Abstract
β-Catenin, a key component of the canonical Wnt pathway is also regulated by tyrosine phosphorylation that regulates its association to E-cadherin. Previously, we reported its association with the hepatocyte growth factor (HGF) receptor-Met, at the membrane. HGF induced met-β-catenin dissociation and nuclear translocation of β-catenin, which was tyrosine-phosphorylation dependent. Here, we further investigate the met-β-catenin interaction, by selectively mutating several tyrosine residues alone or in combination, in β-catenin. The mutants were subcloned into FLAG-CMV vector & stably transfected into rat hepatoma cells, which were treated with HGF. All single or double mutant-transfected cells continued to show HGF induced nuclear translocation of FLAG-β-catenin except the mutations affecting 654 and 670 simultaneously (Y654/670F), which coincided with the lack of formation of β-catenin-TCF complex and DNA synthesis, in response to the HGF treatment. In addition, the Y654/670F-transfected cells also showed no phosphorylation of β-catenin or dissociation from Met in response to HGF. Thus, intact 654 and 670 tyrosine residues in β-catenin are crucial in HGF-mediated β-catenin translocation, activation, and mitogenesis.
INTRODUCTION
Hepatocyte growth factor (HGF) or scatter factor, is a multifunctional peptide that signals via Met, a tyrosine kinase receptor, which undergoes tyrosine phosphorylation and activation [1–3]. In turn, the signal is transduced downstream, either directly or through its adaptor molecules-Gab1 or Grb2, which trigger activation of signaling such as ERK1/2/MAPK, PLC-γ, PI3K, SHP2 and others [4–9]. Some proteins such as Stat3 and Src bind directly to met [10–13]. These interactions elicit biological responses such as epithelial morphogenesis, angiogenesis, tubulogenesis, proliferation and cell-cell adhesion. In addition, HGF/Met pathway also contributes to survival and growth of tumor cells and plays a role in angiogenesis and metastasis [14]. Owing to these responses, HGF acts as a mitogen, morphogen and motogen for epithelial cells [15, 16]. In liver, HGF/Met axis is shown to be important in hepatocyte proliferation during liver regeneration following hepatectomy [17, 18].
β-Catenin is a key component of the canonical Wnt pathway where it is chiefly regulated by serine-threonine phosphorylation [19–21]. Wnt signaling induces inhibition of the kinases with resulting hypophosphorylation of β-catenin, and binding to TCF/LEF to translocate to the nucleus. In the absence of the Wnts or in the presence of Wnt inhibitors such as frizzled-related proteins, β-catenin is phosphorylated at specific serine and threonine residues, which leads to its ubiquitin-mediated proteosomal degradation. Activation of β-catenin due to mutations in β-catenin gene (CTNNB1) or its degradation components such as axin and APC, is observed in many cancers including colorectal, skin, breast and liver [22–24].
β-Catenin also functions as a bridge between the cytoplasmic domain of cadherins and the actin-containing cytoskeleton [25, 26]. β-Catenin complex with the cytoplasmic tail of E-cadherin is mostly regulated by phosphorylation at tyrosine-654, which induces β-catenin dissociation from E-cadherin, thus negatively influencing cell-cell adhesion [27]. Previously others and we have demonstrated key interactions between β-catenin and Met [28, 29]. We observed a subcellular association of these proteins in hepatocytes, which underwent dissociation following HGF-induced tyrosine phosphorylation of β-catenin [29]. This required an intact tyrosine kinase domain of Met. Another study has shown the importance of the tyrosine residue 142 in this interaction, acting as a switch between the adhesion and transcriptional function [30]. In the present study, we examine the importance of tyrosine residue 142 along with other residues to positively address, which residues are critical in Met-β-catenin interaction in hepatocytes along with any associated biological events.
MATERIALS AND METHODS
Plasmid Constructs
The cDNA for Ctnnb1 (human β-catenin gene) encoding β-catenin protein (nt +211 to +2556) was generated by PCR using human liver cDNA as template and using oligonucleotides with incorporated BamHI sites. The cDNA was inserted downstream of the CMV promoter in the p3XFLAG-CMV™-10 expression Vector (Sigma, Saint Louis, MI). Point mutations Tyr-86→Phe, Tyr-86→Glu, Tyr-142→Phe, Tyr-142→Glu, Tyr-604→Glu, Tyr-654→Phe, Tyr-654→Glu, Tyr-670→Phe, Tyr-670→Glu and Tyr-709→Glu, were introduced using the GeneTailor™ site-directed mutagenesis kit as directed (Invitrogen, Carlsbad, CA). The primers were designed as directed by the protocol. For example, sense primers Tyr-654→Phe was 5′-GGAATGAAGGTGTGGCGACATTTGCAGCTGCTGT-3′; for Tyr-654→Glu was 5′-GGAATGAAGGTGTGGCGACAGAGGCAGCTGCTGT-3′ (changes indicated in bold). After amplification, the products were transformed into merBC wild type (DH5α™-T1R) E. Coli. Three double mutants (Tyr-654→Phe/Tyr-142→Phe, Tyr-670→Phe/Tyr-142→Phe and Tyr-654→Phe/Tyr-670→Phe) were also obtained using this procedure. All sequences of generated constructs (WT, Y86F, Y86E, Y142F, Y142E, Y604F, Y604E, Y654F, Y654E, Y670F, Y670E) and double mutant plasmids (Y654/142F, Y670/142F and Y654/670F) were confirmed.
Cell culture, Transfection and Treatment
The rat hepatoma cell line, JM1, was cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum and Gentamycin (500 mg/ml) at 37 °C in a humidified 5% CO2 atmosphere [31]. This cell line has low endogenous expression of β-catenin and does not have any mutations in its exon-3, a site critical for its phosphorylation and degradation.
Semiconfluent layers of JM1 cells were grown in 35-mm-diameter (6-well) dishes and transfected with FLAG-tagged-WT-β-catenin plasmid or tyrosine mutants by FuGENE 6 transfection kit (Roche, Indianapolis, IN). DNA dilutions were prepared in 100 ml of DMEM for transfection of the cells in 35-mm-diameter dishes. The DNA (1 μg) preparations were subsequently mixed with 97 μl of DMEM containing 3 μl of FuGENE 6 respectively. The transfection solutions were incubated for 30 min at room temperature and applied directly to the cell supernatants. After overnight incubation, the transfection solution was replaced with fresh medium containing 600 μg/ml G418, and maintained in the presence of this drug. Cell lines expressing FLAG-tagged protein were screened by Western blots. The selected stably transfected cell lines were maintained in DMEM supplemented with 10% fetal calf serum, Gentamycin (500 μg/ml) and 200 μg/ml G418 sulfate at 37 °C in a humidified 5% CO2 atmosphere. These cells were serum starved for overnight, followed by treatment with HGF at different concentrations for 15 min (unless stated otherwise) for subsequent experiments.
Preparation of Total Cell Lysates, Nuclear Extracts and Western blotting
Total cell lysates and nuclear extracts were prepared as already reported [29]. Briefly, total cell lysate from stably transfected cells was prepared in RIPA buffer containing protease and phosphatase inhibitors (Sigma Chemical Co.). Nuclear extracts were prepared in HEPES buffer. The concentration of the protein in the lysates was determined by bicinchoninic acid protein assay with BSA as a standard. Aliquots of the samples were stored at −80°C until use.
Gel Electrophoresis and Western Blotting
All experiments were performed in triplicates, and representative data is shown. Fifty-100 μg of total or nuclear proteins were resolved using the mini-PROTEAN-3 electrophoresis module (Biorad, Hercules, CA) [32]. Proteins were transfered to Immobilon-PVDF membranes (Millipore, Bedford, MA). Blots were blocked and incubated with primary antibody followed by a horseradish peroxidase (HRP)-conjugated secondary antibody. Proteins were visualized by Super-Signal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL). Primary antibodies including anti–β-catenin (mouse), anti–E-cadherin (rabbit) were used at 1:200 (Santa Cruz Biotech, Santa Cruz, CA.), anti-FLAG (mouse) were used at 10 μg/mL (Sigma, Saint Louis, MI). The secondary antibodies including HRP-conjugated anti-mouse and anti-rabbit were used at 1:50,000 (Chemicon, Temecula, CA).
Immunoprecipitation
400 μg of lysate were precleared using appropriate IgG and protein A/G agarose (Santa Cruz Biotechnology) and incubated with 8 μg of rabbit Anti-FLAG polyclonal antibody and 20 μl of protein A/G agarose [32]. Pellets were washed in RIPA buffer and resuspended in electrophoresis loading buffer. 30 μl of the samples were resolved on ready gels and transferred as described earlier. The antibodies used for blotting included Met, E-cadherin and PY20 as well as HRP-conjugated secondary antibodies. The change in association of FLAG-β-catenin to E-Cadherin or met or detection of PY20-FLAG-β-catenin was quantified by comparing the intergrated optical density (IOD) following scanning of the western blots and densitometry by the NIH Imager software. The IOD was normalized to the non-HGF treated cells and presented as fold-change or change in percentage.
Immunofluorescence Microscopy
The staining protocol has been described before [29]. Briefly, transfected cells grown on glass coverslips were serum starved and experimental group was treated with HGF (50 ng/ml) for 15 minutes. Cells were fixed in methanol, blocked and incubated with rabbit anti-FLAG polyclonal antibody and detected by Alexa Flou 488 fluorochrome-conjugated secondary antibody. Nuclei were counterstained by DAPI and the slides viewed on Zeiss epifluorescence microscope. Digital images were obtained by Nikon Coolpix camera and collages prepared using Adobe Photoshop 7.0 software.
Cell proliferation
Proliferation of the transfected cells in response to HGF was measured by 3H-thymidine incorporation assay. Cells (1 x 105) were plated in 6 well plates in DMEM with 10% fetal calf serum, serum starved for 24 h, and then pulsed with 3H-thymidine (2.5 μCi/ml) +/− HGF for 48 hour at 50 ng/ml. Cells were washed with 0.9% NaCl, lysed in 0.2% SDS, 1× SSC (150 mM NaCl, 15 mM Na-Citrate pH 7.0), 5 mM EDTA and aliquots used to determine 3H-thymidine incorporation on Beckman Scintillation counter and total DNA using Hoechst dye H33258 and fluorescence meter (Hoefer, DyNA Quant 200).
Electrophoretic Mobility Shift Assay (EMSA) for β -Catenin-TCF Binding
The WT- TCF probe was prepared by annealing - 5′-TGCCGGGCTTTGATCTTTG-3′ and 5′-AGCAAAGATCAAAGCCCGG-3′ deoxyoligonucleotides [33]. Double-stranded probes were end-labeled using [g-32P] ATP and T4 polynucleotide kinase (Life Technologies, Inc), followed by purification on a G-25 Sephadex spin column (5 Prime to 3 Prime, Boulder, CO). Binding reactions contained 5–10 μg nuclear protein, 50 ng/ml poly(dI-dC), 10 mM Hepes (pH 7.9), 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, and 12% glycerol. Binding reactions involving non-radiolabeled competitor probes also contained 100-fold molar excess of the competitor. Reactions (excluding radiolabeled probe) were assembled on ice and incubated for 5 min at room temperature. The radiolabeled probe (30,000 cpm) was added and reactions incubated at room temperature for 20 min. Samples were then applied to a 4% polyacrylamide gel in 0.5X TBE at room temperature (3 h, 200 V). Gels were transferred to Whatman paper followed by autoradiography.
Statistical Analysis
For immunofluorescence studies, the numbers of cells showing FLAG-β-catenin in the nuclei were counted at 400X magnification in three random fields. The averages were compared for statistical significance by the student t-test using InStat software and the p<0.001 was considered significant. For thymidine incorporation, the averages (+/−SEM) of counts in response to HGF treatment for each mutant were compared for statistical significant differences using the student t test. P<0.05 was considered significant.
RESULTS
JM1 transfected cells express FLAG-tagged β-catenin fusion protein
We successfully constructed CMV promoter controlled recombinant plasmids: WT β-catenin, tyrosine point mutant β-catenin plasmids (Y86E, Y142E, Y604E, Y654E, Y670E, Y709E, Y86F, Y142F, Y654F, Y670F) and 3 double mutants of β-catenin (Y142F/Y654F, Y142F/670F, Y654F/670F) (Fig. 1).
Figure 1.
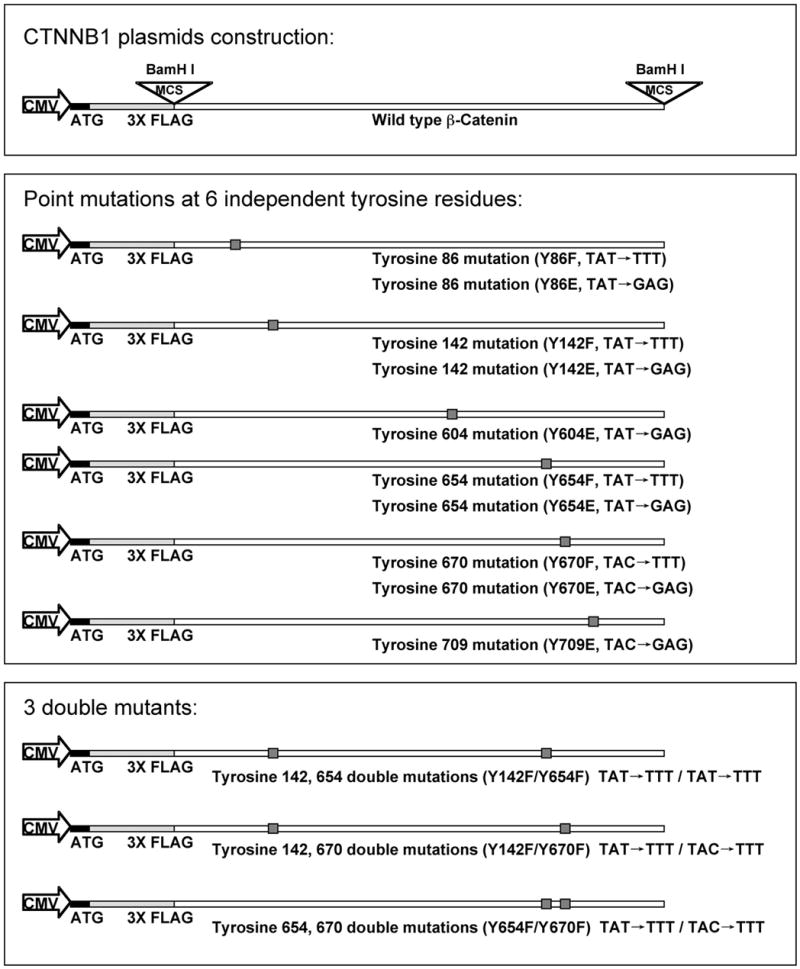
Human β-catenin gene (CTNNB1) plasmid construction and tyrosine residues mutagenesis. Schematic representation of the CTNNB1 cDNA encoding the entire region of human β-catenin protein consisting of 781 residues. Nucleotides +211 to +2556) were inserted in multiple cloning site (BamHI) downstream of the CMV promoter in the p3XFLAG-CMV ™-10 expression vector and labeled as WT-β-catenin plasmid. This was subjected to site-directed mutagenesis to generate the 6 single residue control mutants (Tyrosine to Glutamic acid): Y86E, Y142E, Y604 E, Y654E, Y670E and Y709E; 4 single residue mutants (Tyrosine to Phenylalanine): Y86F, Y142F, Y654F and Y670F; and 3 double mutants (Tyrosine to Phenylalanine): Y142F/Y654F, Y142F/Y670 and Y654F/Y670F. The nucleotides changes were: Tyrosine to Phenylalanine-TAT/TAC to CAG; Tyrosine to Glutamic acid-TAT/TAC to GAG, all shown as small shadowed box.
Transfected JM1 hepatoma cells were examined for FLAG-tagged protein expression by Western blot utilizing an antibody against FLAG. The results confirmed successful generation of stable JM1 cell lines expressing FLAG-tagged WT or tyrosine-mutated β-catenin (Fig. 2A). The selected stable- transfected cell lines were maintained in DMEM medium supplemented with 200 mg/ml G418 and used for all subsequent experiments.
Figure 2.
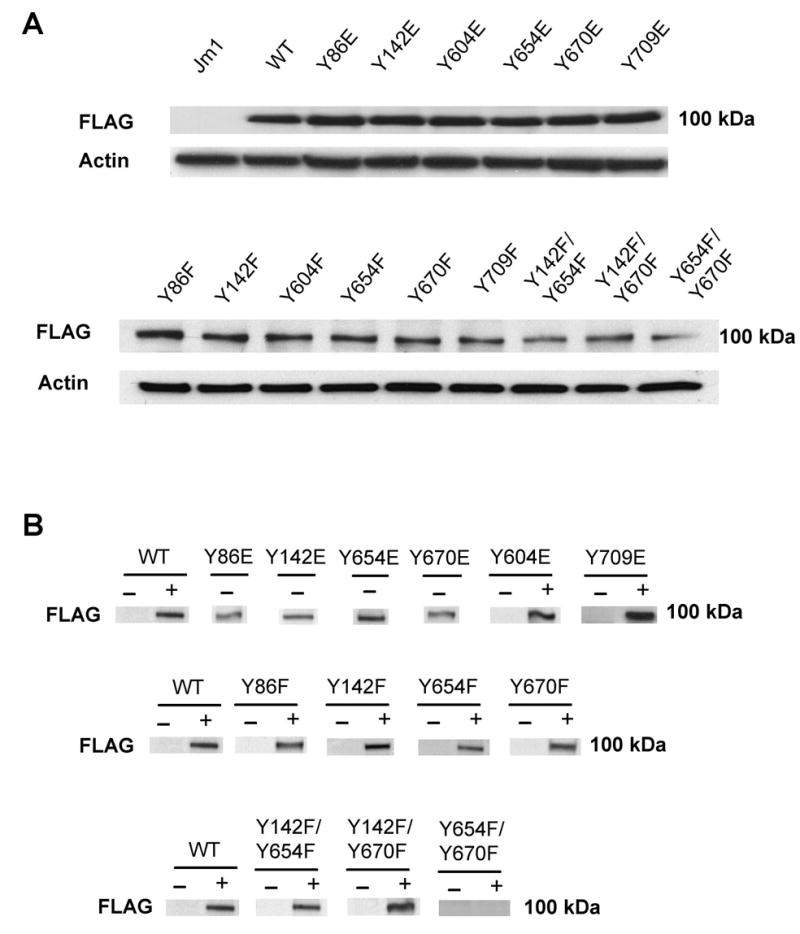
Detection of FLAG-β-catenin using FLAG antibody and changes in nuclear levels of FLAG-β-catenin in response to HGF (50 ng/ml for 15 min) in the cells.
A. FLAG-β-catenin (~100 kDa) is detected in whole cell lysates from FLAG-tagged WT or mutant β-catenin transfected cells.
B. Western blots using nuclear proteins demonstrate presence of FLAG-β-catenin protein in the absence of HGF in Y86E, Y142E, Y654E and Y670E mutants only. HGF induced nuclear translocation of FLAG-β-catenin was observed in WT-, Y604E- and Y709E-transfected cells. Increased nuclear FLAG-β-catenin was observed in response to HGF in Y86F, Y142F, Y654F, Y670F, Y142/654F and Y142F/670F cells and not Y654F/Y670F cells.
HGF increased nuclear translocation of FLAG-tagged β-catenin in all cell lines except Y654/Y670F double mutants
WT or mutant β-catenin transfected cells were cultured in DMEM medium containing 200 mg/ml G418, serum starved for overnight and treated with HGF (50 ng/ml) for 15 minutes. Total and nuclear proteins were isolated for analysis using the anti-FLAG antibody.
For examining the importance of individual tyrosine residues, Y to E mutants were examined for basal nuclear translocation of FLAG-β-catenin in comparison to HGF-induced nuclear translocation of FLAG-WT-β-catenin. HGF increased nuclear FLAG-β-catenin in WT-β-catenin transfected cells as expected, and in Y604E and Y709E cells (Fig. 2B). Spontaneous nuclear FLAG-β-catenin was observed in Y86E, Y142E, Y654E and Y670E-transfected cells only (this mutation imparts negative charge mimicking tyrosine phosphorylation) (Fig. 2B). These findings were consistent with immunofluorescence analysis (Fig. 3A–H).
Figure 3.
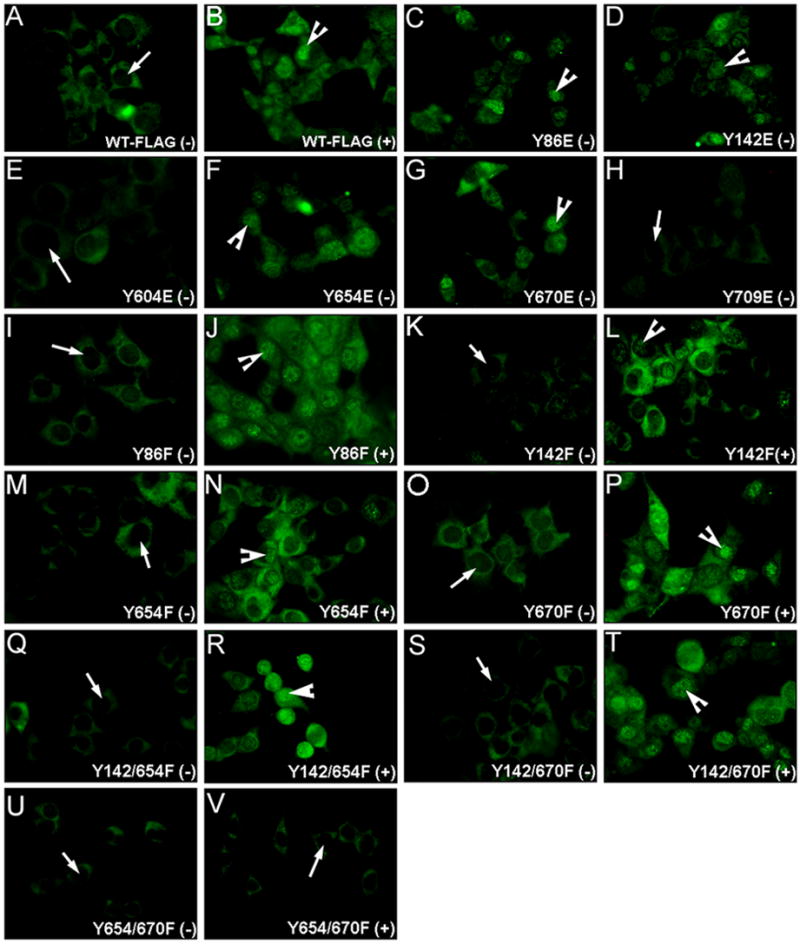
Immunofluorescence using the FLAG antibody reveals nuclear translocation of β-catenin (green) in response to HGF, in all tyrosine to phenylalanine mutants except the Y654/Y670F mutants. (400X). (A)-WT-β-catenin transfected cells show absent nuclear β-catenin without HGF; (B)-HGF induced nuclear translocation in these cells; Nuclear FLAG-β-catenin observed in absence of HGF in (C)-Y86E; (D)-Y142E; (F)-Y654E and; (G)-Y670E-cells and no nuclear FLAG-β-catenin in (E)-Y604E and (H)-Y709E-transfected cells; No nuclear FLAG-β-catenin in absence of HGF in Y86F-cells (I); Y142F-cells (K); Y654-cells (M); Y670F-cells (O); Y142/654F-cells (Q); Y142/670F (S); or Y654/670F-cells (U). Nuclear FLAG-β-catenin detected upon HGF treatment in Y86F-cells (J); Y142F-cells (L); Y654F-cells (N); Y670-cells (P); Y142/654F-cells (R); or Y142/670F-cells (T); (V)-Absent nuclear FLAG-β-catenin in presence of HGF in Y654/670F-cells. (−): without HGF; (+) with HGF.
Based on the above results, Y86F, Y142F, Y654F and Y670F-transfected JM1 cells were treated with HGF. Interestingly, all of the above mutants showed an increase in nuclear FLAG-β-catenin (Fig. 2B), which was also evident in immunofluorescence studies (Fig 3I–P). The numbers of nuclei showing nuclear FLAG-β-catenin were counted as described in methods (400X). A significant increase ranging from 9–16-fold was observed following HGF treatment in all cells (Fig. 4A). This data suggested that there might be multiple tyrosine residues or additional independent tyrosine residues, mediating HGF-induced β-catenin nuclear translocation.
Figure 4.
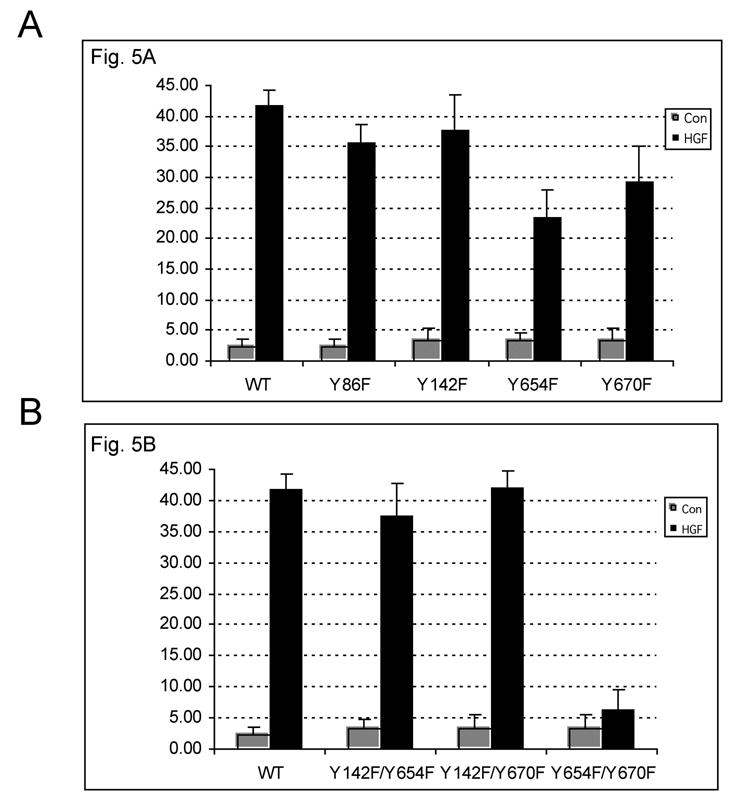
Statistical analysis reveals significant (p<0.001) increase in number of cells showing nuclear FLAG-β-catenin in response to HGF except the Y654/670F-transfected cells.
A. Significant increase in the number of nuclei showing FLAG-β-catenin, following HGF treatment in WT-, Y86F-, Y142F-, Y654F- and Y670F-transfected cells.
B. Significantly higher numbers of nuclei show FLAG-β-catenin, following HGF treatment in WT-, Y142/654F- and Y142F/670F-transfected cells. Nuclei showed no FLAG-β-catenin in presence or absence of HGF in Y654F/Y670F-transfected cells.
Next, double mutants Y142/654F; Y142/670F; and Y654/670F were treated with HGF. WT, Y142/654F and Y142/670F transfected JM1 cells showed nuclear-FLAG-β-catenin in response to HGF treatment by western blot (Fig. 2C), which was also evident by immunofluorescence (Fig. 3Q–T). This increase ranged from 15–17 fold (Fig. 4B). A complete abrogation of nuclear translocation of FLAG-β-catenin was observed Y654/670F-transfected cells in response to HGF (Fig. 2C). This was confirmed by immunofluorescence studies as well (Fig. 3U–V and 4B). Thus intact tyrosine residues 654 and 670 are critical in HGF-mediated nuclear translocation of β-catenin.
Cell proliferation in response to HGF in WT and mutant-transfected JM1 cells
To ascertain any biological relevance of the Met-β-catenin interaction, we examined the various cells for proliferation in response to HGF. DNA synthesis was measured by the 3H-thymidine incorporation assay as described in methods. HGF treatment of WT-β-catenin transfected cells led to 2.5-fold (p<0.001) increase in thymidine incorporation. Similarly Y86E-, Y142E-, Y654E- and Y670E-transfected cells showed around 2-fold (p<0.05) increase in thymidine incorporation even in the absence of HGF (Fig. 5A). A significant (p<0.001) increase ranging from 2–2.5-fold in thymidine incorporation in response to HGF was observed in Y86F-, Y142F-, Y654F- and Y670F cells (Fig. 5B). Also, a significant (p<0.001) and 2.0-fold increase in DNA synthesis in response to HGF was observed in Y142/654F and Y142/670F cells, while no increase occurred in Y654/670F cells (Fig. 5C). This demonstrated the importance of tyrosine residues 654 and 670 in β-catenin, in mediating HGF-induced cell proliferation.
Figure 5.
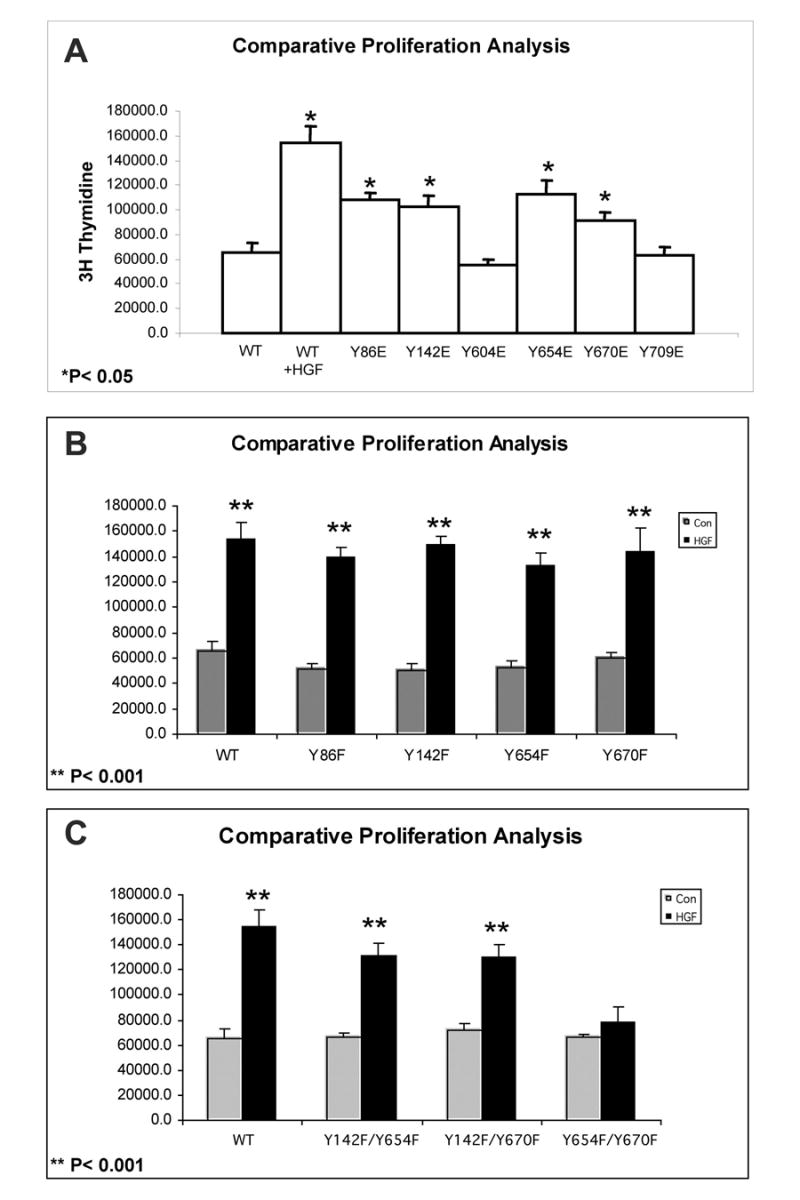
Lack of increase in DNA synthesis following HGF treatment in the Y654F/Y670F-transfected cells.
A. A significant increase in thymidine incorporation was observed in WT-β-catenin transfected cells following HGF treatment and in the Y86E-, Y142E-, Y654E- and Y670E-transfected cells without HGF. No such increase was observed without HGF in Y604E and Y709E-transfected cells.
B. A significant increase in thymidine incorporation occurred in WT-, Y86F-, Y142F-, Y654F- and Y670F-cells following HGF treatment (p<0.001).
C. A significant increase in thymidine incorporation occurred in WT-, Y142/654F- and Y142/670F-transfected cells after HGF treatment (p<0.001), which remained unchanged in Y654/670F-transfected cells.
Met/FLAG-β-Catenin/E-Cadherin association in response to HGF treatment
To directly examine association of FLAG-β-catenin to Met and E-cadherin following HGF treatment, various double mutant-transfected cells were examined by coprecipitation studies as described in methods. HGF induced a decrease in Met-β-catenin association by 3–5 fold in WT-β-catenin-, Y142/654F- and Y142/670F-transfected cells while no change was observed in Y654/670F-transfected cells (Fig. 6A and 6B). At this time and dose of HGF, all cells showed a small but consistent decrease in E-cadherin-β-catenin association ranging from 15–30% (Fig. 6A and 6C). To determine the overall tyrosine phosphorylation status of FLAG-β-catenin in response to HGF, we examined the immunoprecipitate for PY20, which showed a dramatic increase by 2-5-4-fold in WT-, Y142/654F- and Y142/670F-transfected JM1 cells after HGF treatment (Fig. 6A and 6D). No tyrosine phosphorylation was identified in Y654/670F-transfected cells after HGF treatment (Fig. 6A and 6D). This demonstrates that indeed tyrosine residues 654 and 670 undergo tyrosine phosphorylation following HGF/met activation.
Figure 6.
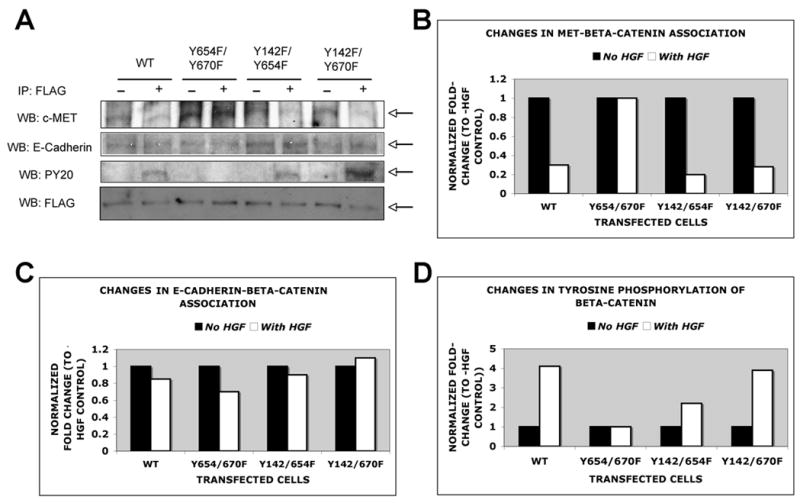
No change in Met/β-catenin association or tyrosine phosphorylation of β-catenin following HGF treatment in Y654/670F-transfected. Total cell lysates from WT or double mutant-transfected cells cultured with or without HGF were immunoprecipitated with anti-FLAG antibody and blotted for Met, E-cadherin or PY20.
A. A representative western blot analysis reveals decrease in Met-β-catenin coprecipitation in WT-, Y142/654F- and Y142/670F-transfected cells in response to HGF, while no change was evident in Y654/670F-cells. A less dramatic difference was observed in E-cadherin-β-catenin association in all cells. Also phosphotyrosine antibody detected β-catenin tyrosine phosphorylation after HGF treatment in WT-, Y142/654F, Y142/670F and not in Y654/670F cells. Comparative pull down of FLAG-β-catenin in all conditions was confirmed by western with anti-FLAG antibody.
B. A representative analysis of studies done in triplicates demonstrates a decrease in Met-β-catenin association ranging from 3–5 fold in all cells except Y654/670F-transfected cells.
C. A small but consistent decrease in E-cadherin-β-catenin association ranging from 15–30% was observed following HGF treatment in all cells.
D. A 2.5–4.0-fold increase in PY20-FLAG-β-catenin was observed in response to HGF in all cells except Y654/670F-transfected cells.
Lack of β-catenin-TCF complex formation in response to HGF in the Y654/670F-transfected JM1 cells
Nuclear translocation of β-catenin along with its association with TCF/LEF factors are the key steps in transactivation of β-catenin target genes, which further go on to regulate events such as cell proliferation. Next, we examine β-catenin–TCF binding in the WT-β-catenin and double-mutant-β-catenin-transfected JM1 cells following HGF treatment by gel mobility shift assay as elaborated in the methods. β-Catenin-TCF binding was observed in WT-β-catenin-, Y142/654F- or Y142/670F-transfected JM1 cells only after HGF treatment (Fig. 7). However no β-catenin-TCF binding was observed in the presence of HGF in the Y654/670F-transfected cells (Fig. 7), indicating the importance of 654 and 670 tyrosine residues for successful HGF/Met mediated β-catenin tyrosine phosphorylation, nuclear translocation and activation.
Figure 7.
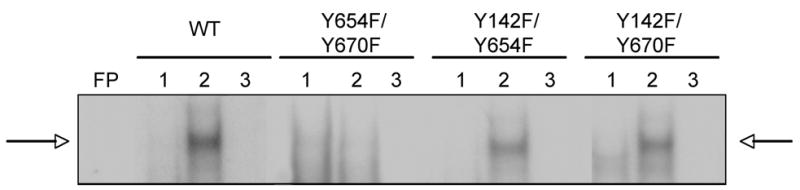
EMSA revealed lack of β-catenin-TCF complex formation in Y654/670F-transfected cells following HGF treatment. 10 μg of nuclear extracts from WT- or double mutant-transfected cells cultured in the absence (1) or presence (2) of HGF were examined for β-catenin-TCF (cyclin-D1) binding. β-Catenin-TCF complex was observed in response to HGF in the WT-, Y142/654F- and Y142/670F-transfected cells, which was successfully competed off with cold probe (3). No complex was observed in Y654/670F-transfected cells after HGF treatment.
DISCUSSION
Tyrosine phosphorylation-dependent regulation of β-catenin is being increasingly recognized for its role in not only modulating cell-cell adhesion but also as a means of β-catenin activation. This broadens the overall role of β-catenin from just being a downstream effector of the canonical Wnt pathway. β-Catenin has now been shown to be tyrosine phosphorylated in response to a number of tyrosine kinases, which in turn leads to its activation. Furthermore, different protein tyrosine kinases can modify specific tyrosine residues in β-catenin, which might dictate the ensuing downstream signaling and eventual biological function. For instance, Src, has been shown to phosphorylated tyrosine 86 in β-catenin [27]. Similarly Fer modifies β-catenin at the tyrosine residue 142 [34].
E-Cadherin-β-catenin association is well characterized at the cell membrane [35, 36]. Tyrosine 654 has been shown to play a role in regulating association of β-catenin to E-cadherin and correlates with the disassembly of adherens junctions and decreased cell adhesion observed in invasion and metastasis in various tumors [27]. Conversely, inhibition of tyrosine phosphorylation can lead to reassociation of β-catenin to E-cadherin in rat-transformed cells [34, 37]. More recently, tyrosine-142 was shown to be phosphorylated in response to HGF/met in MDCK cells, where it acted as a switch between the transcription and adhesive function via interaction with BCL9-2 [30].
In hepatocytes, short-term HGF treatment induced an insignificant change in β-catenin-E-cadherin association while the Met-β-catenin complex undergoes rapid dissociation. A concomitant increased in c-myc has been observed suggesting a more mitogenic consequence of this interaction [29].
Based on previous studies and close proximity to the region of armadillo repeats, we selected tyrosine residues 86, 142, 604, 654, 670 and 709 in β-catenin for additional analysis. The tyrosine 670 was of particular interest since none of the crystal structures of β-catenin encompassed this area and it was close to residue 654, which is a known functional residue. Tyrosine (Y) to glutamate (E) mutation imparts a negative charge and thus mimics tyrosine phosphorylation. Spontaneous nuclear translocation was observed only in 86, 142, 654 and 670 mutants thus identifying their functionality. As mentioned before these residues upon phosphorylation will clearly induce nuclear translocation and perhaps activation of β-catenin. However particular upstream effectors might affect specific residues and this specificity might dictate the eventual response. Additional analysis will be essential in mapping these interactions to examine the contribution of β-catenin in morphogenesis, mitogenesis, motogenesis or other biological responses in response to various proteins. Although tyrosine 709 was also selected for analysis, we were aware that it was not phosphorylated and anticipated its use as a negative internal control [27]. To block phosphorylation, these tyrosine residues were mutated to phenylalanine Interestingly, we failed to identify a single tyrosine residue, whose mutation could block the HGF-induced nuclear translocation and activation of β-catenin suggesting the role of two or more tyrosine residues or the capacity of some of these residues to be redundant.
To address this, we mutated two tyrosine residues simultaneously in β-catenin protein. Based on the importance of tyrosine 142 in HGF-mediated β-catenin phosphorylation we also mutated this site along with tyrosine residues 654 or 670 to phenylalanine [30]. In addition, we concomitantly mutated 654 and 670 residues. These cell lines were examined for their responsiveness to HGF in terms of nuclear translocation and activation of β-catenin as well as for biological relevance by examining proliferation. HGF induced all these events in all double mutants except the Y654/670F-transfected JM1 cells. While Y142/654F and Y142/670F continued to show tyrosine phosphorylation as observed in the PY20 western blot, there was a complete abrogation of tyrosine phosphorylation of β-catenin in response to HGF. This indicated that an intact 654 or 670 residue at any given time is sufficient to respond to HGF/Met stimulation. In addition, there was a lack of β-catenin nuclear translocation as well as β-catenin-TCF complex formation in response to HGF, highlighting the importance of these two neighboring tyrosine residues in HGF-mediated β-catenin regulation. β-Catenin-Met association was lost in response to HGF in all cells except the Y654/670F cells, again demonstrating the importance of these residues in β-catenin dissociation. On the other hand E-cadherin-β-catenin association was mildly affected in all cells in response to HGF. We can speculate that E-cadherin-β-catenin complex in liver cells is more stable in response to the specific dose and duration of HGF treatment, whereas the Met-β-catenin succumbs rapidly. This prevents any negative effect on cell-cell adhesion at this time, but dissociated β-catenin does translocate to the nucleus and begins an increase in expression of pro-proliferative genes such as c-myc [29]. In agreement with this, HGF induced DNA synthesis occurred in all single and double mutants of β-catenin-transfected JM1 cells except the Y654/670Fcells, indicating a definite role of tyrosine phosphorylation-dependent β-catenin activation in HGF-induced mitogenesis. Tyrosine-142 alone or in combination with 654 or 670 does not seem to play a role in HGF-mediated response in liver cells. This might be mostly due to tissue specific differences as the previous work was done in the COS-7 cells [30]. This cell line is in fact kidney fibroblasts and thus is inherently different from liver cells. Additional differences might be due to the dissimilarity in the dose and duration of HGF treatment. However, in the liver cells, tyrosine residues 654 and 670 appear to be critical in HGF-induced nuclear translocation and activation of β-catenin.
While we are sure of the importance of these two tyrosine residues as downstream effectors of HGF/met response, we are unsure of the structural feasibility of this interaction. Although crystal structure like the ones described previously would be needed to determine this, we examined the amino acid sequence around the residues 654 and 670 [36, 38]. We found a proline residue between these two residues, which has previously been shown to be responsible to turn over the helix structure [39]. Such an event would place tyrosine residues 654 and 670 more closely, for appropriate interaction. Additional analysis would be needed to determine such configuration. From the amino acid sequence of the region around tyrosine 670, it appears to lie in a hydrophilic region close to surface of the molecule and to the active site. For tyrosine 670 residue, the amino acid sequence shows Y-1 has an acidic residue (Aspartic acid) as does Y-5 (Aspartic acid) and Y-6 (Glutamic acid). Acidic residues are found close to tyrosine residues in other proteins, which can be phosphorylated in vitro [40, 41]. However, there are also 3 basic amino acids in its Y+1 to Y+3, which some believe, might inhibit tyrosine phosphorylation [27]. Overall, it has been speculated that the N-terminal sequence of the autophosphorylation site of some proteins need to be optimal for both recognition and phosphate acceptance [42]. However, additional secondary and higher order structure may be more important than primary structure for the selection for tyrosine phosphorylation sites.
This report, thus divulges another important aspect of β-catenin signaling, which is tyrosine phosphorylation dependent, putting this protein under the list of HGF/met effectors. Independent of the canonical Wnt pathway, β-catenin in hepatocytes is capable to regulating proliferation in a tyrosine-dependent manner via its association with met. Thus, examining tyrosine-phosphorylation of β-catenin in cancers especially hepatocellular cancer might reflect not just an invasive or metastatic phenotype, but rather a proliferative one as well.
Footnotes
Grant Support: Funded in part by ACS, Grant # RSG-03-141-01-CNE to SPSM and NIH, 1RO1DK62277 to SPSM, Rango’s Fund for Enhancement of Pathology Research and Cleveland Foundation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Naldini L, Vigna E, Narsimhan RP, Gaudino G, Zarnegar R, Michalopoulos GK, Comoglio PM. Hepatocyte growth factor (HGF) stimulates the tyrosine kinase activity of the receptor encoded by the proto-oncogene c-MET. Oncogene. 1991;6:501–4. [PubMed] [Google Scholar]
- 2.Naldini L, Weidner KM, Vigna E, Gaudino G, Bardelli A, Ponzetto C, Narsimhan RP, Hartmann G, Zarnegar R, Michalopoulos GK. Scatter factor and hepatocyte growth factor are indistinguishable ligands for the MET receptor. Embo J. 1991;10:2867–78. doi: 10.1002/j.1460-2075.1991.tb07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vigna E, Naldini L, Tamagnone L, Longati P, Bardelli A, Maina F, Ponzetto C, Comoglio PM. Hepatocyte growth factor and its receptor, the tyrosine kinase encoded by the c-MET proto-oncogene. Cell Mol Biol (Noisy-le-grand) 1994;40:597–604. [PubMed] [Google Scholar]
- 4.Bardelli A, Longati P, Gramaglia D, Stella MC, Comoglio PM. Gab1 coupling to the HGF/Met receptor multifunctional docking site requires binding of Grb2 and correlates with the transforming potential. Oncogene. 1997;15:3103–11. doi: 10.1038/sj.onc.1201561. [DOI] [PubMed] [Google Scholar]
- 5.Leshem Y, Gitelman I, Ponzetto C, Halevy O. Preferential binding of Grb2 or phosphatidylinositol 3-kinase to the met receptor has opposite effects on HGF-induced myoblast proliferation. Exp Cell Res. 2002;274:288–98. doi: 10.1006/excr.2002.5473. [DOI] [PubMed] [Google Scholar]
- 6.Osawa M, Itoh S, Ohta S, Huang Q, Berk BC, Marmarosh NL, Che W, Ding B, Yan C, Abe J. ERK1/2 associates with the c-Met-binding domain of growth factor receptor-bound protein 2 (Grb2)-associated binder-1 (Gab1): role in ERK1/2 and early growth response factor-1 (Egr-1) nuclear accumulation. J Biol Chem. 2004;279:29691–9. doi: 10.1074/jbc.M309371200. [DOI] [PubMed] [Google Scholar]
- 7.Ponzetto C, Bardelli A, Maina F, Longati P, Panayotou G, Dhand R, Waterfield MD, Comoglio PM. A novel recognition motif for phosphatidylinositol 3-kinase binding mediates its association with the hepatocyte growth factor/scatter factor receptor. Mol Cell Biol. 1993;13:4600–8. doi: 10.1128/mcb.13.8.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponzetto C, Bardelli A, Zhen Z, Maina F, dalla Zonca P, Giordano S, Graziani A, Panayotou G, Comoglio PM. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell. 1994;77:261–71. doi: 10.1016/0092-8674(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 9.Wada S, Sasaki Y, Horimoto M, Ito T, Ito Y, Tanaka Y, Toyama T, Kasahara A, Hayashi N, Hori M. Involvement of growth factor receptor-bound protein-2 in rat hepatocyte growth. J Gastroenterol Hepatol. 1998;13:635–42. doi: 10.1111/j.1440-1746.1998.tb00702.x. [DOI] [PubMed] [Google Scholar]
- 10.Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio PM. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391:285–8. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- 11.Faletto DL, Kaplan DR, Halverson DO, Rosen EM, Vande Woude GF. Signal transduction in c-met mediated motogenesis. Exs. 1993;65:107–30. [PubMed] [Google Scholar]
- 12.Runge DM, Runge D, Foth H, Strom SC, Michalopoulos GK. STAT 1alpha/1beta, STAT 3 and STAT 5: expression and association with c-MET and EGF-receptor in long-term cultures of human hepatocytes. Biochem Biophys Res Commun. 1999;265:376–81. doi: 10.1006/bbrc.1999.1681. [DOI] [PubMed] [Google Scholar]
- 13.Zhang YW, Wang LM, Jove R, Vande Woude GF. Requirement of Stat3 signaling for HGF/SF-Met mediated tumorigenesis. Oncogene. 2002;21:217–26. doi: 10.1038/sj.onc.1205004. [DOI] [PubMed] [Google Scholar]
- 14.Gao CF, Vande Woude GF. HGF/SF-Met signaling in tumor progression. Cell Res. 2005;15:49–51. doi: 10.1038/sj.cr.7290264. [DOI] [PubMed] [Google Scholar]
- 15.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–25. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 16.Birchmeier W, Brinkmann V, Niemann C, Meiners S, DiCesare S, Naundorf H, Sachs M. Role of HGF/SF and c-Met in morphogenesis and metastasis of epithelial cells. Ciba Found Symp. 1997;212:230–40. 240–6. doi: 10.1002/9780470515457.ch15. [DOI] [PubMed] [Google Scholar]
- 17.Pediaditakis P, Lopez-Talavera JC, Petersen B, Monga SP, Michalopoulos GK. The processing and utilization of hepatocyte growth factor/scatter factor following partial hepatectomy in the rat. Hepatology. 2001;34:688–93. doi: 10.1053/jhep.2001.27811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pediaditakis P, Monga SP, Mars WM, Michalopoulos GK. Differential mitogenic effects of single chain hepatocyte growth factor (HGF)/scatter factor and HGF/NK1 following cleavage by factor Xa. J Biol Chem. 2002;277:14109–15. doi: 10.1074/jbc.M112196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barker N, Morin PJ, Clevers H. The Yin-Yang of TCF/beta-catenin signaling. Adv Cancer Res. 2000;77:1–24. doi: 10.1016/s0065-230x(08)60783-6. [DOI] [PubMed] [Google Scholar]
- 20.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Es JH, Barker N, Clevers H. You Wnt some, you lose some: oncogenes in the Wnt signaling pathway. Curr Opin Genet Dev. 2003;13:28–33. doi: 10.1016/s0959-437x(02)00012-6. [DOI] [PubMed] [Google Scholar]
- 22.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 23.Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis--a look outside the nucleus. Science. 2000;287:1606–9. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 24.Pennisi E. How a growth control path takes a wrong turn to cancer. Science. 1998;281:1438–9. 1441. doi: 10.1126/science.281.5382.1438. [DOI] [PubMed] [Google Scholar]
- 25.Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J Cell Sci. 1994;107:3655–63. doi: 10.1242/jcs.107.12.3655. Pt 12. [DOI] [PubMed] [Google Scholar]
- 26.Lewis JE, Wahl JK, 3rd, Sass KM, Jensen PJ, Johnson KR, Wheelock MJ. Cross-talk between adherens junctions and desmosomes depends on plakoglobin. J Cell Biol. 1997;136:919–34. doi: 10.1083/jcb.136.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roura S, Miravet S, Piedra J, Garcia de Herreros A, Dunach M. Regulation of E-cadherin/Catenin association by tyrosine phosphorylation. J Biol Chem. 1999;274:36734–40. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- 28.Hiscox S, Jiang WG. Association of the HGF/SF receptor, c-met, with the cell-surface adhesion molecule, E-cadherin, and catenins in human tumor cells. Biochem Biophys Res Commun. 1999;261:406–11. doi: 10.1006/bbrc.1999.1002. [DOI] [PubMed] [Google Scholar]
- 29.Monga SP, Mars WM, Pediaditakis P, Bell A, Mule K, Bowen WC, Wang X, Zarnegar R, Michalopoulos GK. Hepatocyte growth factor induces Wnt-independent nuclear translocation of beta-catenin after Met-beta-catenin dissociation in hepatocytes. Cancer Res. 2002;62:2064–71. [PubMed] [Google Scholar]
- 30.Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W. Essential role of BCL9-2 in the switch between beta-catenin’s adhesive and transcriptional functions. Genes Dev. 2004;18:2225–30. doi: 10.1101/gad.317604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novicki DL, Jirtle RL, Michalopoulos G. Establishment of two rat hepatoma cell strains produced by a carcinogen initiation, phenobarbital promotion protocol. In Vitro. 1983;19:191–202. doi: 10.1007/BF02618059. [DOI] [PubMed] [Google Scholar]
- 32.Monga SP, Pediaditakis P, Mule K, Stolz DB, Michalopoulos GK. Changes in WNT/beta-catenin pathway during regulated growth in rat liver regeneration. Hepatology. 2001;33:1098–109. doi: 10.1053/jhep.2001.23786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze’ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–7. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piedra J, Miravet S, Castano J, Palmer HG, Heisterkamp N, Garcia de Herreros A, Dunach M. p120 Catenin-associated Fer and Fyn tyrosine kinases regulate beta-catenin Tyr-142 phosphorylation and beta-catenin-alpha-catenin Interaction. Mol Cell Biol. 2003;23:2287–97. doi: 10.1128/MCB.23.7.2287-2297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aberle H, Schwartz H, Kemler R. Cadherin-catenin complex: protein interactions and their implications for cadherin function. J Cell Biochem. 1996;61:514–23. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C514::AID-JCB4%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 36.Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 37.Kinch MS, Clark GJ, Der CJ, Burridge K. Tyrosine phosphorylation regulates the adhesions of ras-transformed breast epithelia. J Cell Biol. 1995;130:461–71. doi: 10.1083/jcb.130.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing Y, Clements WK, Kimelman D, Xu W. Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev. 2003;17:2753–64. doi: 10.1101/gad.1142603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couch VA, Cheng N, Nambiar K, Fink W. Structural Characterization of alpha-Helices of Implicitly Solvated Poly-Alanine. J Phys Chem B Condens Matter Mater Surf Interfaces Biophys. 2006;110:3410–9. doi: 10.1021/jp055209j. [DOI] [PubMed] [Google Scholar]
- 40.Dekowski SA, Rybicki A, Drickamer K. A tyrosine kinase associated with the red cell membrane phosphorylates band 3. J Biol Chem. 1983;258:2750–3. [PubMed] [Google Scholar]
- 41.Schaffhausen B, Benjamin TL. Comparison of phosphorylation of two polyoma virus middle T antigens in vivo and in vitro. J Virol. 1981;40:184–96. doi: 10.1128/jvi.40.1.184-196.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMurray JS, Budde RJ, Ke S, Obeyesekere NU, Wang W, Ramdas L, Lewis CA. Cyclic peptides as probes of the substrate binding site of the cytosolic tyrosine kinase, pp60c-src. Arch Biochem Biophys. 1998;355:124–30. doi: 10.1006/abbi.1998.0707. [DOI] [PubMed] [Google Scholar]


