathway, extracellular signaling, and implications for neurologic disease. J. Biomed. Sci. 2005;12:961–974. doi: 10.1007/s11373-005-9026-x. [DOI] [PubMed] [Google Scholar]