Abstract
In addition to serving as a biomarker of oxidative/nitrative stress, elevated levels of nitrotyrosine have been shown to cause DNA damage or trigger apoptosis. Whether the body is equipped with mechanisms for protecting against the potentially harmful nitrotyrosine remains unknown. The present study was designed to investigate the possibility that sulfation serves as a pathway for the metabolism/regulation of nitrotyrosine. Using metabolic labelling, nitrotyrosine O-[35S]sulfate was found to be produced and released into the medium of HepG2 human hepatoma cells labelled with [35S]sulfate in the presence of nitrotyrosine. To identify the enzyme(s) responsible for nitrotyrosine sulfation, a systematic study of all eleven known human cytosolic SULTs (sulfotransferases) was performed. Of the 11 enzymes tested, only SULT1A3 displayed sulfating activity toward nitrotyrosine. The pH-dependence and kinetic constants of SULT1A3 with nitrotyrosine or dopamine as substrate were determined. To examine whether the sulfation of nitrotyrosine occurs in the context of cellular physiology, HepG2 cells labelled with [35S]sulfate were treated with SIN-1 (morpholinosydnonimine), a peroxynitrite generator. Increments of nitrotyrosine O-[35S]sulfate were detected in the medium of HepG2 cells treated with higher concentrations of SIN-1. To gain insight into the physiological relevance of nitrotyrosine sulfation, a time-course study was performed using [3H]tyrosine-labelled HepG2 cells treated with SIN-1. The findings confirm that the bulk of free [3H]nitrotyrosine inside the cells was present in the unconjugated form. The proportion of sulfated [3H]nitrotyrosine increased dramatically in the medium over time, implying that sulfation may play a significant role in the metabolism of free nitrotyrosine.
Keywords: dopamine, nitrotyrosine, sulfation, sulfotransferase, SULT1A3
Abbreviations: Caps, 3-(cyclohexylamino)propane-1-sulfonic acid; Ches, 2-(N-cyclohexylamino)ethanesulfonic acid; DTT, dithiothreitol; MEM, minimal essential medium; M-form, monoamine form; NMWL, nominal molecular-mass limit; PAPS, adenosine 3′-phosphate 5′-phosphosulfate; SIN-1, 3-morpholinosydnonimine; SULT, sulfotransferase; TLE, thin-layer electrophoresis
INTRODUCTION
Nitration of tyrosine, in both protein-bound form and free amino acid form, can readily occur during oxidative/nitrative stress under which oxides of nitrogen, e.g. NO (nitrogen monoxide or nitric oxide) and NO2 (nitrogen dioxide), ONOO− (peroxynitrite) and NO2Cl (nitrylchloride), are formed and react as nitrating agents [1]. Peroxynitrite-mediated nitrotyrosine formation has been detected in a wide range of pathophysiological states including atherosclerosis [2,3], stroke [4], pulmonary injury and other lung diseases [5], and chronic hepatitis [6,7]. It has been reported that tyrosine nitration of proteins may prevent or stimulate the subsequent phosphorylation of the tyrosine residue and change the rate of their proteolytic degradation, possibly leading to disease initiation and progression [1,8–10]. It has been suggested that tyrosine-nitrated proteins may be degraded by proteasome, resulting in the release of free nitrotyrosine [11]. Studies have shown that free nitrotyrosine can induce oxidative DNA damage [12] or trigger apoptosis in cultured cells [13,14]. Using the mouse as a model, it was demonstrated that free nitrotyrosine could elicit neurotoxic effects, leading to striatal neurodegeneration in vivo [15]. Whether the body is equipped with mechanisms for protecting against the potentially harmful effects of nitrotyrosine, however, remains unknown.
In mammals, sulfate conjugation as catalysed by the cytosolic SULTs (sulfotransferases), has been shown to be involved in the biotransformation/excretion of xenobiotics as well as endogenous compounds such as steroid and thyroid hormones, catecholamines, cholesterol and bile acids [16–18]. The cytosolic SULTs catalyse the transfer of a sulfonate group from the active sulfate, PAPS (adenosine 3′-phosphate 5′-phosphosulfate), to an acceptor substrate compound containing either hydroxy or amino groups [19]. Sulfate conjugation by SULT enzymes may lead to the inactivation of these compounds and/or increase their water-solubility, thereby facilitating their removal from the body [16–18]. Our previous studies demonstrated that the human SULT1A3 [previously called the monoamine (M)-form phenol sulfotransferase or catecholamine-preferring phenol sulfotransferase] may sulfonate not only dopamine and other monoamines, but also dopa and tyrosine isomers [20–23]. In view of its structural similarity to tyrosine, we speculated that nitrotyrosine generated under oxidative/nitrative stress conditions may also undergo sulfation.
In the present paper, we report the discovery of the sulfation of nitrotyrosine using HepG2 human hepatoma cells as a model. A systematic study using all 11 known human cytosolic SULTs was carried out and revealed SULT1A3 to be the only enzyme that was capable of sulfating nitrotyrosine. The pH-dependence and the kinetics of the sulfation of nitrotyrosine by SULT1A3 were also elucidated. Moreover, it was demonstrated that sulfated nitrotyrosine was generated and released by HepG2 cells in response to treatment with SIN-1 (morpholinosydnonimine), a peroxynitrite generator.
EXPERIMENTAL
Materials
L-p-tyrosine, DL-m-tyrosine, 3-nitro-L-tyrosine, L-dopa, dopamine, aprotinin, thrombin, ATP, PAPS, SDS, sodium succinate, sodium acetate, Mes, Mops, Hepes, Taps, Ches [2-(N-cyclohexylamino)ethanesulfonic acid], Caps [3-(cyclohexylamino)propane-1-sulfonic acid], Trizma base, DTT (dithiothreitol), DMSO and arylsulfatase (Type IV) from Patella vulgata were obtained from Sigma Chemical Co. Carrier-free sodium [35S]sulfate was from ICN Biomedicals, and L-[3,5-3H]tyrosine was from Amersham Biosciences. Cellulose TLC plates were from EMD Chemicals, and SIN-1 was from Calbiochem. Complete™ Mini protease inihibitor cocktail was from Roche Diagnostics, and Ultrafree-MC 5000 NMWL (nominal molecular-mass limit) filter units were products of Millipore. HepG2 human hepatoma cell line (ATCC HB 8065) was from the American Type Culture Collection (Manassas, VA, U.S.A.). All other chemicals were of the highest grade commercially available.
Preparation of purified human cytosolic SULTs
Recombinant human simple phenol (P)-form (SULT1A1 and SULT1A2) and M-form (SULT1A3) phenol SULTs, thyroid hormone SULT (SULT1B2), two SULT1Cs (designated #1 and #2), oestrogen SULT (SULT1E1), DHEA (dehydroepiandrosterone) SULT (SULT2A1), two SULT2B1s (designated a and b), and a neuronal SULT (SULT4A1), expressed using a pGEX-2TK or pET23c prokaryotic expression system, were prepared as described previously [24–28].
Enzymatic assay
The sulfating activity of the recombinant human cytosolic SULTs was assayed using PAP[35S] as the sulfonate group donor. The standard assay mixture, in a final volume of 25 μl, contained 50 mM Mops buffer at pH 7.0, 1 mM DTT and 14 μM PAP[35S]. Stock solutions of the substrates, prepared in water or alkaline (pH 13) water, were used in the enzymatic assay. The substrate, at 10 times the final concentration was diluted to 50 μM in the assay mixture, was added after Mops buffer and PAP[35S]. A control with water alone was also prepared. The reaction was started by the addition of the enzyme (1.25 μg), allowed to proceed for 3 min at 37 °C, and stopped by placing the thin-walled tube containing the assay mixture on a heating block, pre-heated to 100 °C, for 2 min. The precipitates were cleared by centrifugation at 13000 g for 1 min, and the supernatant was subjected to analysis of [35S]sulfated product using the previously established TLE (thin-layer electrophoresis) procedure [29], with 7.8% ethanoic (acetic) acid/2.5% methanoic (formic) acid (pH 1.9) as the electrophoresis buffer. For two-dimensional thin-layer analysis, the TLE described above was followed in the second dimension by TLC using n-butanol/propan-2-ol/88% methanoic acid/water (2:1:1:2, by vol.) as the solvent system [29]. To examine the pH-dependence of the sulfation of nitrotyrosine or dopamine, different buffers (50 mM succinate at pH 3.5, 4.0 or 4.5; sodium acetate at pH 4.5, 5.0 or 5.5; Mes at pH 5.5, 6.0 or 6.5; Mops at pH 6.5, 7.0 or 7.5; Hepes at pH 7.0, 7.5 or 8.0; Taps at pH 7.5, 8.0, 8.5 or 9.0; Ches at pH 9.0, 9.5 or 10.0; and Caps at pH 10.0, 10.5, 11.0 or 11.5), instead of 50 mM Mops (pH 7.0), were used in the reactions with 100 μM nitrotyrosine or 5 μM dopamine as the substrate. For the kinetic studies on the sulfation of nitrotyrosine, dopamine and L-p-tyrosine, various concentrations of these substrate compounds and 50 mM Mops buffer at pH 7.0 were used. To examine the inhibitory effects of L-p-tyrosine on the sulfation of nitrotyrosine or vice versa, the kinetic studies on the sulfation of nitrotyrosine (or L-p-tyrosine) were performed in the presence of fixed concentrations of L-p-tyrosine (or nitrotyrosine). To study the effects of pH on the kinetics of the sulfation of nitrotyrosine and dopamine, the kinetic studies were performed using different buffer systems at different pH values. Each experiment was performed in triplicate, together with a control without enzyme. The results obtained were calculated and expressed in nmol of sulfated product formed/min per mg of purified enzyme.
Metabolic labelling of HepG2 human hepatoma cells with [35S]sulfate
HepG2 cells were routinely maintained, under a 5% CO2 atmosphere at 37 °C, in MEM (minimum essential medium) supplemented with 10% (v/v) foetal bovine serum, 30 μg/ml penicillin G and 50 μg/ml streptomycin sulfate. Confluent HepG2 cells grown in individual wells of a 24-well culture plate, pre-incubated in sulfate-free (prepared by omitting streptomycin sulfate and replacing magnesium sulfate with MgCl2) MEM for 4 h, were labelled with 0.25 ml aliquots of the same medium containing [35S]sulfate (0.3 mCi/ml) and different concentrations of nitrotyrosine (ranging from 5 to 50 μM) or SIN-1 (ranging from 0.5 to 2.5 mM). At the end of an 18 h labelling period, the media were collected, spin-filtered and subjected to the TLE or two-dimensional thin-layer analysis for nitrotyrosine O-[35S]sulfate based on the procedures described above.
Metabolic labelling of HepG2 cells with [3H]tyrosine in the presence of SIN-1
Confluent HepG2 cells grown in individual wells of a 24-well culture plate and pre-incubated in tyrosine- and phenylalanine-deficient MEM for 4 h were labelled with 0.25 ml aliquots of a low-tyrosine (0.5% of normal) and low-phenylalanine (5% of normal) MEM containing 0.1 mCi/ml [3H]tyrosine plus 2.5 mM SIN-1. Upon incubation for 1, 3 or 18 h, the media were collected and the cells were lysed in 0.25 ml aliquots of a RIPA buffer (containing 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1% sodium cholate and 1% protease inhibitor cocktail). The media and cell lysates collected were spin-filtered using Ultrafree-MC 5000 NMWL filter units. Aliquots of the filtrates were individually mixed with 1 μl of a carrier nitrotyrosine (1.25 mg/ml) solution or 1 μl of a carrier nitrotyrosine O-sulfate (0.6 mg/ml) solution, spotted on to cellulose TLC plates and analysed for [3H]nitrotyrosine and [3H]nitrotyrosine O-sulfate based on the TLE procedure described above. Upon completion of the electrophoresis, the plates were air-dried and sprayed with a ninhydrin solution (0.5% in acetone). The ninhydrin-stained spots of nitrotyrosine and nitrotyrosine O-sulfate were excised and the 3H radioactivity therein was determined using a scintillation counter.
Miscellaneous methods
PAP[35S] was synthesized from ATP and carrier-free [35S]sulfate using the bifunctional human ATP sulfurylase/adenosine 5′-phosphosulfate kinase, and its purity was determined as previously described [30]. The PAP[35S] synthesized was adjusted to the required concentration and a specific activity of 15 Ci/mmol at 1.4 mM by the addition of unlabelled PAPS. SDS/PAGE was performed on 12% polyacrylamide gels using the method of Laemmli [31]. Protein determination was based on the method of Bradford with BSA as the standard [32]. Tyrosine O-sulfate standard was synthesized based on the procedure developed by Jevons [33]. Nitrotyrosine O-sulfate standard was synthesized enzymatically using non-radioactive PAPS as the sulfonate donor under the conditions described in the enzymatic assay section. Desulfation of [35S]sulfate- or [3H]tyrosine-labelled sulfated nitrotyrosine by arylsulfatase (0.1 k-unit/ml) was performed at 37 °C for 30 min with 50 mM Mops buffer (pH 7.5). Analysis of the nitration of tyrosine O-[35S]sulfate by SIN-1 was carried out in PBS in the presence or absence of 26.2 mM sodium bicarbonate (the same concentration as in MEM for HepG2 cell culture). The reaction was started by the addition of SIN-1, allowed to proceed for 1 or 24 h in a CO2 incubator. Afterwards, the samples were collected and subjected to analysis of tyrosine O-[35S]sulfate and nitrotyrosine O-[35S]sulfate using the TLE procedure mentioned above.
RESULTS AND DISCUSSION
The objective of the present study was to investigate the involvement of sulfate conjugation in the metabolism of nitrotyrosine using HepG2 human hepatoma cells as a model. We also sought to identify and characterize the cytosolic SULT enzyme(s) responsible for nitrotyrosine sulfation.
Generation and release of free nitrotyrosine O-[35S]sulfate by HepG2 cells metabolically labelled with [35S]sulfate in the presence of nitrotyrosine
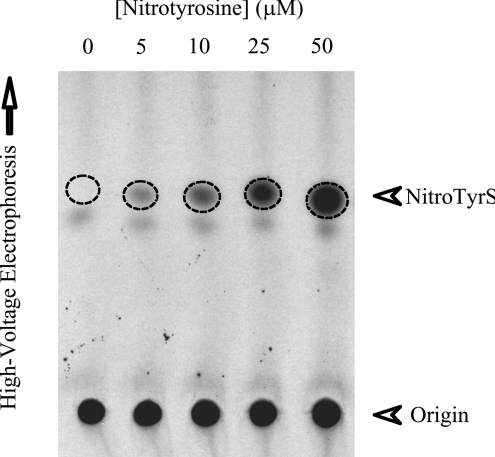
We first attempted to investigate, using HepG2 cells as a model, whether mammalian cells are equipped with enzyme(s) capable of catalysing the sulfation of free nitrotyrosine. HepG2 cells were metabolically labelled with [35S]sulfate in the presence of different concentrations of nitrotyrosine. At the end of an 18 h incubation, the labelling media were collected and analysed for the production and release of nitrotyrosine O-[35S]sulfate. As shown in Figure 1, the autoradiograph taken from the TLC plate used for the analysis of the spent media of HepG2 cells, labelled with [35S]sulfate in the presence of 5, 10, 25 or 50 μM nitrotyrosine, revealed a 35S-radioactive spot in a dose-dependent manner. To verify the authenticity of this [35S]sulfated compound being nitrotyrosine O-[35S]sulfate, a metabolic labelling experiment using [3H]tyrosine was carried out. The 3H-labelled compound migrating at the same position as that of the [35S]sulfate-labelled species, upon TLC, was eluted and subjected to hydrolysis by arylsulfatase (Type IV) from P. vulgata. The hydrolysate was spin-filtered and the filtrate was subjected to TLC analysis together with carrier non-radioactive nitrotyrosine. The results of this analysis clearly showed the co-migration of the 3H-radioactive species with non-radioactive carrier nitrotyrosine (results not shown). Taken together, the findings provide clear evidence that HepG2 cells, and possibly other mammalian cells as well, are equipped with enzyme(s) capable of catalysing the sulfation of nitrotyrosine.
Figure 1. Identification of free nitrotyrosine O-[35S]sulfate generated and released by HepG2 human hepatoma cells metabolically labelled with [35S]sulfate in the presence of nitrotyrosine.
Autoradiograph taken from the TLC plate used for the TLE analysis. Confluent HepG2 cells were incubated in the labelling media containing 0, 5, 10, 25 or 50 μM nitrotyrosine for 18 h as indicated. The broken-line circles indicate the position of nitrotyrosine O-[35S]sulfate (NitroTyrS).
Identification of the human cytosolic SULT(s) responsible for the sulfation of nitrotyrosine
We have previously cloned, expressed and purified all 11 known human cytosolic SULTs: SULT1A1, SULT1A2, SULT1A3, SULT1B2, SULT1C#1, SULT1C#2, SULT1E1, SULT2A1, SULT2B1a, SULT2B1b and SULT4A1 [24–28]. These purified recombinant human cytosolic SULTs were examined for sulfating activities towards nitrotyrosine and other tyrosine isomers/derivatives. Seven (SULT1A2, SULT1B2, SULT1C#1, SULT2A1, SULT2B1a, SULT2B1b and SULT4A1) of the 11 SULTs exhibited no detectable activities towards any of the compounds that we tested as substrates. Three other SULTs (SULT1A1, SULT1C#2 and SULT1E1) displayed weak sulfating activities towards dopamine, but not to other tyrosine isomers/derivatives. Interestingly, only SULT1A3 exhibited significant sulfating activity toward nitrotyrosine (Table 1). The autoradiograph taken from the TLC plate used for the two-dimensional thin-layer analysis of the assay mixture of SULT1A3 clearly revealed a radioactive spot corresponding to nitrotyrosine O-[35S]sulfate generated during the enzymatic reaction (results not shown). These results therefore indicate that SULT1A3 may have a broader involvement in mammalian physiology, being involved in the homoeostasis of not only dopamine and other catecholamines, but also un-modified tyrosine, Dopa and, as demonstrated in the present study, nitrotyrosine.
Table 1. Specific activities of human SULT1A3 with tyrosine isomers and derivatives as substrates.
Results are means±S.D. for three determinations.
| Substrate | Specific activity (nmol/min per mg of protein) |
|---|---|
| L-Nitrotyrosine | 2.11±0.07 |
| L-p-Tyrosine | 0.01±0.01 |
| DL-m-Tyrosine | 0.85±0.01 |
| L-Dopa | 2.06±0.04 |
| Dopamine | 15.92±0.11 |
Characterization of the nitrotyrosine-sulfating activity of human SULT1A3
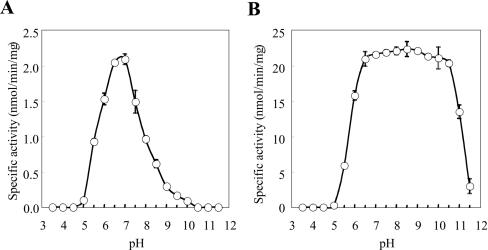
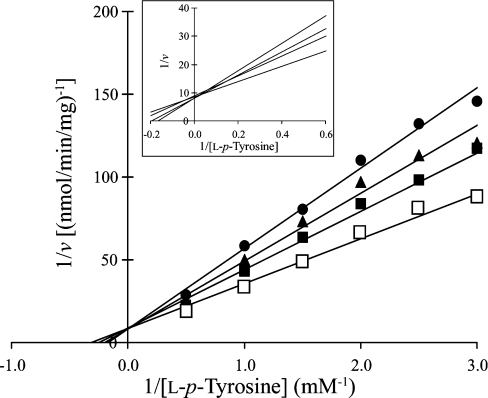
Since a primary role for SULT1A3 in the homoeostasis of dopamine has been suggested [34], we were interested in investigating the sulfating activity of SULT1A3 towards nitrotyrosine in comparison with its activity towards dopamine. To determine the pH-dependence of the sulfation of nitrotyrosine or dopamine by SULT1A3, enzymatic assays were performed under standard conditions described in the Experimental section using buffers at different pH values. As shown in Figure 2, a distinct pH optimum of 7.0 was detected with nitrotyrosine as substrate, whereas a broad pH optimum spanning 6.5–10.5 was observed with dopamine as substrate. Whether these different pH-dependence profiles were due to distinct charge properties between dopamine and nitrotyrosine remain to be clarified. The kinetic parameters of SULT1A3 in catalysing the sulfation of nitrotyrosine and dopamine were subsequently determined. The enzymatic assays were performed at pH 7.0 with various concentrations of these two compounds as substrates. Data obtained were processed using the Excel program to generate the best-fitting trendlines for the Lineweaver–Burk double-reciprocal plots. Table 2 shows the kinetic constants determined for the sulfation of nitrotyrosine and dopamine. It is interesting to note the much higher Km and lower Vmax (130.5 μM and 4.2 nmol/min per mg of protein respectively) determined with nitrotyrosine as substrate in comparison with dopamine as substrate (6.5 μM and 52.7 nmol/min per mg of protein respectively). The catalytic efficiency of SULT1A3, as reflected by the Vmax/Km, therefore appeared to be approx. 250-fold lower with nitrotyrosine as substrate than with dopamine. As mentioned above, SULT1A3 is thought to play a role in the homoeostasis of dopamine and other monoamines [34]. The level of dopamine in vivo is normally within the submicromolar to micromolar concentration range [35]. It is therefore conceivable that a lower Km would be required in order for SULT1A3 to rid excess dopamine and other monoamines through sulfation. In view of their differential charge properties, an important question relates to the effects of pH on the kinetics of the sulfation of nitrotyrosine and dopamine. Kinetic constants were therefore determined at different pH values. Based on the pH-dependence profiles of the sulfation of nitrotyrosine and dopamine (Figure 2), pH 5.5, 7.0 and 9.0 were chosen for studying the sulfation of nitrotyrosine, and pH 7.0, 9.0 and 11.0 were chosen for studying the sulfation of dopamine. The results obtained are shown in Table 3. For the sulfation of nitrotyrosine, the Km values showed only small variations at the three pH values tested, whereas the Vmax value was much higher at pH 7.0 than at pH 5.5 or 9.0. The catalytic efficiency of SULT1A3, as reflected by the Vmax/Km, therefore appeared to be much higher at pH 7.0 than at pH 5.5 or 9.0. For the sulfation of dopamine, while the Vmax value was considerably lower at pH 7.0, the Km value was 5–6-fold lower at pH 7.0 than at pH 9.0 or 11.0. Based on these data, SULT1A3 appeared to be catalytically active for the sulfation of dopamine at all three pH values tested. Another issue is whether L-p-tyrosine may act as an inhibitor for the sulfation of nitrotyrosine. To address this question, kinetic constants of SULT1A3 for the sulfation of nitrotyrosine were determined in the presence of L-p-tyrosine. As shown in Table 2, addition of high concentrations of L-p-tyrosine to the reaction mixtures did not affect significantly either the Km or the Vmax value. These results are consistent with the high Km value (1.95 mM) of SULT1A3 for L-p-tyrosine as determined in our previous study [20]. At the concentration normally present in culture medium (200 μM) or in circulation in mammals (<0.1 mM) [36], L-p-tyrosine would not be expected to exert significant inhibitory effects on the sulfation of nitrotyrosine. The effect of nitrotyrosine on the sulfation of L-p-tyrosine by SULT1A3 was also examined. As revealed in the Lineweaver–Burk double-reciprocal plots shown in Figure 3, the sulfation of L-p-tyrosine was clearly inhibited in the presence of nitrotyrosine. It was noted that the lines corresponding the sulfation of L-p-tyrosine in the presence of various concentrations of nitrotyrosine appeared to converge within a narrow region on the y-axis. These data are therefore indicative of a competitive-type of inhibition of the sulfation of L-p-tyrosine by nitrotyrosine.
Figure 2. pH-dependence of the sulfating activity of human SULT1A3 with (A) nitrotyrosine and (B) dopamine as substrates.
The enzymatic assays were carried out under standard assay conditions as described in the Experimental section, using different buffer systems. Results are means±S.D. for three experiments.
Table 2. Kinetic constants of the sulfation of nitrotyrosine and dopamine by human SULT1A3.
Results are means±S.D. for three determinations.
| Substrate | Vmax (nmol/min per mg of protein) | Km (μM) | Vmax/Km |
|---|---|---|---|
| L-Nitrotyrosine (without L-tyrosine) | 4.2±0.5 | 130.5±15.3 | 0.032 |
| L-Nitrotyrosine (with 250 μM L-tyrosine) | 3.7±0.2 | 117.3±6.1 | 0.032 |
| L-Nitrotyrosine (with 500 μM L-tyrosine) | 3.8±0.6 | 129.4±26.9 | 0.029 |
| Dopamine | 52.7±4.1 | 6.5±0.7 | 8.11 |
Table 3. Kinetic constants of the sulfation of nitrotyrosine and dopamine by human SULT1A3 at different pH values.
Results are means±S.D. for three determinations.
| Vmax (nmol/min per mg) | Km (μM) | Vmax/Km | |
|---|---|---|---|
| Sulfation of nitrotyrosine | |||
| pH 5.5 | 0.9±0.2 | 144.4±30.3 | 0.006 |
| pH 7.0 | 4.2±0.5 | 130.5±15.3 | 0.032 |
| pH 9.0 | 0.3±0.1 | 164.1±31.6 | 0.002 |
| Sulfation of dopamine | |||
| pH 7.0 | 52.7±4.1 | 6.5±0.7 | 8.11 |
| pH 9.0 | 130.6±8.7 | 38.1±2.6 | 3.43 |
| pH 11.0 | 101.3±18.5 | 31.6±12.0 | 3.21 |
Figure 3. Lineweaver–Burk double-reciprocal plots of SULT1A3 with L-p-tyrosine as substrate in the absence or presence of different concentrations of nitrotyrosine.
Concentrations of nitrotyrosine tested were 0 (□), 125 (■), 250 (▲) and 500 (●) μM. Concentrations of L-p-tyrosine were in mM and velocities were expressed as nmol of sulfated product formed/min per mg of protein. Results are means for three determinations. The inset shows an enlargement of the region where the lines converge in the plot.
Generation and release of free nitrotyrosine O-[35S]sulfate by [35S]sulfate-labelled HepG2 cells upon treatment with SIN-1
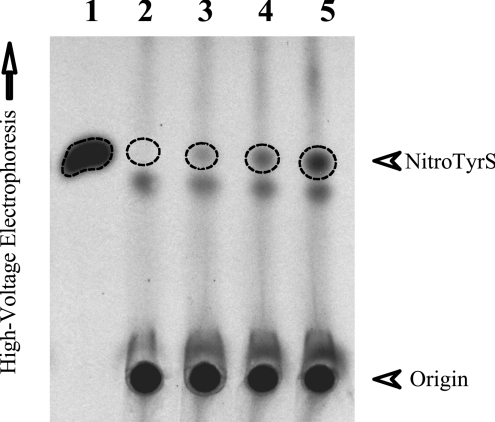
To examine whether the sulfation of nitrotyrosine occurs in the context of cellular physiology, HepG2 cells incubated in media containing [35S]sulfate were treated with different concentrations of SIN-1, a peroxynitrite generator [37–39]. The assumption underlying this study was that, if sufficient amounts of nitrotyrosine were produced upon stimulation by SIN-1, sulfation as catalysed by SULT1A3 (which is known to be expressed in HepG2 cells) may be operative to convert excess nitrotyrosine into its sulfated form. As shown in Figure 4, in the absence of SIN-1, virtually no nitrotyrosine O-[35S]sulfate was produced (lane 2). Upon stimulation by increasing concentrations of SIN-1, there was an increased production and release of nitrotyrosine O-[35S]sulfate by HepG2 cells (as indicated by broken-line circles on lanes 3–5). These results therefore clearly indicate that sulfation of nitrotyrosine may occur in cells under nitrative stress. It is noted that a separate radioactive spot (below the broken-line circle) was present in both untreated HepG2 cells and cells treated with different concentrations of SIN-1. To clarify the identity of the compound present at this radioactive spot, the HepG2 labelling medium that contained 2.5 mM SIN-1, mixed with synthetic nitrotyrosine O-sulfate and tyrosine O-sulfate, was subjected to two-dimensional thin-layer separation. As shown in Figure 5, the autoradiograph taken from the TLC plate used for the two-dimensional thin-layer analysis clearly revealed its identity as tyrosine O-sulfate. It should be pointed out that tyrosine O-sulfate had been demonstrated in our previous studies as a normal metabolite derived from either the de novo sulfation of excess tyrosine or the degradation of tyrosine-sulfated proteins [22]. Based on the intensity of the radioactive spot corresponding to tyrosine O-[35S]sulfate (Figure 4), it appeared that the level of tyrosine O-sulfate, in contrast with that of nitrotyrosine O-sulfate, produced and released by HepG2 cells was little affected by the SIN-1 treatment. An in vitro experiment was performed to test whether tyrosine O-sulfate could be nitrated by SIN-1 in the presence or absence of CO2, a phenomenon previously reported for unsulfated tyrosine [40]. Under the conditions described in the Experimental section, we found that tyrosine O-sulfate, in contrast with unconjugated tyrosine, could not be nitrated by SIN-1 (results not shown).
Figure 4. Generation and release of free nitrotyrosine O-[35S]sulfate by HepG2 human hepatoma cells labelled with [35S]sulfate in the presence of SIN-1.
Autoradiograph taken from the TLC plate used for the TLE analysis. Confluent HepG2 cells were incubated for 18 h in the labelling media containing 0, 0.5, 1 and 2.5 mM SIN-1 (lanes 2–5 respectively). Lane 1 shows the nitrotyrosine O-[35S]sulfate generated in the enzymatic assay. The broken-line circles indicate the position of nitrotyrosine O-[35S]sulfate (NitroTyrS).
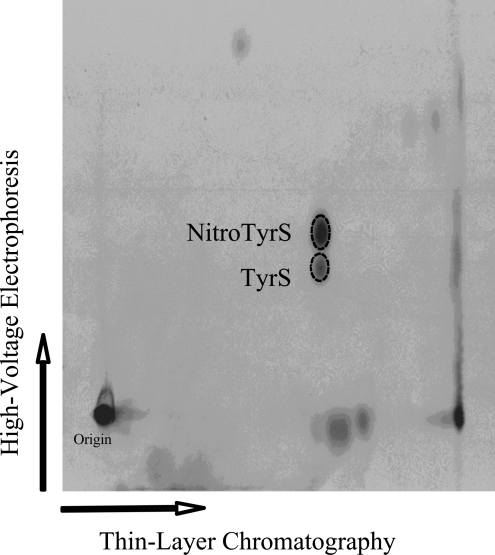
Figure 5. Identification of free nitrotyrosine O-[35S]sulfate and tyrosine O-[35S]sulfate generated and released by HepG2 human hepatoma cells.
Autoradiograph taken from the TLC plate used for the two-dimensional thin-layer analysis of the labelling medium sample. Confluent HepG2 cells were incubated for 18 h in the labelling media containing 2.5 mM SIN-1. The broken-line circles correspond to the positions of synthetic nitrotyrosine O-sulfate (upper) and tyrosine O-sulfate (lower) as revealed by ninhydrin staining.
Kinetics of the generation of free [3H]nitrotyrosine and [3H]nitrotyrosine O-sulfate by [3H]tyrosine-labelled HepG2 cells treated with SIN-1
To gain insight into the physiological relevance of the sulfation of nitrotyrosine, a time-course study was performed using [3H]tyrosine-labelled HepG2 cells treated with SIN-1. As shown in Table 4, the bulk (90.4–94.8%) of the [3H]nitrotyrosine generated upon SIN-1 treatment was present in an unconjugated form inside the HepG2 cells at all three time points studied. In the medium, however, the proportion of the sulfate-conjugated [3H]nitrotyrosine increased (from 23.2% to 59.2%) over time during the period that we monitored. These results indicate that, while the occurrence of other conjugation reactions, e.g. glucuronidation, may still be operational, sulfation may indeed play a significant role in the metabolism of free nitrotyrosine generated under nitrative stress. The question, nevertheless, remains as to the mechanism underlying the generation of nitrotyrosine O-sulfate in vivo. Possible reactions/pathways that may lead to the generation of free nitrotyrosine O-sulfate include (i) the sulfation of free nitrotyrosine formed de novo under nitrative stress, (ii) the sulfation of free nitrotyrosine released upon degradation of tyrosine-nitrated proteins, (iii) the sulfation of tyrosine-nitrated proteins followed by their degradation to release free nitrotyrosine O-sulfate, and (iv) the nitration of free tyrosine O-sulfate, a normal metabolite shown previously to be derived from either the sulfation of excess tyrosine or the degradation of tyrosine-sulfated proteins [22]. Work that extends the present study is warranted in order to elucidate the relative contributions of these various reactions/pathways.
Table 4. Amounts of free [3H]nitrotyrosine and [3H]nitrotyrosine O-sulfate generated by HepG2 human hepatoma cells labelled with [3H]tyrosine in the presence of SIN-1.
Results shown represent the counts (c.p.m.×10−3) corresponding to free [3H]nitrotyrosine or [3H]nitrotyrosine O-sulfate present in the 250 μl cell lysate or labelling medium samples. Results shown are calculated means±S.D. for three experiments. Percentage values in the parentheses indicate the ratio between free [3H]nitrotyrosine and [3H]nitrotyrosine O-sulfate present in cell lysate or medium samples at individual time points.
| [3H]Nitrotyrosine | [3H]Nitrotyrosine O-sulfate | |
|---|---|---|
| Cell lysate | ||
| 1 h | 29.2±4.4 (94.8%) | 1.6±0.5 (5.2%) |
| 3 h | 28.3±1.2 (93.4%) | 2.0±0.1 (6.6%) |
| 18 h | 25.4±3.2 (90.4%) | 2.7±0.8 (9.6%) |
| Medium | ||
| 1 h | 88.9±6.8 (76.8%) | 26.9±4.2 (23.2%) |
| 3 h | 91.8±25.8 (74.9%) | 30.8±1.4 (25.1%) |
| 18 h | 33.5±15.9 (40.8%) | 48.7±4.0 (59.2%) |
In summary, the present study clearly demonstrates the occurrence of the sulfation of nitrotyrosine in HepG2 cells and confirms that the responsible enzyme is the human SULT1A3. The fact that nitrotyrosine can be sulfated implies a role of sulfation in the metabolism, regulation and/or removal of nitrotyrosine generated in vivo under nitrative stress. More work is warranted in order to fully elucidate the physiological relevance of nitrotyrosine sulfation. From a different perspective, elevated levels of nitrotyrosine have in recent years been proposed as a predictor of disease risk and burden or as a potential biomarker for atherosclerosis [2,3], stroke [4], pulmonary fibrosis and other lung diseases [5], and chronic hepatitis [6,7]. Our findings demonstrating the sulfation of nitrotyrosine suggest that sulfated and other potential conjugated form(s) of nitrotyrosine may need to be considered in order to fully appreciate the compound as a biomarker for diseases.
Acknowledgments
This work was supported in part by a Grant-in-Aid (#0555067Y) from the American Heart Association, Texas Affiliate (M.-C.L.) and a Program Project Grant (#PO1 HL076406) from NIH (National Institutes of Health) (S.I.).
References
- 1.Ischiropoulos H. Biological tyrosine nitration: a pathophysiological function of nitric oxide and reactive oxygen species. Arch. Biochem. Biophys. 1998;356:1–11. doi: 10.1006/abbi.1998.0755. [DOI] [PubMed] [Google Scholar]
- 2.Beckmann J. S., Ye Y. Z., Anderson P. G., Chen J., Accavitti M. A., Tarpey M. M., White C. R. Extensive nitration of protein tyrosines in human atherosclerosis detected by immunohistochemistry. Biol. Chem. Hoppe Seyler. 1994;375:81–88. doi: 10.1515/bchm3.1994.375.2.81. [DOI] [PubMed] [Google Scholar]
- 3.Cromheeke K. M., Kockx M. M., De Meyer G. R., Bosmans J. M., Bult H., Beelaerts W. J., Vrints C. J., Herman A. G. Inducible nitric oxide synthase colocalizes with signs of lipid oxidation/peroxidation in human atherosclerotic plaques. Cardiovasc. Res. 1999;43:744–754. doi: 10.1016/s0008-6363(99)00148-0. [DOI] [PubMed] [Google Scholar]
- 4.Forster C., Clark H. B., Ross M. E., Iadecola C. Inducible nitric oxide synthase expression in human cerebral infarcts. Acta Neuropathol. 1999;97:215–220. doi: 10.1007/s004010050977. [DOI] [PubMed] [Google Scholar]
- 5.Andreadis A. A., Hazen S. L., Comhair S. A., Erzurum S. C. Oxidative and nitrosative events in asthma. Free Radical Biol. Med. 2003;35:213–225. doi: 10.1016/s0891-5849(03)00278-8. [DOI] [PubMed] [Google Scholar]
- 6.Cuzzocrea S., Zingarelli B., Villari D., Caputi A. P., Longo G. Evidence for in vivo peroxynitrite production in human chronic hepatitis. Life Sci. 1998;63:PL25–PL30. doi: 10.1016/s0024-3205(98)00252-5. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Monzon C., Majano P. L., Zubia I., Sanz P., Apolinario A., Moreno-Otero R. Intrahepatic accumulation of nitrotyrosine in chronic viral hepatitis is associated with histological severity of liver disease. J. Hepatol. 2000;32:331–338. doi: 10.1016/s0168-8278(00)80080-x. [DOI] [PubMed] [Google Scholar]
- 8.Turko I. V., Murad F. Protein nitration in cardiovascular diseases. Pharmacol. Rev. 2002;54:619–634. doi: 10.1124/pr.54.4.619. [DOI] [PubMed] [Google Scholar]
- 9.Gow A. J., Farkouh C. R., Munson D. A., Posencheg M. A., Ischiropoulos H. Biological significance of nitric oxide-mediated protein modifications. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004;287:L262–L268. doi: 10.1152/ajplung.00295.2003. [DOI] [PubMed] [Google Scholar]
- 10.Ischiropoulos H., Gow A. Pathophysiological functions of nitric oxide- mediated protein modifications. Toxicology. 2005;208:299–303. doi: 10.1016/j.tox.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 11.Souza J. M., Choi I., Chen Q., Weisse M., Daikhin E., Yudkoff M., Obin M., Ara J., Horwitz J., Ischiropoulos H. Proteolytic degradation of tyrosine nitrated proteins. Arch. Biochem. Biophys. 2000;380:360–366. doi: 10.1006/abbi.2000.1940. [DOI] [PubMed] [Google Scholar]
- 12.Murata M., Kawanishi S. Oxidative DNA damage induced by nitrotyrosine, a biomarker of inflammation. Biochem. Biophys. Res. Commun. 2004;316:123–128. doi: 10.1016/j.bbrc.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 13.Peluffo H., Shacka J. J., Ricart K., Bisig C. G., Martinez-Palma L., Pritsch O., Kamaid A., Eiserich J. P., Crow J. P., Barbeito L., Estevez A. G. Induction of motor neuron apoptosis by free 3-nitro-L-tyrosine. J. Neurochem. 2004;89:602–612. doi: 10.1046/j.1471-4159.2004.02363.x. [DOI] [PubMed] [Google Scholar]
- 14.Blanchard-Fillion B., Prou D., Polydoro M., Spielberg D., Tsika E., Wang Z., Hazen S. L., Koval M., Przedborski S., Ischiropoulos H. Metabolism of 3-nitrotyrosine induces apoptotic death in dopaminergic cells. J. Neurosci. 2006;26:6124–6130. doi: 10.1523/JNEUROSCI.1038-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mihm M. J., Schanbacher B. L., Wallace B. L., Wallace L. J., Uretsky N. J., Bauer J. A. Free 3-nitrotyrosine causes striatal neurodegeneration in vivo. J. Neurosci. 2001;21:RC149. doi: 10.1523/JNEUROSCI.21-11-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulder G. J., Jakoby W. B. Sulfation in conjugation reactions. In: Mulder G. J., Jakoby W. B., editors. Drug Metabolism. London: Taylor and Francis; 1990. pp. 107–161. [Google Scholar]
- 17.Falany C., Roth J. A. Properties of human cytosolic sulfotransferases involved in drug metabolism. In: Jeffery E. H., editor. Human Drug Metabolism: from Molecular Biology to Man. Boca Raton: CRC Press; 1993. pp. 101–115. [Google Scholar]
- 18.Weinshilboum R., Otterness D. Sulfotransferase enzymes in conjugation– deconjugation reactions. In: Kaufmann F. C., editor. Drug Metabolism and Toxicity. Berlin: Springer-Verlag; 1994. pp. 45–78. [Google Scholar]
- 19.Lipmann F. Biological sulfate activation and transfer. Science. 1958;128:575–580. doi: 10.1126/science.128.3324.575. [DOI] [PubMed] [Google Scholar]
- 20.Sakakibara Y., Suiko M., Liu M.-C. De novo sulfation of L-tyrosine in HepG2 human hepatoma cells and its possible functional implication. Eur. J. Biochem. 1994;226:293–301. doi: 10.1111/j.1432-1033.1994.tb20053.x. [DOI] [PubMed] [Google Scholar]
- 21.Sakakibara Y., Suiko M., Nakajima H., Liu M.-C. Sulphation of L-tyrosine in mammalian cells: a comparative study. Biochem. J. 1995;305:993–998. doi: 10.1042/bj3050993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suiko M., Sakakibara Y., Nakajima H., Sakaida H., Liu M.-C. Enzymic sulphation of dopa and tyrosine isomers by HepG2 human hepatoma cells: stereoselectivity and stimulation by Mn2+ Biochem. J. 1996;314:151–158. doi: 10.1042/bj3140151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakakibara Y., Katafuchi J., Takami Y., Nakayama T., Suiko M., Nakajima H., Liu M.-C. Manganese-dependent Dopa/tyrosine sulfation in HepG2 human hepatoma cells: novel Dopa/tyrosine sulfotransferase activities associated with the human monoamine-form phenol sulfotransferase. Biochim. Biophys. Acta. 1997;1355:102–106. doi: 10.1016/s0167-4889(96)00166-8. [DOI] [PubMed] [Google Scholar]
- 24.Suiko M., Sakakibara Y., Liu M.-C. Sulfation of environmental estrogen-like chemicals by human cytosolic sulfotransferases. Biochem. Biophys. Res. Commun. 2000;267:80–84. doi: 10.1006/bbrc.1999.1935. [DOI] [PubMed] [Google Scholar]
- 25.Pai T. G., Sugahara T., Suiko M., Sakakibara Y., Xu F., Liu M.-C. Differential xenoestrogen-sulfating activities of the human cytosolic sulfotransferases: molecular cloning, expression, and purification of human SULT2B1a and SULT2B1b sulfotransferases. Biochim. Biophys. Acta. 2002;1573:165–170. doi: 10.1016/s0304-4165(02)00416-6. [DOI] [PubMed] [Google Scholar]
- 26.Sakakibara Y., Takami Y., Nakayama T., Suiko M., Liu M.-C. Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form phenol sulfotransferases. J. Biol. Chem. 1998;273:6242–6247. doi: 10.1074/jbc.273.11.6242. [DOI] [PubMed] [Google Scholar]
- 27.Sakakibara Y., Yanagisawa K., Katafuchi J., Ringer D. P., Takami Y., Nakayama T., Suiko M., Liu M.-C. Molecular cloning, expression, and characterization of novel human SULT1C sulfotransferases that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J. Biol. Chem. 1998;273:33929–33935. doi: 10.1074/jbc.273.51.33929. [DOI] [PubMed] [Google Scholar]
- 28.Sakakibara Y., Suiko M., Pai T. G., Nakayama T., Takami Y., Katafuchi J., Liu M.-C. Highly conserved mouse and human brain sulfotransferases: molecular cloning, expression, and functional characterization. Gene. 2002;285:39–47. doi: 10.1016/s0378-1119(02)00431-6. [DOI] [PubMed] [Google Scholar]
- 29.Liu M.-C., Lipmann F. Decrease of tyrosine-O-sulfate-containing proteins found in rat fibroblasts infected with Rous sarcoma virus or Fujinami sarcoma virus. Proc. Natl. Acad. Sci. U.S.A. 1984;81:3695–3698. doi: 10.1073/pnas.81.12.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanagisawa K., Sakakibara Y., Suiko M., Takami Y., Nakayama T., Nakajima H., Takayanagi K., Natori Y., Liu M.-C. cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. Biosci. Biotechnol. Biochem. 1998;62:1037–1040. doi: 10.1271/bbb.62.1037. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 33.Jevons F. R. Tyrosine O-sulphate in fibrinogen and fibrin. Biochem. J. 1963;89:621–624. doi: 10.1042/bj0890621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falany C. N. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 35.Snider S. R., Kuchel O. Dopamine: an important neurohormone of the sympathoadrenal system. Significance of increased peripheral dopamine release for the human stress response and hypertension. Endocr. Rev. 1983;4:291–309. doi: 10.1210/edrv-4-3-291. [DOI] [PubMed] [Google Scholar]
- 36.Herbert J. D., Coulson R. A., Hernandez T. Free amino acid pools and their role in regulation. Comp. Biochem. Physiol. 1966;17:583–598. doi: 10.1016/0010-406x(66)90589-5. [DOI] [PubMed] [Google Scholar]
- 37.Hogg N., Darley-Usmar V. M., Wilson M. T., Moncada S. Production of hydroxyl radicals from the simultaneous generation of superoxide and nitric oxide. Biochem. J. 1992;281:419–424. doi: 10.1042/bj2810419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saeki M., Maeda S. p130cas is a cellular target protein for tyrosine nitration induced by peroxynitrite. Neurosci. Res. 1999;33:325–328. doi: 10.1016/s0168-0102(99)00019-x. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto T., Bing R. J. Nitric oxide donors. Proc. Soc. Exp. Biol. Med. 2000;225:200–206. doi: 10.1046/j.1525-1373.2000.22525.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H., Joseph J., Feix J., Hogg N., Kalyanaraman B. Nitration and oxidation of a hydrophobic tyrosine probe by peroxynitrite in membranes: comparison with nitration and oxidation of tyrosine by peroxynitrite in aqueous solution. Biochemistry. 2001;40:7675–7686. doi: 10.1021/bi002958c. [DOI] [PubMed] [Google Scholar]