Abstract
To identify estrogen responsive genes in mammary glands, microarray assays were performed. Twenty genes were found to be up-regulated while 16 genes were repressed in the 9h estrogen treated glands. The induction of GAS6, one of the genes up-regulated by estrogen, was confirmed by RNase protection assay. Furthermore, GAS6 was also demonstrated to be induced by estrogen in ER positive breast cancer cells. Analysis of GAS6 promoter revealed that GAS6 promoter was regulated by estrogen. An estrogen response element (ERE) was identified in the GAS6 promoter. Electrophoretic mobility shift assay revealed that ERα interacted with the ERE in the GAS6 promoter. Chromatin immunoprecipitation demonstrated that ERα was recruited to the GAS6 promoter upon estrogen stimulation. These results suggested that GAS6 is an estrogen target gene in mammary epithelial cells.
INTRODUCTION
Estrogen plays an important role in the multi-step development of mammary glands (1, 2). During puberty, the accelerated ductal growth which finally fills the whole fat pad is stimulated by estrogen. Estrogen is also required for the maintenance of mammary ductal structure, as evidenced by the epithelial atrophy and increased apoptosis occurring in the breast during and after menopause. In addition, estrogen stimulates the early lobuloalveolar proliferation during pregnancy along with progesterone (3). Estrogen manifests its effect through estrogen receptor (ER), an inducible transcription factor belonging to the nuclear receptor superfamily (4, 5). There are two forms of ER: ERα (5) and ERβ (6,7). Studies carried out with the mouse models deficient in either ERα or ERβ demonstrated that that ERα is responsible for regulating the development of mammary glands (8, 9). In addition to its essential roles in normal mammary gland development, estrogen is also involved in breast cancer development (10). Overexposure to estrogen is associated with the increased risk of breast cancer and the anti-estrogen agent tamoxifen has been shown to significantly decrease the incidence of breast cancer (11). About 70% of breast cancers are ER positive and half of the ER positive breast cancers are responsive to anti-estrogen therapy (12, 13).
GAS6 (Growth arrest specific gene 6) protein is a 75-KDa secreted protein which bears significant homology at the amino acid level to Protein S, a negative regulator of the coagulation cascade (14). GAS6 was originally identified as a gene of which the expression increased by serum starvation and contact inhibition (14). GAS6 binds as a ligand to the receptor tyrosine kinases Axl (ARK, Ufo, Tyro7), Sky(Rse, Tyro3, Dtk, Etk, Brt, Tif) and Mer (c-Mer, Eyk, Nyk) by its carboxy-terminal globular G domain (15–17). GAS6 activates the kinase activity of each of the receptors. Coexpression of GAS6 protein and its receptors Axl, Sky and Mer are detected in reproductive, neural, lymphoid, vascular tissues and also in primary or tumor cell lines derived from these sources (18–20). The cellular functions of GAS6/Axl/Sky/Mer pathway include cell adhesion, migration, and inhibition of apoptosis (20).
To understand the role of estrogen in normal mammary gland development and tumorigenesis, it is essential to identify the estrogen responsive genes. Here we report the identification of GAS6 as an estrogen-inducible gene in mammary glands.
MATERIALS AND METHODS
Antibodies and plasmids
Anti-ER monoclonal antibody was purchased from Santa Cruz Biotechnology. PcDNA3.1-ER was described elsewhere (21) RNA isolation and hybridization-8-week old wild-type mice (C57/BL6) were ovariectomized. Two weeks later, the mice were injected with 17β-estrodiol (5 ug/kg body weight) intraperitoneally. The mice were then sacrificed and the No. 3–5 mammary glands were harvested for isolation of total RNA by TRIZOL (Invitrogen). Total mammary gland RNA was purified with RNeasy (Qiagen). The integrity of RNA was confirmed by the presence of sharp 28S and 18S bands on a denaturing agarose gel.
Five micrograms of purified cDNA was reversely transcribed using Enzo BioArray RA transcript labeling kit (Affymetrix) and the product was purified with RNeasy spin colums (Qiagen). According to instructions from Affymetrix, 20 μg of cRNA was fragmented. A 300 μl volume of hybridization mixture with 0.1 mg/ml herring sperm DNA, 0.5 mg/ml acetylated bovine serum albumin, and 2X MES hybridization buffer was added to the 20μg of fragmented cRNA. The mouse genome 430 2.0 array (Affymetrix) was incubated with the hybridization mixture for 16 hours at 45°C, followed by washing, signal-amplification, and staining according to the instructions from Affymetrix. The Chips were scanned to obtain the hybridization values using an Affymetrix scanner. Microarray data analysis was performed with the Affymetrix microarray software. Difference in the fluorescent spot intensities between the matched oligonucleotides and their mismatches were analyzed to determine the presence or absence of gene expression and the relative level of gene expression. We excluded genes with absent expression and further applied a threshold of 2.5-fold change in expression level between ligand-treated samples and controls for identifying estrogen responsive genes.
RNase protection assay
Fragments of Mouse GAS6 and GAPDH were PCR amplified and subcloned into pGEM-T vector. The linearized vectors were in vitro transcribed with T7 RNA polymerase in the presence of α-32P-UTP. Unlabeled UTP was added into GAPDH probe labeling reaction to decrease corresponding probe intensity. Total RNAs (10 μg each) from mouse mammary glands were hybridized overnight at 42°C with equal amounts of gel-purified antisense probes. Hybrids were digested with RNase A and RNase T1 according to the instructions of the manufacturer (RPAII Kit, Ambion, Inc.). The protected fragments (GAS6 194 bp; GAPDH 258 bp) were separated by 5% acrylamide/8 M urea PAGE gel and autoradiographed.
Semi-quantitative RT-PCR
Inguinal glands were collected from wild type C57BL/6J mice and pooled. Mammary epithelial cells were separated from stromal cells by collagenase digestion and Percoll gradient centrifugation method (22). Total RNAs were isolated by TRizol (Invitrogen) method. RT-PCR was performed using SuperScript one-step RT-PCR kit (Invitrogen, CA). For each reaction, 40 μl of amplified product was taken out at cycles 25. Sequences for primer pairs used were: mouse GAS6, 5′-TCTTCTCACACTGTGCTGTTGCG-3′ and 5′-GGTCAGGCAAGTTCTGAACACAT-3′; mouse whey acidic protein (WAP), 5′-CCTGACACCGGTACCATGCG-3′ and 5′-ATGTTCTCTCTGGATCCACG-3′; 18S ribosomal RNA: 5′-TCGCCATCACTGCCATTAAGGG-3′ and 5′-GAACCTGGCTGTACTTCCCATCC-3′.
Quantitative real-time PCR (qPCR)-RNA was extracted from MCF-7 cells treated with estrogen or control vehicle using TRIzol® reagent (Invitrogen). qPCR was performed on ABI 7300 (Applied Biosystems) by using the SYBR Green Supermix (Applied Biosystems) according to the manufacturer's protocol. The expression levels of GAS6 or pS2 were normalized against 18S ribosomal RNA. The following PCR primers were used: Human GAS6, 5′-CATCAACAAGTATGGGTCTCCGT-3′ and 5′-GTTCTCCTGGCTGCATTCGTTGA-3′; human pS2 5′-GCTTCTTACCTGTGCACTTTCAG-3′ and 5′-CTCTGGGACTAATCACCGTGCT-3′.
Electrophoretic mobility shift assay (EMSA)
MCF-7 cells nuclear extracts were prepared as described (23). 5 μg of nuclear extracts were incubated for 20 min at room temperature with a 32P-labeled oligonucleotide derived from the human Gas6 promoter region containing the potential sequence for ERα binding site (shown in bold): 5′-CCGCCAGGACGGGATGACCGGAGCCT-3′. The double-stranded GAS6 probe was end-labeled using T4 polynucleotide kinase and [γ-32P] ATP according to standard protocols. DNA-protein complexes were resolved on 6% TBE polyacrylamide gel. Gels were dried and autoradiographed at −80 °C. For supershift experiments, nuclear extracts were incubated with anti-ERα monoclonal antibody in binding buffer for 45 min at room temperature before 32P-labeled probe was added to the binding mixture.
Chromatin Immunoprecipitation
MCF-7 cells were maintained in phenol red-free modified Eagle's medium supplemented with 10% charcoal-dextran-stripped fetal bovine serum. Cells were treated with E2 or control vehicle for 45 min, and cross-linked with 1% formaldehyde for 10 min at room temperature. After the cells were collected, nuclei were prepared by incubating the cells in cell lysis buffer (10 mM Tris, pH 8.0, 10 mM NaCl, 0.2% Nonidet P-40) containing protease inhibitors (phenylmethylsulfonyl fluoride, pepstatin, aprotinin, and leupeptin) for 10 min on ice. Nuclei were precipitated, and lysed with nuclear lysis buffer (50 mM Tris, pH 8.1, 10 mM EDTA, 1% SDS, phosphatase and protease inhibitors) for 10 min on ice. The chromatin was sheared by sonication to an average size of 500–1000 base pairs. Soluble chromatin was diluted 10-fold with dilution buffer (16.7 mM Tris-Cl, pH 8.1, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl) and precleared with salmon sperm DNA-blocked preimmune IgG protein A beads. Immunoprecipitation was performed by incubating the precleared cell lysate with specific antibodies at 4 °C for 12 h. Immune complexes were collected by binding to protein A-beads and washed sequentially for 10 min with washing buffer I (0.1% SDS, 1% Triton-X100, 2 mM EDTA, 20 mM Tris-Cl, pH 8.1, 150 mM NaCl), buffer II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl, pH 8.1, 500 mM NaCl), and buffer III (0.25 M LiCl, 1% Nonidet P-40, 1% deoxycholate, 10 mM Tris-Cl, pH 8.1). DNAs were extracted from precipitates and used as templates in PCR amplifications. The primer pairs for PCR reactions are: pS2, 5′-CCAACCTGACCTTAATCCA-3′ and 5′-CTGCTTTGGCCTCCCAAA-3′; GAS6, 5′-CCGCGATGTGACCTTCAG-3′ and 5′-CGAGAGCGAAGGGGCCAT-3′. To ensure specificity of ChIP experiment, additional control PCR primers were designed to amplify DNA sequences existing about 2000bp up stream or down stream of identified EREs. Sequences for these control primers are: GAS6 upstream control, 5′-ATGGAGACATCCTCCATTGC-3′ and 5′-TCTGGACCCCACTCTCATTC-3′; GAS6 downstream control, 5′-GTGCTGGTGATGGGAGTTG-3′ and 5′-CAGGTAGGGGACTGACTTGG-3′; pS2 upstream control, 5′-CGGACACCACTCACACACTG-3 and 5′-GCATTCTTCTGTCGGAACAGG-3′; pS2 downstream control, 5′-CATCTCCAGTATAACTGTGCAACG-3′ and 5′-AGGGTGGCATGGACTGACC-3′. In all cases, PCR was performed with a serial dilution of input and various cycles (29–35 cycles) to ensure that amplification was maintained in the linear range. Construction of GAS6 luciferase reporter Plasmid and luciferase assays—GAS6-luciferase fusion genes were constructed in the pGL3-Basic vector (Promega, Madison, WI). The GAS6 promoter region was amplified from mouse genomic DNA by PCR using the forward primer 5′-AGCTAGGGTACCTTCTCCAGAGAGGAGTATCC-3′ and the reverse primer 5′-CTAGCTAGATCTAGAGCGAAGGGGCCATGGCG-3′. The PCR product was digested with KpnI and BglII and was inserted into pGL3-Basic KpnI/BglII sites to generate GAS6 promoter-LUC.
CV-1 cells (1 × 105) were plated in DMEM containing 10% fetal calf serum in six-well plates and were cultured for 24 hr before transfection. Transfections were performed using Lipofectamine 2000 reagent (Invitrogen) with β-galactosidase expression vector pCMV-β as an internal control for transfection efficiency. Cell extracts were prepared 36 hr after transfection and were assayed for luciferase and β-galactosidase activities.
RESULTS and DISCUSSION
The induced or repressed genes in the 9h estrogen treated mammary glands
Ovariectomized mice were treated with 17β-estradiol for 9 hours and mammary glands were then isolated. Total RNA was prepared from estrogen-treated mammary glands or control mammary glands for microarray analysis. We observed that 20 genes were significantly induced following the 9 hour treatment (Table 1). WISP-2 and amphiregulin, which have been shown to be estrogen-responsive genes, were induced in mammary glands (24). However, several well-characterized estrogen-inducible genes from breast cancer cell lines, including pS2 (or trefoil factor 1) (24), were not induced at all in mammary glands, indicating that the breast cancer cells may respond differently to estrogen compared to the normal mammary epithelial cells. Microarray analysis also revealed that sixteen genes were significantly down-regulated 9h after the treatment of estrogen (Table 2).
Table 1.
Genes significantly induced after 9h-estrogen treatment
| GeneBank No. | Gene Name | Fold |
|---|---|---|
| M27960 | interleukin-4 receptor (secreted form) | 2.5 |
| M15131 | Interleukin 1 beta | 2.6 |
| AF045887 | Angiotensinogen | 2.8 |
| J04596 | GRO1 oncogene | 3 |
| AF036893 | circadian clock protein (Per2) | 3 |
| X70058 | Small inducible cytokine A7 | 3.5 |
| M19681 | Small inducible cytokine A2 | 3.6 |
| L34570 | Arachidonate 15-lipoxygenase | 3.6 |
| K02236 | Metallothionein 2 | 3.8 |
| AF100778 | connective tissue growth factor related protein WISP-2 (Wisp2) | 3.8 |
| U16959 | FK506 binding protein 5 | 4.1 |
| X59846 | GAS6 | 4.2 |
| AB021861 | apoptosis signal-regulating kinase 2 | 4.2 |
| M88694 | thioether S-methyltransferase | 4.3 |
| L41352 | Amphiregulin | 5.2 |
| V00835 | Metallothionein 1 | 5.2 |
| D12619 | Proprotein convertase subtilisin/kexin type 5 | 5.3 |
| AF001871 | guanine nucleotide exchange factor and integrin binding protein homolog GRP1 | 5.6 |
| X83601 | Pentaxin related gene | 5.8 |
| U50712 | Small inducible cytokine A12 | 16.0 |
Table 2.
Genes significantly repressed after 9h estrogen treatment
| Genebank No. | Gene Name | Fold |
|---|---|---|
| U65091 | Melanocyte specific gene | 0.08 |
| M64228 | Potassium voltage gated channel, Shab-related subfamily, member 1 | 0.21 |
| M76124 | EGP314 precursor | 0.23 |
| U02602 | Thyroid stimulating hormone receptor | 0.24 |
| X05546 | Gag related peptide | 0.26 |
| AF029791 | UDP-Gal-betaglcnac beta 1,3-galactosyltranferase-II (b3gt2) | 0.28 |
| D78175 | Natriuretic peptide receptor 3 | 0.28 |
| D10204 | Prostaglandin E receptor EP3 subtype | 0.29 |
| Y12713 | Endogenous retroviral sequence muerv-L gag, pol and dutpase | 0.31 |
| AB016248 | Sterol-C5-desaturase | 0.32 |
| X95503 | Zinc finger protein (cell line att20) | 0.36 |
| D90225 | Pleiotrophin | 0.38 |
| AB007848 | Osteomodulin | 0.38 |
| L25274 | Transmembrane glycoprotein (DM-GRASP) | 0.40 |
| U88623 | Aquaporin 4 | 0.40 |
| X95504 | Zinc finger protein | 0.40 |
GAS6 is induced by estrogen in normal mammary epithelial cells and breast cancer cells
Among the genes up-regulated by estrogen, GAS6 (growth arrest specific gene 6, Genebank No. X59846) attracted our special attention. As a secreted protein, GAS6 plays an important role in sustaining hematopoietic progenitor cells growth (25) and can induce mesangial cell proliferation (26). GAS6 could be involved in mediating the action of estrogen in mammary glands. At first we performed RNase protection assay to confirm the microarray data. RNase protection assay revealed that GAS6 mRNA expressions were increased in mammary glands treated with estrogen for 9 hour or 24 hour (Fig. 1a).
Fig. 1.

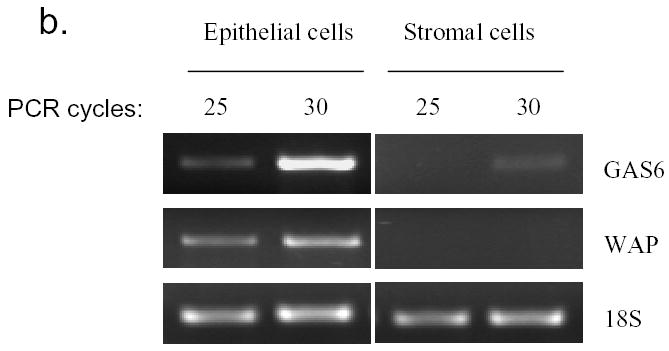
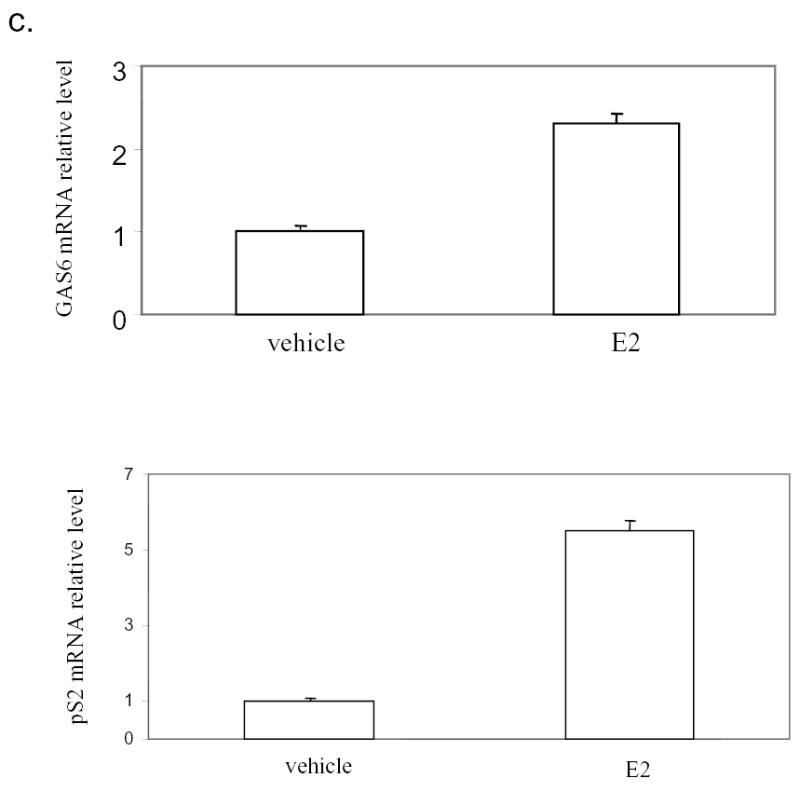
Enhancement of GAS6 mRNA expression in normal mammary epithelial cells and ER positive breast cancer cells by estrogen. a). Induction of GAS6 by estrogen in mammary glands. Ovariectomized mice were treated with 17β-estradiol (E2) for 9 hours or 24 hours and mammary glands were then isolated for the preparation of total RNA. Total RNA (10 μg) was hybridized with radiolabeled anti-sense GAS6 probe generated by in vitro transcription. Glyceraldehyde-3-phosphate dehydrogenase probe was included in each hybridization reaction as control. Nuclear resistant fragments were resolved on polyacrylamide-urea gels. b). GAS6 is mainly expressed in mammary epithelial cells. RT-PCR was performed with total RNA prepared from the mammary epithelial cells and stromal cells, which had been separated by collagenase digestion and Percoll gradient centrifugation. c). GAS6 is induced by 17β-estradiol (E2) in MCF-7 cells. Total RNA was made form MCF-7 cell treated with E2 or control vehicle for 3 hours. The GAS6 and pS2 mRNA levels were quantified by SYBR Green real-time PCR and normalized to 18S ribosomal RNA.
To find if the stromal cells or mammary epithelial cells express GAS6, the mammary epithelial cells and stromal cells were separated by collagenase digestion and Percoll gradient centrifugation. RT-PCR revealed that whey acidic protein (WAP), which is a marker for mammary gland epithelium, was only amplified from the total RNA made from epithelial cells, confirming the well-separation of stromal and epithelial cells. Semi-quantitative RT-PCR showed that GAS6 mRNA was mostly present in the epithelial cells (Fig. 1b).
As the normal mammary epithelial cells and breast cancer cells might respond differently to estrogen, we tested if GAS6 is also induced in ER positive breast cancer cells by estrogen. After MCF-7 cells were treated with 17β-estradiol for 3 hour, the pS2 gene, another estrogen inducible gene, was revealed to be induced by estrogen as expected (Fig.1c). Real-time PCR also demonstrated that GAS6 mRNA was induced by estrogen although to a lesser extent compared with pS2 gene (Fig. 1c).
The promoter of GAS6 is regulated by estrogen
The rapid increase of gene expression after estrogen administration suggested that GAS6 could be a direct target gene of estrogen receptor. We analyzed whether GAS6 promoter activity is regulated by estrogen. We transfected CV-1 cells with GAS6 promoter-luciferase construct, together with human ERα expression plasmid, and treated with estrogen only or with a combination of estrogen and its antagonist ICI182,780. As shown in Fig. 2, the promoter activity was increased up to 4-fold in the presence of estrogen alone. The induction of promoter activity by estrogen was blocked with the addition of ICI182,780, suggesting that the GAS6 promoter carries a functional estrogen response element (ERE).
Fig. 2.
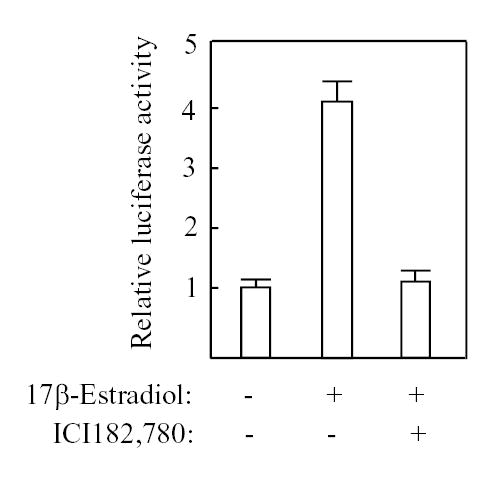
Regulation of GAS6 promoter by estrogen. CV-1 cells were cotransfected with 1.5 μg of GAS6 promoter-LUC, 20 ng of PCMV-ERα, and 0.1 μg of PCMV-β. The cells were treated with 0.01 μM 17-estradiol and 1μM ICI182,780 as indicated. The luciferase and β-galactosidase activities were measured. The results represent the average of three independent transfections normalized with the internal control β-galactosidase activity.
GAS6 is a direct target of estrogen receptor
The result from the promoter analysis promoted us to look for estrogen response elements (ERE) in GAS6 promoter region. By examining the sequence upstream of GAS6 gene using ERE sequence analysis software, Dragon ERE Finder program, we identified an estrogen response element (ERE) spanning −72~-89 bp from the translation start site (Fig.3a). To test authenticity of the ERE site found in GAS6 gene, we performed electrophoretic mobility shift assay (EMSA). As shown in figure 3b, the 32P labeled probe containing the putative ERE formed a DNA-protein complex with MCF-7 cell nuclear extract. The complex could be competed away by corresponding cold probe but not by non-specific irrelevant probe. Furthermore, addition of anti-ERα antibody to MCF-7 nuclear extract before incubation with 32P labeled probe significantly reduced the formation of the complex (Fig.3b), demonstrating that the ERE sequence from GAS6 promoter can bind ERα.
Fig. 3.


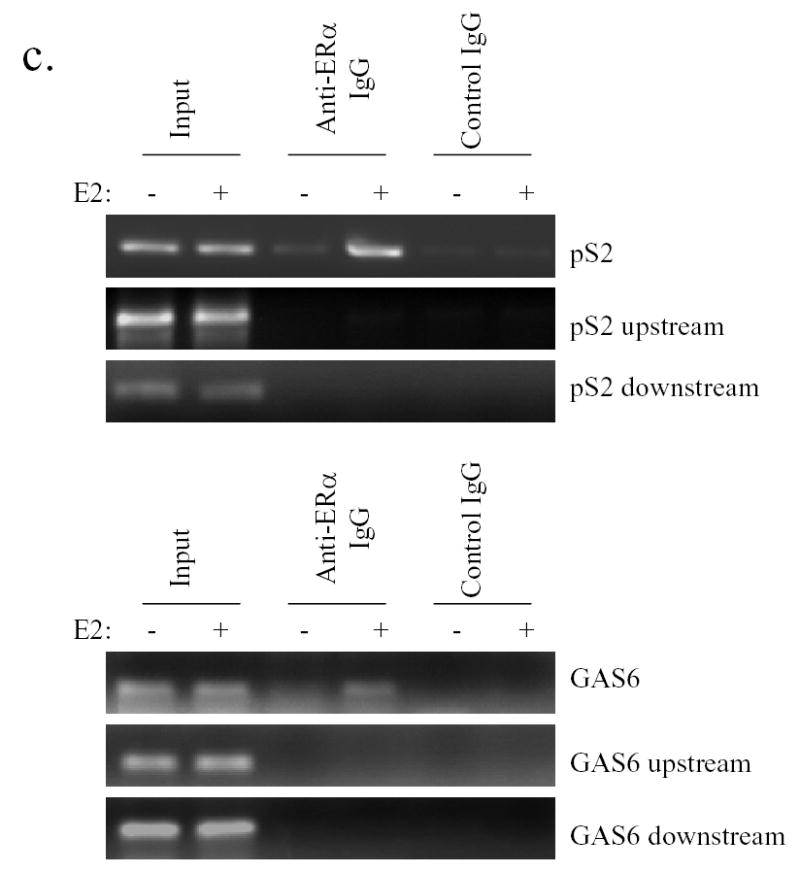
GAS6 promoter contain an ERE (estrogen response element) interacting with ERα. a). An ERE site was identified in the GAS6 promoter by DRAGON ERE FINDER. The underlined ERE site along with adjacent sequence is shown. b).Binding of ERα to the ERE of GAS6 gene in vitro. The GAS6 ERE oligonucleotide as a probe was incubated with 10 μg of nuclear extracts prepared from MCF-7 cells in the presence of unlabeled specific or non-specific competitors. The nuclear extracts were incubated with anti-ERα antibody as indicated or control IgG before being added to the probe. Specific DNA-protein complex is indicated by an arrow. c). ERα binds to GAS6 promoter in vivo. Chromatin extracts were made from MCF-7 cells following treated with or without 10nM E2 for 45 min and ChIP assays were performed using an antibody against ERα, or control IgG. DNA fragments were extracted from immunoprecipitates. The GAS6 and pS2 promoter regions containing EREs were amplified.
To further test this result in vivo, we performed chromatin immunoprecipitation assay in MCF-7 cells. The well-characterized pS2 ERE region was served as a positive control. After 45min of estrogen treatment, monoclonal antibody against ERα could efficiently precipitate the estrogen-responsive region of pS2 (Fig. 3c). Similarly, GAS6 ERE region was also amplified from anti-ERα immunoprecipitant while non-specific regions from both upstream and downstream of pS2 and GAS6 ERE sites generated no PCR products (Fig. 3c), showing that ERα directly binds to the GAS6 gene promoter region containing the ERE site in MCF-7 cells.
The exact down-stream signals regulated by estrogen and responsible for estrogen-stimulated growth in mammary glands remain elusive. Although amphiregulin is induced by estrogen and can stimulate the proliferation of mammary epithelial cells (27), mammary glands from the mouse model with amphiregulin null mutation can still respond to estrogen (28), indicating that there are other factors mediating the action of estrogen. Given the role of GAS6 in stimulating the proliferation of other cells (25, 26) and its induction in mammary glands by estrogen, GAS6 could be one of the factors involved in the estrogen-stimulated ductal growth of mammary glands.
Acknowledgments
We thank Dr. Janardan K. Reddy for his advice and support.
The abbreviations used are
- GAS6
growth arrest specific gene 6
- ER
estrogen receptor
Footnotes
This work was supported by National Institutes of Health Grant CA 88898 (To Y. J. Z).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hennighausen L, Robinson GW. Think globally, act locally: the making of a mouse mammary gland. Genes Dev. 1998;12:449–455. doi: 10.1101/gad.12.4.449. [DOI] [PubMed] [Google Scholar]
- 2.Topper YJ, Freeman CS. Multiple hormone interactions in the developmental biology of the mammary gland. Physiol Rev. 1980;60:1049–1106. doi: 10.1152/physrev.1980.60.4.1049. [DOI] [PubMed] [Google Scholar]
- 3.Brisken C. Hormonal control of alveolar development and its implications for breast carcinogenesis. J Mammary Gland Biol Neoplasia. 2002;7:39–48. doi: 10.1023/a:1015718406329. [DOI] [PubMed] [Google Scholar]
- 4.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 6.Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci U S A. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosselman S, Polman J, Dijkema R. ER beta: identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- 8.Korach KS, Couse JF, Curtis SW, Washburn TF, Lindzey J, Kimbro KS, Eddy EM, Migliaccio S, Snedeker SM, Lubahn DB, Schomberg DW, Smith EP. Estrogen receptor gene disruption: molecular characterization and experimental and clinical phenotypes. Recent Prog Horm Res. 1996;51:158–159. 186. [PubMed] [Google Scholar]
- 9.Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, Mahler JF, Sar M, Korach KS, Gustafsson JA, Smithies O. Generation and reproductive phenotypes of mice lacking estrogen receptor beta. Proc Natl Acad Sci U S A. 1998;95:15677–15682. doi: 10.1073/pnas.95.26.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nass SJ, Davidson NE. The biology of breast cancer. Hematol Oncol Clin North Am. 1999;13:311–332. doi: 10.1016/s0889-8588(05)70058-7. [DOI] [PubMed] [Google Scholar]
- 11.Murphy LC, Watson P. Steroid receptors in human breast tumorigenesis and breast cancer progression. Biomed Pharmacother. 2002;56:65–77. doi: 10.1016/s0753-3322(01)00157-3. [DOI] [PubMed] [Google Scholar]
- 12.Baum M. Tamoxifen--the treatment of choice. Why look for alternatives? Br J Cancer. 1998;78:1–4. doi: 10.1038/bjc.1998.753. Suppl 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan VC, Wolf MF, Mirecki DM, Whitford DA, Welshons WV. Hormone receptor assays: clinical usefulness in the management of carcinoma of the breast. Crit Rev Clin Lab Sci. 1988;26:97–152. doi: 10.3109/10408368809106860. [DOI] [PubMed] [Google Scholar]
- 14.Manfioletti G, Brancolini C, Avanzi G, Schneider C. The protein encoded by a growth arrest-specific gene (gas6) is a new member of the vitamin K-dependent proteins related to protein S, a negative coregulator in the blood coagulation cascade. Mol Cell Biol. 1993;13:4976–4985. doi: 10.1128/mcb.13.8.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stitt TN, Conn G, Gore M, Lai C, Bruno J, Radziejewski C, Mattsson K, Fisher J, Gies DR, Jones PF, et al. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell. 1995;80:661–670. doi: 10.1016/0092-8674(95)90520-0. [DOI] [PubMed] [Google Scholar]
- 16.Ohashi K, Nagata K, Toshima J, Nakano T, Arita H, Tsuda H, Suzuki K, Mizuno K. Stimulation of sky receptor tyrosine kinase by the product of growth arrest-specific gene 6. J Biol Chem. 1995;270:22681–22684. doi: 10.1074/jbc.270.39.22681. [DOI] [PubMed] [Google Scholar]
- 17.Godowski PJ, Mark MR, Chen J, Sadick MD, Raab H, Hammonds RG. Reevaluation of the roles of protein S and Gas6 as ligands for the receptor tyrosine kinase Rse/Tyro 3. Cell. 1995;82:355–358. doi: 10.1016/0092-8674(95)90424-7. [DOI] [PubMed] [Google Scholar]
- 18.Sun W, Fujimoto J, Tamaya T. Coexpression of Gas6/Axl in human ovarian cancers. Oncology. 2004;66:450–457. doi: 10.1159/000079499. [DOI] [PubMed] [Google Scholar]
- 19.Sainaghi PP, Castello L, Bergamasco L, Galletti M, Bellosta P, Avanzi GC. Gas6 induces proliferation in prostate carcinoma cell lines expressing the Axl receptor. J Cell Physiol. 2005;204:36–44. doi: 10.1002/jcp.20265. [DOI] [PubMed] [Google Scholar]
- 20.Hafizi S, Dahlback B. Signalling and functional diversity within the Axl subfamily of receptor tyrosine kinases. Cytokine Growth Factor Rev. 2006;17:295–304. doi: 10.1016/j.cytogfr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM, Zhu YJ. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J Biol Chem. 2002;277:28624–28630. doi: 10.1074/jbc.M201053200. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Richards J, Guzman R, Imagawa W, Nandi S. Sustained growth in primary culture of normal mammary epithelial cells embedded in collagen gels. Proc Natl Acad Sci U S A. 1980;77:2088–2092. doi: 10.1073/pnas.77.4.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carroll JS, Brown M. Estrogen receptor target gene: an evolving concept. Mol Endocrinol. 2006;20:1707–1714. doi: 10.1210/me.2005-0334. [DOI] [PubMed] [Google Scholar]
- 25.Dormady SP, Zhang XM, Basch RS. Hematopoietic progenitor cells grow on 3T3 fibroblast monolayers that overexpress growth arrest-specific gene-6 (GAS6) Proc Natl Acad Sci U S A. 2000;97:12260–12265. doi: 10.1073/pnas.97.22.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanagita M, Arai H, Ishii K, Nakano T, Ohashi K, Mizuno K, Varnum B, Fukatsu A, Doi T, Kita T. Gas6 regulates mesangial cell proliferation through Axl in experimental glomerulonephritis. Am J Pathol. 2001;158:1423–1432. doi: 10.1016/S0002-9440(10)64093-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Lacaci I, Saceda M, Plowman GD, Johnson GR, Normanno N, Salomon DS, Dickson RB. Estrogen and phorbol esters regulate amphiregulin expression by two separate mechanisms in human breast cancer cell lines. Endocrinology. 1995;136:3983–3992. doi: 10.1210/endo.136.9.7649107. [DOI] [PubMed] [Google Scholar]
- 28.Luetteke NC, Qiu TH, Fenton SE, Troyer KL, Riedel RF, Chang A, Lee DC. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]


