Abstract
Introduction
Long-term storage of biological materials is a critical component of any epidemiological study. In designing specimen repositories, efforts need to balance future needs for samples with logistical constraints necessary to process and store samples in a timely fashion.
Objectives
In the Norwegian Mother and Child Cohort Study (MoBa), the Biobank was charged with long-term storage of more than 380,000 biological samples from pregnant women, their partners and their children for up to 100 years.
Methods
Biological specimens include whole blood, plasma, DNA and urine; samples are collected at 50 hospitals in Norway. All samples are sent via ordinary mail to the Biobank in Oslo where the samples are registered, aliquoted and DNA extracted. DNA is stored at −20 °C while whole blood, urine and plasma are stored at − 80 °C.
Results
As of July 2006, over 227,000 sample sets have been collected, processed and stored at the Biobank. Currently 250–300 sets are received daily. An important part of the Biobank is the quality control program.
Conclusion
With the unique combination of biological specimens and questionnaire data, the MoBa Study will constitute a resource for many future investigations of the separate and combined effects of genetic, environmental factors on pregnancy outcome and on human morbidity, mortality and health in general.
Keywords: Automation, Biobank, Birth Cohort Study, DNA, Plasma, Quality Control
Introduction
Collection and long-term storage of biological specimens are becoming an integral part of many human studies [1]. When designing longitudinal studies with biological specimens, researchers are faced with the question of how to perform sampling and store a vast number of samples in a way that allows the greatest number of analytical options in the future [1–4]. Issues of cost, feasibility, sample size and volume, sample processing, storage and retrieval of samples all contribute to the decisions that affect the scientific questions.
The Norwegian Mother and Child Cohort Study (MoBa) is a prospective birth cohort enrolling pregnant women throughout Norway from 1999–2008. Biological material is to be collected from women in 100,000 pregnancies. In addition cord blood from their newborn infants and samples from 80,000 fathers may result in more than 380,000 unique sample sets for long-term storage. In addition to biological materials, detailed questionnaire data are collected during pregnancy and as the child ages. Additional health outcome information is available through linkage to Norwegian national health registries.
The overall aim is to study the effects of genetic and environmental factors on pregnancy outcomes and later health of the children as well as the parents. The study includes collection of questionnaires and registry information, and aims at having DNA for all participants as well as biological samples from pregnant women to assess environmental exposures. With this unique combination of biological specimens, DNA, and questionnaire data, the MoBa Study will constitute a resource for many future investigations of the effects of genetic and environmental factors on pregnancy outcome and on human morbidity, mortality and health in general. Here we present the chosen methods for sampling of biological material, sample processing, storage, and retrieval.
Material and methods
Study population
The target population consists of all women who give birth in Norway. The study is described in detail elsewhere [5]. Briefly, pregnant women and their partners are recruited by mail prior to their ultrasound appointment at 17th week’s gestation; 99% of pregnant women in Norway have routine ultrasounds. Direct participation contact occurs at the ultrasound appointment and at delivery. The opportunity for collection of biological material exists only on these occasions. Table 1 shows the progression of the study enrolment over time; currently 50 of 52 hospitals that perform deliveries in Norway are participating.
Table 1.
Number of participating hospitals in the MoBa Study and number of sample sets registered and processed at the Biobank (Numbers per 07/07/06)
| Year | Number of participating hospitals | Number of sample sets from mothers | Number of sample sets |
|---|---|---|---|
| 1999 | 3 | 298 | 304 |
| 2000 | 5 | 3,121 | 4,117 |
| 2001 | 25 | 9,515 | 15,209 |
| 2002 | 45 | 18,100 | 32,403 |
| 2003 | 46 | 21,649 | 41,276 |
| 2004 | 49 | 24,754 | 47,462 |
| 2005 | 50 | 29,150 | 56,105 |
| 2006 | 50 | 15,687 | 30,629 |
| Total | 50 | 122,274 | 227,505 |
Sampling
Biological specimens are collected from the mothers at 17–18 weeks of gestation and at the time of delivery. Blood samples from the fathers are collected at the ultrasound appointment; recruitment of fathers began in 2000. Cord blood samples are drawn at delivery or, if cord blood is unavailable, a capillary sample from the newborn child is collected at the time of phenylketonuria (PKU) screening (3–4 days after birth). Over the course of the study, the specimens collected have changed as research interests have grown. Figure 1 presents a timeline of the specimen collection activities.
Figure 1.
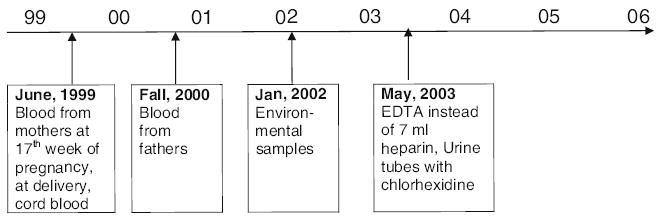
Timeline of sample collection strategies in MoBa.
Initially, only two 7-ml EDTA tubes (Becton-Dickinson (BD), Plymouth, UK) were drawn from women at the ultrasound appointment. One of the EDTA tubes is spun following sample collection in the hospital laboratory and plasma is pulled o3 before shipping; the other is shipped in the vacutainer directly to the Biobank. In 2002, based on particular interests in environmental toxins, the sample set was extended to include an additional 7-ml anticoagulant tube for plasma analysis and one plastic 3-ml EDTA tube (BD, Plymouth, UK) of blood for trace metal analysis. Additionally, 8-ml of urine is collected in a urine cup and transferred to a urine transport tube. (Becton-Dickinson (BD), Franklin Lakes, NJ, USA). Urine was initially shipped in tubes without bacterostatic additive (BD, Plymouth, UK), but most of the urine samples showed bacterial growth upon receipt in the Biobank. We therefore tested the impact of urine preservation methods on potential environmental analyses of interest (e.g., phthalates, bisphenol A) [6]. As a result of this evaluation we changed the protocol in 2003 to ship urine samples in tubes with chlorhexidine (UAP Vacutainers, BD, Franklin Lakes, NJ, USA). From fathers, the umbilical cord and mothers one day after delivery two 7-ml EDTA tubes are drawn.
Sample processing at the Biobank
Samples are shipped overnight from hospital laboratories to the Biobank. A majority of samples are received the day after collection. Samples are processed the day of receipt. A specially designed computer program (The Mother–Child program) supports all processing at the Biobank. This Laboratory Information Management System (LIMS) controls all steps where sample identity can be mixed-up, e.g. transfer of samples from one tube to another. Specimen tubes from hospitals are labelled with the woman’s name and national identity number; upon registration in the Biobank the sample sets get a unique Biobank ID which is linked to the MoBa participant ID for this pregnancy. At registration the LIMS records maternal ID, date of sampling, date of receipt, collection hospital, and sample status at arrival (e.g., incomplete volume, haemolysed, coagulated). Bar codes with the unique Biobank identifier are automatically printed for further sample processing. Our LIMS also stores locations for all aliquots for each Biobank ID (which freezer, rack, plate, and position on the 96 well plate). Access to the Mother-Child program is highly restricted, and a tape back-up of the database is made each night and stored in a fire-protected area.
Aliquoting of samples
Whole blood is aliquoted into two polypropylene deep-well plates (930 μl in each, ABgene, Surrey, UK). For the plasma pulled off at the hospital laboratories before shipment, a total of 1.8-ml is aliquoted onto 6 different polypropylene microtitre plates (300 μl each, ABgene, Surrey, UK) using Tecan pipetting robots (three Genesis RSP 200 and one Freedom Evo, Tecan Ltd, Männedorf, Switzerland). The plates are sealed with heat-sealing foil sheets (Easy Peel, ABgene, Surrey, UK). Maternal plasma collected for environmental analyses is spun at the Biobank and aliquoted into individually labelled vials on three Matrix™ plates (TrackMates tubes, Hudson, NH, individual nanobar code; e.g. 2D bar code on each tube, 930 μl in each). Urine is divided into 930 μl aliquots on six Matrix plates.
DNA extraction
The 930 μl volume was selected because this is the maximum efficient volume for the Tecan machine; larger volumes require a much greater aliquoting time and thus all samples are stored in 930 μl or smaller volumes. DNA is extracted manually using the FlexiGene kit (Qiagen, Hilden, Germany). Quality control is performed on all DNA samples using a spectrophotometer (Spectramax 190, Molecular Devices, Sunnyvale, CA) where optical density (OD) is tested in triplicates. Stringent parameters must be adhered to for DNA to be accepted. These parameters include a DNA purity of 1.6–2.0 260/280 ratio, a DNA concentration in excess of 20 ng/μl, and OD at zero for the negative control included in each batch of 24 samples (e.g. extracted at the same time). All indications of quality outside the requirement are marked in our LIMS. The DNA concentrations are subsequently normalised to 100-ng/μl using a Tecan pipetting (Generis 2000, Ma¨ nnedorf, Switzerland) robot; which also tracks the amount of DNA aspirated throughout the process. DNA normalisation is based on four different computer programs that communicate with each other (Softmax Pro 3.0 for the spectrophotometer, Gemini for the Tecan machine, MCS (specially made for communication between Softmax Pro 3.0 and Gemini). DNA is then aliquoted into four to ten 1.4-ml deep-well plates (930 μl in each) dependent on the DNA yield, the volume in each well which is not totally filled is calculated and recorded, and the file from the Gemini program of the Tecan robot is imported to the Mother–Child program.
Storage
DNA is stored at −20 °C while whole blood, urine and plasma are stored at −80 °C. Plates containing aliquots from the same sample are stored in two different chest freezers. The 3-ml EDTA tube for metal analysis is stored at −20 °C. All freezers are connected to an external alarm system, and temperatures are logged routinely. An emergency electrical system is connected to all freezers to provide backup power.
Retrieval
We plan to retrieve specified samples using automated equipment for all samples stored in the 96-well format. Study investigators will identify subjects and samples of interest; these personal identification numbers will be linked to the encrypted Biobank IDs and pull lists will be generated. Once the samples to be retrieved are identified, the retrieval file from the Mother–Child program will be transferred to a MultiPROBE II robot (Perkin Elmer, Wellesley, MA). Up to 100 plates are loaded into the MultiPROBE II robot for automatic retrieval and transfer to a delivery plate. The retrieved volume from a certain position is tracked in the Mother–Child program.
Quality Assurance and Quality Control (QA/QC) procedures
Conduct of such a large study over such a long period of time requires a comprehensive quality assurance program to ensure the ability to track changes over time. In addition to standard operating procedures (SOPs) which are constantly updated, the Biobank is being ISO-certified (ISO 9001). As part of the QA, we keep five samples from each lot of materials used in the study; this includes vacutainers, pipettes, tips and plates. Because a majority of sample collection and processing occurs at the 50 hospitals, annually we conduct a written survey of all hospitals involved to assess collection, processing, and storage procedures in order to identify variations in protocols throughout the study period. We are conducting an evaluation of changes in different markers in plasma and urine during repeatedly freeze-thaw cycles and long-term storage. Sodium, cholesterol, triglycerides, free fatty acids, vitamin E and aspartic aminotransferase were analysed in plasma while sodium and creatinine were analysed in urine [7]; results will be presented in a future manuscript.
Results
Status to Date
As of July 07, 2006, a total of 227,510 sample sets have been received, processed and stored at the Biobank (representing 68,385 pregnancies, 47,663 fathers and 57,573 children). The tubes, the shipment, as well as the processing and storage are shown in Figure 2, illustrated by the sample set from mothers at 17th–18th week of pregnancy. The number of received sample sets and sample types are shown in Table 2. Currently, 250–300 sample sets are received and processed daily by 14 full-time technicians.
Figure 2.
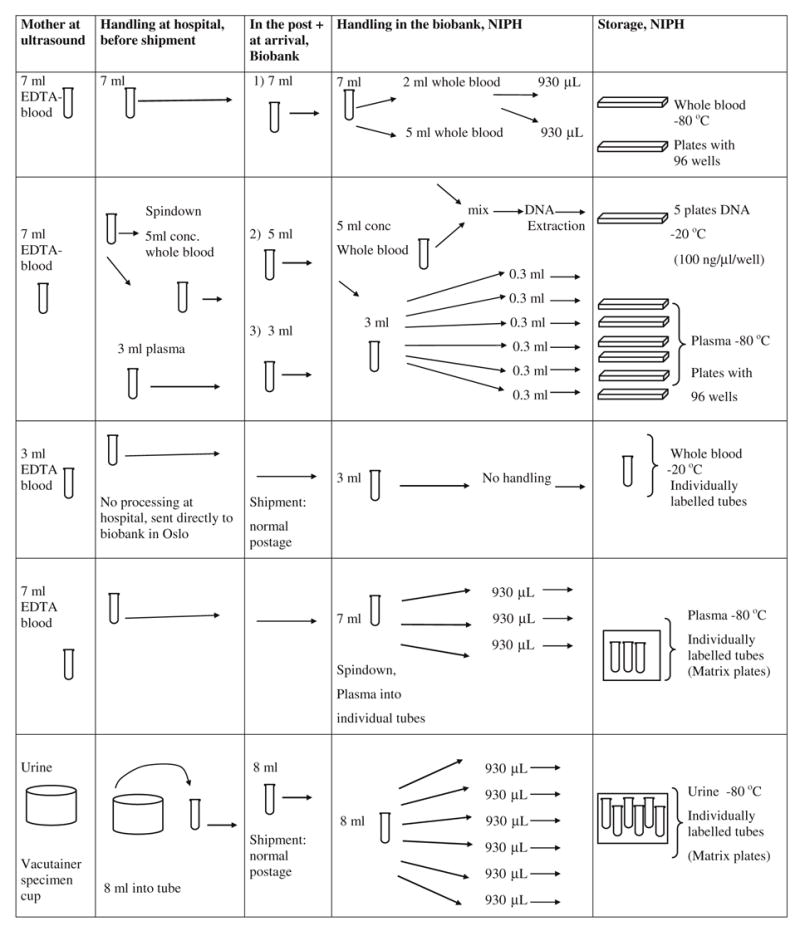
Sample set from mothers at 17th–18th week of pregnancy.
Table 2.
Number of received sample sets and sample types stored at the Biobank
| Stored sample types, ready for retrieval in the MoBa study as of July 2006
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of sets | sample | Whole blood | Plasma microtitre plates | Plasma Matrix Heparin | Plasma Matrix EDTA | Whole blood for metals (FE3) | Urine Matrix without chlorhexidine | Urine Matrix with chlorhexidine | DNAf |
| M11a | 17,131 | 16,561 | 16,883 | 13,542 | |||||
| M12b | 51,254 | 49,630 | 50,308 | 10,116 | 40,050 | 48,460 | 9,488 | 40,020 | 33,475 |
| M2c | 53,889 | 52,878 | 53,437 | 39,444 | |||||
| Cd | 57,573 | 34,737 | 56,679 | 39,581 | |||||
| Fe | 47,663 | 47,108 | 47,296 | 33,432 | |||||
| Total | 227,510 | 200,914 | 224,603 | 10,116 | 40,050 | 48,460 | 9,488 | 40,020 | 159,474 |
M11 = Mother 1 sample, blood sample set from mothers at 17th week of pregnancy, two 7-ml EDTA tubes drawn.
M12 = Mother 1 sample, blood sample set from mothers at 17th week of pregnancy, including tubes for environmental factors.
M2 = Mother 2 sample, blood sample set from mothers at delivery.
F = Father, blood sample set from fathers at 17th week of pregnancy.
C = Cord, blood sample from the babies umbilical cord or a capillary sample from the PKU screening. 1.8% of samples from babies are obtained in this way.
The deviation in number of registered samples and DNA samples is that not all DNA samples have been adjusted for concentration, which is needed for DNA aliquoting, and to give location on plates for storage.
The QA/QC program is ongoing and will be incorporated into analyses using the biological specimens. This information will facilitate investigators’ decisions regarding what samples to use for a specific analysis; for example, if the analyte is stable over multiple freeze thaw cycles, it will be less critical which aliquot to select than if the analyte is changed as a result of thawing and refreezing.
Discussion
In determining the logistics of a biological specimen repository, issues of optimal processing and storage of samples need to be balanced by the cost and feasibility of these methods.
Biological specimen collection
Key factors that affect the stability of biological samples include anticoagulants, stabilising agents, temperature, elapsed time from collection to initial processing and endogenous degrading properties (enzymes, cell death) [1–4]. The effects of each of these on known factors have been carefully considered together with the simplest logistics and lowest costs. EDTA works well for DNA-based assays; citrate stabilised blood gives a higher yield of lymphocytes for culture, while heparin is the anticoagulant usually used for immediate analysis and is regarded as the best for trace element analysis. Since MoBa never planned to isolate lymphocytes or perform Epstein-Barr virus transformation of peripheral mononuclear cells, EDTA was chosen as the anticoagulant. Citrate has been regarded as the only anticoagulant for analysis of coagulation factors, but recently a new enzyme immunoassay for measuring immunologically intact fibrinogen in EDTA samples has been developed [8]. To make the procedure for blood sampling as easy and standardised as possible we ended up collecting only EDTA tubes for whole blood, plasma and DNA. Another large cohort study collecting biological specimens, the UK Biobank study, will also be using EDTA plasma for almost all analytes (UK Biobank www.ukbiobank.ac.uk, see Sample Handling and Storage subgroup protocol and recommendations, Appendix 3).
The temperature of the sample during transport is a critical factor in ensuring integrity and flexibility in future analysis [1–4]. The appropriate temperature depends on the biomolecules of interest and, ideally, the MoBa Biobank would need to take into account the temperature requirements for the stability of each biomarker. This was not feasible, partly due to the extra work it would require of the hospital laboratory personnel and partly because of the increased postage costs to ship on ice. MoBa samples are therefore sent by ordinary mail, as is also the case for many blood samples for clinical testing in Norway and elsewhere. In our freeze-thaw program, samples were tested before freezing and then each time for 30 individual freeze-thaw cycles for six analytes. The biggest difference in test levels was found between the test before freezing and after the first thawing. This will be true for all samples in the Biobank, because all analysis will be performed after at least one thawing.
DNA extraction
Many archives of biological material store whole blood and extract DNA later based on a nested case-control design [1]. Because many outcomes in the MoBa study are anticipated to occur soon after enrolment (e.g., pregnancy outcomes), it was a priority to extract DNA from all samples at time of collection, even though the cost of this option was high. To facilitate DNA sample delivery, DNA concentration adjustment is also performed at the time of extraction. Thus, retrieval only has to consider volume to determine the amount of required DNA. Commercial companies use this strategy as well [9, 10].
Sample materials
Microtitre plates, although facilitating storage (both costs and space), may result in challenges because each time a certain sample is required, all the 95 other samples on that plate will be thawed as well. It was therefore decided as part of the collaborative project with National Institute of Environmental Health Sciences to store maternal plasma also on Matrix plates. The cost per sample is approximately 10 times higher compared to microtitre plates (1.8 compared to 0.15 US $), but allows the flexibility of being able to retrieve individual vials still working within the 96-well format allowing for automated aliquoting of samples.
Samples available at the Biobank
As of July 7, 2006, a total number of 227,510 sample sets are registered and processed at the Biobank. We continue to receive 1200–1300 sample sets per week (five working days) and will soon begin retrieving samples for nested case-control studies. Researchers who would like to collaborate on specific research questions should consult the “guidelines for access to data” published on the website of NIPH (www.fhi.no/tema/morogbarn). Managing such diverse activities present challenges for the Biobank, but given the forethought in the sample collection activities, analysts should be able to take full advantage of this resource to explore the factors associated with successful pregnancy out-come and health among Norwegian families.
Acknowledgments
The Biobank of the MoBa study is supported by NIH/NIEHS (grant no. N01-ES-85433), NIH/NINDS (Autism Birth Cohort U01 NS 047537), EU/EARNEST (grant no. 007036), Norwegian Research Council/FUGE (grant no. 151918/S10) and the Norwegian Government. We thank the International Advisory Committee: Elaine Gunter, Richard Jones, Mads Melbye, Egil Jellum, and Anne-Lise Børresen-Dale for their helpful advice for design and logistics of the Biobank, the MoBa group in Bergen for recruitment and tracking of study participants, and all the laboratory technicians at the Biobank for their efforts.
Abbreviations
- OD
Optical Density
- LIMS
Laboratory Information Management System
- MoBa
The Norwegian Mother and Child Cohort Study
- PKU
Phenylketonuria
- QA, QC
Quality Assurance, Quality Control
- SOP
Standard Operating Procedure
References
- 1.Smith GD, Ebrahim S, Lewis S, Hansell AL, Palmer LJ, Burton PR. Genetic epidemiology and public health: Hope, hype, and future prospects. Lancet. 2005;22:1484–1498. doi: 10.1016/S0140-6736(05)67601-5. [DOI] [PubMed] [Google Scholar]
- 2.Gunter EW. Biological and environmental specimental banking at the Centers for Disease Control and Prevention. Review Chemosphere. 1997;34:1945–1953. doi: 10.1016/s0045-6535(97)00056-8. [DOI] [PubMed] [Google Scholar]
- 3.Ollier W, Sprosen T, Peakman T. UK Biobank: from concept to reality. Pharmacogenomics. 2005;6:639–646. doi: 10.2217/14622416.6.6.639. [DOI] [PubMed] [Google Scholar]
- 4.Eskenazi B, Gladstone EA, Berkowitz GS, et al. Methodological and logistic issues in conducting longitudinal birth cohort studies: Lessons learned from the Center for Children’s Environmental Health and Disease Prevention Research. Environmental Health Perspectives. 2005;113:1419–1429. doi: 10.1289/ehp.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magnus P, Irgens LM, Haug K, Nystad W, Skjærven R Stoltenberg and the MoBa Study Group. Cohort profile: The norwegian mother and child cohort study (MoBa) Int J Epidemiol. doi: 10.1093/ije/dyl170. (in press) [DOI] [PubMed] [Google Scholar]
- 6.Hoppin AJ, Ulmer R, Calafat AM, Barr DB, Baker SV, Meltzer HM, Rønningen KS. Impact of urine preservation methods and duration of storage on measured levels of environmental contaminants. Journal of Exposure Analysis and Environmental Epidemiology. 2006;16:39–48. doi: 10.1038/sj.jea.7500435. [DOI] [PubMed] [Google Scholar]
- 7.Paltiel L, Meltzer HM, Hoppin JA, Skjerden T, Baker S, Magnus P, Rønningen KS. QA/QC in the Norwegian Mother and Child Cohort study – Assessment of freeze-thaw cycles on markers in plasma. Presented at the ISBER (International Society for Biological and Environmental Repositories); Perugia-Italy. October 17th–20th; 2004. p. 20. [Google Scholar]
- 8.Sobel JH, Wu HQ, Canfield RE. The development of assays for the detection of fibrin(ogen)olysis based on COOH-terminal A alpha chain epitopes. Blood. 1994;84:535–546. [PubMed] [Google Scholar]
- 9.Mahan S, Ardlie KG, Krenitsky KF, Walsh G, Clough G. Collaborative design for automated DNA storage that allows for rapid, accurate, large-scale studies. ASSAY and Drug Development Technologies. 2004;2:683–689. doi: 10.1089/adt.2004.2.683. [DOI] [PubMed] [Google Scholar]
- 10.Potera C. GenVault system improves DNA sample storage. High-throughput archive system and management of large collection of DNA samples. Genetic Engineering News. 2004;24:30. [Google Scholar]


